Abstract
The transcriptional modulator Cited2 is induced by various biological stimuli including hypoxia, cytokines, growth factors, lipopolysaccharide (LPS) and flow shear. In this study, we report that Cited2 is required for mouse fetal liver development. Cited2−/− fetal liver displays hypoplasia with higher incidence of cell apoptosis, and exhibits disrupted cell-cell contact, disorganized sinusoidal architecture, as well as impaired lipid metabolism and hepatic gluconeogenesis. Furthermore, we demonstrated the physical and functional interaction of Cited2 with liver-enriched transcription factor HNF4α. Chromatin immunoprecipitation (ChIP) assays further confirmed the recruitment of Cited2 onto the HNF4α-responsive promoters and the reduced HNF4α binding to its target gene promoters in the absence of Cited2. Taken together, this study suggests that fetal liver defects in mice lacking Cited2 result, at least in part, from its defective coactivation function for HNF4α.
Keywords: Cited2, coactivator, HNF4α, liver development
Introduction
Hepatogenesis proceeds through multiple developmental stages. During embryonic development, the induction of hepatic fate depends on the reciprocal interactions between ventral foregut endoderm and adjacent mesenchymal tissues (Zaret, 2002). At around embryonic day 8 (E8) in the mouse, the ventral wall of foregut endoderm initiates its development toward a hepatic fate in response to the inductive growth factor signaling, including fibroblast growth factor (FGF) signaling from the adjacent cardiogenic mesoderm and bone morphogenic proteins (BMPs) signaling from nearby septum transversum (st) mesenchyme (Jung et al, 1999; Rossi et al, 2001; Duncan, 2003). Then cells within the foregut endoderm start to proliferate and differentiate, leading to the formation of the liver bud at E9. Once the liver bud is generated and the hepatoblasts delaminate from the foregut and migrate as cords into the septum transversum mesenchyme, the cells within the embryonic liver environment must reorganize to generate the complex hepatic architecture that is crucial for normal liver function (Zhao and Duncan, 2005). This requires extensive differentiation of the hepatocytes, organization of the extracellular matrix, development of the biliary tract, maturation of sinusoidal capillaries and hepatic vasculature and formation of a polarized epithelium (Lemaigre and Zaret, 2004). While the embryonic liver lacks most of the liver functions in the adult as the center of metabolism, the liver acquires those functions during the perinatal and postnatal stages.
Over the last decade, analyses of transcriptional regulation of liver development have identified several liver-enriched transcription factors that are involved in liver development, differentiation and function, including C/EBP family, HNF1α and β, HNF3α, β and γ, HNF4α and HNF6 (Duncan, 2000; Costa et al, 2003). The cumulative data also strongly suggest that HNF4α acts as a central regulator of hepatogenesis, through the activation of a cascade of transcription factors that ultimately define the gene expression profile of the mature hepatocytes (Watt et al, 2003). Genetic studies in mice have also revealed that numerous signaling molecules are required for continued fetal liver growth. Although the combined use of molecular genetics, molecular biology and embryology has allowed us to realize the proceedings in unraveling of the mechanisms that control hepatogenesis, the blueprint regarding hepatic development remains largely unknown.
Cited2 is a founding member of a new family of non-DNA-binding transcriptional coregulators (Shioda et al, 1997; Dunwoodie et al, 1998). Members of the Cited family share a well-conserved C-terminal acidic domain (the conserved region 2, CR2) that binds to the transcriptional coactivators CBP/p300 with high affinity (Bhattacharya et al, 1999; Leung et al, 1999). On this basis, Cited proteins are thought to target specific transcriptional responses through interactions with other transcription factors, and to modify these responses through binding with CBP/p300. Indeed, several lines of evidence supported the view that Cited2 associates with various DNA-binding transcription factors and acts as a transcriptional cofactor to regulate gene expression mediated by these proteins, including the LIM domain-containing DNA-binding protein Lhx2 (Glenn and Maurer, 1999), the transcription factor AP-2 (TFAP-2) (Braganca et al, 2003), the nuclear receptors peroxisome proliferator-activated receptors (PPAR) α and γ (Tien et al, 2004), and the TGFβ-signaling mediators Smad2/3 (Chou et al, 2006). An additional role has been proposed for Cited2 as a negative regulator of DNA-binding protein hypoxia-inducible factor-1α (HIF-1α) (Bhattacharya et al, 1999; Freedman et al, 2003). Collectively, these initial in vitro studies underscore the potential roles of Cited2 in different biological processes.
Cited2 plays an essential role in mouse embryonic development based on knockout studies (Bamforth et al, 2001, 2004; Martinez-Barbera et al, 2002; Yin et al, 2002; Weninger et al, 2005; Withington et al, 2006). During early mouse embryogenesis, Cited2 expression is clearly evident in the cardiac mesenchyme and septum transversum mesenchyme, which gives rise to aspects of the mature liver (Dunwoodie et al, 1998), suggesting its potential role in hepatogenesis. In this study, we characterized fetal liver defects of Cited2−/− embryos, which are represented by hepatic hypoplasia and dysfunction of hepatocytes. These phenotypes are accompanied by the downregulation of a subset of genes involved in hepatogenesis. Moreover, we demonstrated that Cited2 interacts with HNF4α and coactivates HNF4α-mediated transcription. Therefore, we propose that fetal liver defects observed in Cited2−/− embryos result, at least in part, from its defective function as an HNF4α coactivator.
Results
Expression of Cited2 in the developing mouse liver
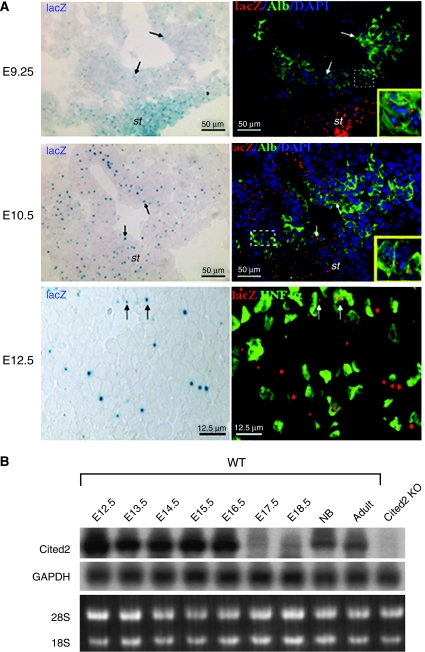
During early mouse embryogenesis (8- to 12-somite stage), Cited2 transcripts are localized to the myocardium and septum transversum mesenchyme (Dunwoodie et al, 1998). By E9.25, we found Cited2 continued to express in specified hepatic endodermal cells within the liver bud as well as in the surrounding septum transversum mesenchyme, as demonstrated by the coexpression of hepatocyte-specific marker albumin and β-galactosidase from the Cited2-lacZ allele in heterozygous embryos (Figure 1A). At E10.5, the lacZ- and albumin-double-positive cells could be found in the hepatic primordia, as well as in the mesenchymal portion of the developing liver (Figure 1A). At E12.5, the lacZ-positive staining was detected in hepatocytes (HNF4α-positive cells) as well as in non-hepatocytes (Figure 1A). The Northern blot analysis of developing fetal liver (E12.5–E18.5), new-born (NB) liver and adult liver (8-week-old) further showed that Cited2 transcripts are highly expressed in fetal livers from E12.5 through E16.5, compared with the low expression levels in E17.5 and E18.5 livers and intermediate expression levels in newborn and adult livers (Figure 1B). The RNA in situ hybridization analysis also confirmed the high expression of Cited2 in E14.5 liver (Supplementary Figure 1). Taken together, the expression pattern of Cited2 in developing mouse liver supports the idea that Cited2 may play an important role in liver development and function.
Figure 1.
Cited2 expression in developing liver. (A) Cited2 expression in liver (E9.25, E10.5 and E12.5) by lacZ staining. Arrows indicate that lacZ-positive cells are also positive for albumin or HNF4α. The lacZ staining (blue) was pseudocolorized to red for the ease in visualization. st, septum transversum mesenchyme. (B) Northern blot analysis of Cited2 expression in developing liver. Cited2−/− fetal liver (KO) was used as a negative control. NB, newborn.
Liver hypoplasia in Cited2-deficient embryos
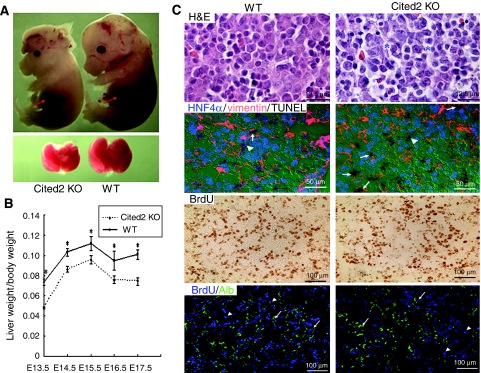
To test whether Cited2 plays a role in liver development, we examined the gross morphology of fetal livers from wild-type and Cited2−/− embryos. Compared with wild-type littermate controls, Cited2−/− embryos could be recognized by their growth retardation and hypoplastic liver starting from E13.5. Although smaller in size, liver from Cited2−/− embryos had the correct number of lobes and appeared red, suggesting that the initial steps of liver development are intact in Cited2−/− embryos (Figure 2A). However, liver weight/body weight ratio was significantly decreased in Cited2-null embryos from E13.5 to E17.5 when compared with wild-type controls (Figure 2B). Analysis of hematoxylin and eosin (H&E)-stained liver sections from both E14.5 wild-type and Cited2-null littermates revealed the distorted liver architecture with reduced cellularity, increased sinusoidal space and dissociated parenchymal cells in Cited2−/− fetal liver (Figure 2C). Three possibilities could account for the observed hypocellularity and smaller sizes of Cited2-null livers: (i) increased cell death, (ii) reduced cell proliferation and (iii) disrupted cell–cell interaction. We examined apoptosis by the TUNEL assay and cell proliferation by the BrdU incorporation assay for E14.5 wild-type and Cited2-null liver sections, followed by immunostaining of cell type-specific markers such as vimentin for mesenchymal cells and albumin or HNF4α for hepatocytes. The TUNEL assay showed increased apoptosis in Cited2−/− livers (12.8±1.2%), whereas the wild-type littermate controls had 3.9±1.5% apoptotic cells (Figure 2C). The immunostaining showed that majority of the apoptotic cells were mesenchymal cells, although some were hepatocytes (Figure 2C). The BrdU incorporation assay showed that 37.8±1.8 and 36.9±1.5% of cells in normal and Cited2−/− livers were BrdU positive, respectively, suggesting that wild-type and Cited2-knockout embryos were not significantly different in the proportion of liver cells that are undergoing proliferation (Figure 2C). The proliferating cells were both hepatocytes (albumin positive) and non-hepatocytes (Figure 2C).
Figure 2.
Liver hypoplasia in Cited2-deficient embryos. (A) Hypoplasia of Cited2−/− embryonic liver compared with a wild-type littermate control. (B) Liver weight/body weight ratio of wild-type versus Cited2−/− embryos from E13.5 to E17.5. The data represent the mean±s.e.m.; n=4–6 mice for each group. *P-value from Student's t-test <0.05 when comparing wild-type with Cited2-null mice. (C) H&E-stained E14.5 sagittal liver sections showed that Cited2−/− embryonic liver has a loosened liver structure with enlarged sinusoidal space and dissociated parenchymal cells (asterisks). TUNEL staining indicated an elevated number of apoptotic cells in E14.5 Cited2−/− liver. Majority of the apoptotic cells (brown) are mesenchymal cells (vimentin positive, arrows), although a few of apoptotic cells are hepatocytes (HNF4α positive, arrowheads). The HNF4α staining (green) was pseudocolorized to blue. The percentage of the apoptotic cells was evaluated from 3–5 sections (5000 cells/section) obtained from three each of wild-type and Cited2−/− embryos. BrdU incorporation assay indicated normal hepatic proliferation in E14.5 Cited2−/− fetal liver. Some of the BrdU-positive cells are hepatocytes (albumin positive, arrows) while some are non-hepatocytes (arrowheads). BrdU staining (brown) was pseudocolorized to blue. The percentage of the BrdU-positive cells was evaluated from 3–5 sections (2000 cells/section) obtained from three each of wild-type and Cited2−/− embryos.
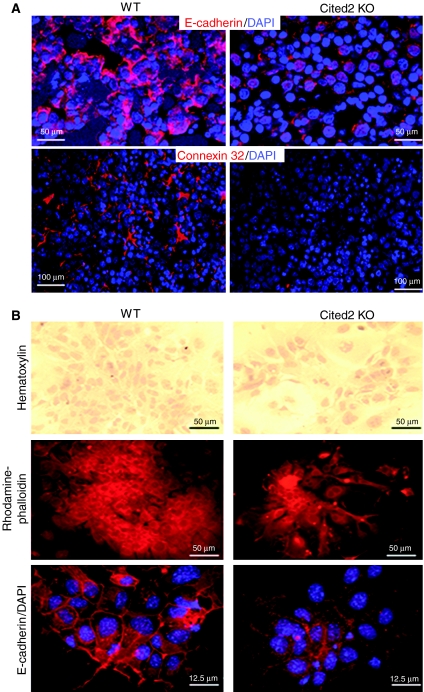
To address whether hypoplastic fetal liver phenotype in Cited2−/− embryos results from the impaired cell–cell contact, we investigated cell–cell interaction by examining the localization and expression levels of cell junction markers, E-cadherin and connexin 32, in E14.5 livers. Immunostaining showed that E-cadherin and connexin 32 were primarily membranous in wild-type liver, but the staining was much weaker or even absent from a substantial proportion of cells in Cited2−/− liver, which is suggestive of defective cell–cell interaction (Figure 3A). Furthermore, Western blot analysis of E-cadherin and connexin 32 expression levels confirmed the above observation (Supplementary Figure 2). These preliminary results suggested that mutant hepatocytes might have altered adhesive properties. We therefore cultured E14.5 hepatocytes from normal and mutant livers to directly test cell adhesion. Hematoxylin staining showed that wild-type hepatocytes were able to adhere and form good epithelial sheets on the extracellular matrix, collagen, whereas Cited2−/− hepatocytes failed to adhere to the substrate (Figure 3B). Unlike cells from wild type, most cells from Cited2−/− liver were small and round and adhered loosely to the few cells that attached to the substrate (Figure 3B). Viability of these cells was monitored using trypan blue staining and the results indicated that the small round-shaped cells were not dead but were non-adherent (data not shown). Rhodamine–phalloidin staining showed that normal hepatocytes primarily expressed cortical actin bundles, further confirming the epithelial nature of these cells (Figure 3B). However, Cited2−/− hepatocytes contained multiple stress fibers (Figure 3B). E-cadherin was detected at the site of juxtaposed plasma membrane of wild-type hepatocytes, but the expression was much weaker in Cited2−/− hepatocytes (Figure 3B). Furthermore, adenovirus-mediated exogenous Cited2 expression could rescue the ability of Cited2−/− hepatocytes to adhere to the substrate (Supplementary Figure 3).
Figure 3.
Disruption of cell–cell interaction in Cited2−/− fetal liver. (A) Immunostaining showed that the membranous expression of E-cadherin and connexin 32 was intense in E14.5 wild-type liver but much weaker or even absent in a substantial proportion of cells in Cited2−/− littermate liver. (B) Perturbation of cellular adhesion in E14.5 Cited2−/− hepatocytes. Hematoxylin staining, rhodamine–phalloidin staining and E-cadherin immunostaining indicated defective adhesion of Cited2−/− hepatocytes.
Disorganized sinusoidal architecture, impaired lipid homeostasis and hepatic gluconeogenesis in Cited2−/− fetal liver
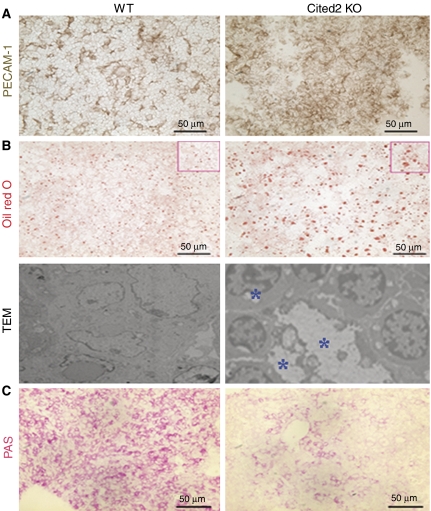
Since well-developed sinusoidal architecture is necessary for liver function, we examined if the sinusoidal architecture is normal in Cited2−/− liver. PECAM-1 immunostaining for E16.5 liver sections showed that wild-type liver had very organized sinusoidal architecture, with PECAM-1-positive endothelial cells surrounding large vessels as well as sinusoids, which were distributed throughout the parenchyma (Figure 4A). In contrast, the sinusoidal structure of Cited2−/− liver was greatly disrupted, exhibiting disorganized sinusoidal capillaries (Figure 4A).
Figure 4.
Disorganized sinusoidal architecture, impaired lipid homeostasis and hepatic gluconeogenesis in Cited2−/− fetal liver. (A) Disorganized sinusoidal architecture was identified by PECAM-1 immunostaining (brown) in E16.5 Cited2−/− fetal liver. (B) Oil red O staining showed that abundant fine- to medium-sized lipid storage droplets accumulated diffusely in E17.5 Cited2−/− liver, whereas lipid storage droplets were rarely observed in wild-type fetal liver. Ultrastructural analysis showed abundant lipid droplets accumulated in the sinusoidal space in E17.5 Cited2−/− hepatocytes (asterisks). Original magnification: × 6000. (C) PAS staining revealed impaired glycogen storage in E17.5 Cited2−/− fetal liver, compared with wild type.
Proper formation of the intricate sinusoidal vascular network provides all hepatocytes direct access to the blood plasma and efficient lipoprotein metabolism (Postic et al, 2004). Therefore, the disrupted sinusoidal architecture observed in Cited2−/− liver could affect the ability of the liver to properly function in lipid homeostasis. Oil red O staining showed abundant fine- to medium-sized lipid storage droplets accumulated diffusely in Cited2−/− liver, whereas lipid storage droplets were rarely observed in wild-type liver (Figure 4B). Electron microscopic analysis further showed numerous lipid droplets being accumulated in the sinusoidal space in E17.5 Cited2−/− liver, whereas normal liver contained less lipid droplets (Figure 4B).
In addition, periodic acid–Schiff (PAS) histochemistry revealed that E17.5 wild-type liver exhibited robust levels of glycogen accumulation. In contrast, accumulation of intracellular glycogen was greatly reduced in E17.5 Cited2−/− liver, which displayed only sparse and weaker glycogen staining than wild-type liver (Figure 4C).
Gene expression profiling in Cited2−/− fetal liver
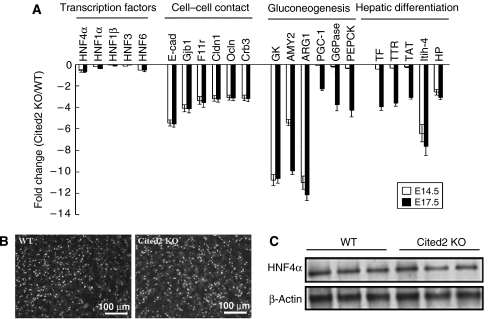
To determine whether loss of Cited2 alters the gene expression profile in developing liver, we performed Affymetrix mouse oligonucleotide gene array analysis, which identified 160 and 238 genes whose expression was downregulated and upregulated⩾2-fold (P⩽0.05) in E14.5 Cited2-null liver when compared with wild-type liver, respectively. To validate microarray data, we compared steady-state mRNA levels of differentially expressed genes identified by microarray in sorted wild-type and Cited2-null hepatocytes by real-time RT–PCR. We isolated the hepatocyte populations (albumin+CD45−Ter119−) from wild-type and Cited2−/− fetal livers (E14.5 and E17.5) by flow cytometry. The percentage of hepatocytes (albumin+CD45−Ter119−) in Cited2-null embryos (16.66% in E14.5 liver and 20.03% in E17.5 liver) was slightly lower than that in wild-type (18.10% in E14.5 liver and 21.07% in E17.5) littermate controls. The RNA samples extracted from sorted wild-type and Cited2-null hepatocytes were used for real-time RT–PCR. The downregulated genes were categorized based on their biological functions. Figure 5A shows that in the absence of Cited2, the steady-state mRNA levels of a subset of genes which are involved in cell–cell contact assembly (E-cadherin (E-cad), gap junction protein β1 (Gjb1, also known as connexin 32), claudin 1 (Cldn1), occludin (Ocln), F11 receptor (F11r), crumb 3 (Crb3)), hepatic carbohydrate homeostasis (amylase 2 (AMY2), arginase 1 (ARG1), PPARγ coactivator-1α (PGC-1α), glucose-6-phosphatase (G6Pase), phosphoenolpyruvate carboxykinase (PEPCK), glucokinase (GK)) and several well-characterized hepatic differentiation markers (transferrin (TFN), transthyretin (TTR), tyrosine amino transferase (TAT), inter-α-trypsin inhibitor-4 (Itih-4), haptoglobin (HP)) were downregulated. However, the mRNA expression levels of several hepatic regulators such as HNF4α, HNF1α, HNF1β, HNF3 and HNF6 did not show significant difference between sorted wild-type and Cited2−/− hepatocytes (Figure 5A). Notably, most of these genes with altered expression profiles are putative HNF4α target genes when compared with the array data of fetal liver-specific HNF4α deficient mouse (Battle et al, 2006; Supplementary Table 1). It is also worth noting that this downregulation of HNF4α target genes by Cited2 deficiency does not appear to be due to decreased expression of HNF4α mRNA, since real-time RT–PCR did not show statistical difference (<2-fold) in HNF4α expression levels between wild-type and Cited2-null livers (Figure 5A). The immunostaining (Figure 5B) and Western blot analysis (Figure 5C) of HNF4α protein levels in wild-type and Cited2−/− liver also did not show significant difference. Rather, these results are consistent with the observation that Cited2 coactivates HNF4α through direct physical association and augmentation of its transcriptional activity (see below).
Figure 5.
Gene expression profiling in Cited2−/− fetal liver. (A) Compared with wild type, a subset of genes involved in cell junction assembly (E-cadherin, Gjb1, Cldn1, Ocln, F11r, Crb3), hepatic carbohydrate metabolism (AMY2, ARG1, PGC-1, G6Pase, PEPCK, GK), as well as several well-characterized hepatic differentiation markers (TF, TTR, TAT, Itih-4, HP) were downregulated in Cited2−/− hepatocytes. The steady-state mRNA levels of HNF4α, HNF1α, HNF1β, HNF3 and HNF6 in wild-type and Cited2-null hepatocytes did not show significant difference (<2-fold). Bars represent the mean±s.e.m.; n=3–5 RNA samples from different mice for each group. The immunofluorescence (B) and western blot analysis (C) of HNF4α proteins in wild-type and Cited2−/− liver did not show significant difference. The immunofluorescence of HNF4α was presented as grayscale.
Cited2 acts as a coactivator for HNF4α
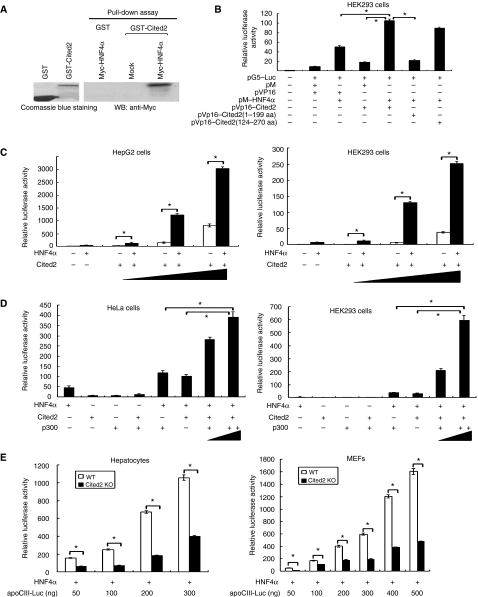
Cited2 is a transcriptional cofactor that in vitro can act as either a positive or a negative regulator of transcription (Bhattacharya et al, 1999; Glenn and Maurer, 1999; Braganca et al, 2003; Tien et al, 2004; Chou et al, 2006). Therefore, one mechanism that may underlie the abnormalities observed in Cited2−/− fetal liver is defective coactivation of a transcription factor that is critical for liver development and function. Because Cited2-deficient fetal liver displays overlapping phenotypes with those of fetal liver-specific HNF4α knockout, we hypothesized that Cited2 could serve as a coactivator for HNF4α and regulate hepatic gene expression during liver development and differentiation. To test this possibility, we first examined subcellular localization of Cited2 and HNF4α proteins by confocal microscopic analysis, which showed that these two proteins colocalized in the nuclei (Supplementary Figure 4). The in vitro GST pull-down assay showed that GST-Cited2, but not GST, efficiently pulled down Myc-HNF4α protein (Figure 6A), indicating the interaction of Cited2 with HNF4α in vitro. Mammalian two-hybrid analysis further showed that HNF4α failed to interact with the Cited2 mutant missing its C-terminal region, but was still able to bind to the mutant without the N-terminal amino acids (Figure 6B), indicating that Cited2 C-terminal region is essential for interacting with HNF4α.
Figure 6.
Physical and functional interaction of Cited2 with HNF4α. (A) Interaction of Cited2 with HNF4α in vitro. GST or GST-Cited2 was immobilized on glutathione-conjugated Sepharose beads and incubated with lysates from cells transfected with a plasmid expressing Myc-HNF4α protein or vector alone (mock). After extensive washing, coprecipitated Cited2 was detected by anti-Myc western blotting (WB). (B) Cited2 C-terminal region is essential for interacting with HNF4α. All the luciferase reporter assays in (C–E) were performed as described in Materials and methods. (C) Cited2 enhances HNF4α-mediated apoCIII transcription in a dose-dependent manner. (D) Coactivator p300 synergizes with Cited2 to enhance HNF4α transcriptional activity in a dose-dependent manner. The data represent the mean±s.e.m.; n=3–5 samples for each group in panels B–D. *P-value from Student's t-test <0.05 when comparing the two indicated bars. (E) Loss of Cited2 abrogates HNF4α-mediated transactivation activity. HNF4α and various amounts of apoCIII promoter/reporter plasmids were cotransfected into wild-type and Cited2-null cells, followed by luciferase reporter assays. The data represent the mean±s.e.m.; n=3–5 samples for each group. *P-value from Student's t-test <0.05 when comparing wild-type with Cited2-null cells.
The functional significance of the identified interaction between Cited2 and HNF4α was further assessed in the luciferase reporter assay. HepG2 cells (with endogenous HNF4α expression), or HeLa and HEK293 cells (without endogenous HNF4α expression) were cotransfected with the plasmid expressing HNF4α, apoCIII promoter/luciferase reporter plasmid harboring HNF4α-responsive elements, together with varying amounts of the plasmid expressing Cited2 proteins. As previously reported, HNF4α was able to activate apoCIII transcription, while Cited2 only had marginal transactivation activity (Figure 6C). However, substantial activation was observed when Cited2 was cotransfected with HNF4α in a dose-dependent manner upon cotransfection of various amounts of Cited2 (Figure 6C). More importantly, coactivator p300 synergized with Cited2 to enhance HNF4α transcriptional activity in a dose-dependent manner (Figure 6D). Furthermore, the reporter assay showed that the combination of Cited2, HNF4α and p300 resulted in synergistic coactivation of the E-cadherin gene promoter, which was entirely dependent on the presence of Cited2 in a dose-dependent manner (Supplementary Figure 5). Taken together, these data demonstrate that Cited2 acts as a coactivator to potentiate the transactivation activity of HNF4α.
To further explore the role of endogenous Cited2 in transactivation by HNF4α, we investigated the transactivation activity of HNF4α in primary hepatocytes and murine embryonic fibroblasts (MEFs) derived from E14.5 Cited2-null embryos to ascertain its function in the absence of Cited2. The transcriptional activity of HNF4α was evaluated by cotransfection of HNF4α and apoCIII promoter/reporter plasmid into Cited2-null cells. As shown in Figure 6E, although HNF4α did show transcriptional activity in Cited2−/− hepatocytes and MEFs, the activity of the reporter gene in Cited2-null cells was greatly reduced, indicating that HNF4α function in these cells was defective and the full activation of HNF4α target genes requires endogenous Cited2. Taken together, these loss-of-function approaches complement the overexpression studies and demonstrate that Cited2 serves as a coactivator for HNF4α to regulate the expression of HNF4α target genes, which is consistent with the downregulation of HNF4α target genes seen in Cited2−/− fetal liver (Figure 5A).
Recruitment of Cited2 onto the HNF4α-responsive promoters and reduced HNF4α occupancy to its responsive promoters in Cited2−/− fetal liver
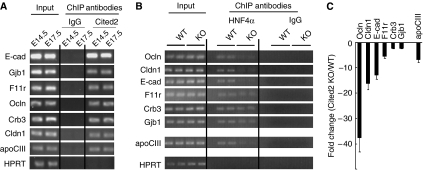
To demonstrate that Cited2 serves as an HNF4α coactivator for endogenous gene expression, we performed chromatin immunoprecipitation (ChIP) assay to determine whether Cited2 is recruited onto native, chromosomally integrated genomic sequences, which contain the HNF4α-binding sites in the context of mouse fetal liver. Recent studies demonstrated that HNF4α binds to the regulatory elements within many of the genes encoding cell junction and adhesion proteins during embryonic liver development, such as E-cadherin, Cldn1, Crb3, Eppk1, F11r, Gjb1, Lgals9 and Ocln (Battle et al, 2006). Since we observed disrupted cell junction assembly as well as the downregulated expression of genes functioning in cell–cell contact assembly in Cited2−/− fetal liver, and proposed that Cited2 acts as a coactivator for HNF4α, we tested whether Cited2 interacts with the same HNF4α-binding regions that are known to be occupied by HNF4α. Formaldehyde-crosslinked protein–chromatin complexes isolated from mouse embryonic livers (E14.5 and E17.5) were immunoprecipitated with a Cited2-specific antibody to isolate the fraction of DNA associated with Cited2, or preimmune rabbit IgG to control for nonspecific immunoprecipitation of DNA. We identified sequences enriched in the pool of anti-Cited2-precipitated liver chromatin compared with the negative controls using PCR with oligonucleotides flanking HNF4α-binding sites identified by Battle et al (2006). We showed that Cited2 was recruited onto the previously identified HNF4α-binding sites in six promoters out of eight cell junction genes assayed: E-cadherin, Gjb1, Cldn1, Crb3, F11r, Ocln, as well as apoCIII, whereas we failed to detect Cited2 recruitment to chromatin from the coding region of exon 9 of the mouse hypoxanthine phosphoribosyltransferase (HPRT) gene, which lacks HNF4α-binding sites (Figure 7A), indicating that the observed Cited2 chromatin association is gene specific and not due to a nonspecific activity of the antibody used. Collectively, these data demonstrate that Cited2 coactivates HNF4α on the HNF4α target gene promoters through HNF4α-responsive elements.
Figure 7.
Recruitment of Cited2 onto HNF4α-responsive promoters and reduced occupancy of HNF4α to its responsive promoters in Cited2−/− fetal liver. (A) Crosslinked chromatin from E14.5 and E17.5 wild-type fetal livers was incubated with antibodies against Cited2, or preimmune rabbit IgG (negative control). Immunoprecipitates were analyzed by PCR using primers specific for HNF4α-responsive elements in tested HNF4α target gene promoters. A sample representing 1% of total chromatin was included in the PCR (input). (B) The binding of HNF4α to its responsive promoters was reduced in chromatins from Cited2-null mice compared with wild-type controls. Crosslinked chromatin from E14.5 wild-type and Cited2-null livers was incubated with antibodies against HNF4α, or preimmune rabbit IgG (negative control). Immunoprecipitates were analyzed by real-time PCR for quantitative analysis. A sample representing 1% of total chromatin was included in the PCR (input). (C) The reduction of HNF4α occupancy to its responsive promoters in chromatins from Cited2-null mice was evaluated by quantitative PCR. The data represent the mean±s.e.m.; n=3–5 reactions for each group. *P-value from Student's t-test <0.05.
In the absence of Cited2, the HNF4α transactivation activity is diminished (Figure 6E), consistent with a cooperative role of Cited2 in HNF4α activity. Quantitative ChIP assays were performed to determine the effect of Cited2 depletion on the HNF4α occupancy onto its target gene promoters. As shown in Figure 7B, in the absence of Cited2, the binding of HNF4α to the target gene promoters was reduced when compared with wild-type controls, ranging from 2- to 38-fold (Figure 7C). The ablation of Cited2 results in the reduced HNF4α occupancy to the responsive promoters, suggesting that Cited2 can activate HNF4α-mediated transcription via enhancing the DNA binding of HNF4α to its responsive promoters.
Discussion
The results presented in this study establish a new role for Cited2 in hepatogenesis, in part through its coactivation of HNF4α, a master regulator for hepatic gene expression, liver development, differentiation and function (Li et al, 2000; Hayhurst et al, 2001; Inoue et al, 2002; Parviz et al, 2003; Rhee et al, 2003). We showed that Cited2 is preferentially expressed in developing liver and characterized fetal liver defects when Cited2 gene is absent during development (Figures 1, 2, 3 and 4). We have also shown that Cited2 acts as an HNF4α coactivator capable of regulating its target genes by a variety of in vitro biochemical and cellular approaches (Figures 6 and 7). The physical interaction of Cited2 with HNF4α and the functional cooperation report here provide supporting evidence that Cited2 functions as a coactivator for HNF4α to transduce cellular signals triggered by physiologic and developmental cues to the transcriptional control of liver development, differentiation and function by HNF4α, which further corroborates with the overlapping phenotypes of Cited2−/− livers and mice with liver-specific HNF4α deficiency. Based on our analyses of Cited2−/− fetal liver, it is likely that loss of Cited2 activity in adult liver will have pleiotropic consequences that grossly affect liver function, which can be best determined by generating hepatocyte-specific Cited2-knockout mice. Furthermore, in clinical practice, we would speculate that impaired activation of HNF4α due to inactivation/mutations of Cited2 might be involved in the etiology of certain patients with HNF4α mutation-associated maturity-onset diabetes of the young 1 (MODY1) (Ellard and Colclough, 2006) and type 2 diabetes mellitus (Hani et al, 1998). Hence, the modulation of Cited2 docking to HNF4α may provide a novel therapy to manage liver dysfunctions in these diseases.
Transactivation activity of HNF4α and other nuclear receptors is regulated by the orchestrated recruitment and assembly of several coactivators into multisubunit protein complexes on target gene promoters (Rosenfeld et al, 2006). The coactivators stimulate transcription either through direct interactions with the basal transcription machinery or by inducing local chromatin remodeling through the intrinsic histone acetyltransferase (HAT) activity (Rosenfeld et al, 2006). Since Cited2 lacks an obvious catalytic domain for HAT, which is responsible for coactivation properties for most coactivators, the underlying mechanisms for Cited2 to coactivate HNF4α transcriptional activity remain undefined. However, Cited2 may serve as an adaptor molecule, either by recruiting or stabilizing promoter-specific coregulator complexes. In this regard, based on the fact that nearly all cellular Cited2 molecules bind physically with HAT-containing proteins CBP/p300 with high affinity (Bhattacharya et al, 1999) and the mechanism of HNF4α-mediated transcriptional activation involves the interaction with CBP/p300 (Soutoglou et al, 2000), we speculate that the contribution of Cited2 to HNF4α transcriptional activity occurs through the recruitment of CBP/p300 to exert chromatin-remodeling activity. It is worth noting that the reporter assay showed a synergistic effect of p300 on HNF4α coactivation by Cited2, suggesting an involvement of p300 (and probably CBP as well) in HNF4α coactivation by Cited2 (Figure 6D; Supplementary Figure 5).
Although Cited2 has no intrinsic HAT activity, its recruitment to the HNF4α target genes may also promote increased histone acetylation through its interaction with CBP/p300, thus indirectly altering the local chromatin structure. Consistent with this speculation, we observed that Cited2−/− hepatocytes have abnormal nuclei containing large areas of heterochromatin accumulation (Figure 4B), suggesting that the absence of Cited2 may lead to the suppressed chromatin structure. More detailed studies will be necessary to clarify the potential involvement of Cited2 in remodeling chromatin structure and the related mechanisms. It is also worth noting that by the ChIP assay Cited2 is recruited to only certain but not all HNF4α-responsive promoters (6 out of 8 genes encoding cell junction proteins assayed), suggesting gene-specific coactivation of Cited2 for HNF4α. A genome-wide screen by ChIP-on-chip strategy could further identify specific target genes coactivated by HNF4α and Cited2. In addition, quantitative ChIP assays showed that diminished occupancy of HNF4α to its responsive promoters varied to some extent (Figure 7C), and did not correlate with observed decreases in the expression levels of target RNA (Figure 5A). This could be due to the differential HNF4α binding affinity for response elements found in various promoters. It could also result from the different composition and/or configuration of transcriptional protein complexes formed around DNA-bound HNF4α, thereby reflecting the contribution of other factors working in concert with HNF4α. In this regard, it has been proposed that DNA acts as an allosteric modulator that, when bound by transcription factors, could change the conformation of transcription factors, resulting in altered DNA binding affinity and transcriptional activity (Lefstin and Yamamoto, 1998; Wu and Chiang, 2007). In turn, DNA-induced changes in transcription factor conformation were predicted to alter transcription factor affinity for other ‘ligands' such as coactivators or corepressors (Lefstin and Yamamoto, 1998; Wu and Chiang, 2007). The quantitative ChIP data in this study indicate that HNF4α target genes are subjected to diverse modes of regulation. Although this is likely due to different modes of HNF4α interaction with DNA, it is also possible that Cited2 contributes significantly to this diversity, namely, the nature of target gene promoters influences the relative ability of Cited2 to stimulate HNF4α-dependent gene transcription. Another important consideration is that several HNF4α targets are not affected in Cited2-null mice. Although the mechanism for this selective effect of Cited2 on HNF4α target genes is unknown, Cited2 may confer a subtle conformational change upon its partner transcription factor that affects its ability to bind certain DNA sequences. Alternatively, small differences in response elements in promoters may determine whether a productive interaction occurs between a bound transcription factor and a coactivator.
Taken together, our data suggest that Cited2 actions in liver development are in part mediated by its function as a transcriptional coactivator for HNF4α. Our findings provide new insights for the molecular mechanisms underlying liver development, differentiation and function, which may explain the etiology of liver dysfunction and resulting complications in human diseases. More importantly, the understanding of molecular mechanisms that underlie the Cited2 coactivation function for HNF4α in a variety of physiological and pathophysiological situations will offer novel targets for potential therapeutic intervention, hence contributing to drug discovery and new clinical applications.
Materials and methods
Mouse line and genotyping
Cited2+/− mice were genotyped by Southern blot analysis (Yin et al, 2002) and backcrossed at least 13 generations into C57BL/6J genetic background. The Cited2-lacZ mouse line (Martinez-Barbera et al, 2002) was used for Cited2 expression studies.
Histology, lacZ staining, immunohistochemistry, immunofluorescence and electron microscopic analysis
LacZ staining was performed on sections from E9.25, E10.5 and E12.5 Cited2 heterozygous (Cited2+/lacZ) embryos as described (Bort et al, 2006). The same sections were double stained with antibodies against albumin or HNF4α. H&E-stained sections were used for histological analysis. Cell apoptosis was analyzed using a terminal deoxynucleotidyl transferase biotin-dUTP nick-end labeling (TUNEL) kit according to the manufacturer's manual (Chemicon), followed by immunostaining for vimentin and HNF4α. BrdU incorporation was detected by immunohistochemistry analysis using anti-BrdU-POD antibodies (clone BMG-6H8) according to the protocol provided by the manufacturer (Roche), and further stained by anti-albumin antibodies. Immunostaining of vimentin, albumin, HNF4α, E-cadherin, connexin 32, and PECAM-1 was performed using indirect antibody immunostaining procedures with appropriate fluorescence-labeled secondary antibodies. Frozen liver sections were stained with Oil red O (Sigma Chemical Co.). PAS staining was performed on paraffin-embedded sections according to the manufacturer's protocol (Sigma Chemical Co.). The TUNEL/vimentin/HNF4α, E-cadherin and connexin 32-stained sections were examined under a confocal microscope. Fetal livers were processed for resin embedment using standard procedures. The ultra-thin sections were examined under a JEOL 100CX transmission electron microscope.
Microarray analysis
Total RNA (15 μg) isolated from three each of wild-type and Cited2-null E14.5. fetal livers using Qiagen RNeasy kit (Valencia, CA) was used to prepare biotinylated cRNA according to the protocol described in Affymetrix Expression Analysis Technical Manual. We hybridized a total of six GeneChip Mouse Genome 430A 2.0 arrays (Affymetrix), three for controls and three for experimental samples. Images were acquired by using a GeneChip Scanner 3000 (Affymetrix). GENECHIP Operating Software (GCOS) from Affymetrix was used to analyze the data. The microarray data have been submitted to the ArrayExpress database under accession number E-MEXP-1240.
Northern blotting and real-time RT–PCR
Total RNAs from liver tissues were extracted by using Trizol reagent (Invitrogen) following the manufacturer's instructions. A 10-μg aliquot of each RNA sample was applied for NORTHERN blot analysis by hybridization with 32P-labeled cDNA probes (Cited2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) according to standard protocols. A 100-ng weight of total RNAs extracted from sorted wild-type and Cited2-null hepatocytes were reverse transcribed to cDNA using the SuperScript™ First-Strand Synthesis System (Invitrogen). Real-time PCR was performed with iQ™ SYBR Green Supermix (Bio-Rad) on an iCycler™ Thermal Cycler (Bio-Rad) according to the manufacturer's instructions. The expression levels of genes tested were normalized to the expression levels of a housekeeping gene HPRT.
Cell culture, transient transfection and luciferase assay
Primary hepatocytes were isolated and inoculated onto collagen-coated cell culture dishes at a cell density of 4 × 105 cells/cm2 as described (Matsui et al, 2002). Primary MEFs were isolated and cultured according to the standard protocol. Primary cells, HepG2, HeLa and HEK293 cells were transfected with FuGENE 6 Transfection Reagent (Roche) according to the manufacturer's protocol. The luciferase activity was assessed by using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol. As an internal control, SV40 promoter driven Renilla luciferase control vector (pRL-SV40) was cotransfected to normalize transfection efficiency. All experiments were performed in triplicate (mean±s.e.m.).
GST pull-down assay and western blot analysis
GST and GST fused-Cited2 proteins (GST-Cited2) were expressed in Escherichia coli (BL21, DE3) and purified according to the manufacturer's protocol (Amersham Biosciences). For GST pull-down assay, 20–50 μl of glutathione beads containing approximately 0.5 μg of GST or GST-Cited2 proteins in pull-down buffer (150 mM sodium acetate, 25 mM Hepes pH 7.2, 2 mM EDTA, 0.25% BSA, 0.1% Nonidet P-40, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride) were incubated with equal amounts of lysates from HEK293 cells transfected with Myc-HNF4α or vector alone (mock). After extensive washing, the bound proteins were eluted with 50 μl of 2 × SDS–PAGE loading buffer. The immunoprecipitated products were fractionated by SDS–PAGE and transferred to polyvinylidene difluoride membranes (PVDF-plus; Osmonics Inc.). After blocking with 1% BSA in Tris-buffered saline/0.1% Tween 20 (TBST), the membranes were incubated with anti-Myc monoclonal antibodies (A14; Santa Cruz Biotechnology). After washing, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies and followed by extensive washing with 1 × TBST, membranes were processed for enhanced chemiluminescence detection (ECL) (Amersham Pharmacia Biotech).
Mammalian two-hybrid system
Protein–protein interactions in intact cells were examined by mammalian two-hybrid analysis (Promega). In HEK293 cells, VP16-fused wild-type or mutant Cited2 (pVP16-Cited2, pVP16-Cited2 (1–199 aa) and pVP16-Cited2 (124–270 aa)) were cotransfected with GAL4 DNA-binding domain (BD)-fused HNF4α (pM-HNF4α) and reporter plasmid pG5-Luc, which contains five consensus GAL4-binding sites driving the expression of firefly luciferase reporter gene.
ChIP assay
ChIP assays were performed with fetal livers by using the Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology, CA) according to the manufacturer's protocol. Immunoprecipitation was performed with 2 μg of either anti-HNF4α (SC-6556; Santa Cruz Biotechnology) or anti-Cited2 (SC-21795; Santa Cruz Biotechnology) antibodies for each aliquot (1.2 ml). For each preparation, an additional mock immunoprecipitation with rabbit preimmune IgG (Santa Cruz Biotechnology) was performed in parallel. Immunoprecipitated chromatin was detected by PCR amplification. For quantitative ChIP, PCR amplification was real-time monitored and allowed to proceed in the exponential phase on an iCycler Thermal Cycler (Bio-Rad). Experimental quantitative ChIP–PCR values were normalized against values obtained by a standard curve constructed by input DNA with the same primer set.
Statistical analysis
Statistical analyses were performed using Student's t-test. P⩽0.05 was considered statistically significant.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Drs Iannis Talianidis and Michele Battle for providing us with the protocol for ChIP assay in fetal liver, and to Dr Michele Battle for sharing the microarray data of HNF4α fetal liver-specific knockout mouse. We thank Dr Hitoshi Shimano for providing HNF4α constructs, Ms Midori Hitomi for technical assistance with electron microscopic analysis, Dr Shi Gu for help in depositing microarray data and Dr Cheng-Ming Chiang for comments on the manuscript. This work was supported by NIH grants RO1-HL075436–02 (to YCY and MW), RO1-HL076919-07 (to YCY), and NHMRC grant 404805 (to SLD). SLD is a Pfizer Foundation Australia Senior Research Fellow.
References
- Bamforth SD, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, Kranc KR, Farza H, Henderson DJ, Hurst HC, Bhattacharya S (2001) Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet 29: 469–474 [DOI] [PubMed] [Google Scholar]
- Bamforth SD, Braganca J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, Brown NA (2004) Cited2 controls left–right patterning and heart development through a Nodal–Pitx2c pathway. Nat Genet 36: 1189–1196 [DOI] [PubMed] [Google Scholar]
- Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA (2006) Hepatocyte nuclear factor 4 {alpha} orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci USA 103: 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Mitchels CL, Leung MK, Arany ZP, Kung AL, Livingston DM (1999) Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev 13: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bort R, Signore M, Tremblay K, Martinez-Barbera JP, Zaret KS (2006) Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev Biol 290: 44–56 [DOI] [PubMed] [Google Scholar]
- Braganca J, Eloranta JJ, Bamforth SD, Ibbitt JC, Hurst HC, Bhattacharya S (2003) Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J Biol Chem 278: 16021–16029 [DOI] [PubMed] [Google Scholar]
- Chou YT, Wang H, Chen Y, Danielpour D, Yang YC (2006) Cited2 modulates TGF-β-mediated upregulation of MMP9. Oncogene 25: 5547–5560 [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AX, Wang X (2003) Transcription factors in liver development, differentiation, and regeneration. Hepatology 38: 1331–1347 [DOI] [PubMed] [Google Scholar]
- Duncan SA (2000) Transcriptional regulation of liver development. Dev Dyn 219: 131–142 [DOI] [PubMed] [Google Scholar]
- Duncan SA (2003) Mechanisms controlling early development of the liver. Mech Dev 120: 19–33 [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Rodriguez TA, Beddington RSP (1998) Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev 72: 27–40 [DOI] [PubMed] [Google Scholar]
- Ellard S, Colclough K (2006) Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum Mutat 27: 854–869 [DOI] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZY, Kung AL, France DS, Wagner G, Eck MJ (2003) Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2. Nat Struct Biol 10: 504–512 [DOI] [PubMed] [Google Scholar]
- Glenn DJ, Maurer RA (1999) MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone α-subunit gene expression. J Biol Chem 274: 36159–36167 [DOI] [PubMed] [Google Scholar]
- Hani EH, Suaud L, Boutin P, Chevre JC, Durand E, Philippi A, Demenais F, Vionnet N, Furuta H, Velho G, Bell GI, Laine B, Froguel P (1998) A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. J Clin Invest 101: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ (2001) Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Hayhurst GP, Inoue J, Mori M, Gonzalez FJ (2002) Defective ureagenesis in mice carrying a liver-specific disruption of hepatocyte nuclear factor 4α (HNF4α): HNF4α regulates ornithine transcarbamylase in vivo. J Biol Chem 277: 25257–25265 [DOI] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, Zaret KS (1999) Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science 284: 1998–2003 [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR (1998) Allosteric effects of DNA on transcriptional regulators. Nature 392: 885–888 [DOI] [PubMed] [Google Scholar]
- Lemaigre F, Zaret KS (2004) Liver development update: new embryo models, cell lineage control, and morphogenesis. Curr Opin Genet Dev 14: 582–590 [DOI] [PubMed] [Google Scholar]
- Leung MK, Jones T, Mitchels CL, Livingston DM, Bhattacharya S (1999) Molecular cloning and chromosomal localization of the human CITED2 gene encoding p35srj/Mrg1. Genomics 61: 307–313 [DOI] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA (2000) Mammalian hepatocyte differentiation requires the transcription factor HNF4α. Genes Dev 14: 464–474 [PMC free article] [PubMed] [Google Scholar]
- Martinez-Barbera JP, Rodriguez TA, Greene ND, Weninger WJ, Simeone A, Copp AJ, Beddington RSP, Dunwoodie SL (2002) Folic acid prevents exencephaly in Cited2 deficient mice. Hum Mol Genet 11: 283–293 [DOI] [PubMed] [Google Scholar]
- Matsui T, Kinoshita T, Morikawa Y, Tohya K, Katsuki M, Ito Y, Kamiya A, Miyajima A (2002) K-Ras mediates cytokine-induced formation of E-cadherin-based adherens junctions during liver development. EMBO J 21: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA (2003) Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34: 292–296 [DOI] [PubMed] [Google Scholar]
- Postic C, Dentin R, Girard J (2004) Role of the liver in the control of carbohydrate and lipid homeostasis. Diabetes Metab 30: 398–408 [DOI] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM (2003) Regulation of hepatic fasting response by PPARgamma coactivator-1 alpha (PGC-1): requirement for hepatocyte nuclear factor 4 alpha in gluconeogenesis. Proc Natl Acad Sci USA 100: 4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428 [DOI] [PubMed] [Google Scholar]
- Rossi JM, Dunn RN, Hogan BLM, Zaret KS (2001) Distinct mesodermal signals, including BMPs from septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev 15: 1998–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T, Fenner MH, Isselbacher KJ (1997) MSG1 and its related protein MRG1 share a transcription activating domain. Gene 204: 235–241 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Katrakili N, Talianidis I (2000) Acetylation regulates transcription factor activity at multiple levels. Mol Cell 5: 745–751 [DOI] [PubMed] [Google Scholar]
- Tien ES, Davis JW, Heuvel JP (2004) Identification of the CREB-binding protein/p300-interacting protein CITED2 as a peroxisome proliferator-activated receptors α coregulator. J Biol Chem 279: 24053–24063 [DOI] [PubMed] [Google Scholar]
- Watt AJ, Garrison WD, Duncan SA, Phil D (2003) HNF4: a central regulator of hepatocyte differentiation and function. Hepatology 37: 1249–1253 [DOI] [PubMed] [Google Scholar]
- Weninger WJ, Floro KL, Bennett MB, Withington SL, Preis JI, Barbera JP, Mohun TJ, Dunwoodie SL (2005) Cited2 is required both for heart morphogenesis and establishment of the left–right axis in mouse development. Development 132: 1337–1348 [DOI] [PubMed] [Google Scholar]
- Withington SL, Scott AN, Saunders DN, Lopes FK, Preis JI, Michalicek J, Maclean K, Sparrow DB, Barbera JP, Dunwoodie SL (2006) Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev Biol 294: 67–82 [DOI] [PubMed] [Google Scholar]
- Wu SY, Chiang CM (2007) The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282: 13141–13145 [DOI] [PubMed] [Google Scholar]
- Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, Yuan S, Prabhakar NR, Herrup K, Conlon RA, Hoit BD, Watanabe M, Yang YC (2002) The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci USA 99: 10488–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS (2002) Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet 3: 499–512 [DOI] [PubMed] [Google Scholar]
- Zhao R, Duncan SA (2005) Embryonic development of the liver. Hepatology 41: 956–967 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information