Abstract
Protein interacting with c kinase 1 (PICK1) regulates the trafficking of receptors and ion-channels such as AMPA receptors. Traditionally, the PICK1 PDZ domain is regarded as an adaptor capable of binding to receptors trafficked by PICK1, and the lipid-binding BAR domain functions to tether PICK1 directly to membranes. Here, we show that the PICK1 PDZ domain can directly interact with lipid membranes. The PDZ domain and lipid membrane interaction is mediated by both a polybasic amino-acid cluster and a conserved ‘Cys-Pro-Cys' motif located away from the peptide ligand-binding groove. Disruption of the PDZ and lipid membrane interaction totally abolished synaptic targeting of PICK1. Although mutation of the CPC motif did not affect the interaction between PICK1 and AMPA receptors, the mutant PICK1 was unable to cluster the GluR2 subunit of the receptor. In neurons, PICK1 containing the same mutation displayed dramatically compromised capacity in the trafficking of AMPA receptors. Taken together, our findings not only uncovered the novel lipid membrane-binding property of the PICK1 PDZ domain, but also provided direct evidence supporting the functional relevance of the PDZ–lipid interaction.
Keywords: AMPA receptor, membrane binding, PDZ domain, PICK1, protein trafficking
Introduction
Subcellular localization of proteins is crucial for their functions. Targeting proteins to specific subcellular domains is a complex process requiring sophisticated protein trafficking machinery as well as protein–lipid membrane interactions (Pfeffer, 2003). PICK1 (protein interacting with ckinase 1) is a peripheral membrane protein implicated in the trafficking of multiple proteins (Xu and Xia, 2007). PICK1 contains two major domains, an N-terminal PDZ (for PSD-95, Dlg, ZO-1) domain and a central BAR (for Bin, Amphiphysin, Rvs) domain. A number of proteins, most of them membrane proteins, have been reported to interact with the PDZ domain of PICK1. In most cases, PICK1 regulates the subcellular localization, the membrane surface expression, or both of its PDZ domain-binding partners (Xu and Xia, 2007).
One of the well-studied PICK1 PDZ-binding partners is GluR2, a subunit of the AMPA-type glutamate receptors that mediate the majority of excitatory synaptic transmission in the central nervous system. PICK1 was reported to change the subcellular localization of GluR2 by forming co-clusters, regulating the amount of GluR2 at plasma membranes and targeting GluR2 to synapses (Dev et al, 1999; Xia et al, 1999; Perez et al, 2001; Terashima et al, 2004; Jin et al, 2006). PICK1-regulated trafficking of GluR2 was found to be important in synaptic plasticity, which is the change of synaptic transmission strength and is believed to be the cellular basis of learning and memory (Malinow and Malenka, 2002). Disruption of PICK1 and GluR2 interaction by excessive GluR2 C-terminal peptide or PICK1 PDZ domain antibody inhibits the expression of synaptic plasticity (Matsuda et al, 2000; Xia et al, 2000; Kim et al, 2001). Evidence from PICK1-knockout mice, which had deficiencies in AMPA receptor trafficking and synaptic plasticity, further supported PICK1's indispensable role in these two processes (Gardner et al, 2005; Steinberg et al, 2006).
The central part of PICK1 is occupied by a BAR domain, a protein module frequently found in proteins involved in membrane trafficking. BAR domains, such as those from amphiphysin and endophilins, are known to form banana-shaped dimers capable of sensing and bending lipid membrane bilayers (Peter et al, 2004). The BAR domain of PICK1 was found to bind to liposomes containing phosphoinositides (Jin et al, 2006). Mutations of the critical lysine residues of the BAR domain eliminated PICK1's lipid binding, and consequently compromised PICK1's synaptic targeting, indicating that the BAR domain-mediated lipid binding of PICK1 is important for its subcellular localization. PICK1's lipid-binding capability is also critical for the trafficking of AMPA receptors and synaptic plasticity, as lipid-binding-deficient PICK1 could no longer cluster GluR2 and caused reduced synaptic localization of AMPA receptors. When perfused into neurons, lipid-binding-deficient PICK1 inhibits the expression of synaptic plasticity. The physiological significance of the BAR domain-mediated lipid binding was further confirmed in PICK1-knockout mice. Whereas the wild-type PICK1 rescued the deficiency in synaptic plasticity, lipid-binding-deficient PICK1 failed to do so (Steinberg et al, 2006).
As one of the most abundant protein modules in metazoan genomes, PDZ domains are generally viewed as protein–protein interaction modules. The most frequent interaction mode of PDZ domains is to recognize a short stretch of amino-acid residues at the extreme C-termini of target proteins, although other modes of interactions are also possible (Sheng and Sala, 2001; Zhang and Wang, 2003). Recently, PDZ domains were suggested to bind to phosphatidylinositol (PI) lipids (Zimmermann et al, 2002; Mortier et al, 2005; Yan et al, 2005). As many PDZ domain proteins are known to regulate the membrane surface expression and/or the clustering of diverse receptors and ion-channels (Craven and Bredt, 1998; Sheng and Sala, 2001), direct interactions between PDZ domains and lipid membranes are likely to be significant, as such interactions can not only regulate the membrane localization of PDZ domain proteins, but may also provide a potential mechanism for sensing phosphoinositide signaling. In contrast to a number of well-characterized PI lipid-binding modules such as PH, FYVE, FERM, and PX domains, the interaction mechanism between PDZ domains and lipid membranes is poorly understood. Additionally, evidence supporting the physiological significance of PDZ domain–lipid membrane interactions is still lacking.
In this work, we determined the three dimensional structure of PICK1 PDZ domain in complex with the carboxyl tail peptide of GluR2. The structure of the PDZ-GluR2 tail peptide complex not only provides mechanistic insights into the interaction between PICK1 and GluR2, but also explains why the PICK1–GluR2 interaction is not regulated by phosphorylation of Ser(−3) in the GluR2 tail. We further discovered that the PDZ domain of PICK1 can directly bind to lipid membranes containing phosphoinositides. We found that, in addition to several positively charged residues, a conserved ‘Cys-Pro-Cys' motif at the bottom of its peptide ligand-binding groove is absolutely required for PICK1 to bind to lipid membranes. We demonstrate that the PDZ domain-mediated lipid membrane binding of PICK1 is essential for synaptic targeting of PICK1 and PICK1-regulated trafficking of AMPA receptors.
Results and discussion
Structure of the PICK1–GluR2 tail peptide complex
The PDZ domain of PICK1 has unique target protein-binding properties, as it can recognize both the type I (e.g., the GluR2 tail peptide) and II (e.g., the mGluR5 tail peptide) peptide ligands (Madsen et al, 2005; Dev, 2007; Xu and Xia, 2007). Additionally, phosphorylation of Ser(−3) of the GluR2 tail disrupts the interaction between GluR2 and the PDZ45 tandem from GRIP1, but does not alter the interaction between GluR2 and PICK1 PDZ (Chung et al, 2000; Perez et al, 2001; Seidenman et al, 2003). To elucidate the molecular mechanism governing PICK1 PDZ's peptide ligand recognition, we sought to determine the solution structure of the ligand-free form of the PDZ domain by NMR. However, the ligand-free PICK1 PDZ was prone to aggregation and readily precipitated shortly after purification, indicating that the protein is not amenable for high-resolution structural determination by NMR spectroscopy. Addition of a synthetic peptide comprised of the last 9 amino-acid residues of GluR2 (the GluR2 peptide) improved the stability of the PDZ domain. But the 1H-15N HSQC spectrum of the complex displayed poor homogeneity, most likely due to the unfavorable chemical exchange between the free and the GluR2-peptide-bound forms of the PDZ domain (data not shown). To obtain a stable PICK1 PDZ–GluR2 peptide complex, we covalently linked the GluR2 peptide to the C-terminal end of the PICK1 PDZ domain with a peptide fragment consisting of the cleavage sequence of protease 3C. The purified fusion protein was monomeric and highly stable in solution (data not shown). The protein also displayed high quality 1H-15N HSQC spectrum suitable for NMR-based 3D structure determination. Cleavage of the fusion protein by protease 3C did not change the overall HSQC spectrum of the protein (Supplementary Figure 1), indicating that the covalent linker did not alter the conformation of the PDZ-GluR2 peptide complex.
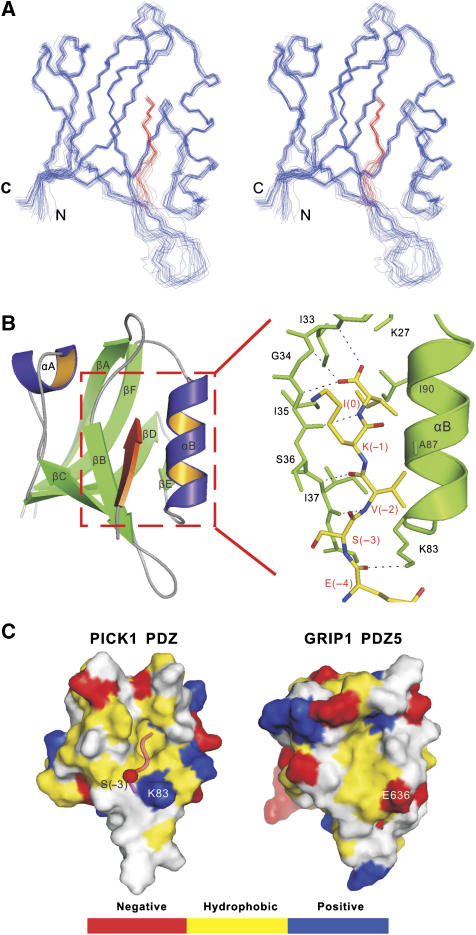
The structure of PICK1 PDZ-GluR2 peptide fusion protein was determined to high resolution by NMR spectroscopy (Figure 1A and B and Supplementary Table 1). PICK1 PDZ adopts a canonical PDZ domain fold consisting of a partially opened β-barrel with six β-strands (βA to βF) and two α-helices capping the opening sides of the β-barrel (Sheng and Sala, 2001; Zhang and Wang, 2003). The GluR2 peptide binds to the αB/βB-groove of PICK1 PDZ by augmenting the βB-strand of the PDZ domain in an antiparallel manner (Figure 1B). The side chain of Ile(0) of the GluR2 peptide inserts into the hydrophobic pocket at the end of the αB/βB groove. The carboxyl group of the peptide forms hydrogen bonds with the backbone amides of Ile33, Gly34, and Ile35 (the so-called ‘GLGF-motif' in PDZ domains) of PICK1 PDZ. The side chain of Lys(−1) makes minimal contact with the PDZ domain, explaining that the amino-acid residue in the −1 position in all known PICK1 PDZ-binding peptides is highly diverse (Madsen et al, 2005; Dev, 2007; Xu and Xia, 2007). As a type II PDZ ligand, the hydrophobic Val at the −2 position plays an important role in binding to PICK1 PDZ. The side chain of Val(−2) is sandwiched between the aliphatic side chain of Lys83 at the αB1 position and the methyl group of Ala87 one helical turn above Lys83 (Figure 1B). The PICK1 PDZ–GluR2 complex is further stabilized by the hydrogen bond between the amino group of Lys83 and backbone carbonyl of Glu(−4) in the GluR2 peptide, and charge–charge interaction between the side chains of Glu(−4) and Lys83. The PICK1 PDZ–GluR2 peptide complex structure determined here is similar to the crystal structure of the PICK1 PDZ in complex with an ephrin B1 tail peptide (amino-acid sequence of ‘−YYKV') from a structural genomic effort (Elkins et al, 2007).
Figure 1.
Structure of the PICK1–GluR2 tail peptide complex. (A) Stereo view showing the backbones of 20 superimposed NMR structures of PICK1 PDZ–GluR2 peptide complex. The structures are superimposed against the averaged structure using residues 18–40 and 47–103 of PICK1 PDZ including the last five residues of the GluR2 peptide. The GluR2 peptide is drawn in red. (B) Ribbon diagram of a representative NMR structure of the PICK1 PDZ–GluR2 peptide complex. The GluR2 peptide is shown in red. The secondary structures are labeled following the scheme of the canonical PDZ domains. The boxed region represents the GluR2 peptide-binding groove of the PDZ domain, and the detailed interaction between the GluR2 peptide and the PDZ domain is drawn in explicit atomic model. (C) Comparison of the surface structure PICK1 PDZ with that of GRIP1 PDZ5 (PDB code 1P1D). In this presentation, the hydrophobic amino-acid residues are drawn in yellow, the positively charged residues in blue, the negatively charged residues in red, and the uncharged polar residues in gray. The GluR2 peptide in the PICK1 PDZ–GluR2 complex is shown in worm model.
The GluR2 subunit is known to interact with PDZ domains from PICK1 and GRIP1 (also known as ABP) (Dong et al, 1997; Xia et al, 1999). The interaction of GluR2 with the 5th PDZ domain of GRIP1 is known to be disrupted by phosphorylation of Ser(−3) of GluR2 tail by PKC. In contrast, the interaction between GluR2 and PICK1 PDZ is not sensitive to the phosphorylation of Ser(−3) (Chung et al, 2000; Perez et al, 2001). Comparison of the 3D structure of GRIP1 PDZ5 (Feng et al, 2003) with that of PICK1 PDZ provides a mechanistic explanation for the above difference (Figure 1C). In the PICK1 PDZ–GluR2 peptide complex, the side chain of Ser(−3) is solvent exposed and in the vicinity of Lys83 of PICK1 PDZ. Phosphorylation of Ser(−3) of GluR2 may even strengthen the interaction between GluR2 and the PICK1 PDZ domain by adding a favorable charge–charge interaction mediated by phosphor-Ser(−3) and Lys83. In contrast, the amino-acid residue in GRIP1 PDZ5, corresponding to the position of Lys83 in PICK1 PDZ, is a negatively charged Glu (Glu636 in rat GRIP1). Phosphorylation of Ser(−3) of GluR2 is expected to introduce unfavorable charge–charge repulsions between the GluR2 tail and GRIP1 PDZ5.
The PDZ domain of PICK1 binds to phosphoinositol-lipid membranes
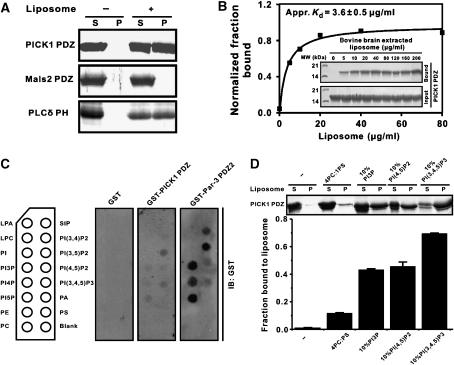
As some PDZ domains were recently shown to bind to lipid membranes (Zimmermann et al, 2002; Mortier et al, 2005; Yan et al, 2005), we tested the possibility whether the PDZ domain of PICK1 can directly bind to lipid membranes. We used the PICK1 PDZ–GluR2 peptide fusion protein for sedimentation-based membrane-binding assay, as the isolated PICK1 PDZ formed aggregates and interfered with the assay. Indeed, the PICK1 PDZ domain binds to liposomes prepared from total bovine brain lipids, and the binding capacity of PICK1 PDZ to brain liposomes is comparable to that of the PH domain from phospholipase Cδ (Figure 2A). An unrelated PDZ domain from rat Mals2 (also known as Lin-7b) showed no detectable liposome binding, demonstrating that the lipid binding of PICK1 PDZ is specific. We further showed that PICK1 PDZ binds to brain liposomes in a concentration-dependent manner (Figure 2B). To rule out the possibility that the lipid binding of PICK1 PDZ was an artifact of the fusion of the GluR2 peptide to the PDZ domain, we prepared a B1 domain of streptococcal protein G (GB1)-fused PICK1 PDZ domain. The GB1-PDZ sample was stable and highly soluble, and therefore suitable for the centrifugation-based membrane-binding assay (Supplementary Figure 2). The peptide ligand-free form of PICK1 also binds robustly to lipid membranes. The lipid-binding specificity of PICK1 PDZ was probed using membrane-immobilized lipid strips. Of the 15 lipids tested, PICK1 PDZ only weakly recognized PI lipids (Figure 2C). However, the stereo-specificity among different PIPs is not high. Finally, we repeated assays of the binding between PICK1 PDZ and synthetic phosphatidylcholine (PC)/phosphatidylserine (PS) liposomes containing 10% of selected PIPs. In agreement with the brain liposome sedimentation- and lipid strip-based assays, PICK1 PDZ robustly binds to synthetic liposomes containing PI3P, PI(4,5)P2, and PI(3,4,5)P3 (Figure 2D). Although PICK1 PDZ binds to PI(3,4,5)P3 with somewhat higher affinity than to PI(4,5)P2, the much higher concentration of PI(4,5)P2 in living cells may offset this binding-affinity difference. The relatively weak binding between PICK1 PDZ and immobilized PIPs (rather than PIPs embedded in the membrane bilayers) indicated that both the head groups of PIPs and the membrane bilayers are required for binding to the PICK1 PDZ domain. Taken together, the above data clearly demonstrate that PICK1 PDZ binds to membrane bilayers containing PIPs.
Figure 2.
The PDZ domain of PICK1 binds to phosphoinositol-lipid membranes. (A) Sedimentation assay of bindings of PICK1 PDZ domain with liposomes prepared from bovine brain lipid extracts. Fractions labeled with ‘S' and ‘P' represent proteins present in supernatants and pellets after centrifugation. In this assay, the well-characterized PH domain from rat PLCδ served as the positive lipid binder, and an unrelated PDZ domain from mouse Mals2 was used as a negative control. (B) Dose-dependent interaction of PICK1 PDZ with the bovine brain liposomes (liposome concentrates were varied from 0 to 200 μg/ml in each reaction). An apparent binding affinity of PICK1 PDZ binding to bovine brain liposomes was derived by fitting the dose-dependent binding curve (Kd∼3.6±0.5 μg/ml). (C) The lipid strip-based binding assay of PICK1 PDZ and the Par-3 PDZ2 domain, respectively. The data also indicate that the Par-3 PDZ2 domain has higher PIP-binding affinity than PICK1 PDZ. (D) Interaction of PICK1 PDZ with various PIPs reconstituted into defined PC/PS liposomes. Values are mean±s.d. of three different experiments. Note that the concentration of PIPs was kept at 10% to mimic the total PIP concentration found in plasma membranes.
Characterization of the PICK1 PDZ lipid membrane interaction
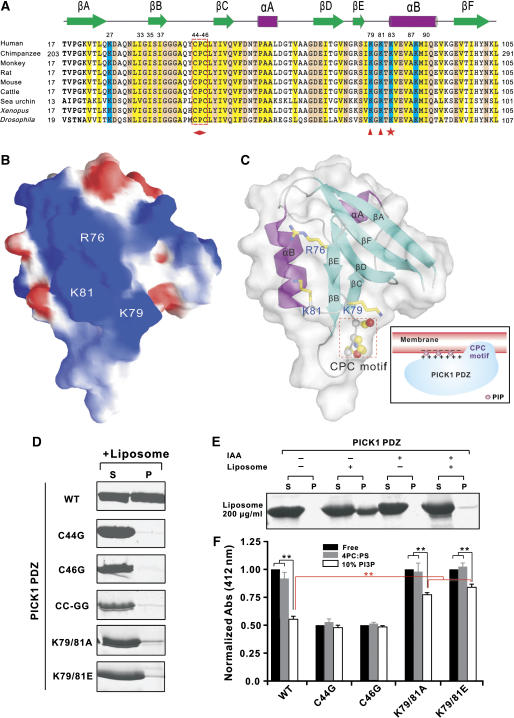
Analysis of the surface charge properties of the PICK1 PDZ structure showed that the protein contains a continuous patch of positively charged residues composed of Arg76, Lys79, and Lys81 at the opposite side of the peptide ligand groove (Figure 3A and B). We reasoned that this positively charged surface is likely to play a role in binding to negatively charged lipid membrane surfaces. To test this hypothesis, we substituted two of these positively charged residues (Lys79 and Lys81 in the βE/αB-loop) with neutral Ala or with negatively charged Glu and tested the lipid-binding capacities of these mutants, and found that these led to near complete abolishment of the lipid binding (Figure 3D). We conclude that the clustered positively charged residues in PICK1 PDZ play a critical role in binding to negatively charged phospholipid membrane surfaces.
Figure 3.
Characterization of the interaction between PICK1 PDZ and lipid membrane. (A) Structure-based sequence alignment of PICK1 PDZ from different species. Conserved hydrophobic residues are shown in yellow, positively charged residues in blue and the rest of the highly conserved residues in pink. The residues forming the positive charge cluster important for membrane interaction between βE and αB are highlighted with red triangles. The Lys residue in the αB1 position is highlighted with a red star. The absolutely conserved CPC motif is boxed in red and further highlighted with a diamond shape at the bottom. (B) Surface charge representation of PICK1 PDZ. In this diagram, the positive charge potential is drawn in blue, and the negative charge potential is in red. The positions of Arg76, Lys79, and Lys81 are labeled. (C) Combined surface and ribbon diagram representations of the PICK1 PDZ illustrating the position of the positive charge cluster and the orientation of the CPC motif. (D) Analysis of the roles of the positively charged residues (Lys79&81) and CPC motif (Cys44&46) in lipid membrane binding by sedimentation-based lipid-binding assay. Lanes labeled with ‘S' and ‘P' stand for proteins present in supernatants and pellets after centrifugation. (E) Centrifugation-based lipid membrane-binding assay of iodoacetic acid modified PICK1 PDZ. (F) The Ellman assay-based measurements of the solvent accessibilities of sulfhydryl groups in the wild-type PICK1 PDZ and its various Cys to Gly substitution mutants. The absorption values are normalized to the value of the wild-type PICK1 PDZ without addition of liposomes. The data represent mean±s.d. of three different experiments. The stars indicate the significant differences between the indicated bars (**P<0.01).
Efficient membrane bindings of a number of protein domains or polybasic peptide fragments are known to require additional structural elements (often hydrophobic residues) in addition to positively charged amino acids (DiNitto et al, 2003; McLaughlin and Murray, 2005; Heo et al, 2006). Careful analysis of the PICK1 PDZ domain structure revealed that the protein contains an absolutely conserved ‘Cys44-Pro45-Cys46' motif in the βB/βC-loop (Figure 3A and C). This hydrophobic CPC motif is solvent accessible and in the vicinity of the positively charged surface comprising of Arg76, Lys79, and Lys81 (Figure 3C). It is known that the P1B-type family ATPases also contain a CPC motif, and this CPC motif is directly embedded in the membrane bilayers (Arguello, 2003). We reasoned that the hydrophobic CPC motif may function together with the neighboring positive charges in mediating lipid membrane binding of PICK1 PDZ. To test this hypothesis, we chemically modified the two Cys residues (Cys44 and Cys46, the only two Cys in PICK1 PDZ) using iodoacetic acid. Conversion of Cys to S-carboxymethylcysteine completely abolished lipid membrane binding (Figure 3E), indicating that the Cys residues are critical for PICK1 PDZ–membrane interaction. We further demonstrated that both Cys residues are important for the membrane binding, as substitutions of either Cys with a Gly abolished the lipid membrane binding of the PDZ domain (Figure 3D). As expected, substitutions of both Cys residues with Gly also eliminated the membrane binding of the domain. NMR experiments showed that substitution of Cys residues with Gly introduced chemical shift changes limited to the CPC-motif-containing loop of the PDZ domain, indicating the mutations did not alter its overall conformation (Supplementary Figure 3A and B). Consistent with the above structural data, ligand-binding experiments also showed that the Cys to Gly mutations of PICK1 PDZ did not change the GluR2 peptide-binding affinity of the domain (Supplementary Figure 4).
It is possible that the hydrophobic CPC motif facilitates PICK1 PDZ membrane binding by directly inserting into membrane bilayers. To test this hypothesis, we probed the Cys accessibility of PICK1 PDZ using the Ellman's Reagent (5,5′-dithiobis-(2-nitrobenzoic acid, DTNB). Without addition of liposomes, the two Cys residues can be quantitatively modified by DTNB (Figure 3F). Addition of reconstituted PC/PS (4:1 ratio) liposomes to PICK1 PDZ has limited impact on the surface accessibility of the Cys residues, and this is consistent with the very weak basal level interaction of the PC/PS liposomes with the PDZ domain (Figure 2C). In contrast, binding of PC/PS/PI(4,5)P2 (70/20/10%) liposomes to PICK1 PDZ significantly reduced the Cys accessibility, presumably due to the insertion of the Cys residues into the membrane bilayers. In agreement with the centrifugation-based liposome-binding assay, mutation of either Cys44 or Cys46 to Gly caused complete accessibility of the remaining Cys in the mutant proteins. Mutation of Lys79&81 also increased the Cys accessibility of PICK1 PDZ, although the increase is not as significant as the Cys mutations. The increased Cys accessibility of the Lys79&81 mutant of PICK1 PDZ correlates well with the weakened liposome binding of the mutants shown in Figure 3D. Summarizing all structural and biochemical data above, a schematic model is depicted to describe the interaction of PICK1 PDZ with PIP-containing lipid membranes (Figure 3C). In this model, the positively charged surface of PICK1 PDZ interacts with negatively charged membranes nonspecifically. Presence of PIPs in membranes increases the negative charge density of the membranes, thereby increasing the binding affinity of PICK1 PDZ to the membranes (McLaughlin and Murray, 2005; Heo et al, 2006). In addition to the polybasic residues, the hydrophobic CPC motif directly penetrates into the hydrophobic core of the membrane bilayers, although the molecular mechanism of the CPC-motif-mediated membrane insertion seen in PICK1 PDZ may not be the same as that in the P1B-ATPases. Increasing examples demonstrate that combined interactions of hydrophobic residues and polybasic cluster from proteins with membrane bilayers are a common feature for membrane association of a diverse range of proteins (DiNitto et al, 2003; Lemmon, 2003; McLaughlin and Murray, 2005; Heo et al, 2006). In the case of PICK1 PDZ domain, the DTNB-based Cys accessibility assay suggested that the CPC motif plays an even more critical role than the positively charged residues in the PICK1 PDZ and lipid membrane interaction.
The PDZ domain-mediated lipid membrane binding regulates subcellular localization of PICK1
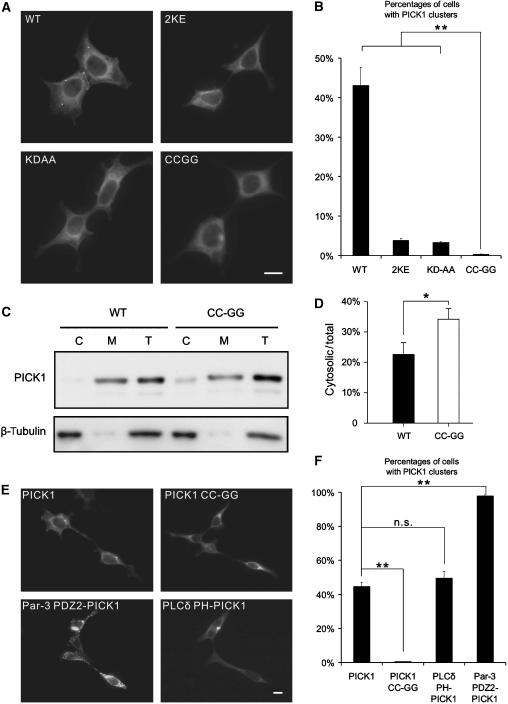
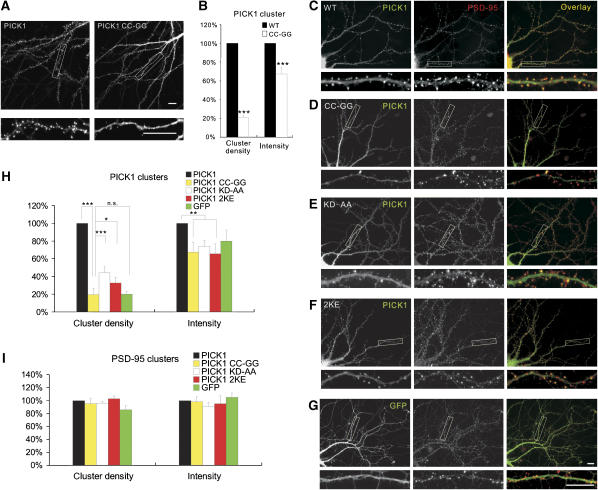
Binding to lipid membranes is critical for PICK1 to regulate protein trafficking (Xu and Xia, 2007). In addition to the BAR domain, our biochemical data demonstrated that the PDZ domain of PICK1 also functions as a lipid membrane-binding domain. Several experiments were designed to test the physiological roles of the PICK1 PDZ's lipid membrane-binding capacity. As the CPC motif is absolutely required for PICK1 PDZ to bind to membranes, we created a PICK1 mutant where both Cys residues in its PDZ domain were substituted with Gly (referred to as PICK1 CC-GG). Compared to the wild-type full-length PICK1, the PICK1 CC-GG mutant displayed significantly diminished lipid membrane binding (Supplementary Figure 5A and B). We also constructed a Lys79&81 to Glu mutant of PICK1, and this mutant has a modest impact on lipid membrane binding (data not shown). As substitutions of the two Cys residues with Gly do not change the peptide ligand-binding property of the PICK1 PDZ domain, we reasoned that PICK1 CC-GG is an ideal lipid-binding-deficient mutant to study the cellular role of the PDZ–lipid membrane interaction of the protein. First, we investigated whether the CC-GG mutation affects PICK1's subcellular localization in human embryonic kidney 293T (HEK293T) cells. Wild-type PICK1 formed some small clusters in the cytosol, as previously reported (Jin et al, 2006). The PICK1 CC-GG mutant, on the other hand, was mainly diffused in the perinuclear regions. The quantification results indicated that the wild-type PICK1 formed clusters in 43±4.6% of transfected cells. In contrast, the CC-GG mutant formed clusters in only 0.35±0.1% of cells (Figure 4A and B, n=3, P<0.01 comparing WT PICK1 and PICK1 CC-GG). We performed a subcellular fractionation experiment to assess the effect of the CC-GG mutation in membrane association of PICK1. As expected, the majority of the wild-type PICK1 is associated with membranes. Mutation of the two Cys residues in the CPC motif significantly increased cytosolic fraction of PICK1 (Figure 4C and D). We also compared the subcellular distribution of the PICK1 CC-GG mutant to PICK1 2KE, a PICK1 lipid-binding-deficient mutant with mutations in its BAR domain (Jin et al, 2006). Consistent with our earlier report, PICK1 2KE formed clusters in about 3.8±0.48% of cells, a percentage significantly lower than the wild-type PICK1, but still considerably higher than the PICK1 CC-GG-transfected cells (Figure 4A and B, P<0.01 comparing PICK1 CC-GG to PICK1 2KE). We also measured the clustering capacity of PICK1 KD-AA, another PICK1 mutant with deficiency in its PDZ domain-mediated peptide ligand binding (Xia et al, 1999). PICK1 KD-AA mutant also has reduced clustering, likely due to the loss of interactions with membrane proteins. However, PICK1 KD-AA still has significantly more clusters than PICK1 CC-GG (Figure 4A and B, PICK1 KD-AA=3.3±0.23%, P<0.01 comparing PICK1 CC-GG to PICK1 KD-AA). The PICK1 CC-GG mutant has the least number of clusters among all the PICK1 mutants tested here and those reported in the literature. The above data suggest that the PDZ domain-mediated lipid membrane binding is a fundamental determinant of PICK1's self-clustering and its association with membrane structures.
Figure 4.
The PDZ domain-mediated lipid membrane binding regulates subcellular localization of PICK1. (A) WT PICK1 was shown to form clusters in cells, but not the PICK1 mutants. Scale bar=10 μm. (B) Percentage of clustered cell was calculated by counting the number of cells that had at least one cluster divided by total number of transfected cells. Results were averaged from three independent experiments. At least 2000 cells were quantified for each experiment. Error bars represent s.e.m. (C) Subcellular fractionation of PICK1 and PICK1 CC-GG. HEK293T cells transfected with GFP-PICK1 or GFP-PICK1 CC-GG were lysed. The total ‘T', cytosolic ‘C', and membrane ‘M' fractions were prepared for both proteins. Equal amounts of proteins were analyzed by SDS-PAGE and detected by Western blot using the anti-GFP antibody or anti-β-tubulin antibody. (D) The cytosolic/total ratios of PICK1 and PICK1 CC-GG were quantified. There are significant more PICK1 CC-GG in cytosolic fraction (33.54±3.9% of total) compared to the WT PICK1 (22.38%±4.1 of total). N=4, *P<0.05. (E) WT PICK1 and PLCδ PH-PICK1 formed some clusters in cells, whereas Par-3 PDZ2-PICK1 formed numerous clusters. Scale bar=10 μm. (F) Quantification results showed that the cell numbers with PICK1 clusters were similar between the group transfected with PLCδ PH-PICK1 and the wild-type PICK1 (n=3 P>0.05). Par-3 PDZ2-PICK1 formed significant higher percentage of clusters in transfected cells than the wild-type PICK1 (n=3, **P<0.01).
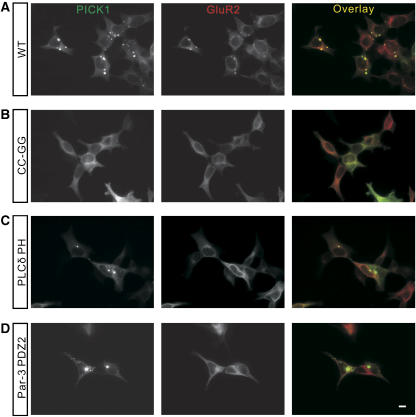
If the CC-GG mutation-mediated disruption of the PDZ domain's lipid membrane binding is solely responsible for the loss of PICK1's self-clustering, restoring of the PICK1 PDZ's lipid binding should rescue its self-clustering. To test the above prediction, we created a PICK1 chimera by replacing its PDZ domain with the PH domain of PLCδ, which is a well-characterized lipid-binding protein module (Lemmon and Ferguson, 2000). We transfected PLCδ PH-PICK1 into HEK293T cells and found that the chimera PICK1 formed clusters in 49.6±4.1% of transfected cells, a number similar to the wild-type PICK1 (44.8±2.4%), supporting our conclusion that the PDZ domain-mediated membrane binding is essential for the formation of PICK1 clusters (Figure 4E). To consolidate this conclusion further, we replaced the PDZ domain of PICK1 with the second PDZ domain of Par-3 (referred to as Par-3 PDZ2-PICK1). The Par-3 PDZ2 was recently discovered to bind to PIP lipid-containing membranes with higher affinities than the PDZ domain of PICK1 (Figure 2C and our unpublished results). However, the Par-3 PDZ2 and the PICK1 PDZ share no overlap in their peptide ligand binding (H Wu and M Zhang, unpublished data). As shown in Figure 4F, Par-3 PDZ2-PICK1 formed even higher percentage of clusters in transfected cells (97.9±0.6% for Par3 PDZ2-PICK1 versus 44.8±2.4% for the wild-type PICK1), perhaps due to the higher lipid-binding affinity of the Par-3 PDZ2. Taken together, the above chimera data strongly suggest that the PDZ domain-mediated lipid binding is absolutely required for PICK1 to form clusters in heterologous cells.
In neurons, PICK1 is localized at synapses. We have previously reported that the lipid-binding capacity of the PICK1's BAR domain is critical for its synaptic targeting. We wondered whether the PDZ domain-mediated lipid membrane binding may also be involved in the synaptic targeting of PICK1. To test this, we transfected PICK1 CC-GG to cultured hippocampal neurons and compared its localization with the wild-type PICK1. PICK1 CC-GG is completely absent from dendritic spines. This is in stark contrast to the wild-type PICK1, which is highly enriched at synapses (Figure 5A). Quantification results indicate that both the number and intensity of PICK1 synaptic clusters were drastically reduced in the PICK1 CC-GG mutant (Figure 5B, the wild-type PICK1 is normalized to 100% for the cluster density of dendrite, PICK1 CC-GG=20.8±2.8%, for intensity, PICK1 CC-GG=67.3±7.0%, n=30, P<0.001).
Figure 5.
PICK1 CC-GG mutant has minimal synaptic targeting capability. (A) The wild-type PICK1 is highly clustered at synapses but not the PICK1 CC-GG mutant. Scale bar=10 μm. (B) Quantification results indicated that both the density of dendrite and intensity of PICK1 synaptic clusters were significantly reduced in the PICK1 CC-GG mutant. (n=30, ***P<0.001). To confirm that the trasfection of PICK1 CC-GG does not affect synapse formation of neurons, hippocampal neurons were infected with GFP-PICK1 (C), GFP-PICK1 CC-GG (D), GFP-PICK1 KD-AA (E), GFP-PICK1 2KE (F) and GFP alone (G) and stained by antibody against PSD-95. Wild-type PICK1 is highly clustered and colocalized with PSD-95 well. GFP-PICK1 KD-AA and 2KE mutants were shown to form less clusters than the wild-type PICK1. PICK1 CC-GG and GFP are missing from PSD-95 clusters compared to the wild-type PICK1, KD-AA, and 2KE mutants. Scale bar=10 μm. (H) Quantification results indicated PICK1 CC-GG formed significantly less clusters numbers than PICK1 KD-AA (n=10, *P<0.05) and 2KE mutants (n=10, ***P<0.001), but no difference between PICK1 CC-GG and GFP vector alone (n=10, P>0.05). Intensity of PICK1 synaptic clusters were also significantly reduced in the PICK1 CC-GG, KD-AA, and 2KE mutant (n=10, **P<0.01). (I) GFP-PICK CC-GG did not significantly change the number or intensity of PSD-95 clusters (n=10, P>0.05).
The disappearance of PICK1 CC-GG clusters is likely due to the failure of PICK1 CC-GG to localize at synapses. Alternatively, it may simply result from the elimination of synapses by the mutant. To exclude the later possibility, we examined the effect of PICK1 CC-GG on the number of synapses by staining neurons with PSD-95, a well-characterized marker of excitatory synapses. As shown in Figure 5C, neurons transfected with the wild-type PICK1 formed numerous clusters and these clusters colocalized well with PSD-95, indicating that the PICK1 clusters are synaptic. In PICK1 CC-GG-transfected neurons, PSD-95 clusters were essentially the same as those from neurons transfected with the wild-type PICK1 (Figure 5C, D, H and I, wild-type PICK1 was normalized to 100% for the PSD-95 cluster density of dendrite, PICK1 CC-GG=98.1±8.4%, P>0.05). Therefore, the PICK1 CC-GG mutation does not affect formation of synapses.
We also compared the synaptic targeting of PICK1 CC-GG to that of PICK1 2KE and PICK1 KD-AA, two mutants previously reported to have reduced synaptic localization (Jin et al, 2006). We found that PICK1 CC-GG showed significantly less clustering than either PICK1 2KE or PICK1 KD-AA (Figure 5D–F, H and I; wild-type PICK1 was normalized as 100% for PICK1 cluster density, PICK1 CC-GG=19.5±6.8%, PICK1 KD-AA=45.5±6.4%, PICK1 2KE=32.8±6.0, n=10, P<0.001 comparing PICK1 CC-GG to PICK1 KD-AA and P<0.05 comparing PICK1 CC-GG to PICK1 2KE). Finally, we compared the synaptic targeting of PICK1 CC-GG with the GFP vector alone and found that there is no difference between PICK1 CC-GG and GFP (Figure 5D and G–I; for PICK1 cluster density, PICK1 CC-GG=19.5±6.8 versus GFP=20.1±2.9%), indicating that PICK1 CC-GG has zero preference for synaptic localization. The above results indicate that compared to the BAR domain-mediated lipid binding and the PDZ domain–peptide ligand interactions the PDZ domain-mediated lipid binding plays a dominant role in the synaptic localization of PICK1.
The lipid-binding property of the PDZ domain is required for PICK1-mediated clustering and synaptic targeting of AMPA receptors
PICK1 clusters and targets AMPA receptors to synapses (Xia et al, 1999; Jin et al, 2006). To investigate the function of the PICK1 PDZ's lipid-binding capacity in AMPA receptor trafficking, we first examined whether the CC-GG mutation of PICK1 could affect PICK1's capability in clustering AMPA receptors. When the wild-type PICK1 was transfected to HKE293T cells together with the AMPA receptor subunit GluR2, they form numerous co-clusters (Figure 6A). In contrast, when PICK1 CC-GG and GluR2 were co-transfected into 293T cells, no co-clusters could be found at all, indicating that the PDZ domain-mediated lipid binding is required for PICK1-induced co-clustering of AMPA receptors (Figure 6B). As expected, the PICK1-mediated AMPA receptor clustering also requires direct interaction of the PICK1 PDZ domain with the GluR2 tail, as neither PLCδ PH-PICK1 nor Par-3 PDZ2-PICK1 could restore the clustering of AMPA receptors (Figure 6C and D). As both PLCδ PH-PICK1 and Par-3 PDZ2-PICK1 formed self-clusters efficiently, we concluded that the lipid-binding and peptide ligand-binding properties of the PICK1 PDZ domain can be dissociated. Mutations of PICK1 that can specifically disrupt either of the two functions are likely to be valuable for future mechanistic dissections of PICK1-mediated protein trafficking.
Figure 6.
Lipid binding of PICK1's PDZ domain is required for PICK1-mediated clustering of GluR2. The wild-type PICK1 and GluR2 formed co-clusters in the cells (A), but the PICK1 CC-GG mutant did not form any clusters with GluR2 (B). PLCδ PH-PICK1 (C) and Par-3 PDZ2-PICK1 (D) formed many self-clusters, but none of these clusters contain GluR2. Scale bar=10 μm.
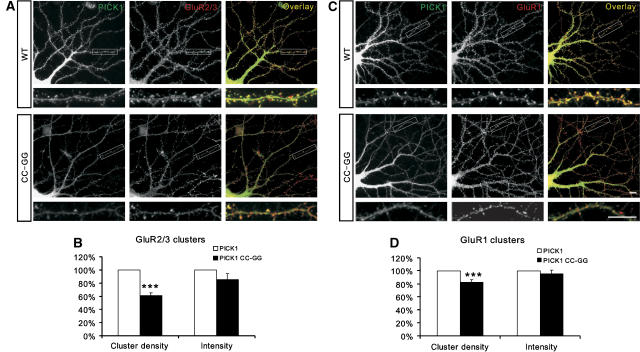
Synaptic targeting of AMPA receptors is critical for neuronal plasticity. Both the PDZ domain and the BAR domain have been reported to be involved in the PICK1-mediated synaptic targeting of AMPA receptors (Jin et al, 2006). To determine specifically the role of the PICK1 PDZ domain's lipid binding in synaptic targeting of AMPA receptors, we transfected both the wild-type and the CC-GG mutant of PICK1 into neurons, respectively, and then compared the synaptic localization of the AMPA receptor subunit GluR2/3. As expected, the wild-type PICK1 colocalized well with GluR2/3 at synapses. In contrast, PICK1 CC-GG-transfected neurons had much less synaptic GluR2/3 (Figure 7A). Quantification results indicated that both the intensity and number of GluR2 clusters were significantly reduced in PICK1 CC-GG transfected neurons (Figure 7B, for GluR2/3 cluster density, PICK1 wild-type was normalized to 100%, PICK1 CC-GG=61.0±4.1%, n=13, P<0.001). Although the majority of GluR1 are in complex with GluR2 in neurons, it has been reported that GluR1 may traffic differently than GluR2 (Shi et al, 2001). To test whether the lipid binding of the PICK1 PDZ domain may have different effect on the GluR1 and GluR2 trafficking, we examined the synaptic localization of GluR1 in PICK1 CC-GG-transfected neurons. We found that the PICK1 CC-GG mutant also reduced the number of GluR1 clusters at synapses (Figure 7C and D, for GluR1 cluster density, PICK1 wild-type was normalized to 100%, PICK1 CC-GG=82.3±3.4%, n=12, P<0.001). Taken together, our data strongly indicate that the lipid-binding capacity of the PICK1 PDZ domain is critical for synaptic targeting of AMPA receptors.
Figure 7.
Lipid binding of PICK1's PDZ domain is critical for synaptic targeting of AMPA receptors. (A) The GluR2/3 clusters colocalized with the PICK1 clusters at synapses. Neurons infected with the wild-type PICK1 had substantial more synaptic clusters of GluR2/3 comparing to neurons infected with the PICK1 CC-GG mutant. (B) The intensity and density of the GluR2/3 clusters were quantified form multiple experiments. The number of the GluR2 clusters form GFP-PICK1-infected neurons were significantly higher than neurons infected with the GFP-PICK1 CC-GG mutant (n=13, ***P<0.001). (C) The wild-type PICK1-infected neurons had more GluR1 clusters compared with the PICK1 CC-GG mutant-infected neurons. Scale bar=10 μm. (D) Quantification result showed that the density of GluR1 clusters was significantly higher in wild-type PICK1-infected neurons than PICK1 CC-GG mutant infected neurons (n=12, ***P<0.001).
In summary, the structure of the PICK1 PDZ-GluR2 peptide complex determined in this work not only reveals the recognition mechanism of the PICK1 PDZ domain for the GluR2 AMPA receptor subunit, but also provides an explanation as to why the bindings of PICK1 and GRIP1 to GluR2 are differentially regulated by the phosphorylation of the receptor subunit at Ser880. We discovered that, in addition to binding to well-documented peptide ligands, the PICK1 PDZ domain is capable of binding directly to lipid membranes. The interaction between PICK1 PDZ and membrane bilayers requires both the conserved, hydrophobic CPC motif and a polybasic cluster neighboring the CPC motif in the domain. We further demonstrated that the PDZ domain-mediated membrane binding plays a dominant role for both self-clustering of PICK1 and PICK1-mediated synaptic trafficking of AMPA receptors. The data presented in this work also provide direct evidence to support the notion that PDZ domain–lipid membrane interaction may play diverse cellular functions.
Materials and methods
Protein purification
The coding sequences of the PICK1 PDZ domain (18–110), the PDZ domain of Mals2 (94–173) and the PH domain of PLCδ (22–132) were PCR amplified from the full-length rat PICK1, mouse Mals2 and rat PLCδ cDNAs, respectively, and cloned into a modified pET32a vectors (Long et al, 2003). For the PICK1 PDZ-3C-GluR2 construct, the C-terminal 9 amino acid (VYGIESVKI) was linked to the PDZ domain with a protease 3C cleavage site. The full-length PICK1 protein was expressed with an N-terminal maltose-binding protein tag. The PICK1 PDZ domain and its mutants were expressed in N-terminal GB1-fused or His6-tagged forms. Recombinant proteins were expressed in BL21 (DE3) Escherichia coli cells and purified using Ni2+-nitrilotriacetic acid agarose affinity chromatography followed by size-exclusion chromatography. Uniformly isotope-labeled PICK1 PDZ were prepared by growing bacteria in M9 minimal medium using 15NH4Cl as the sole nitrogen source or 15NH4Cl and 13C6-glucose (Cambridge Isotope Laboratories Inc.) as the sole nitrogen and carbon sources, respectively.
NMR spectroscopy
The protein samples for NMR studies were concentrated to ∼0.2 mM for titration experiments and ∼1.0 mM for structural determinations in 100 mM potassium phosphate at pH 6.5. NMR spectra were acquired at 30°C on Varian Inova 500 or 750 MHz spectrometers. Backbone and side-chain resonance assignments were achieved by combination of standard heteronuclear correlation experiments including HNCO, HNCACB, CBCA(CO)NH, and HCCH-TOCSY using 15N/13C-labeled protein samples, and 1H 2D TOCSY and NOESY experiments (Wüthrich, 1986; Bax and Grzesiek, 1993). Approximate interproton distance restraints were derived from 2D 1H-NOESY, 3D 15N-seperated NOESY, and 3D 13C-seperated NOESY spectra, and followed the standard methods as we have used earlier (e.g., see Long et al, 2005). Structures were calculated using the program CNS (Daniels et al, 1998). The figures were prepared using the programs MOLSCRIPT (Kraulis, 1991), PyMOL (http://pymol.sourceforge.net/), MOLMOL (Koradi et al, 1996), and GRASP (Nicholls, 1992).
Liposome preparation and sedimentation assay
Interactions between proteins and liposomes prepared from total bovine brain lipid extracts (Folch fraction I, Sigma B1502) were assayed as described earlier by Yan et al (2005). Interactions of PICK1 PDZ with various PIP lipids immobilized on membrane strips (Echelon Biosciences, P-6001) also followed our earlier method (Yan et al, 2005). Briefly, each protein sample (∼10 μg) was incubated with 1 mg/ml liposomes in 40 μl of assay buffer (40 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM DTT) for 15 min at room temperature. The mixture was then spun at 80 000 g for 15 min at 4°C in a Beckman TLA100.1 rotor. Proteins existing in the supernatant and pellet were analyzed by SDS–PAGE. Defined liposomes were reconstituted from synthetic L-α-phosphatidylcholine and L-α-phosphatidylserine (Avanti Polar Lipids) with or without specified phosphoinositides (Echelon Biosciences). Lipids dissolved in chloroform were mixed in a glass tube at an appropriate ratio, and the solvent was evaporated under a stream of N2 gas at 4°C. Buffer composed of 40 mM HEPES (pH 7.4) and 100 mM NaCl was added to the dried lipid mixtures to yield a final lipid concentration of 2–10 mg/ml. The lipid mixture was rigorously vortexed for 5 min, followed by 10 cycles of freeze-and-thaw with liquid N2. For sedimentation assay, the lipid mixtures were sonicated in a water-bath for ∼5 min to form suitable liposomes or passed several 10–20 times through an extruder with a filtration membrane with expected molecular size (Avanti Polar Lipids).
The Ellman's assay and cysteine modification
Purified and fully reduced PICK1 PDZ and its mutants were dissolved in 40 mM HEPES buffer in the presence of 100 mM NaCl (pH 7.4), and pre-incubated with reconstituted liposomes at 4°C for 1 h. The protein samples were then subjected to the Ellman's assay by adding 2–3 molar ratio amount of DTNB (Sigma Aldrich) to the total free sulfhydryls of each protein. Absorption values at 412 nm of each reaction mixture were measured 5 min after the initiation of the reaction. In parallel, absorptions at 412 nm of the same reaction mixture, except without addition of the corresponding protein sample, were recorded as blank values of the Ellman's reaction. For iodoacetic acid modification of Cys residues in PICK1 PDZ, purified PICK1 PDZ-GluR2C fusion protein was thoroughly reduced by incubating with 20 mM DTT. The remaining DTT was removed by passing the reaction mixture through a PD-10 desalting column, and protein sample was concomitantly exchanged into the reaction buffer (50 mM Tris-Cl, 100 mM NaCl, pH 7.5). Each protein sample was reacted with 5 molar ratio of iodoacetic acid to the total sulfhydryls for 1 h at room temperature. The excess iodoacetic acid was removed by dialysis.
HEK293T cell culture, transfection, and immunostaining
HEK293T cells were cultured in MEM media (Invitrogen, Grand Island, NY) plus fetal bovine serum. For immunostaining, HEK293T cells were grown on coverslips coated with 0.2% gelatin. cDNA constructs of GluR2, green fluorescent protein (GFP)-tagged wild-type PICK1, or mutant PICK1 were transfected into the HEK293T cells by calcium phosphate coprecipitation method. The cells were fixed 36–48 h after transfection by 4% paraformaldehyde and 4% sucrose in PBS for 20 min at room temperature. The cells were then permeabilized by 0.2% Triton X-100 in PBS for 10 min at room temperature. After blocking with 10% normal donkey serum (NDS) in PBS for 1 h, the cells were incubated with affinity-purified rabbit anti-GluR2/3 antibody in 3% NDS for 1 h at room temperature, followed by 1 h of incubation with Red-X-conjugated fluorescence anti-rabbit secondary antibody (Jackson ImmunoResearch, West Grove, PA). After washing with PBS, the coverslips were mounted with Permafluor (Immunon, Pittsburgh, PA). The cells were observed with a Nikon Eclipse TE2000 (Nikon, Tokyo, Japan) inverted fluorescence microscope. Pictures were taken by a monochrome low noise cooled CCD camera (SPOT-RT; Diagnostic Instruments, Sterling Heights, MI) controlled by MetaMorph imaging acquisition software (Universal Imaging, West Chester, PA). Images were processed with Adobe Photoshop (Adobe Systems, San Jose, CA) to adjust intensity and contrast, to select the region of interest, and to overlay two images. All images were taken in monochrome gray scale and artificially colored for presentation.
Subcellular fractionation of PICK1 and PICK1 CC-GG
GFP-PICK1 or GFP-PICK1 CC-GG was transfected into the 293T cells. Cells were collected in a HEPES buffer (20 mM HEPES, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 5 mM sucrose,1 mM PMSF, pH 7.5) 48 h after transfection, and homogenized with 10 complete strokes with a glass Teflon homogenizer. A postnuclear supernatant was obtained by spinning the cell extract at 600 g for 10 min. The resultant supernatant was then subjected to centrifugation at 100 000 g for 1 h to separate cytosol and membrane fractions. Equivalent amounts of the cytosol and membrane fractions were analyzed by Western Blot. Anti- GFP and β-tubulin antibodies were used to detect the PICK1 and β-tubulin levels. PICK1 WT and PICK1 CC-GG expression level were quantified by densitometry.
Generation of Sindbis viruses
The cDNA encoding GFP-tagged wild-type and mutant PICK1 proteins were subcloned into the pSinRep5 viral vector (Invitrogen, Carlsbad, CA). The pSinRep5 constructs and DH26S helper DNA (Invitrogen) were then subjected to restriction enzyme linearization, DNA purification, and in vitro transcription using an SP6 promotor in vitro transcription kit (Ambion, Austin, TX). For production of viruses, mRNAs were transfected into baby hamster kidney cells by Lipofectamine 2000 (Invitrogen). Culturing media were removed 36–48 h after transfection and centrifuged at 2000 g for 10 min at 4°C to remove all cell debris. The supernatants were further centrifuged at 20 000 g at 4°C for 4 h to harvest the virus particles. The viruses were aliquoted and stored at −80°C. To infect the neurons, the viruses were directly added to the culture media at a titration determined for each batch of viruses. One day after infection, the neurons were fixed and stained for imaging analysis.
Neuronal culture, staining, and quantification
Cultured hippocampal or cortical neurons were prepared from embryonic day 18 Sprague–Dawley rats and grown on coverslips coated with poly-L-lysine (Sigma). The hippocampal neurons were infected with Sindbis viruses between days 16 and 18. For staining, neurons were fixed by 4% paraformaldehyde plus 4% sucrose in PBS for 15 min at 4°C. The neurons were further treated with −20°C methanol for 10 min, washed three times with 0.03% Triton X-100, and then permeabilized by 0.2% Triton X-100 for 10 min at 4°C. After blocking with 10% normal donkey serum for >2 h at room temperature, the neurons were incubated with primary antibody in 3% NDS at 4°C overnight to stain their endogenous proteins or 1 h at room temperature for overexpressed proteins. After washing, coverslips were mounted and observed under a fluorescence microscope. Quantitative immunofluorescence was performed under a × 60 Plan Apochromatic oil lens (1.4 NA; Nikon Instech). Data were acquired and quantified using MetaMorph acquisition and analysis software (Universal Imaging, West Chester, PA). Synaptic clusters were determined by a threshold set at twice the average dendritic gray value. Cluster density was defined as the number of clusters per 100 μm of dendrite. All quantification experiments were performed in a blinded fashion.
Coordinates
The coordinates of the PICK1 PDZ in complex with the GluR2 peptide have been deposited in the Protein Data Bank under the accession codes of 2PKU.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by grants from the Research Grants Council of Hong Kong to MZ (HKUST6125/04M, 6419/05M, and 6442/06M) and JX (HKUST6510/03M and 6130/04M). We thank Mr Anthony Zhang for critical reading of the manuscript. The NMR spectrometers used in this work were purchased with funds donated to the Biotechnology Research Institute by the Hong Kong Jockey Club. MZ was a recipient of the Croucher Foundation Senior Research Fellow Award.
References
- Arguello JM (2003) Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J Membr Biol 195: 93–108 [DOI] [PubMed] [Google Scholar]
- Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26: 131–138 [Google Scholar]
- Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL (2000) Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci 20: 7258–7267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven SE, Bredt DS (1998) PDZ proteins organize synaptic signaling pathways. Cell 93: 495–498 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Cohen AR, Anderson JM, Brunger AT (1998) Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat Struct Biol 5: 317–325 [DOI] [PubMed] [Google Scholar]
- Dev KK (2007) PDZ domain protein–protein interactions: a case study with PICK1. Curr Top Med Chem 7: 3–20 [DOI] [PubMed] [Google Scholar]
- Dev KK, Nishimune A, Henley JM, Nakanishi S (1999) The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology 38: 635–644 [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Cronin TC, Lambright DG (2003) Membrane recognition and targeting by lipid-binding domains. Sci STKE 2003: re16. [DOI] [PubMed] [Google Scholar]
- Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL (1997) GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature 386: 279–284 [DOI] [PubMed] [Google Scholar]
- Elkins JM, Papagrigoriou E, Berridge G, Yang X, Phillips C, Gileadi C, Savitsky P, Doyle DA (2007) Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions. Protein Sci 16: 683–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Shi Y, Li M, Zhang M (2003) Tandem PDZ repeats in glutamate receptor-interacting proteins have a novel mode of PDZ domain-mediated target binding. Nat Struct Biol 10: 972–978 [DOI] [PubMed] [Google Scholar]
- Gardner SM, Takamiya K, Xia J, Suh JG, Johnson R, Yu S, Huganir RL (2005) Calcium-permeable AMPA receptor plasticity is mediated by subunit-specific interactions with PICK1 and NSF. Neuron 45: 903–915 [DOI] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T (2006) PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314: 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Ge WP, Xu J, Cao M, Peng L, Yung W, Liao D, Duan S, Zhang M, Xia J (2006) Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J Neurosci 26: 2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Chung HJ, Lee HK, Huganir RL (2001) Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc Natl Acad Sci USA 98: 11725–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Lemmon MA (2003) Phosphoinositide recognition domains. Traffic 4: 201–213 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM (2000) Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J 350 (Part 1): 1–18 [PMC free article] [PubMed] [Google Scholar]
- Long JF, Feng W, Wang R, Chan LN, Ip FC, Xia J, Ip NY, Zhang M (2005) Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem. Nat Struct Mol Biol 12: 722–728 [DOI] [PubMed] [Google Scholar]
- Long JF, Tochio H, Wang P, Fan JS, Sala C, Niethammer M, Sheng M, Zhang M (2003) Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J Mol Biol 327: 203–214 [DOI] [PubMed] [Google Scholar]
- Madsen KL, Beuming T, Niv MY, Chang CW, Dev KK, Weinstein H, Gether U (2005) Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J Biol Chem 280: 20539–20548 [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC (2002) AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25: 103–126 [DOI] [PubMed] [Google Scholar]
- Matsuda S, Launey T, Mikawa S, Hirai H (2000) Disruption of AMPA receptor GluR2 clusters following long-term depression induction in cerebellar Purkinje neurons. EMBO J 19: 2765–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Murray D (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438: 605–611 [DOI] [PubMed] [Google Scholar]
- Mortier E, Wuytens G, Leenaerts I, Hannes F, Heung MY, Degeest G, David G, Zimmermann P (2005) Nuclear speckles and nucleoli targeting by PIP(2)–PDZ domain interactions. EMBO J 24: 2556–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A (1992) GRASP: Graphical Representation and Analysis of Surface Properties. New York: Columbia University [Google Scholar]
- Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB (2001) PICK1 targets activated protein kinase C{alpha} to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci 21: 5417–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Pfeffer S (2003) Membrane domains in the secretory and endocytic pathways. Cell 112: 507–517 [DOI] [PubMed] [Google Scholar]
- Seidenman KJ, Steinberg JP, Huganir R, Malinow R (2003) Glutamate receptor subunit 2 Serine 880 phosphorylation modulates synaptic transmission and mediates plasticity in CA1 pyramidal cells. J Neurosci 23: 9220–9228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C (2001) Pdz domains and the organization of supramolecular complexes. Annu Rev Neurosci 24: 1–29 [DOI] [PubMed] [Google Scholar]
- Shi S, Hayashi Y, Esteban JA, Malinow R (2001) Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell 105: 331–343 [DOI] [PubMed] [Google Scholar]
- Steinberg JP, Takamiya K, Shen Y, Xia J, Rubio ME, Yu S, Jin W, Thomas GM, Linden DJ, Huganir RL (2006) Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron 49: 845–860 [DOI] [PubMed] [Google Scholar]
- Terashima A, Cotton L, Dev KK, Meyer G, Zaman S, Duprat F, Henley JM, Collingridge GL, Isaac JT (2004) Regulation of synaptic strength and AMPA receptor subunit composition by PICK1. J Neurosci 24: 5381–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K (1986) NMR of Proteins and Nucleic Acids. New York: John Wiley [Google Scholar]
- Xia J, Chung HJ, Wihler C, Huganir RL, Linden DJ (2000) Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron 28: 499–510 [DOI] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL (1999) Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron 22: 179–187 [DOI] [PubMed] [Google Scholar]
- Xu J, Xia J (2007) Structure and function of PICK1. Neurosignals 15: 190–201 [DOI] [PubMed] [Google Scholar]
- Yan J, Wen W, Xu W, Long JF, Adams ME, Froehner SC, Zhang M (2005) Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin. EMBO J 24: 3985–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang W (2003) Organization of signaling complexes by PDZ-domain scaffold proteins. Acc Chem Res 36: 530–538 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J (2002) PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell 9: 1215–1225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data