Abstract
Vinculin links integrin receptors to the actin cytoskeleton by binding to talin. Vinculin is held in an inactive, closed-clamp conformation through hydrophobic interactions between its head and tail domains, and vinculin activation has long been thought to be dependent upon severing the head–tail interaction. Talin, α-actinin, and the invasin IpaA of Shigella flexneri sever vinculin's head–tail interaction by inserting an α-helix into vinculin's N-terminal four-helical bundle, provoking extensive conformational changes by a helical bundle conversion mechanism; these alterations in vinculin structure displace its tail domain, allowing vinculin to bind to its other partners. IpaA harbors two juxtaposed α-helical vinculin-binding sites (VBS) in its C-terminus. Here, we report that the lower affinity VBS of IpaA can also bind to the adjacent C-terminal four-helical bundle of vinculin's head domain through a helix addition mechanism. These hydrophobic interactions do not alter the conformation of this helical bundle, and the architecture of the complex suggests that IpaA can simultaneously interact with both of the four-helical bundle domains of vinculin's N-terminus to stabilize vinculin–IpaA interactions.
Keywords: crystal structure, helical bundle conversion, Shigella flexneri's invasin IpaA, talin, vinculin
Introduction
Vinculin is a 117 kDa ubiquitously expressed cytoskeletal protein that is localized to cell–matrix junctions (focal adhesions) mediated by integrin–talin interactions and to cell–cell junctions (adherens junctions) that are directed by interactions of cadherin receptors and catenins (Critchley, 2004; Ziegler et al, 2006). Vinculin provides essentials links for these receptors to the actin cytoskeleton by directly binding to talin in focal adhesions and to α-actinin and α-catenin in adherens junctions (Critchley, 2004). Accordingly, vinculin is essential for embryonic development and vinculin-null cells display chaotic cell migration with few focal adhesions (Xu et al, 1998). Furthermore, vinculin displays tumor suppressor functions, where its overexpression impairs cell motility and tumor cell metastasis (Rodriguez Fernandez et al, 1992), whereas suppression of vinculin augments cell motility and tumorigenesis (Rodriguez Fernandez et al, 1993).
Vinculin is comprised of three seven-helical bundle α-catenin-like domains (Vh1, Vh2, and Vh3) that contain two four-helical bundle subdomains (Izard et al, 2004), and a four-helical bundle (Vt2) domain that is connected to a five-helical bundle tail (Vt) domain via a proline-rich hinge region (Bakolitsa et al, 2004; Borgon et al, 2004). The first four domains N-terminal to the proline-rich hinge comprise the globular vinculin head (VH) domain observed by electron microscopy. Vinculin's binding partners have been shown to bind to vinculin's N-terminal Vh1 domain, its proline-rich region, and to the Vt domain (Critchley, 2004). Extensive hydrophobic interactions between vinculin's Vh1 and Vt domains clamp vinculin in a closed, inactive conformation and severing this interaction is necessary for vinculin to bind to some of its partners, for example for the binding of F-actin to the Vt domain (Johnson and Craig, 1995; Gilmore and Burridge, 1996). The physiological triggers that sever the Vh1–Vt interaction have been a source of controversy and include the binding of phosphatidylinositol-4,5-bisphosphate (PIP2) that binds to the Vt domain (Gilmore and Burridge, 1996; Bakolitsa et al, 2004; Weis, 2004; Ziegler et al, 2006), and the amphipathic α-helical vinculin-binding sites (VBS) that reside in the central rod domains of talin and α-actinin, which bind to vinculin's Vh1 domain (McGregor et al, 1994; Bass et al, 2002; Critchley, 2004; Gingras et al, 2005). Although vinculin mutants that cannot bind to PIP2 still localize to focal adhesions (Chandrasekar et al, 2005), biochemical and structural studies have demonstrated that the VBS of talin and α-actinin are sufficient to provoke extensive conformational changes in Vh1's N-terminal helical bundle subdomain that create an entirely new structure by a mechanism coined helical bundle conversion (Izard et al, 2004; Izard and Vonrhein, 2004; Papagrigoriou et al, 2004; Bois et al, 2005; Fillingham et al, 2005; Gingras et al, 2005), which distorts the head–tail interface and thus displaces Vt, and which allows vinculin to bind to F-actin (Bois et al, 2006).
Helical bundle conversion occurs specifically within the N-terminal four-helical bundle subdomain of vinculin's Vh1 domain, where the VBSs of talin or α-actinin, or of the IpaA invasin of S. flexneri (Izard et al, 2006), burrow between and alter the structures of Vh1 helices α1 and α2, and the structure of helix α3. These changes displace Vt yet leave the structure of the C-terminal four-helical bundle subdomain of Vh1 unaltered (Izard et al, 2004; Izard and Vonrhein, 2004; Bois et al, 2005). However, the binding of talin to vinculin and the localization of vinculin to sites of cell–matrix junctions were originally mapped to vinculin residues 167–207 that reside in the C-terminal helical bundle subdomain of Vh1 (Bendori et al, 1989; Jones et al, 1989), suggesting that this represents another site in vinculin that interacts with talin.
Thus far, about a dozen crystal structures have been determined for the Vh1 domain in complex with various VBSs (Izard et al, 2004, 2006; Izard and Vonrhein, 2004; Papagrigoriou et al, 2004; Bois et al, 2005; Fillingham et al, 2005; Gingras et al, 2005, 2006). All of these structures show remarkably similar alterations in the N-terminal four-helical bundle subdomain of Vh1, and the binding of these VBSs has heretofore not been detected in any other region of vinculin. The invasin IpaA protein of S. flexneri harbors two high-affinity VBSs for vinculin, IpaA-VBS and IpaA-VBS2 (Izard et al, 2006). Here, we present the crystal structure of vinculin's Vh1 domain in complex with two molecules of IpaA-VBS2. Our structural and biochemical studies show that this VBS of IpaA can also bind to vinculin in a conformation reminiscent of its closed, inactive conformation through hydrophobic interactions with the C-terminal helical bundle subdomain of Vh1, and that this occurs through a helix addition mechanism. Furthermore, biochemical studies suggest that at least some of the VBSs of talin can bind to vinculin in a fashion akin to the helix addition mechanism described here for the vinculin-IpaA-VBS2 interaction, without activating vinculin. Studies in cells have established the significance of this second binding site for localization of vinculin to adhesion plaques (Bendori et al, 1989; Jones et al, 1989), and together with our data presented herein, suggest a model in which the interactions of vinculin with its binding partners is stabilized by simultaneously binding of VBSs to its two N-terminal four-helical bundles.
Results
Crystal structure of human vinculin in complex with two molecules of IpaA-VBS2
The crystal structure of vinculin's Vh1 domain in complex with IpaA-VBS2 peptide was solved to 3.2 Å resolution (Tables I and II). The structure revealed that one IpaA-VBS2 molecule bound to Vh1's N-terminal helical bundle subdomain, where it provoked extensive conformational changes in its structure typical of helical bundle conversion (Izard et al, 2004; Izard and Vonrhein, 2004; Bois et al, 2005), where the four α-helices within this bundle completely rearrange to incorporate IpaA-VBS2 and form a new five-helical bundle structure. As seen with other VBSs, the IpaA-VBS2 has extensive hydrophobic interactions with Vh1 helices α1, α2, and α3 (Figure 1A).
Table 1.
Data reduction statistics of the Vh1:IpaA-VBS2 data set
Table 2.
Crystallographic refinement statistics of the Vh1:IpaA-VBS2 structure
| Resolution | 77.1–3.2 Å |
| Last Shell | 3.39–3.2 Å |
| No. of reflections (working set) | 10 834 |
| No. of reflections (test set) | 1070 |
| R-factora (overall) | 0.2426 |
| R-factora (last shell) | 0.2519 |
| Rfreeb (overall) | 0.2697 |
| Rfreeb (last shell) | 0.2981 |
| No. of amino-acid residues | 302 |
| No. of protein atoms | 2359 |
| Average B-factor (main chain) | 73.2 Å2 |
| Average B-factor (side chain) | 75.4 Å2 |
| R.m.s.d. from ideal geometry: | |
| Covalent bond lengths | 0.005 Å |
| Bond angles | 0.903° |
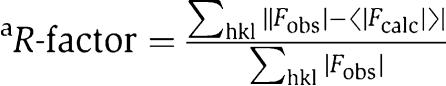 where 〈∣Fcalc∣〉 is the expectation of ∣Fcalc∣ under the error model used in maximum-likelihood refinement. where 〈∣Fcalc∣〉 is the expectation of ∣Fcalc∣ under the error model used in maximum-likelihood refinement. | |
| bThe free R-factor is a cross-validation residual calculated by using 9% of the native data, which were randomly chosen and excluded from the refinement. | |
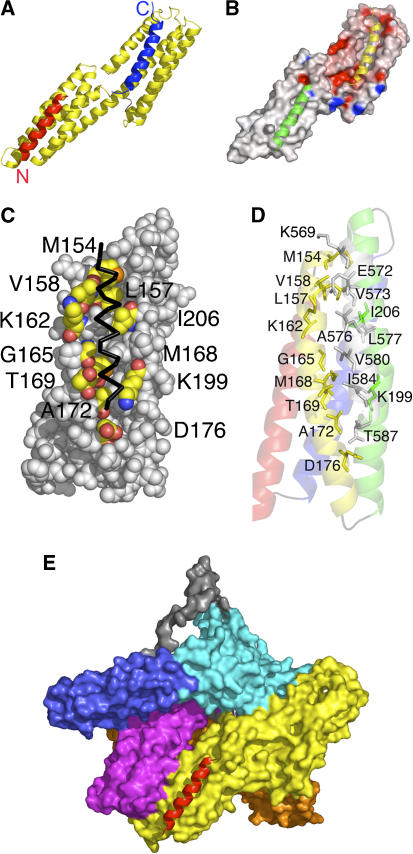
Figure 1.
The crystal structure of human vinculin bound to Shigella's IpaA-VBS2. (A) Cartoon representation of vinculin (Vh1, residues 1–258, yellow) bound to two IpaA-VBS2 molecules (red and blue). The binding of IpaA-VBS2 (blue) to the N-terminal helical bundle of Vh1 causes helical bundle conversion from a four-helical vinculin bundle to a five-helical bundle, and is very similar to the structures that have been observed for the binding of talin's or α-actinin's VBSs to Vh1 (Izard et al, 2004; Izard and Vonrhein, 2004; Papagrigoriou et al, 2004; Bois et al, 2005, 2006; Fillingham et al, 2005; Gingras et al, 2006). The C-terminus of IpaA-VBS2 is indicated. The binding of a second IpaA-VBS2 (red) to the C-terminal helical bundle is unprecedented and its binding rather occurs via a helix addition mechanism. The N- and C-termini of IpaA-VBS2 are indicated. (B) Electrostatic surface potential (red, negative; blue, positive) of <−20 to >+20 kbT e−1, where kb is the Boltzmann constant and T is the temperature of the Vh1 structure in its IpaA-VBS2-bound state. IpaA-VBS2 bound to the N-terminal or C-terminal helical bundles are shown as a cartoon in yellow and green, respectively. (C) View of the surface vinculin residues (labeled; carbon, yellow; oxygen, red; nitrogen, blue; sulfur, orange) of the C-terminal helical bundle of Vh1 in contact with IpaA-VBS2 (back, Cα trace). (D) Cartoon representation of the C-terminal helical bundle of Vh1 (residues 130–150, red; 154–181, yellow; 185–218, green; 223–251, blue) in complex with IpaA-VBS2 (gray). Residues involved in binding are labeled. (E) Superposition of the C-terminal helical Vh1 bundle of the Vh1:IpaA-VBS2 structure onto the full-length structure of vinculin in its inactive conformation (PDBID 1TR2; Borgon et al, 2004) reveals no changes in the structure of this bundle upon binding to IpaA-VBS2, and also establishes that this new binding site is exposed; therefore, IpaA can bind to a conformation reminiscent of inactive vinculin. Vinculin domains Vh1, Vh2, Vh3, Vt2, and Vt are colored in yellow, orange, magenta, blue, and cyan, respectively, and the disordered proline-rich region was modeled and is shown in gray. IpaA-VBS2 bound to the C-terminal helical bundle of Vh1 is shown in red.
In all of the Vh1:VBS crystal structures defined thus far, the binding of VBSs has only been seen to vinculin residues 8–126 within the N-terminal helical bundle subdomain. Surprisingly, however, the vinculin-IpaA-VBS2 crystal structure also identified a second IpaA-VBS2 molecule bound to the C-terminal four-helical bundle subdomain of Vh1 (Figure 1A). Furthermore, an entirely new binding mode is seen for the interaction of IpaA-VBS2 and the C-terminal helical bundle of Vh1, which occurs via a simple helix addition of IpaA-VBS2 onto a hydrophobic patch that lies between helices α2 and α3 of this bundle, with no interactions with helices α1 or α4 (Figures 1A–D and 2). Specifically, residues K569, A570, V573, A576, L577, V580, I584, and T587 of IpaA make hydrophobic contacts with residues M154, L157, V158, K162, G165, M168, T169, and A172 of the C-terminal Vh1 helix α2 and with residues D176, K199, I206, and K210 of the C-terminal Vh1 helix α3 (Figures 1C, D and 2; Supplementary Table). Polar interactions are also seen between IpaA residues E572 and A570 with Vh1 residues K162 and K210, respectively (Figure 1D; Supplementary Table). Notably, the structure of the C-terminal helical bundle does not change upon binding of IpaA-VBS2 when superimposed onto the structure of full-length, inactive vinculin (Figure 1E), where the Cα of the C-terminal helical bundle (residues 130–258) can be superimposed with root mean square deviation (r.m.s.d.) of 0.74 Å.
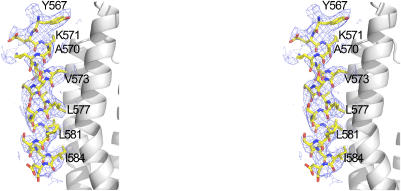
Figure 2.
Stereo view of the final 2Fo–Fc electron density map for IpaA-VBS2 bound to the C-terminal helical bundle of Vh1. The contour level of the electron density map is 0.6 σ and the resolution is 3.2 Å. Key residues of IpaA-VBS2 that interact with the α2 and α3 helices of the C-terminal helical bundle of vinculin's Vh1 domain are indicated.
Helix addition in adhesion signaling
The interaction of the IpaA-VBS2 of IpaA with the C-terminal helical subdomain of vinculin's Vh1 domain occurs via a helix addition mechanism, which is new to vinculin signaling. However, other interactions involved in adhesion signaling use this mechanism, where the α-helical LD2 and LD4 motifs of paxillin make hydrophobic contacts with either helices α2 and α3, or with helices α1 and α4, of the FAT domain of FAK, without altering the structure of this domain (Hayashi et al, 2002; Hoellerer et al, 2003; Gao et al, 2004). We therefore speculated that the structure of the C-terminal four-helical bundle of Vh1 bound to IpaA-VBS2 might share structural similarities with the FAK–paxillin interactions. Indeed, the 113 Cα of the C-terminal helical bundle of Vh1 can be superimposed onto the FAT domain of FAK with r.m.s.d. of 2.5 Å. However, in the crystal structures of FAT bound to LD2 or LD4 (Hoellerer et al, 2003), these α-helices are in an orientation opposite to that seen for the binding of IpaA-VBS2, which is parallel to helix α3 and antiparallel to helix α2 (Figure 3A–C).
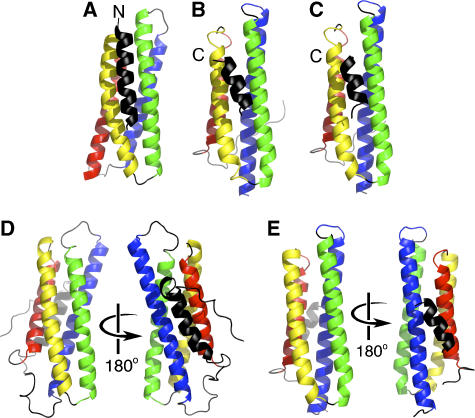
Figure 3.
The helix addition mechanism in vinculin. (A) Architecture of the four-helical bundle of the C-terminal bundle subdomain of Vh1 (helices are colored from the N-terminus: red, yellow, green, and blue) when bound by the IpaA-VBS2 (black) α-helix. The N-terminus of IpaA-VBS2 is indicated. While similar in terms of their helix addition mechanism of interaction, the crystal structures of FAK:LD2 (B) or FAK:LD4 (C), PDBID 1OW8 and 1OW7, respectively, show binding of the paxillin helix in the opposite orientation compared to binding of IpaA-VBS2 to the C-terminal helical bundle of the Vh1 domain of vinculin (A). The C-termini of paxillin are indicated. (D) The Vt domain of vinculin (PDBID 1QKR) is also a five-helical bundle that has an architecture that is somewhat similar to that of the Vh1:IpaA-VBS2 complex, with an additional fifth helix on its N-terminus (black), interacting with Vt helices α1 (red) and α4 (blue). (E) Paxillin-LDs also bind to the interface of helices α1 (red) and α4 (blue), PDBID 1OW7 (Hoellerer et al, 2003), similar to the Vt structure.
The vinculin tail domain Vt is a five-helical bundle (Bakolitsa et al, 2004; Borgon et al, 2004) and a comparison of its structure to that of the C-terminal helical bundle of Vh1 bound to IpaA-VBS2 also revealed similarities in their architecture. Specifically, the 92 Cα positions of the four helices of this helical bundle when bound to IpaA-VBS2 can be superimposed with r.m.s.d. of 3.2 Å. In the Vt structure, helix α0 interacts with helices α1 and α4 (Figure 3D), while IpaA-VBS2 interacts with helices α2 and α3 of the C-terminal helical bundle of Vh1 (Figure 3A).
In the paxillin-FAK crystal structure (Figure 3B, C, and E), the LD2 and LD4 helices of paxillin can simultaneously bind to the two sides of the four-helix bundle of the FAT domain of FAK (Hayashi et al, 2002; Hoellerer et al, 2003; Gao et al, 2004). In contrast, binding to the hydrophobic patch that lies between helices α2 and α3 of the C-terminal helical bundle of the Vh1 domain in full-length vinculin is sterically hindered from binding to an additional α-helix by the juxtaposed Vh2 domain. Therefore, unlike FAK, the C-terminal helical bundle of Vh1 can only bind to one helix in the conformation as seen for full-length inactive vinculin.
Vinculin activation mechanism
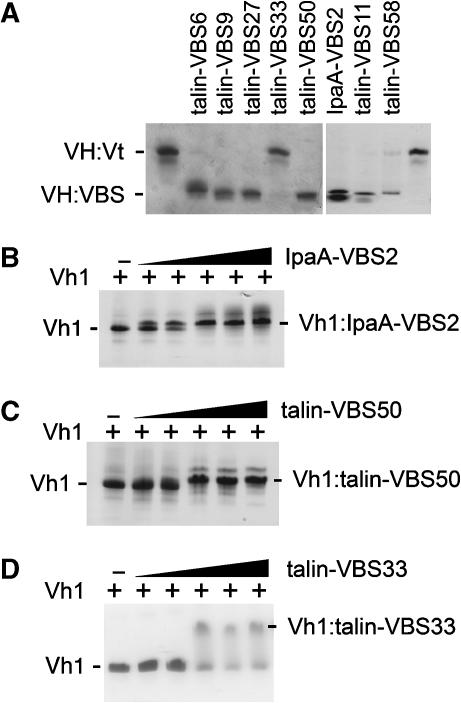
The structure of the C-terminal helical bundle is unaltered following binding to IpaA-VBS2. Indeed, the novel IpaA-VBS2-binding site found on the C-terminal helical bundle of Vh1 is equally available for binding to IpaA in the full-length vinculin structure (Figure 1E). This suggests that, unlike the helical bundle conversion mechanism, the helix addition mechanism does not activate vinculin. To test this hypothesis, the functionality of talin's many VBSs were evaluated. Originally, three talin VBSs were identified in its rod domain (Bass et al, 1999, 2002) and more recently eight additional talin VBSs have been proposed (Gingras et al, 2005). To test whether these eight new talin-VBSs were indeed capable of disrupting the head–tail interaction, we performed vinculin VH:Vt displacement assays (Figure 4A). Surprisingly, in these assays two of the newly proposed talin-VBSs, talin-VBS33 and talin-VBS36, failed to displace Vt from pre-existing VH:Vt complexes (Figure 4A and data not shown), although they were capable of binding to a portion of the VH domain that contains the Vh1, Vh2, and Vh3 domains (residues 1–714) in solid-phase SPOT-peptide arrays (Gingras et al, 2005). The crystal structure of the Vh1:talin-VBS36 complex is known (Gingras et al, 2005), confirming that talin-VBS36 can indeed bind to the Vh1 domain.
Figure 4.
Displacement of Vt from pre-existing VH:Vt complexes by talin's VBSs. (A) Five of the new, recently identified talin-VBSs (Gingras et al, 2005) lack crystal structure information and four of these, talin-VBS6, talin-VBS9, talin-VBS27, and talin-VBS50, activate vinculin by displacing the Vt domain to form a new VH:talin-VBS complex detectable in native gel analyses. In contrast, talin-VBS33 failed to displace the Vt domain from pre-existing VH:Vt complexes. The Vh1:talin-VBS crystal structures of three of these additional talin-VBSs (talin-VBS11, talin-VBS36, and talin-VBS58) have been determined (PDBID 1ZVZ, 1ZW2, and 1ZW3, Gingras et al, 2005). While talin-VBS11 and talin-VBS58 do indeed activate vinculin by displacing the Vt domain, talin-VBS36 aggregated with VH:Vt, and prevented it from running into the gel (data not shown). Consistent with its ability to provoke helical bundle conversion of the N-terminal bundle subdomain of Vh1 (Figure 1A), IpaA-VBS2 displaced the Vt domain from the VH:Vt complex. Titration of (B) IpaA-VBS2, (C) talin-VBS50, and (D) talin-VBS33 to Vh1 as analyzed by native PAGE of free Vh1 (lanes 1) and of Vh1 bound to the VBSs (lanes 2–6). Free Vh1 and Vh1:VBS are indicated. A slower migrating band above the indicated Vh1:VBS complex corresponds to a complex that contains two VBSs bound to Vh1 (seen in lanes 4–6 for IpaA-VBS2 (B) or talin-VBS50 (C) but not for talin-VBS33 (D)).
Surface plasmon resonance analyses also suggested that, similar to IpaA-VBS2, talin-VBS50 has two binding sites in vinculin (unpublished data). We therefore tested by native gel shift assay whether vinculin's Vh1 head domain can bind to two molecules of IpaA-VBS2 or to two molecules of talin-VBS50 in solution (Figure 4B and C). Titration of either IpaA-VBS2 (Figure 4B) or talin-VBS50 (Figure 4C) to Vh1 resulted in the appearance of a slower migrating Vh1:VBS complex, yet at higher VBS concentrations a second, slower migrating Vh1:VBS:VBS complex was also evident, consistent with the notion that two molecules of these VBSs can simultaneously bind to one molecule of Vh1. In contrast, titration of talin-VBS33 resulted in the appearance of only one slower migrating Vh1:VBS complex (Figure 4D). Further, consistent with a weaker secondary binding site on the C-terminal helical bundle of Vh1, talin-VBS33 never fully saturates Vh1 (Figure 4D), whereas Vh1 binding by IpaA-VBS2 or talin-VBS50 was saturable (Figure 4B and C). Given the failure of talin-VBS33 to displace the VH:Vt complex (Figure 4A), these findings suggested that talin-VBS33 might only bind to the C-terminal helical bundle of Vh1.
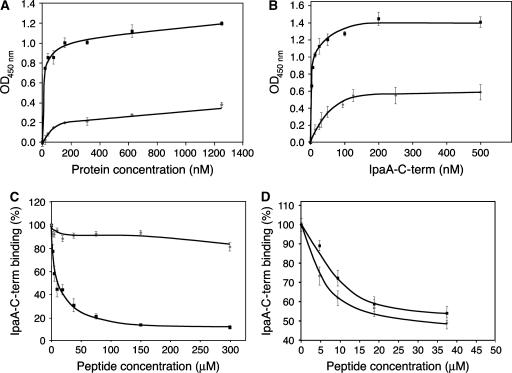
The vinculin-binding domain of IpaA of S. flexneri, IpaA-C-term, contains two VBSs (IpaA-VBS and IpaA-VBS2; Izard et al, 2006), and the crystal structure of the Vh1:IpaA-VBS2 complex established that this VBS also binds to the C-terminal helical bundle subdomain (Figure 1). We therefore further assessed the binding properties of IpaA-C-term using purified recombinant Vh1 or Vh1Δ126, a truncated derivative of Vh1 that lacks the N-terminal four-helical bundle subdomain of Vh1, but which retains the C-terminal helical bundle where the second-site binding of IpaA-VBS2 was observed. Indeed, when IpaA-C-term was incubated in wells coated with increasing concentrations of Vh1 or Vh1Δ126, the binding of both Vh1 and Vh1Δ126 was saturable, with maximal binding at concentrations above 200 nM for both (Figure 5A). Similarly, when wells were coated at a fixed concentration of 200 nM of Vh1 or Vh1Δ126 and were then incubated with increasing concentrations of IpaA-C-term, saturable binding was also observed, with a IpaA-C-term half-maximal binding concentration of 3.1±0.6 nM for Vh1, and of 30.2±4.5 nM for Vh1Δ126 (Figure 5B). Finally, consistent with Vh1 bearing two sites for binding to IpaA-C-term and Vh1Δ126 containing only one, the saturation binding for Vh1 occurs at IpaA-C-term levels that were twice those of Vh1Δ126. Therefore, the second-site binding of IpaA-VBS2 to the C-terminal helical bundle subdomain of vinculin's Vh1 domain detected in the crystal structure can indeed direct binding to the VBSs present in the C-terminal vinculin-binding domain of IpaA.
Figure 5.
Binding of the IpaA-VBS2 and talin-VBS33 to the C-terminal helical bundle subdomain of Vh1. Solid-phase binding assays were performed with the indicated proteins; values presented correspond to the average of three independent experiments, performed in duplicate, and the s.e.m. are indicated. (A) Wells were coated with Vh1 (solid squares) or with Vh1Δ126 (gray diamonds) proteins at concentrations ranging from 19 to 1200 nM. IpaA-C-term was used at a concentration of 200 nM. (B) Wells were coated with 200 nM Vh1 (solid squares) or with 200 nM Vh1Δ126 (gray diamonds) protein and were then incubated with IpaA-C-term at concentrations ranging from 3.1 to 500 nM. (C) Wells were coated with 200 nM Vh1 and incubated with 30 nM IpaA-C-term in the presence of the IpaA-VBS2 (solid squares) or talin-VBS33 (gray diamonds) peptides at concentrations ranging from 2.3 to 75 μM. (D) Wells were coated with 200 nM Vh1Δ126 protein and were incubated with 150 nM IpaA-C-term in the presence of the IpaA-VBS2 (solid squares) or talin-VBS33 (gray diamonds) peptides at concentrations ranging from 2.3 to 37.5 μM.
Talin-VBS33 peptide bound to Vh1, but was not able to sever the VH–Vt interaction that activates vinculin (Figure 4A and D). We therefore tested the effects of talin-VBS33 versus the IpaA-VBS2 on the binding properties of IpaA-C-term to Vh1. IpaA-C-term binding to Vh1 was inhibited by IpaA-VBS2 (IC50=6.8±0.8 μM; Figure 5C). In contrast, talin-VBS33 had no significant effects on IpaA-C-term binding to Vh1 even at concentrations as high as 300 μM (Figure 5C), corresponding to a 2000-fold excess over the IpaA-C-term probing concentration. Collectively, these data are consistent with the presence of one binding site present in the N-terminal four-helical bundle and with another in the C-terminal four-helical bundle of Vh1, and with the binding of IpaA-VBS2, but not of talin-VBS33, to both of these sites.
To test the hypothesis that talin-VBS33 could only bind to the C-terminal helical bundle of vinculin's Vh1 domain, we assayed the ability of talin-VBS33 to displace IpaA-C-term bound to either Vh1 or Vh1Δ126, and compared its activity to that of IpaA-VBS2, which was used as a positive control. Notably, talin-VBS33 displaced IpaA-C-term from Vh1Δ126 with an efficiency that was similar to that of IpaA-VBS2 (Figure 5D). Therefore, talin-VBS33 only binds to the C-terminal helical bundle, which remains in an inactive conformation even when vinculin is activated (Izard et al, 2004; Bois et al, 2005, 2006). By contrast, binding by IpaA-VBS2 provokes helical bundle conversion when bound to the N-terminal helical bundle subdomain, yet is also capable of second-site binding on the C-terminal Vh1 subdomain as shown in the crystal structure (Figure 1).
Discussion
A novel helix addition mechanism also contributes to vinculin's interactions
The Vh1:IpaA-VBS2 crystal structure establishes that vinculin can interact with VBSs using at least two distinct mechanisms of α-helix-helical bundle interactions. First, intimate burrowing of VBSs into the N-terminal helical bundle subdomain provokes helical bundle conversion, as seen in all Vh1:VBS structures (Izard et al, 2004; Izard and Vonrhein, 2004; Papagrigoriou et al, 2004; Bois et al, 2005; Fillingham et al, 2005). In contrast, the binding of IpaA-VBS2 to the C-terminal helical bundle of Vh1 establishes that VBSs can also interact with vinculin through a helix addition mechanism that does not alter the structure of this domain as it is found in inactive vinculin.
Superposition of the Vh1:IpaA-VBS2 structure reported here with that of the FAK:paxillin complex revealed similarities in these interactions, and establishes that helix addition is a common mechanism for four-helical bundle interactions in cytoskeletal proteins. Specifically, like the interaction of IpaA-VBS2 with the C-terminal helical bundle of vinculin's Vh1 domain, the binding of paxillin's LD2 or LD4 motifs does not alter the structure of the four-helical bundle of the FAT domain of FAK (Hayashi et al, 2002; Hoellerer et al, 2003; Gao et al, 2004).
The fact that IpaA-VBS2 interacts with vinculin by using at least two mechanisms prompted a reassessment of the functionality and binding of talin's 11 proposed VBSs, which are localized throughout its central rod domain (Bass et al, 1999; Gingras et al, 2005). The structures and biochemical analyses of the most studied vinculin-talin-VBS interactions, those of talin-VBS1, -VBS2, and -VBS3, have shown that all three provoke helical bundle conversion (Izard et al, 2004; Izard and Vonrhein, 2004; Papagrigoriou et al, 2004; Fillingham et al, 2005; Gingras et al, 2006), and that they are sufficient to alter the conformation and to activate full-length vinculin, for example in triggering the latent F-actin binding potential of vinculin (Bois et al, 2006). Our further assessment of the eight recently reported talin-VBSs (Gingras et al, 2005), however, led to some surprises, where talin-VBS33 and talin-VBS36 failed to sever vinculin's head–tail interactions. The assignment of these peptides of talin as VBSs was based upon their ability to bind to a portion of vinculin's globular head domain in peptide SPOT-array analyses, and this was also confirmed in the crystal structure of the Vh1:talin-VBS36 complex (Gingras et al, 2005). Collectively, these results suggest that the affinity of talin-VBS33 and talin-VBS36 is lower than that of the head–tail intramolecular interaction. Furthermore, talin-VBS33 cannot displace IpaA-C-term from Vh1 in solid-phase assays (Figure 5C), but does bind to the C-terminal four-helical bundle of vinculin (i.e., Vh1Δ126; Figure 5B). Therefore, some α-helical VBSs (e.g., IpaA-VBS2 and talin-VBS50) have the potential to bind to helical bundles by disparate mechanisms, whereas others (e.g., talin-VBS33) appear restricted in the types of interactions they can undergo.
The interaction of IpaA-VBS2 with the C-terminal helical bundle of vinculin's Vh1 domain does not alter the structure of this domain, as it is found in vinculin's closed, inactive conformation. We therefore reasoned that a similar scenario might also apply to some of the many VBSs of talin, and indeed talin-VBS50 and talin-VBS33 fit these criteria. Specifically, like IpaA-VBS2, talin-VBS50 is capable of displacing vinculin's head–tail interactions yet also binds to the C-terminal helical bundle of Vh1 (Figure 4 and data not shown). Furthermore, talin-VBS33 only shows binding to the C-terminal helical bundle in solid-phase assays (Figure 5), and accordingly this VBS of talin would be predicted to also bind to inactive pools of vinculin.
Dual, simultaneous binding of VBSs to vinculin
The severing of vinculin's head–tail interaction is required for binding to some of its partners, and this has been most clearly established for the binding of F-actin to the Vt domain in full-length vinculin (Johnson and Craig, 1995; Gilmore and Burridge, 1996; Bois et al, 2006). The crystal structure of the Vh1:IpaA-VBS2 complex presented herein now shows that, in addition to activating vinculin, this VBS of Shigella's invasin IpaA protein can also bind to a conformation of vinculin that is seen in the inactive conformation of vinculin. The crystal structure establishes that this interaction is unique, in that it occurs through a region of vinculin that directs protein–protein interactions, the C-terminal helical bundle subdomain of vinculin's Vh1 domain, which is freely accessible in the inactive vinculin structure (Borgon et al, 2004).
The fact that IpaA-VBS2 can engage in both mechanisms, helical bundle conversion and helix addition, suggests that the sequence of a given VBS does not necessarily direct mechanism. Rather, the inherent structure of the helical bundle that binds to VBSs appears to specify mechanism where the N-terminal helical bundle of Vh1 requires less unfolding and rearrangement for a VBS insertion, compared to what would be required for such an insertion in the C-terminal helical bundle. Indeed, the insertion site resides at the N-terminus of the Vh1 domain, between helices α1 and α2, whereas the latter binding site is formed by helices α2 and α3 of the C-terminal helical bundle.
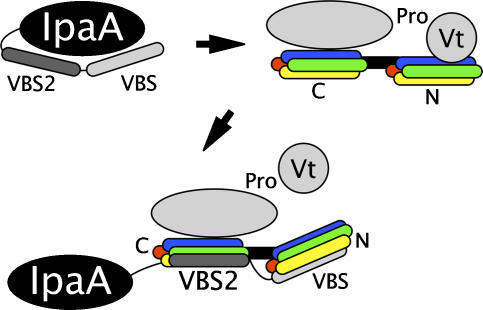
Superposition of the Vh1:IpaA-VBS crystal structure (Izard et al, 2006) onto the Vh1:IpaA-VBS2 crystal structure presented here also suggests that the two VBSs of IpaA can simultaneously bind to vinculin (Figure 6). One possible scenario is that the addition of the IpaA-VBS2 helix to the C-terminal helical bundle could serve as a guide for positioning the insertion IpaA-VBS helix to its correct site in the N-terminal helical bundle subdomain, in a fashion akin to that described for the interactions of gelsolin with F-actin, where binding of gelsolin's G2 domain to the side of F-actin positions its G1 domain to sever actin–actin contacts (Irobi et al, 2003 and references therein). However, our deletion studies have shown that loss of IpaA-VBS compromises the entry and spread of S. flexneri, whereas loss of IpaA-VBS2 selectively impairs localization of vinculin to internalized bacterium in the infected cell (Izard et al, 2006). Such findings are more consistent with the (60-fold) higher affinity IpaA-VBS (residues 611–633) first binding to and altering the structure of the N-terminal helical bundle of Vh1 (as shown in blue for IpaA-VBS2 in Figure 1A), leaving IpaA-VBS2 (residues 565–587) then free to bind to the C-terminal helical bundle of Vh1 (as shown in red in Figure 1A). In accord with this notion, this superposition places residue 585 of IpaA-VBS2 when bound to the C-terminal helical bundle of Vh1 within 25 Å of residue 610 of IpaA-VBS bound to the N-terminal helical bundle, allowing the unstructured residues 586–609 to connect IpaA-VBS and IpaA-VBS2 without any steric hindrance (Figure 6). Finally, we crystallized the Vh1:IpaA-VBS2 complex by adding an excess of IpaA-VBS2 to Vh1, and initially only observed binding of IpaA-VBS2 to the N-terminal helical bundle (Izard et al, 2006), again suggesting that this site is occupied first. We propose that this scenario would stabilize the IpaA:vinculin complex, particularly if IpaA needs to unfurl before it can bind to vinculin, as has been suggested for talin (Fillingham et al, 2005; Gingras et al, 2006).
Figure 6.
Proposed mechanism of activation of vinculin by Shigella's invasin IpaA. The hydrophobic surface of the VBSs of IpaA are proposed to require unfurling from their buried conformation within IpaA to allow them to activate vinculin. The two VBSs of IpaA are spatially separated enough to allow the C-terminal IpaA-VBS (light gray) to bind to the N-terminal four-helical bundle subdomain of vinculin's Vh1 domain (red, yellow, green, blue of bundle labeled ‘N') and for IpaA-VBS2 (dark gray) to simultaneously bind to the C-terminal four-helical bundle subdomain (red, yellow, green, blue of bundle labeled ‘C'). The former interaction occurs via helical bundle conversion, whereas the latter interaction via helix addition. Simultaneous binding of IpaA-VBS and IpaA-VBS2 to Vh1 would stabilize the vinculin–IpaA interaction. The severing of the Vh1–Vt interaction by helical bundle conversion of the N-terminal helical bundle of Vh1 upon binding of IpaA-VBS is indicated. Vinculin's remaining domains, Vh2, Vh3, and Vt2 are shown as a gray oval and the proline-rich region connecting the head and tail is indicated.
Of course oligomerization of IpaA and/or vinculin could allow for other levels of IpaA–vinculin interactions as well. For example, dimerization of IpaA might facilitate the binding of IpaA-VBS2 to both N- and C-terminal helical bundles of Vh1, as seen in the crystal structure presented here. However, we feel this is unlikely given the significantly higher affinity of IpaA-VBS versus IpaA-VBS2 for vinculin's Vh1 domain. More likely would be scenarios where IpaA oligomerization would facilitate the bridging of vinculin molecules. Given the high degree of similarity of IpaA-VBS and IpaA-VBS2, it seems likely that IpaA-VBS, like IpaA-VBS2, could bind to both N- and C-terminal helical bundles of Vh1.
The talin-binding site in vinculin was initially localized to vinculin residues 167–207 by analyses of deletion mutants that span this domain (Johnson et al, 1998). Strikingly, this is the precise region that comprises the second helix addition binding site in the C-terminal helical bundle of our Vh1:IpaA-VBS2 structure (Figures 1 and 2; Supplementary Table). Moreover, expression of vinculin residues 1–398 directs localization to cell–matrix junctions, whereas vinculin (1–398)Δ167–207 fails to localize to focal adhesions. Collectively, this indicated that vinculin residues 167–207 contained a bonafide talin-binding site as well as a focal contact binding site (Bendori et al, 1989; Johnson et al, 1998). More recently, yeast two-hybrid assays have shown that the binding of talin-VBS3 to vinculin's Vh1 domain was reduced to 70% for vinculin residues 1–131 and to 79% for vinculin residues 1–167 compared to the entire domain (vinculin residues 1–258, Bass et al, 2002). Along with our biochemical data, this suggests that at least some of the VBSs of talin, like those of IpaA, can simultaneously bind to both four-helical bundles of Vh1, with a higher affinity for binding to the N-terminal four-helical bundle subdomain by a helical bundle conversion mechanism and a lower binding affinity for the C-terminal helical bundle subdomain by a helix addition mechanism. Such a scenario would stabilize the activated and unfurled talin (Fillingham et al, 2005) in the vinculin:talin complex at sites of focal adhesions.
Materials and methods
Protein purification and crystallization
VH:Vt was co-purified as described previously (Bois et al, 2006). Human talin-VBSs and Shigella's IpaA-VBS2 peptides were synthesized and HPLC purified. Shigella IpaA-C-term (residues 559–633) was purified as described previously (Izard et al, 2006). An N-terminally hexa-histidine tagged recombinant form of human vinculin Vh1Δ126 (residues 127–258) was cloned into the pET3 expression vector (Novagen). Human vinculin Vh1Δ126 protein was expressed in Escherichia coli BL21(DE3) and purified using a chelating nickel affinity column by elution over a gradient to 0.5 M imidazole pH 8, followed by an anion exchange column. The protein was dialyzed into PBS.
We first obtained cubic crystals (space group I432) of Vh1 in complex with the minimal IpaA-VBS2-binding site (residues 565–587), but were unable to improve their diffraction beyond 7–8 Å Bragg spacings at the Advanced Photon Source (19ID and 22ID beamlines). We therefore synthesized longer IpaA-VBS2 peptides by adding three- or six-residue extensions on either terminus. This resulted in rhombohedral Vh1:IpaA-VBS2 (559–587) or Vh1:IpaA-VBS2 (562–590) crystals (space group R32), cubic Vh1:IpaA-VBS2 (565–591) crystals (space group I213), or cubic crystals (space group I432) for native and seleno-methionine-substituted Vh1:IpaA-VBS2, or of native Vh1 in complex with mutated IpaA-VBS2s (V-590-SeMet or L-581-SeMet). We eventually obtained diffraction to about 4 Å Bragg spacings, but the selenium derivative did not show a usable anomalous signal for phasing. We solved four structures of various IpaA-VBS2 lengths in space groups R32 and I432 (Izard et al, 2006) by molecular replacement and eventually obtained Vh1:IpaA-VBS2 (558–587) crystals (space group P4332) that diffracted to 3.2 Å Bragg spacings, which were used for the structure described here.
These Vh1:IpaA-VBS2 crystals were obtained by hanging drop vapor diffusion based on the initial crystallization conditions identified by the Hauptman-Woodward Institute (Buffalo, NY). The best conditions obtained were from 2.1 M NaCl and 100 mM citric acid (pH 5).
X-Ray data collection, structure determination, and refinement
The Vh1:IpaA-VBS2 X-ray data were collected at the Advanced Photon Source (SCB-CAT 19ID beamline) and processed using MOSFLM (Collaborative Computational Project, 1994). The Vh1:IpaA-VBS1 crystals belong to space group P4332, with unit-cell parameters a=b=c=154.7 Å and a packing density, VM (Matthews, 1968), of 4.9 Å3/Da corresponding to a solvent content of about 74%. A total of 304 649 observations were merged to 10 867 reflections with an overall Rmerge of 0.11 and an Rmerge of 0.365 in the highest resolution (3.37–3.2 Å) shell. Data reduction statistics are provided in Table I. The Vh1:IpaA-VBS structure (PDBID 2GWW) was used as a search model. The helices were then refined as rigid bodies followed by extensive model building using the O (Jones et al, 1991) interactive graphics program, and positional and B-factor refinement using BUSTER/TNT (Tronrud et al, 1987; Bricogne, 1997). A Ramachandran plot analysis using the program PROCHECK (Collaborative Computational Project, 1994) indicates that 94.9% of all the residues lie in most favorable regions and that 5.1% are in additional allowed regions, and that all stereochemical parameters are better than expected at the given resolution. Refinement statistics are provided in Table II. Full-length human vinculin, VH (residues 1–740), Vh1, and Vt domains were purified as described previously (Izard et al, 2004; Bois et al, 2006).
Vinculin head and tail displacement assays
All binding assays were performed in PBS buffer. IpaA-VBS2 or the talin VBSs were added to the VH:Vt complex. The VH:Vt and newly formed VH:VBS complexes were resolved on homogeneous 7.5 PhastGel native polyacrylamide gels and stained with Commassie blue. Displaced Vt is not detectable in these assays because of its basic pI (Izard et al, 2004).
Solid-phase binding assays
Ninety-six-microtiter-well plates were coated with human vinculin domains Vh1 (residues 1–258), Vh1Δ126 (residues 127–258), or the S. flexneri IpaA domain IpaA-C-term (residues 559–633), at the indicated concentrations for 16 h at 4°C in 50 mM Tris pH 7.5, 50 mM NaCl (TN buffer). All subsequent steps were carried out at 21°C. Samples were blocked in TN buffer containing 2% BSA for 90 min. Wells were washed twice in TN buffer and incubated with IpaA-C-term at the indicated concentrations for 120 min with gentle shaking. Samples were washed twice and bound IpaA-C-term was detected by immuno-enzymatic detection using anti-GST monoclonal Ab (Upstate Biotechnology), at a final concentration of 0.5 μg/ml and anti-mouse IgG Ab coupled to horseradish peroxidase (Amersham Biosciences), at dilution of 1:2000. Samples were developed using 3,3,5,5-tetramethylbenzidine in 100 mM citric acid, 0.1% H2O2, pH 3.9. Reactions were stopped by adding H2SO4 to 1 N and absorbance at 450 nm was determined using an MR4000 plate reader (Dynatech). The values were obtained by subtracting the background values obtained from wells coated with BSA alone. The linearity of the reaction was tested in standard curve using purified IpaA-C-term at coating concentrations ranging from 2 to 187 pM. The Supplementary Table lists the residues involved in IpaA binding to the C-terminal helical bundle of Vh1.'
Note added in proof
Our recently performed surface plasmon resonance (SPR) experiments have independently confirmed that IpaA-VBS2 displays two-site binding to vinculin (Kd values of 7.8 and 510 nM). Furthermore, talin-VBS50, as well as talin-VBS1, -VBS2, and -VBS3, also display two-site binding to vinculin by SPR (with Kd values of 5.4 and 7.2 nM, 77 nM and 8.5 μM, 530 nM and 60 μM, and 74 nM and 74 μM, respectively). Therefore, in a manner akin to IpaA-VBS2, these four VBSs of talin appear to be also capable of binding to vinculin in its closed conformation, a scenario that would stabilize talin–vinculin interactions.
Supplementary Material
Supplementary Table
Acknowledgments
We are indebted to John Cleveland and Philippe Bois (Scripps-Florida) for helpful discussions and for critical review of the manuscript. Thanks also to the St Jude Children's Research Hospital (SJ) Hartwell Center for peptide synthesis; Charles Ross (SJ) for maintaining the computing facilities; Gerard Bricogne and Michael Roberts (Global Phasing, Ltd; GPhL) for substantial help with the crystallography; Clemens Vonrhein (GPhL) for continuous advice; the Hauptman-Woodward Institute for determining the initial crystallization conditions; and Megan Ellis, Christen Gregory, and Van Morris (all from SJ) for technical assistance. Finally, sincere thanks to the staff at the Advanced Photon Source (19ID and 22ID beamlines) for synchrotron support and Hajeung Park (Scripps-Florida) for Vh1Δ126 purification. Some of this work was carried out in Marco Giovaninni's laboratory (Inserm U674, Paris, France). This work was supported by the National Institutes of Health Grants GM071596 and AI067949, the Cancer Center Support (CORE) Grant CA21765, and by the American Lebanese Syrian Associated Charities (ALSAC). This is publication no. 19021 from The Scripps Research Institute.
Data deposition The coordinates have been deposited with the Protein Data Bank (PDB ID code 2IBF).
References
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430: 583–586 [DOI] [PubMed] [Google Scholar]
- Bass MD, Patel B, Barsukov IG, Fillingham IJ, Mason R, Smith BJ, Bagshaw CR, Critchley DR (2002) Further characterization of the interaction between the cytoskeletal proteins talin and vinculin. Biochem J 362: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass MD, Smith BJ, Prigent SA, Critchley DR (1999) Talin contains three similar vinculin-binding sites predicted to form an amphipathic helix. Biochem J 341: 257–263 [PMC free article] [PubMed] [Google Scholar]
- Bendori R, Salomon D, Geiger B (1989) Identification of two distinct functional domains on vinculin involved in its association with focal contacts. J Cell Biol 108: 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois PR, Borgon RA, Vonrhein C, Izard T (2005) Structural dynamics of alpha-actinin-vinculin interactions. Mol Cell Biol 25: 6112–6122 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Bois PR, O'Hara BP, Nietlispach D, Kirkpatrick J, Izard T (2006) The vinculin binding sites of talin and alpha-actinin are sufficient to activate vinculin. J Biol Chem 281: 7228–7236 [DOI] [PubMed] [Google Scholar]
- Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T (2004) Crystal structure of human vinculin. Structure 12: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Bricogne G (1997) Bayesian statistical viewpoint on structure determination: basic concepts and examples. Methods Enzymol 276: 361–423 [DOI] [PubMed] [Google Scholar]
- Chandrasekar I, Stradal TE, Holt MR, Entschladen F, Jockusch BM, Ziegler WH (2005) Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J Cell Sci 118: 1461–1472 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Critchley DR (2004) Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem Soc Trans 32: 831–836 [DOI] [PubMed] [Google Scholar]
- Fillingham I, Gingras AR, Papagrigoriou E, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL (2005) A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure 13: 65–74 [DOI] [PubMed] [Google Scholar]
- Gao G, Prutzman KC, King ML, Scheswohl DM, DeRose EF, London RE, Schaller MD, Campbell SL (2004) NMR solution structure of the focal adhesion targeting domain of focal adhesion kinase in complex with a paxillin LD peptide: evidence for a two-site binding model. J Biol Chem 279: 8441–8451 [DOI] [PubMed] [Google Scholar]
- Gilmore AP, Burridge K (1996) Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4–5-bisphosphate. Nature 381: 531–535 [DOI] [PubMed] [Google Scholar]
- Gingras AR, Vogel KP, Steinhoff HJ, Ziegler WH, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL (2006) Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry 45: 1805–1817 [DOI] [PubMed] [Google Scholar]
- Gingras AR, Ziegler WH, Frank R, Roberts GC, Critchley DR, Emsley J (2005) Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J Biol Chem 280: 37217–37224 [DOI] [PubMed] [Google Scholar]
- Hayashi I, Vuori K, Liddington RC (2002) The focal adhesion targeting (FAT) region of focal adhesion kinase is a four-helix bundle that binds paxillin. Nat Struct Biol 9: 101–106 [DOI] [PubMed] [Google Scholar]
- Hoellerer MK, Noble ME, Labesse G, Campbell ID, Werner JM, Arold ST (2003) Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure 11: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Irobi E, Burtnick LD, Urosev D, Narayan K, Robinson RC (2003) From the first to the second domain of gelsolin: a common path on the surface of actin? FEBS Lett 552: 86–90 [DOI] [PubMed] [Google Scholar]
- Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PR (2004) Vinculin activation by talin through helical bundle conversion. Nature 427: 171–175 [DOI] [PubMed] [Google Scholar]
- Izard T, Tran Van Nhieu G, Bois PR (2006) Shigella applies molecular mimicry to subvert vinculin and invade host cells. J Cell Biol 175: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard T, Vonrhein C (2004) Structural basis for amplifying vinculin activation by talin. J Biol Chem 279: 27667–27678 [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW (1995) F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 373: 261–264 [DOI] [PubMed] [Google Scholar]
- Johnson RP, Niggli V, Durrer P, Craig SW (1998) A conserved motif in the tail domain of vinculin mediates association with and insertion into acidic phospholipid bilayers. Biochemistry 37: 10211–10222 [DOI] [PubMed] [Google Scholar]
- Jones P, Jackson P, Price GJ, Patel B, Ohanion V, Lear AL, Critchley DR (1989) Identification of a talin binding site in the cytoskeletal protein vinculin. J Cell Biol 109: 2917–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Matthews BW (1968) Solvent content of protein crystals. J Mol Biol 33: 491–497 [DOI] [PubMed] [Google Scholar]
- McGregor A, Blanchard AD, Rowe AJ, Critchley DR (1994) Identification of the vinculin-binding site in the cytoskeletal protein alpha-actinin. Biochem J 301: 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagrigoriou E, Gingras AR, Barsukov IL, Bate N, Fillingham IJ, Patel B, Frank R, Ziegler WH, Roberts GC, Critchley DR, Emsley J (2004) Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J 23: 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Fernandez JL, Geiger B, Salomon D, Ben-Ze'ev A (1992) Overexpression of vinculin suppresses cell motility in BALB/c 3T3 cells. Cell Motil Cytoskeleton 22: 127–134 [DOI] [PubMed] [Google Scholar]
- Rodriguez Fernandez JL, Geiger B, Salomon D, Ben-Ze'ev A (1993) Suppression of vinculin expression by antisense transfection confers changes in cell morphology, motility, and anchorage-dependent growth of 3T3 cells. J Cell Biol 122: 1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronrud DE, Ten Eyck LF, Matthews BW (1987) An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallographica Section A 43: 489–501 [Google Scholar]
- Weis WI (2004) Cell biology: how to build a cell junction. Nature 430: 513–515 [DOI] [PubMed] [Google Scholar]
- Xu W, Baribault H, Adamson ED (1998) Vinculin knockout results in heart and brain defects during embryonic development. Development 125: 327–337 [DOI] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR (2006) The structure and regulation of vinculin. Trends Cell Biol 16: 453–460 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table