Abstract
Transcription by RNA polymerase II is accompanied by dynamic changes in chromatin, including the eviction/deposition of nucleosomes or the covalent modification of histone subunits. This study examined the role of the histone H3/H4 chaperones, Asf1 and HIR, in histone mobility during transcription, with particular focus on the histone exchange pathway, using a dual histone expression system. The results showed that the exchange of H3/H4 normally occurs during transcription by the histone chaperones. Both Asf1 and HIR are important for histone deposition but have a different effect on histone exchange. While Asf1 mediated incorporation of external H3/H4 and renewal of pre-existing histones, HIR opposed it. The balance of two opposing activities might be an important mechanism for determining current chromatin states.
Keywords: Asf1, HIR, histone exchange, histone H3/H4 chaperone, histone mobility
Introduction
The eukaryotic genome is packed in chromatin as nucleosomes. Nucleosome is the repeating unit containing two copies of the histones H2A, H2B, H3, and H4 (Luger et al, 1997). DNA functions such as transcription, replication, repair, and recombination are strongly influenced by the packaging state of the DNA in chromatin.
The bulk of the nucleosomes are assembled when the DNA is replicated in the S phase through the replication coupled (RC) pathway that is mediated by the histone H3/H4 chaperone, chromatin assembly factor 1 (CAF-1) (Verreault, 2000; Loyola and Almouzni, 2004). Outside of S phase, the histones are deposited into the nucleosome by HIRA (HIR complex in Saccharomyces cerevisiae) via a replication independent (RI) pathway (Henikoff and Ahmad, 2005). Each histone deposition pathway is related to the transcriptionally active or inactive chromatins. HIRA mediates the accumulation of the variant histone H3.3 in the active euchromatic region, while CAF-1 mediates the canonical H3 in the heterochromatic region (Mckittrick et al, 2004; Tagami et al, 2004). In yeast, HIR and CAF-1 are genetically redundant in the nucleosome assembly, although their precise roles are unclear. Another histone H3/H4 chaperone, Asf1, interacts with both CAF-1 and HIR, and affects both the RC and RI pathways (Krawitz et al, 2002; Green et al, 2005).
Nucleosomes normally block the progression of RNA polymerase II (pol II). Therefore, transcription must be accompanied by a large change in chromatin. One such change is the eviction and deposition of histones. Histones are evicted from and deposited onto the pol II track during transcription (Bernstein et al, 2004; Kristjuhan and Svejstrup, 2004; Lee et al, 2004; Schwabish and Struhl, 2004). The histone H2A/H2B chaperone, FACT (SPT16 and POB3 in yeast, and their homologs, hSpt16 and SSRP1, in human) complex, travels with pol II, binds the H2A/H2B dimers, and mediates the disassembly and reassembly of the nucleosomes (Formosa et al, 2002; Kireeva et al, 2002; Beloserkovskaya and Reinberg, 2004). The eviction and deposition process has a potential to provide a chance for histones to be exchanged along the gene. Indeed, H2A/H2B and H3/H4 is actively replaced during transcription, but with different kinetics: H2A/H2B is easily exchanged, but H3/H4 is less frequently exchanged and appears to be more strictly dependent on transcription (Kimura and Cook, 2001; Thiriet and Hayes, 2005; Jamai et al, 2007). In addition, the extent of H3 exchange is not even along the gene. H3 exchange is mainly observed around the promoter (Chow et al, 2005; Dion et al, 2007; Jamai et al, 2007; Mito et al, 2007). However, it also occurs within an entire region of the actively transcribed gene, with a lesser extent, compared to the promoter region (Choi et al, 2005; Schwartz and Ahmad, 2005; Wirbelauer et al, 2005; Daury et al, 2006). These reports indicate that multiple H3/H4 exchange pathways operate simultaneously. Although many factors are supposed to play a role in chromatin dynamics, the precise features of the changes in chromatin, the mobility of the histone subunits, and the responsible factors are not completely understood.
This study examined the role of the H3/H4 chaperones in chromatin dynamics to determine the changes that occur in chromatin during transcription. The episomal expression of histones H3 or H4 under the TFA1 promoter enabled histone mobility to be monitored along the DNA during transcription. Here we report that histone H3/H4 exchange occurs preferentially at the transcription sites and is mediated by Asf1 and HIR. Interestingly, Asf1 mediates the deposition of new histones, while HIR mediates the deposition of old histones. Their balanced activity might be important for maintenance or renewal of chromatin during transcription.
Results
Histones are exchanged during transcription
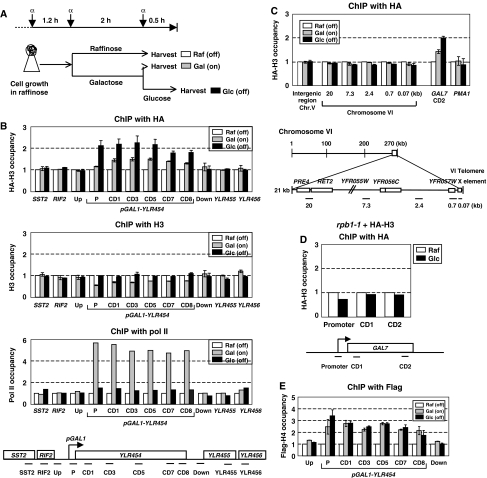
Transcription by pol II on a chromatin template is accompanied by dynamic changes in the chromatin structure, such as the eviction/deposition or exchange of histones. To understand histone mobility and the role of histone chaperones, a slight modification of a dual histone expression strategy was applied (Schermer et al, 2005). HA-H3 or Flag-H4 was produced episomally from the TFA1 promoter, whose expression is not controlled by the cell cycle (Ferea et al, 1999). In this strategy, most of the nontagged endogenous histones are produced in the S phase, and are incorporated into the chromatin mainly through the RC pathway. In contrast, the TFA1 promoter expresses HA-H3 (or Flag-H4) continuously. Therefore, tagged histones can be incorporated into the chromatin through both the RC and RI pathway because there is a ready supply of soluble histones throughout the cell cycle. Hence, if the chromatin histones are exchanged with histones from the soluble histone pool (a source in trans) during transcription, the level of tagged histones should increase comparatively in the transcribed region by replacing pre-existing histones. With this system, the galactose-inducible genes were analyzed by changing the medium from raffinose (off) to galactose (on), and then to glucose (off) to observe dynamic incorporation of tagged histones. The cells were treated with the α-factor to arrest them in the G1 phase before the galactose induction in an attempt to minimize the incorporation of HA-H3 (or Flag-H4) through replication (Figure 1A).
Figure 1.
Histone H3/H4 is exchanged during transcription. (A) Experimental design. Yeast cells carrying plasmids expressing HA-H3 or Flag-H4 were grown in raffinose medium and treated with the α factor as shown during incubation. (B) Histone H3 is actively exchanged during transcription. ChIP with 12CA5 (HA-H3), H3 (total H3), or 8WG16 (pol II) antibodies was performed as described in the Materials and methods with the indicated primer sets at the bottom. Each PCR signal was quantified and normalized to the intergenic control and the input DNA. In order to show the relative fold difference between the samples, the ChIP value obtained in raffinose was arbitrarily set to 1. The data are reported as the mean±s.d. from at least six independent experiments. (C) ChIP to measure the occupancy of HA-H3 in the nontranscribed regions (telomere and intergenic region), nonactivated gene, PMA1, and the galactose-activated gene (the coding region of GAL7). Bottom, the schematic diagram of the PCR primer pairs for the telomeric region from the right end of chromosome VI. (D) The HA-H3 occupancy in GAL7 regions was monitored in rpb1-1 after shifting to 37°C. (E) The Flag-H4 occupancy in yeast carrying the pRS316-TFA1-Flag-H4 was analyzed as described in (A). The data points were obtained from three independent experiments.
The cross-linking of HA-H3 to the GAL1-promoter-linked-YLR454 (∼8 kb gene) increased gradually as the cells were sequentially incubated in raffinose, galactose, and glucose media (Figure 1B, top panel). The expression of HA-H3 under the TFA1 promoter was not altered by the carbon source or the α factor in the media (Supplementary Figure S1). Nevertheless, the occupancy of HA-H3 across pGAL1-YLR454, but not in the upstream or downstream flanking regions, was higher in glucose (off) than in raffinose (off), indicating that HA-H3 from a soluble histone pool was incorporated successfully into the chromatin during transcription via galactose induction (relative value in raffinose equals 1). However, the total level of H3 immunoprecipitated with the anti-H3 antibody was low during transcription, but recovered to the original levels after transcription had been turned off, as reported previously (Figure 1B, the second panel) (Kristjuhan and Svejstrup, 2004; Schwabish and Struhl, 2004). The transcription state of the target gene depending on the carbon source was confirmed by pol II occupancy (Figure 1B, the third panel). This suggests that total histone H3 is rapidly displaced and deposited onto a transcribed region, during which the continuous exchange of H3 also occurs.
We then determined if HA-H3 incorporation was specific to transcription. First, we examined nontranscribed regions such as intergenic and telomeres. The occupancy of HA-H3 in silent regions was not altered by media changes (Figure 1C). Second, constitutively transcribed genes not regulated by galactose, such as SST2, RIF2, YLR455, or YLR456, which are located in the flanking region of pGAL1-YLR454 (Figure 1B), or PMA1, did not show occupancy changes of HA-H3, while GAL7 increased (Figure 1C). The relative fold difference in the HA-H3 occupancy was invisible in these nonactivated genes unless their transcription was induced by galactose. Lastly, we monitored HA-H3 occupancy in GAL7 after transcription was inhibited by shifting the rpb1-1 (yeast mutant expressing a temperature-sensitive largest subunit of pol II) to 37°C for 30 min prior to galactose induction (Nonet et al, 1987). Without transcription, HA-H3 incorporation was not changed (Figure 1D).
Flag-H4 was expressed under the TFA1 promoter to determine if H4 was also exchanged during transcription. Flag-H4 was preferentially exchanged at the same sites of transcription as H3 (Figure 1E), with minor differences in the exchange profile. The cross-linking of Flag-H4 during galactose incubation was observed as high as the glucose sample, although H4 appeared to be evicted and deposited normally (data not shown). In addition, H4 exchange tended to be more biased toward the promoter. All together, increased occupancy of HA-H3 and Flag-H4 in the pGAL1-YLR454 gene upon galactose induction was transcription specific, and certainly due to stable incorporation of H3/H4 from a soluble histone pool.
Asf1 and HIR are important for exchange of H3/H4
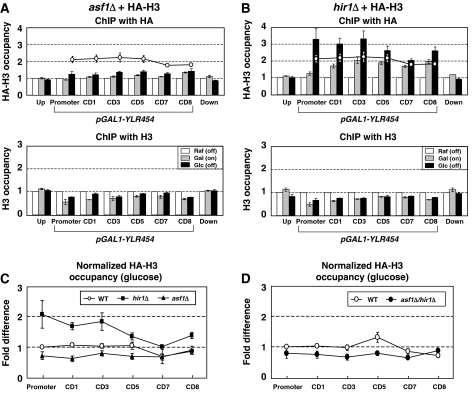
We next examined the role of Asf1 and HIR (consisting of Hir1, Hir2, Hir3, and Hpc2 in budding yeast) in the eviction/deposition and the exchange of histones in our system. Both asf1Δ and hir1Δ responded to the α factor as efficiently as wild type and G1 arrest was stably maintained during experiment as indicated by the flow cytometry analysis of DNA content (Supplementary Figure S2). Interestingly, as shown in Figure 2A (top panel), HA-H3 occupancy was significantly reduced in asf1Δ compared with that of the wild type (∼64% in average, data from wild type in Figure 1B is plotted overlapped). Similarly, the extent of histone deposition (recovery of total H3 occupancy in glucose) was also reduced in asf1Δ as reported (∼85%) (Figure 2A, bottom panel) (Schwabish and Struhl, 2006). However, the reduced deposition of histones could not account for the lower level of HA-H3 incorporation observed in this mutant. The incorporation efficiency of HA-H3 in asf1Δ was still lower than the wild type even after normalization to total H3 levels (Figure 2C). Total level of HA-H3 was unaffected by asf1Δ or hir1Δ (data not shown). In contrast, hir1Δ incorporated HA-H3 more efficiently than the wild type (∼137%), even though the deposition of H3 was apparently reduced to a similar extent as asf1Δ (Figure 2B). Interestingly, the hir1Δ-dependent increase of HA-H3 incorporation was more prominent around the promoter and the 5′ of the coding region (∼191% in this region when normalized to the H3 level) than downstream (Figure 2B and C). This suggests that (1) Asf1 and Hir1 are important for the eviction and deposition of histones during transcription, and are particularly important for restoring H3 levels after transcription is turned off, as reported, and (2) Asf1 and Hir1 play an additional role in the exchange and stable incorporation of new H3 from a soluble histone source. Asf1 appears to enhance the stable incorporation of external HA-H3 into chromatin while Hir1 appears to oppose it. This suggests that H3/H4 chaperones might assist pol II to progress by facilitating the eviction/deposition of histones, and at the same time regulate chromatin states by mediating the exchange of histones through the balance of the two opposing activities. Next, yeast with a double deletion of ASF1 and HIR1 was examined for the H3 exchange to see whether they function equivalently in the same pathway. Figure 2D shows that HA-H3 occupancy in asf1Δ/hir1Δ remained lower than wild type. Thus, Asf1 and HIR might not function independently or equivalently. Histone exchanging pathway could be more dependent on Asf1, such that Asf1 activity is a prerequisite for HIR.
Figure 2.
Asf1- and Hir1-dependent exchange of HA-H3. The exchange and stable incorporation of HA-H3 was decreased in asf1Δ (YC207) (A), while it was increased in hir1Δ (YC199) (B). ChIP was performed and the data were analyzed as described in Figure 1. The experiment was repeated at least five times. The data points of the HA-H3 occupancy obtained in the wild type (glucose sample) was depicted as open circles. The occupancy of total H3 was performed as described in Figure 1. (C, D) The relative occupancy of the HA-H3 obtained from each genetic background (glucose sample) is shown as the fold difference (the value obtained from promoter region of the wild type was arbitrarily set to 1). Each value was obtained after normalization by the level of total H3.
Histone mutations on the Asf1-binding surface lead to an aberrant pattern of histone exchange
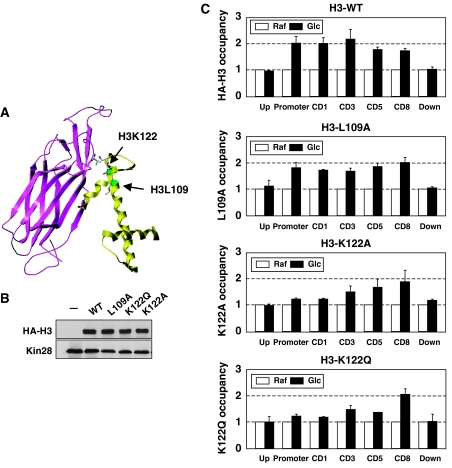
The behavior of the histone H3 mutants with a specific defect in chaperone interaction was examined to confirm the role of histone chaperones in histone exchange. The interaction between H3 and Asf1 has been studied based on a well-defined structure (Antczak et al, 2006; English et al, 2006; Agez et al, 2007; Natsume et al, 2007). The C-terminal α3 helix of H3 (amino acids 122–134) makes direct contact with the N-terminal β strands of Asf1. Consistent with this, H3 K122 is essential for Asf1 binding and develops an asf1Δ-like phenotype (Zeocin sensitivity) when mutated (English et al, 2006). In particular, K122A is defective in PHO5 induction, where Asf1-mediated chromatin disassembly is essential (English et al, 2006). On the other hand, L109 is located on the secondary interaction surface of H3, which is expected to be less important for Asf1 binding (Figure 3A) (Munakata et al, 2000; Mousson et al, 2005; Antczak et al, 2006; English et al, 2006; Agez et al, 2007; Natsume et al, 2007). Unlike asf1Δ, L109A showed no sensitivity to the DNA-damaging agent (MMS; methyl methane sulfonate) or the DNA replication-blocking agents (hydroxyurea and Camptothecine) (Supplementary Figure S3).
Figure 3.
H3 mutation defective in Asf1 interaction leads to the aberrant histone exchange. K122 mutants showed an aberrant pattern of histone exchange. (A) The location of each H3 mutation in the structure of the yAsf1 N (1–164) and Xenopus laevis H3 (60–135) complex (English et al, 2006). Asf1 and H3 are colored in purple and yellow, respectively. The locations of K122 and L109 are colored in green. The residues of L109 (H3), K122 (H3), and V92 (Asf1) are shown as sticks. The diagram was generated using SYBYL 7.3 (Tripos, USA) on the basis of Linux Redllat 4.0. (B) The expression level of the H3 mutants under the TFA1 promoter was similar. The immunoblotting analysis was performed with the whole-cell extract prepared from the wild type (YC73) that harbors the pRS315-TFA1-HA-H3, L109A, K122Q, or K122A. (C) The incorporation of the H3 mutants was analyzed as described in Figure 1. The experiment was repeated at least two times.
The histone exchange pattern of the K122 (K122Q, K122A) and L109A mutants of H3 was examined. Protein level of the H3 mutants under the TFA1 promoter was measured in the wild-type background in cultures grown without the α factor. All H3 mutants were expressed at similar levels, indicating that protein stability was not significantly affected and they can normally be incorporated into chromatin (Figure 3B). The α factor was then added to examine the transcription-coupled incorporation of the H3 mutants through histone exchange, as described in Figure 1A. Chromatin immunoprecipitation (ChIP) from raffinose and glucose were compared. Interestingly, K122A and K122Q, which were predicted to have a defect in the Asf1 interaction, were not normally incorporated into the chromatin, while L109A was able to incorporate as successfully as wild type (Figure 3C). Remarkably, the incorporation of K122A and K122Q showed an aberrant histone exchange profile within the gene. Histone exchange was mainly affected in the promoter and the 5′ coding region, and it was gradually restored toward the 3′ of the gene, which is opposite to what hir1Δ showed in Figure 2B. Although asf1Δ developed a defect in histone exchange within the entire gene region, the H3 mutants defective in Asf1 binding lacked H3 exchange proximal of the promoter region. These data suggest the importance of the Asf1–H3 interaction, especially around the promoter region. The broader defect of asf1Δ than H3 K122 may suggest that Asf1 might adopt multiple ways to mediate histone exchange during different phases of transcription, or that there are alternative histone exchanging pathways that bypass direct interaction between Asf1 and H3. Taken together, Asf1 and HIR are important for the exchange of H3/H4 along the transcription track, particularly during early transcription, by playing opposite roles.
Direct interaction between Asf1–H3/H4 is important for histone exchange
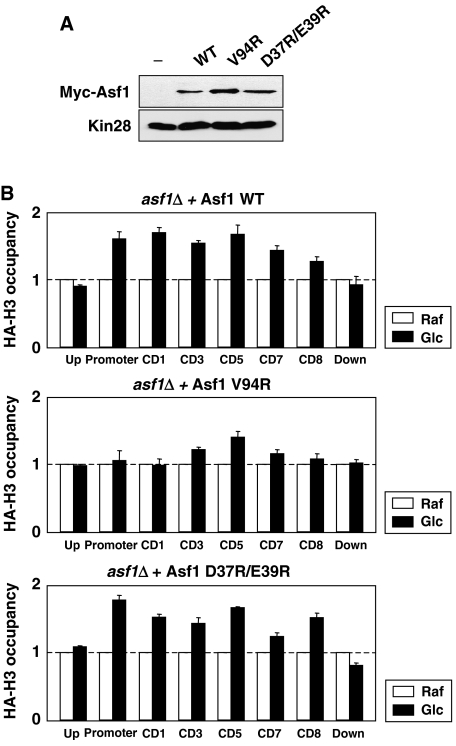
Detailed analysis of Asf1–HIRA and Asf1–H3/H4 structures predicts that Asf1 is able to interact with HIRA and H3/H4 simultaneously to form a ternary complex by bridging them (Antczak et al, 2006; Mousson et al, 2005, Tang et al, 2006). Based on this structural configuration, it was suggested that Asf1 has a potential to disassemble the nucleosome to present H3/H4 to HIR. Next, we further analyzed Asf1–H3 interaction during histone exchange and also asked whether direct interaction between Asf1 and HIR is absolutely required. To do this, two Asf1 mutants were examined; V94R, a mutant defective in H3 binding, and D37R/E39R, a mutant defective in HIR binding (Mousson et al, 2005). V94R carries asf1Δ phenotypes such as DNA damage sensitivity, thermo-sensitive growth, and a defect in gene silencing, whereas D37R/E39R shows a defect only in gene silencing. The experimental difference using H3-K122 and Asf1 V94R is that H3-K122 verifies Asf1's interaction with external H3, whereas V94R verifies its interaction with both internal and external H3. Wild-type and two Asf1 mutants were expressed at similar levels in yeast (Figure 4A). asf1Δ, supplemented with wild-type Asf1, recovered the HA-H3 level via subunit exchange, although the overall efficiency was lower than ASF1 wild-type yeast (Figure 4B, top panel). However, interestingly, asf1Δ supplemented with V94R remained defective (Figure 4B, middle panel). Its pattern was more like asf1Δ rather than H3-K122 (Figure 3C), suggesting that Asf1–H3 interaction is a major step to promote nucleosome disassembly along the gene and this disassembly step of pre-existing nucleosome is necessary prior to subunit exchange. Asf1–Hir1 interaction is important for gene silencing but does not have a role in Asf1-dependent DNA damage repair pathway (Mousson et al, 2005). Interestingly, H3 incorporation in D37R/E39R was as normal as wild type in our assay. It suggests that direct interaction between Asf1 and Hir1 may not be necessary for determining the extent of histone exchange.
Figure 4.
Asf1 in direct contact with H3 is important for histone exchange. (A) Wild type and Asf1 mutants, V94R and D37R/E39R, were expressed in asf1Δ. All Asf1 construct provide 13 Myc tag. (B) HA-H3 occupancy within a target gene was diminished in V94R mutant, but not in D37R/E39R. The incorporation of HA-H3 in Asf1 mutants was analyzed as described in Figure 1. The experiment was repeated at least four times.
Discussion
Chromatin undergoes a dynamic change as pol II transcribes genes through it. Our results show that histone H3/H4 is evicted, deposited, and actively exchanged. This suggests an intermediate step involving the complete or partial unfolding of nucleosomes into subunits. These results are consistent with recent ideas about transcription-dependent exchange of H3/H4 or potential disruption of H3/H4 tetramer by Asf1 (English et al, 2006; Morillon, 2006; Workman, 2006; Kulaeva et al, 2007; Natsume et al, 2007).
Asf1 and HIR mediate dynamic histone exchange while transcription is ongoing. Interestingly, the exchange of pre-existing histones with external histones was decreased in asf1Δ, but increased in hir1Δ. While both Asf1 and Hir1 deposit histones into chromatin, the histone source they utilize might be different. Asf1 catalyzes the incorporation of histones from a source in trans (external free histones), while Hir1 catalyzes the incorporation of histones from a source in cis (original chromatin histones). It resembles the two opposing activities of Spt16 and Pob3 within the FACT complex for the concerted disassembly and reassembly of the nucleosomes during transcription (Formosa et al, 2002; Beloserkovskaya and Reinberg, 2004).
The role of histone chaperones in histone eviction and deposition has been observed mostly in the PHO5 and PHO8 promoters. Asf1 is important for the activation-dependent eviction of the nucleosomes from the promoters of the PHO genes. Moreover, upon repression, Asf1 and HIR are responsible for the deposition of nucleosomes to the promoters (Adkins et al, 2004; Schermer et al, 2005; Korber et al, 2006). We assumed that histone eviction/deposition and subunit exchange must take place simultaneously, such that subunits could be exchanged while nucleosomes are deposited. According to our data, the increase in H3 exchange led by hir1Δ and the decrease in H3 exchange led by asf1Δ are more prominent around the promoter region, indicating that Asf1 and HIR are major histone exchanging partners that operate together in this region. The predominant exchange of H3 at the promoter has been reported by other groups (Chow et al, 2005; Jamai et al, 2007). Our results suggest Asf1 and HIR to be excellent devices that modulate the chromatin states during early transcription. In addition to Asf1-HIR pair, we assume that there must be other pathways that allow continuous histone exchange throughout the coding region. As asf1Δ and V94R diminish overall histone exchange along the gene, Asf1 might play an overlapping role downstream, such as by providing split nucleosomes to other proteins.
Although the role of HIR in the histone exchange pathway is not completely understood, there is some concern as to what the general consequence of their opposing activities is. One possibility is that the renewal of chromatin can be regulated by the extent of histone exchange through the balance between the two activities. If so, histone exchange might allow modification of nucleosome composition or pre-existing histone modification. In this regard, it is interesting that Asf1 is particularly essential for H3 exchange around the promoter and the early-transcribed region through a direct interaction with H3.
Chromatin can be remodeled by substituting conventional histones with variants by their cognate histone chaperones together with chromatin remodeling complexes. HIRA in higher eukaryotes is a histone chaperone specific to H3.3 and is purified in association with Asf1 (Tagami et al, 2004; Green et al, 2005). HIRA and Asf1 cooperate to contribute to nucleosome formation in vitro (Green et al, 2005). Given the activities of the yeast chaperones, Asf1 (in this case, Asf1a, as HIRA preferentially binds Asf1a rather than Asf1b) might participate more actively in the HIRA-dependent RI pathway than has been generally understood. This predicts that the concerted activity of HIRA and Asf1 is essential for both introducing H3.3 into and retaining H3.3 within the chromatin (Mito et al, 2005). Hence, any epigenetic information, either histone modifications or nucleosome composition, can be preserved, while it has a continuous opportunity to change. However, more study will be needed to determine the precise roles of Asf1 and HIR in the dynamic regulation of the chromatin states and gene expression.
Materials and methods
Yeast strain and plasmid construction
Yeast strains used in this study: YC73 (MATa, ura3-1, leu2-3,112, trp1-1, his3-11,15, ade2-1, can1-100, TRP1∷pGAL1-YLR454w), YC199 (as YC73, hir1Δ∷KanMX), YC207 (as YC73, asf1Δ∷KanMX), YC252 (as YC73, asf1Δ∷HIS3, hir1Δ∷KanMX), or Y262 (MATa, ura3-52, rpb1-1; provided by DK Lee).
The deletion mutants were constructed using one-step PCR-mediated disruption, which replaces the entire gene with the KanMX4 cassette. The GAL1-YLR454 strains were constructed by the one-step integration of the GAL1 promoter fragment upstream of the YLR454w coding region. The pRS315-TFA1-HA-H3 or pRS316-TFA1-Flag-H4 (TFA1 promoter driven HA-H3 or Flag-H4) was generated by PCR using pRS315-CEG1 as a starting material, which was provided by S Buratowski. The pRS315-TFA1-HA-H3 K122Q, K122A, or L109A (TFA1 promoter driven HA-H3 mutants) were generated by site-directed mutagenesis. pRS316-ASF1, V94R, or D37R/E39R (13Myc-tagged Asf1 or mutants under the endogenous promoter) was constructed using pRS315-Asf1-13myc, V94R-13myc, or D37R/E39R-13myc as a starting material (provided by F Ochsenbein), respectively. All constructs were confirmed by sequencing.
Growth conditions and analysis
Yeasts were grown at 30°C in either YPD media or in synthetic minimal media that lacked the nutritional supplements required for maintaining plasmids. For the galactose induction experiment, cells were grown to an A600=0.4 in SC-LEU or URA plus 2% raffinose, treated with the α factor (5 μg/ml), and incubated for 1.2 h. The synchronized cells were divided into two parts and incubated in the medium containing 2% raffinose or 2% galactose in the presence of α factor. After 2 h, half of the 2% galactose culture was quickly switched to a medium containing 2% glucose plus the α factor to repress GAL1 and incubated for another 0.5 h. In rpb1-1, yeast cells were grown overnight at 24°C. Before α arrest, the temperature of the culture was shifted to 37°C to inactivate Rpb1.
Immunoblotting
To analyze the expression of tagged histones, yeasts were harvested by centrifugation and boiled in a cracking buffer (8 M urea, 40 mM Tris–HCl, pH 8.0, 5% SDS, 0.1 M EDTA, 0.01% β-mercaptoethanol, 0.001% bromophenol blue supplemented with complete protease inhibitors) for 10 min with occasional vortexing. The resulting lysate was clarified by centrifugation and separated onto a 15% denaturing polyacrylamide gel and analyzed by immunoblotting with 12CA5 (Roche), Myc (Roche), or Kin28 (Covance) antibodies.
ChIP
ChIPs were performed as described previously (Cho et al, 2001; Kim et al, 2005). The PCR signals were quantified using a Phosphoimager (Fujix BAS 2040) and normalized to the input DNA and the intergenic control. PCR primers were (the coordinates are defined relative to the translation initiation site): PMA1 CD2 (+584 to +807), GAL1-YLR454 promoter (−271 to +69), CD1: 1 kb (+951 to +1149), CD3: 3 kb (+2954 to +3150), CD5: 5 kb (+5091 to +5283), CD7: 7 kb (+7096 to +7278), CD8: 8 kb (+7536 to +7866), SST2 (+28 to +196), RIF2 (+561 to +786), upstream region of YLR454 (−755 to −525), downstream (+8741 to +8873), YLR455 (+374 to +546), GAL7 promoter (−296 to −80), CD1 (+59 to +390), CD2 (+570 to +907), intergenic region Chromosome V (position on chromosome, 9716–9863).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Information
Acknowledgments
We thank Stephen Buratowski, Peter D Kaufman, Françoise Ochsenbein, and Dongki Lee for providing yeast strains and plasmids. We thank HJ Park (Medicinal Chemistry Laboratory) and KW Song (Yonsei University) for technical assistance with structure graphics and FACS analysis, respectively. This work was supported by grants from Korea Research Foundation (2005-E00045) and from Basic Research Program of the Korea Science & Engineering Foundation (RO1-2006-000-10707-0) to E-J Cho. While this paper was in progress, Rufiange et al, reported similar results that Asf1 is important for transcription-dependent incorporation of H3 at promoters (Rufiange A, Jacques P-É, Bhat W, Robert F, Nourani A (2007). Mol Cell 27: 393–405).
References
- Adkins W, Howar SR, Tyler JK (2004) Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 14: 657–666 [DOI] [PubMed] [Google Scholar]
- Agez M, Chen J, Guerois R, van Heijenoort C, Thuret JY, Mann C, Ochsenbein F (2007) Structure of the histone chaperone Asf1 bound to the histone H3 C-terminal helix and functional insights. Structure 15: 191–199 [DOI] [PubMed] [Google Scholar]
- Antczak AJ, Tsubota T, Kaufman PD, Berger JM (2006) Structure of the yeast histone H3–ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloserkovskaya R, Reinberg D (2004) Facts about FACT and transcript elongation through chromatin. Curr Opin Genet Dev 14: 139–146 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL (2004) Global nucleosome occupancy in yeast. Genome Biol 5: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S (2001) Opposing effects of Ctk1 kinase and fcp1 phosphatase at ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15: 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ES, Shin JA, Kim HS, Jang YK (2005) Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res 33: 7102–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N (2005) Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep 6: 354–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daury L, Chailleux C, Bonvallet J, Trouche D (2006) Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep 7: 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ (2007) Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK (2006) Structural basis for the histone chaperone activity of Asf1. Cell 127: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferea TL, Botstein D, Brown PO, Rosenzweig RF (1999) Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA 96: 9721–9726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ (2002) Defects in SPT16 or POB3(yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR III, Kaufman PD (2005) Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol 15: 2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K (2005) Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21: 133–153 [DOI] [PubMed] [Google Scholar]
- Jamai A, Imoberdorf RM, Strubin M (2007) Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell 25: 345–355 [DOI] [PubMed] [Google Scholar]
- Kim DH, Shim JS, Kwon HJ (2005) Coordinated transcriptional regulation of calmegin, a testis-specific molecular chaperon, by histone deacetylase and CpG methyltransferase. Exp Mol Med 37: 492–496 [DOI] [PubMed] [Google Scholar]
- Kimura H, Cook PR (2001) Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol 153: 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM (2002) Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell 9: 541–552 [DOI] [PubMed] [Google Scholar]
- Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Hörz W (2006) The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J Biol Chem 281: 5539–5545 [DOI] [PubMed] [Google Scholar]
- Krawitz DC, Kama T, Kaufman PD (2002) Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol Cell Biol 22: 614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjuhan A, Svejstrup JQ (2004) Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J 23: 4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Studitsky VM (2007) Transcription through chromatin by RNA polymerase II:histone displacement and exchange. Mutat Res 618: 116–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G (2004) Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 1677: 3–11 [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mckittrick E, Gafken PR, Ahmad K, Henikoff S (2004) Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA 101: 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2005) Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37: 1090–1097 [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2007) Histone replacement marks the boundaries of cis-regulatory domains. Science 315: 1408–1411 [DOI] [PubMed] [Google Scholar]
- Morillon A (2006) Is histone loss a common feature of DNA metabolism regulation? Biochem Cell Biol 84: 450–462 [DOI] [PubMed] [Google Scholar]
- Mousson F, Lautrette A, Thuret JY, Agez M, Courbeyrette R, Amigues B, Becker E, Neumann JM, Guerois R, Mann C, Ochsenbein F (2005) Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc Natl Acad Sci USA 102: 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M (2000) A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5: 221–233 [DOI] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T (2007) Structure and function of the histone chaperone CIA/ASF1 complexed with histone H3 and H4. Nature 446: 338–341 [DOI] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol 7: 1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermer UJ, Korber P, Hörz W (2005) Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell 19: 279–285 [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K (2004) Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K (2006) Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell 22: 415–422 [DOI] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K (2005) Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R (2006) Structure of a human ASF1a–HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol 13: 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ (2005) Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev 19: 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A (2000) De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev 14: 1430–1438 [PubMed] [Google Scholar]
- Wirbelauer C, Bell O, Schübeler D (2005) Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev 19: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL (2006) Nucleosome displacement in transcription. Genes Dev 20: 2009–2017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Information