Abstract
Reducible polycations represent promising carriers of therapeutic nucleic acids. Oligomers of 2-dimethylaminoethyl methacrylate (DMAEMA) containing terminal thiol groups were synthesized by reversible addition-fragmentation chain transfer (RAFT) polymerization using difunctional chain transfer agent. Reducible poly(DMAEMA) (rPDMAEMA) was synthesized by oxidation of the terminal thiol groups, forming a polymer with disulfide bonds in the backbone. Physico-chemical properties of DNA polyplexes of rPDMAEMA were evaluated by dynamic and light scattering methods, revealing lower structural density and DNA content than control PDMAEMA polyplexes. Cytotoxicity and transfection activity of rPDMAEMA-based DNA polyplexes were evaluated in vitro. In comparison with control PDMAEMA, only minimum toxic effects of rPDMAEMA were observed in a panel of cell lines. Transfection activity was tested in B16F10 mouse melanoma and six human pancreatic cancer cell lines. rPDMAEMA polyplexes showed a comparable or better activity than control PDMAEMA polyplexes.
Keywords: gene delivery, reducible polycations, RAFT polymerization, transfection, cytotoxicity
1. INTRODUCTION
Redox-sensitive polyplexes attract attention because of their promise of a significant improvement of the efficacy of the gene delivery process and reduced toxicity [1]. Such polyplexes are typically prepared by introducing disulfide bonds into the structure of the polycations and show the capability to release the therapeutic nucleic acids selectively in the subcellular reducing space. The easy intracellular reversibility of disulfide bonds in redox-sensitive polyplexes was already proven to be a highly advantageous feature for delivering a variety of nucleic acids, including plasmid DNA, mRNA, antisense oligonucleotides, and siRNA [2-6].
While easy availability and potential inherent biological activity made peptide-based polycations among the most attractive for preparation of redox-sensitive polyplexes [7-9], recent reports also indicate increased interest in new types of synthetic reducible polycations. Synthesis of well-defined analogues of reducible linear polyethylenimine (PEI) by oxidation of α,ω-bisthiol-oligoethylenimines has been described by Park and colleagues [10]. The synthesized reducible PEI show reduced cytotoxicity and comparable transfection activity when compared with control non-reducible PEI. The synthesized reducible PEI represent better defined polymers in comparison with earlier attempts to prepare reducible PEI by random crosslinking of PEI oligomers [11, 12]. In another recent example, reducible poly(amido ethylenimines) (rPAEI) were synthesized by Michael addition reactions of different oligoamines with cystamine bisacrylamide linker by Kim and Feijen [13, 14]. Like the reducible PEI, rPAEI showed promising activity in delivering plasmid DNA as well as siRNA [15].
(Meth)acrylate-based polycations, such as poly(2-dimethylaminoethyl methacrylate) (PDMAEMA), have shown promising transfection activity but their high cytotoxicity and lack of biodegradability limit their potential in gene delivery [16-20]. This study reports, for the first time, a synthesis of reducible PDMAEMA copolymers using reversible addition-fragmentation chain transfer (RAFT) polymerization and their use in gene delivery. The polymers show minimal cytotoxicity and transfection activity comparable to or better than control non-reducible PDMAEMA.
2. MATERIALS AND METHODS
2.1. Materials
2,2′-azobis(isobutyronitrile) (AIBN, Sigma-Aldrich, 98%), hexylamine (Sigma-Aldrich, 99%), 1,4-diisopropenylbenzene (TCI), sulfur (EMD, 99%), anhydrous methanol (EMD, 99.8%), sodium methoxide (Alfa Aesar, 25∼30%), benzyl chloride (Alfa Aesar, 99%), carbon tetrachloride (Sigma-Aldrich, 99%), tetrahydrofuran (VWR, 99%), diethyl ether (EMD, 99%), hexane (EMD, 98.5%), and dithiothreitol (DTT, Fluka, >99%) were used as received. 2-(Dimethylamino)ethyl methacrylate (DMAEMA, Alfa Aesar, 99%) was passed through a column of activated basic alumina to remove the inhibitor and stored at 4°C before use. gWiz™ High-Expression Luciferase plasmid (6.7 kbp) was purchased from Aldevron and used as received. Dulbecco's Modified Eagle Medium (DMEM), Roswell Park Memorial Institute medium (RPMI-1640), fetal bovine serum (FBS), and L-glutamine were from Invitrogen.
2.2. Synthesis
2.2.1 Synthesis of bisfunctional RAFT agent
Bisfunctional RAFT agent, 1,4-bis(2-(thiobenzoylthio)prop-2-yl)benzene (BTBP), was synthesized by a reaction of dithiobenzoic acid (DTBA) with 1,4-diisopropenylbenzene as previously described [21]. First, DTBA was synthesized by reacting elemental sulfur with benzyl chloride in methanolic solution of sodium methoxide [22]. DTBA was obtained as an oil with 38% yield. Then, a mixture of DTBA (14.0 g, 0.091 mol) and 1,4-diisopropenylbenzene (6.5 g, 0.041 mol) in carbon tetrachloride (100 mL) was heated at 80°C for 24 h. The volatiles were removed under reduced pressure and the residue was triturated with 1:4 diethyl ether/hexanes to isolate the BTBP as a pink solid (yield 26%). 1H NMR, δ (ppm): 2.0 (12H, s); 7.3 (m, 4H); 7.47 (m, 6H); and 7.84 (m, 4H). 13C NMR: δ (ppm) in CDCl3: 227, 146.5, 142.6, 132,128.3, 126.5, 126.3, 56.2, 28.2.
2.2.2 RAFT polymerization of DMAEMA
RAFT polymerization of DMAEMA was performed in THF at 60°C using AIBN as the initiator ([BTBP]:[AIBN] > 20). Solution containing DMAEMA (1 g), AIBN (80 mg), and BTBP was added into glass ampoule, thoroughly deoxygenated, sealed under vacuum, and placed in a thermostated water bath at 60°C for 48 h. The polymer, α,ω-dithioester-terminated PDMAEMA (DTE-PDMAEMA), was obtained by precipitation into excess of hexane and isolated by filtration. All the polymers in this study were analyzed by 1H-NMR on Varian spectrometer (400 MHz) using d-chloroform as solvent. The number average (Mn) and weight-average (Mw) molar mass and polydispersity index (PDI, Mw/Mn) of the polymers were determined by size exclusion chromatography (SEC) on Polymer Labs PL gel 5 μm mixed C column. The system was equipped with a 3-angle miniDAWN light scattering detector and Optilab differential refractometer (Wyatt Technology, Inc.). N,N-Dimethylformamide (DMF) was used as an eluent at a flow rate of 1.0 mL/min and temperature of 35°C. SEC data were analyzed using ASTRA software from Wyatt Technology. Refractive index increments of the polymers in DMF were determined by the Optilab differential refractometer and used in SEC analysis.
2.2.3 Synthesis of α,ω-dithiol-PDMAEMA (DT-PDMAEMA)
To synthesize bisthiol-terminated PDMAEMA, the α,ω-dithioester PDMAEMA (1 g) was dissolved in THF (4 mL) containing a few drops of aqueous sodium bisulfite (Na2S2O4), the reaction mixture was purged of oxygen by bubbling with N2 for 30 min, hexylamine (0.2 mL) was added, and the reaction was stirred for 3 h under N2. The reaction mixture was then added dropwise to a 10-fold excess of diethyl ether, and the polymer was collected by filtration.
2.2.4 Synthesis of reducible PDMAEMA (rPDMAEMA)
Reducible PDMAEMA was prepared by one of the following methods: (i) small amount of DMSO was added to solution of DT-PDMAEMA in deionized water and the reaction mixture was stirred for 13 days. Then the water was removed and rPDMAEMA was isolated by precipitation from DMSO; (ii) DMSO (0.2 mL) and hexylamine (0.2 mL) were added to solution of DTE-PDMAEMA in methanol and the reaction mixture was stirred at room temperature under oxygen atmosphere. The solvent was then removed and rPDMAEMA dissolved in THF and precipitated with hexane.
2.2.5 Synthesis of control PDMAEMA
Free radical polymerization of DMAEMA was performed in THF at 60°C using AIBN as the initiator ([DMAEMA]:[AIBN] = 11 for PDMAEMA1, [DMAEMA]:[AIBN] = 45 for PDMAEMA2). Solution containing DMAEMA (1.0 g) and AIBN in THF (2.0 mL) was added into glass ampoule, thoroughly deoxygenated, sealed under vacuum, and placed in a thermostated water bath at 60°C for 20 h. PDMAEMA was obtained by precipitation into excess of hexane and isolated by filtration.
2.3. Formulation of DNA polyplexes
Polyplexes were prepared at a desired N:P ratio by adding a predetermined amount of polycation to the solution of plasmid DNA in 30 mM sodium acetate pH 5.0 (PDMAEMA and rPDMAEMA) and in 10 mM HEPES pH 7.4 (PEI) to achieve a final DNA concentration of 20 μg/mL. Mass of 325 per one phosphate group of DNA was assumed in the calculations.
2.4. Light scattering analysis of the polyplexes
The static light scattering (SLS) measurements were performed with a DAWN EOS light scattering instrument (Wyatt Technology) equipped with a 30 mW linearly polarized GaAs laser. The analysis of the polyplex solutions was conducted at an angular range θ = 22.5°–147° in 20 mL glass scintillation vials at 25°C. The static light scattering data were analyzed by the second-order Debye fit (R(θ)/Kc vs. sin2(θ/2) to obtain weight-average molar mass (Mw) and z-average of the root mean square radius (radius of gyration, RG) of the DNA polyplexes [23]. K is the optical constant which includes the square of the refractive index increment (ν); R(θ) is the Rayleigh ratio, proportional to the intensity of the light scattered from solutions and the polyplex concentration c in g/mL. Extrapolation to zero concentration was not performed due to very low concentrations of the polyplexes (< 10−5 g/mL) [24]. The polyplex concentrations were calculated using 325 g/mol phosphates for DNA and 157.11 g/mol amines for all DMAEMA polycations. Complete complexation between DNA and polycations was assumed at N:P < 1 and 1:1 stoichiometry at N:P > 1. The refractive index increments of the DNA polyplexes were calculated as weight-average values using the same assumptions about the polyplex stoichiometry (νDNA = 0.185, νPDMAEMA = 0.178) [19, 25]. It was assumed that the refractive index increment of rPDMAEMA is identical to that of PDMAEMA.
The determination of hydrodynamic diameters and zeta potentials of the DNA polyplexes was performed by dynamic light scattering (DLS) using ZetaPlus Particle Size and Zeta Potential analyzer (Brookhaven Instruments) equipped with 35 mW solid state laser (658 nm). Scattered light was detected at 90° angle and a temperature of 25°C. Mean hydrodynamic diameters were calculated from size distribution by weight, assuming a lognormal distribution using the supplied algorithm. Zeta potential values were calculated from measured velocities using Smoluchowski equation.
2.5. Cell lines
Mouse melanoma B16F10 cell line and a panel of human pancreatic cancer cell lines (Panc-1, AsPC-1, COLO-357, BxPC-3, MiaPaCa, and Panc-28) were used in this study. The pancreatic cell lines were a kind gift from Dr. Sarkar (Karmanos Cancer Institute). BxPC-3 and Panc-28 are human pancreatic adenocarcinoma cell lines, Panc-1 and MiaPaCa are human ductal pancreatic adenocarcinoma cell lines, AsPC-1 is human pancreatic adenocarcinoma derived from ascites metastatic site, COLO-357 is derived from lymph node metastasis of pancreatic carcinoma. B16F10, Panc-1, AsPC-1, MiaPaCa and Panc-28 were maintained in DMEM, COLO-357 was maintained in DMEM supplemented with 2 mM L-glutamine and BxPC-3 was maintained in RPMI 1640. The above media were supplemented with 4 mM L-alanyl-L-glutamine and 10% FBS.
2.6. Cytotoxicity evaluation
Cytotoxicity of rPDMAEMA and control PDMAEMA in EA.hy926, and MiaPaCa cells was determined by MTS assay using a commercially available kit (CellTiter 96® Aqueous Cell Proliferation Assay, Promega). Twenty thousand cells seeded in a 96-well plate 2 days before the experiment were incubated with increasing concentration of the polycations in 100 μL of DMEM/FBS. The medium was removed after 16 h and replaced with a mixture of 200 μL fresh DMEM and 20 μL MTS reagent solution. The cells were incubated for 1 h at 37°C in CO2 incubator. The absorbance of each sample was then measured at 505 nm to determine cell viability. The results are expressed as mean percentage cell viability relative to untreated cells ± S.D.
2.7. Transfection efficiency in vitro
All transfection studies were performed in 48-well plates with cells plated 24 h before transfection at the seeding density of 60,000 cells per well (pancreatic cancer cell lines) and 30,000 cells per well (B16F10) as described previously [4, 9]. The cells were incubated for 3 h with the polyplexes in 150 μL of complete growth medium with 10% FBS. After 3 h of incubation, the transfection mixture was completely removed and the cells were cultured for additional 24 h in complete growth medium prior to analysis of reporter gene expression. Calibration curve was constructed using recombinant luciferase and the transfection results are expressed as pg of luciferase per mg of cellular protein ± SD of triplicate samples.
3. RESULTS AND DISCUSSION
3.1. Synthesis and characterization of rPDMAEMA
In a search for efficient polymer carriers of nucleic acid therapeutics, methacrylate- and acrylate-based polycations have been widely investigated due to convenient synthesis by free radical polymerization and the possibility to optimize their properties by copolymerization of a number of ionic and non-ionic comonomers [19]. PDMAEMA emerged as one of the most promising polycations in this class [26, 27]. Substantial cytotoxicity, however, impeded PDMAEMA prospects in gene delivery and prompted a development of biodegradable DMAEMA alternatives [16].
Reducible polycations that are degraded in the reducing intracellular environment are known to efficiently deliver nucleic acids. We hypothesize that reducible PDMAEMA would exhibit similar beneficial properties for gene delivery as other previously reported reducible polycations. A widely exploited synthetic approach relies on the oxidation of oligomers containing two terminal sulfhydryl groups [28]. This approach is readily applicable to peptides which can be synthesized with terminal cysteinyl residues. The traditional way of preparing DMAEMA copolymers by free radical polymerization leads to polymers with high polydispersity and with poorly defined end-group chemistries; both of which severely limit the possibility of synthesizing the necessary bis-sulfhydryl precursors of reducible PDMAEMA. The emergence during the past several years of RAFT polymerization as an exceptionally versatile tool for the synthesis of α,ω-functionalized polymers of narrow molar mass distributions has opened new possibilities for the synthesis of previously unattainable (meth)acrylate copolymers [29]. RAFT polymerization is arguably more versatile with respect to monomer choice than other living free radical polymerization techniques as it can be applied to virtually any type of monomers without the necessity of protecting functional groups. RAFT polymerization produces polymers with well-defined reactive end groups and is therefore very effective in preparing block copolymers and other complex polymer structures [30].
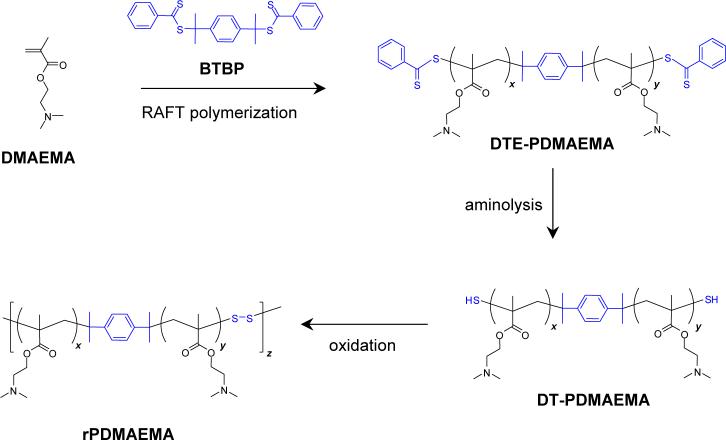
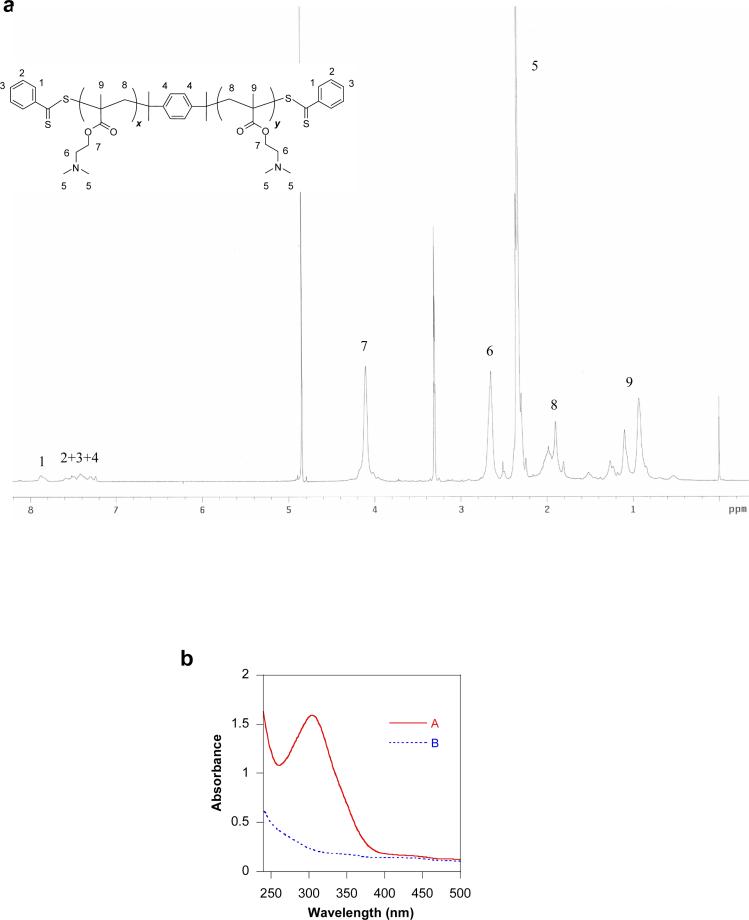
It was recently shown that polymers obtained by using a difunctional RAFT agent will yield macromolecules containing chain transfer residues at both ends of the polymer chain, which can be converted into terminal sulfhydryl groups [21, 31, 32]. Based on these findings, successful synthesis of reducible poly(styrene) was reported by Monteiro and colleagues [33]. Here, we report the synthesis of reducible PDMAEMA (rPDMAEMA) using approach depicted in Scheme 1. Difuctional RAFT agent (BTBP) was synthesized and used in an AIBN-initiated radical polymerization of DMAEMA to obtain α,ω-dithioester-functionalized oligomers (DTE-PDMAEMA). The presence of the terminal dithioesters was confirmed by the presence of the aromatic protons (δ = 7.3∼7.7) in 1H-NMR spectrum and by an absorption at 305 nm (Figure 1). Changing the [DMAEMA]:[BTBP] ratio in the polymerization mixture allowed preparation of polymers with different molar masses (Table 1). Because of the living nature of the RAFT polymerization, the molar mass increases with increasing conversion thus offering another way of controlling the molar mass. The terminal dithioester groups of the DTE-PDMAEMA were converted into sulfhydryl groups by aminolysis with hexylamine (Figure 1b). Final rPDMAEMA was synthesized by oxidation of the terminal sulfhydryl groups of DT-PDMAEMA (Figure 2). DMSO and oxygen were successfully used for the oxidation. The oxidation reaction is a polycondensation type of polymerization and thus the rPDMAEMA1 and rPDMAEMA2 exhibit higher polydispersity indexes than the starting oligomers (Table 1). While the oxidation resulted in a successful formation of reducible polymers, the number of the DT-PDMAEMA blocks connected together in the rPDMAEMA was generally lower than that of the previously reported peptide-based reducible polymers, which consisted of up to 100 oligopeptide blocks [28]. Additional optimization of the RAFT polymerization conditions to enhance the content of dithiol-polymers is required to increase the possibility of further enhancing the molar mass of the rPDMAEMA. Last, reducibility of rPDMAEMA was confirmed by treatment with DTT, which resulted in the decrease of molar mass to values similar to the original dithiol DMAEMA oligomer (Figure 2).
Scheme 1.
Synthesis of rPDMAEMA.
Figure 1.
Characterization of rPDMAEMA. (a) 1H-NMR spectrum of DTE-PDMAEMA oligomer; (b) Absorbance spectrum documenting aminolysis of the terminal dithioester groups of DTE-PDMAEMA (A) into thiol groups in DT-PDMAEMA (B).
Table 1.
Molar mass and buffering capacity of the used polycations
| α,ω-dithiol oligomer | polymer | ||||
|---|---|---|---|---|---|
| Mn | Mw/Mn | Mn | Mw/Mn | buffering capacitya | |
| rPDMAEMA1 | 3,900 | 1.06 | 16,700 | 2.5 | 3.0 |
| rPDMAEMA2 | 5,500 | 1.09 | 53,000 | 4.3 | 2.4 |
| PDMAEMA1 | n.a.b | n.a. | 13,000 | 1.4 | -- |
| PDMAEMA2 | n.a. | n.a. | 26,000 | 1.4 | 3.9 |
...buffering capacity determined as μmol of H+ per mg of polymer required to decrease pH of 0.1 mg/mL polymer solution in water from 7.4 to 5.
...not applicable
Figure 2.
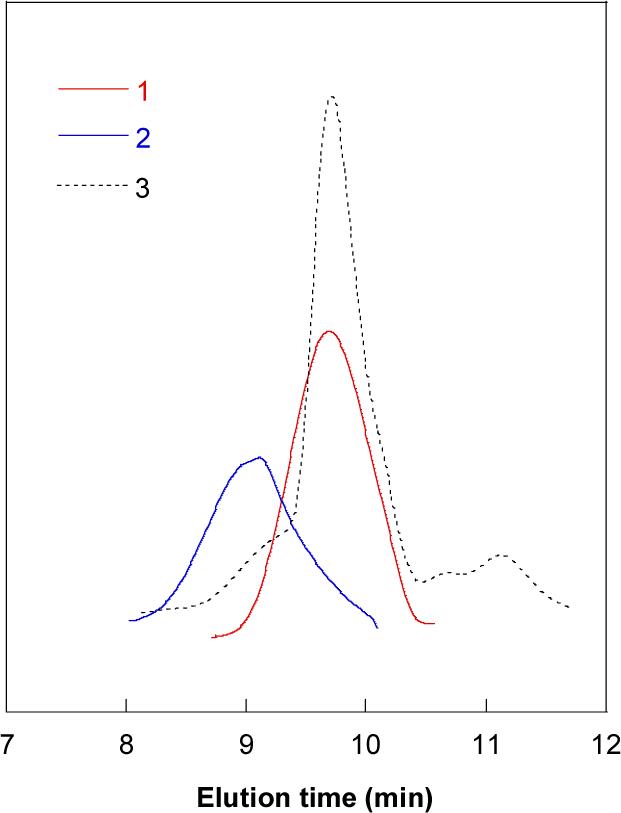
Size exclusion chromatograms documenting reduction of rPDMAEMA. (1) DT-PDMAEMA oligomer; (2) rPDMAEMA; (3) rPDMAEMA treated with 20 mM DTT.
3.2. Physico-chemical properties of rPDMAEMA/DNA polyplexes
The ability of the rPDMAEMA polycations to condense DNA was first confirmed by ethidium bromide exclusion and gel retardation assays (data not shown). No discernible differences were observed between rPDMAEMA and control PDMAEMA as all the tested polycations fully condensed plasmid DNA at N:P ∼ 1.0 when the experiment was performed at pH = 5.0. The sensitivity of the rPDMAEMA/DNA polyplexes to reduction was verified by following the polyplex dissociation with NaCl after reduction with 20 mM DTT using previously described protocol [6].
A combination of static and dynamic light scattering techniques is a powerful tool in analyzing the physico-chemical properties of DNA polyplexes. Combining the information about molar mass and radius of gyration from SLS and hydrodynamic radius and ζ potential from DLS allows one to acquire knowledge about solution properties of polyplexes not available from other methods. Information about polyplex structure (RG/RH), structural density (d), number of DNA molecules per polyplex (n) can all be obtained. Data summarizing all the major studied parameters are shown in Table 2. Representative Debye plots for selected polyplexes are shown for illustration of the quality of the static light scattering measurements (Figure 3). Polyplexes of plasmid DNA and rPDMAEMA and PDMAEMA were evaluated at four N:P ratios. The data reveal that all the polyplexes show an N:P dependent behavior similar to previously reported results [34]. Molar mass and both size parameters (RH, RG) of the polyplexes exhibit a maximum around the equivalence point (N:P ∼1) and steadily decline at N:P > 1. Structural density of the polyplexes shows a similar trend as molar mass and size, with polyplexes having the highest density when formed around N:P ∼1, indicating the most efficient DNA packing due to the lowest intra-molecular charge repulsion. As the N:P increases, the polyplexes become stabilized by excess positive charge leading to reduction in molar mass and size but also to a decrease in density due to intra-polyplex repulsion from the excess positive charges. The structure sensitive parameter (RG/RH) can provide additional information about the polyplex structures. The parameter has a value of 0.775 for solid spheres, 1.3–1.5 for random coils and >2 for elongated structures [35]. The RG/RH ratio was ∼1.2 for polyplexes prepared at N:P 2 and 4 and slightly lower (∼1) for the lower N:P ratios, indicating a formation of more sphere-like structures close to the N:P equivalence.
Table 2.
Comparison of molecular parameters of rPDMAEMA and PDMAEMA polyplexes
| Polycation | N:P | Mw (×10-8) (g/mol) | RG (nm) | RH (nm) | ζ (mV) | d (g/mL)* | n** |
|---|---|---|---|---|---|---|---|
| rPDMAEMA1 | 0.75 | 10.8 | 122 | -- | -30 | 0.236 | 179 |
| 1.2 | 7.40 | 113 | 109 | 46 | 0.203 | 113 | |
| 2 | 2.89 | 98 | 80 | 51 | 0.123 | 44 | |
| 4 | 1.44 | 83 | 70 | 42 | 0.101 | 22 | |
| rPDMAEMA2 | 0.75 | 5.21 | 114 | 109 | -35 | 0.138 | 87 |
| 1.2 | 5.96 | 114 | 116 | 39 | 0.158 | 91 | |
| 2 | 2.35 | 99 | 78 | 48 | 0.095 | 36 | |
| 4 | 1.37 | 87 | 66 | 50 | 0.084 | 21 | |
| PDMAEMA1 | 0.75 | 8.69 | 121 | 132 | -28 | 0.196 | 144 |
| 1.2 | 11.7 | 120 | -- | 35 | 0.268 | 179 | |
| 2 | 3.75 | 100 | 81 | 49 | 0.149 | 57 | |
| 4 | 1.83 | 84 | 66 | 53 | 0.123 | 28 | |
| PDMAEMA2 | 0.75 | 10.5 | 124 | -- | -27 | 0.218 | 175 |
| 1.2 | 7.42 | 112 | 102 | 47 | 0.212 | 113 | |
| 2 | 3.61 | 97 | 79 | 54 | 0.159 | 55 | |
| 4 | 2.01 | 84 | 69 | 44 | 0.137 | 31 |
Calculated from d = 3Mw/(4πNARG3)
Estimated average number of DNA molecules per one polyplex
Figure 3.
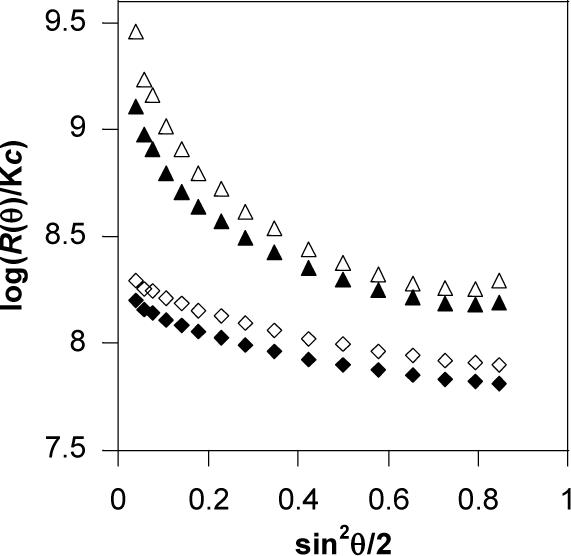
Typical light-scattering spectra angular dependence (Guinier plot) of rPDMAEMA1/DNA and PDMAEMA1/DNA polyplexes. (◇ PDMAEMA1/DNA (N:P 4), ◆ rPDMAEMA1/DNA (N:P 4), △ PDMAEMA1/DNA (N:P 1.2), ▲ rPDMAEMA1/DNA (N:P 1.2)).
Comparison of rPDMAEMA and PDMAEMA polyplexes shows that the presence of disulfide bonds in the rPDMAEMA has only a subtle effect on the physical properties of the DNA polyplexes. Most notably, the density of the rPDMAEMA polyplexes at the higher N:P ratios is consistently lower than that of the PDMAEMA controls. A previous report suggested that reducible polypeptides form more hydrophobic and dense complexes with DNA than non-reducible polypeptides [36]. The observed density of rPDMAEMA polyplexes is therefore more likely associated with the lower charge density of the reducible polycation than its higher hydrophobicity. Higher polyplex compaction is often associated with tendency to aggregation. An important parameter that may affect the efficiency of gene delivery by polyplexes is the number of DNA molecules per polyplex. In this study, n was calculated form polyplex molar mass assuming that the molar mass of the used plasmid DNA was 4.42×106 g/mol (6.7 kbp). As a result of increased density and associated aggregation of primary polyplexes, average PDMAEMA polyplex contains more DNA molecules than comparable rPDMAEMA polyplex.
3.3. Transfection activity
Transfection activity of the rPDMAEMA1 and PDMAEMA1 polyplexes was compared to PEI polyplexes in B16F10 cells at N:P ratios ranging from 15 to 25 (Figure 4). Transfection activity remained almost constant over the range of N:P ratios tested. rPDMAEMA1 polyplexes mediated a modest 3- to 4-fold increase in luciferase reporter gene expression compared to PDMAEMA1 polyplexes (Figure 4a). The control PEI polyplexes mediated ∼10- to 20-fold increased gene expression levels compared to rPDMAEMA1 polyplexes. As suggested by data in Figure 4b, the main contribution to the differences between rPDMAEMA and PEI polyplexes is significantly less efficient endosomal escape of the rPDMAEMA polyplexes. The presence of 100 μM chloroquine in transfection medium increased the levels of luciferase expression of rPDMAEMA polyplexes ∼200-fold while the activity of PEI polyplexes was less sensitive (45–80-fold enhancement). The results also show that rPDMAEMA polyplexes are less sensitive to chloroquine enhancement than control non-reducible PDMAEMA polyplexes (460–580-fold enhancement). This indicates that introduction of disulfide bonds does not negatively affect the crucial step in cellular delivery of polyplexes and can, in fact, have a positive impact. This notion is further confirmed by comparable buffering capacities of the reducible and non-reducible polycations (Table 1). It should be mentioned in this context, however, that the importance of buffering capacity of even structurally close polycations for polyplex activity has been brought into question by numerous researchers [37, 38] while confirmed by others [39]. The transfection activity of rPDMAEMA2 polyplexes was not significantly different from the rPDMAEMA1 (data not shown) and thus only rPDMAEMA1 polyplexes were used in all the transfection studies.
Figure 4.
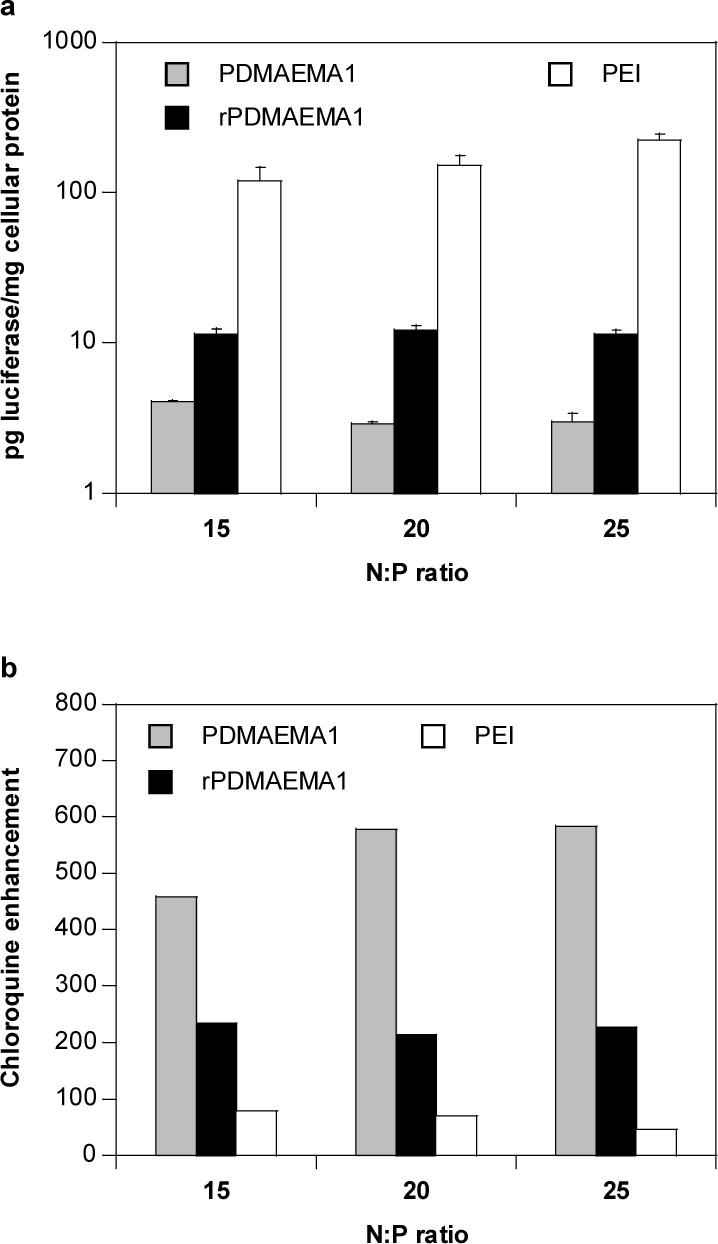
Effect of chloroquine on transfection activity of rPDMAEMA1/DNA, PDMAEMA1/DNA and PEI/DNA polyplexes in B16F10 cells. (a) Luciferase expression in the absence of chloroquine. (b) Enhancement of luciferase expression in the presence of 100 μM chloroquine. All transfection experiments were performed in DMEM supplemented with 10% FBS.
After establishing a promising activity of rPDMAEMA polyplexes in an easy-to-transfect B16F10 cell line, the transfection activity was studied in a panel of six different human pancreatic cancer cell lines. The transfection of rPDMAEMA and PDMAEMA polyplexes was studied at N:P ratios ranging from 5 to 20 and compared to an optimized formulation of PEI polyplexes at N:P 10 (Figure 5). The enhanced activity of rPDMAEMA polyplexes observed in B16F10 cells was not confirmed in the pancreatic cancer cell lines. Overall, the data indicate that rPDMAEMA1 polyplexes mediate comparable levels of transfection to that of PDMAEMA1 polyplexes in all the evaluated cell lines. This observation confirms previous findings indicating the levels of polyplex transgene expression are strongly influenced by multiple cell-line dependent factors. Some of the crucial factors include the cell line itself [40], levels of cellular uptake, mode of cellular uptake [41, 42] and eventually subcellular trafficking of polyplexes. The above factors need to be analyzed before making conclusions regarding transfection activity of the rPDMAEMA1 polyplexes in the pancreatic cancer cell lines. Interestingly, the transfection activity of the DMAEMA-based polyplexes was comparable to that of control PEI polyplexes in five out of the six cell lines (except Panc-1) as opposed to the results with B16F10 where PEI polyplexes showed substantially better transfection activity compared to DMAEMA-based polyplexes.
Figure 5.
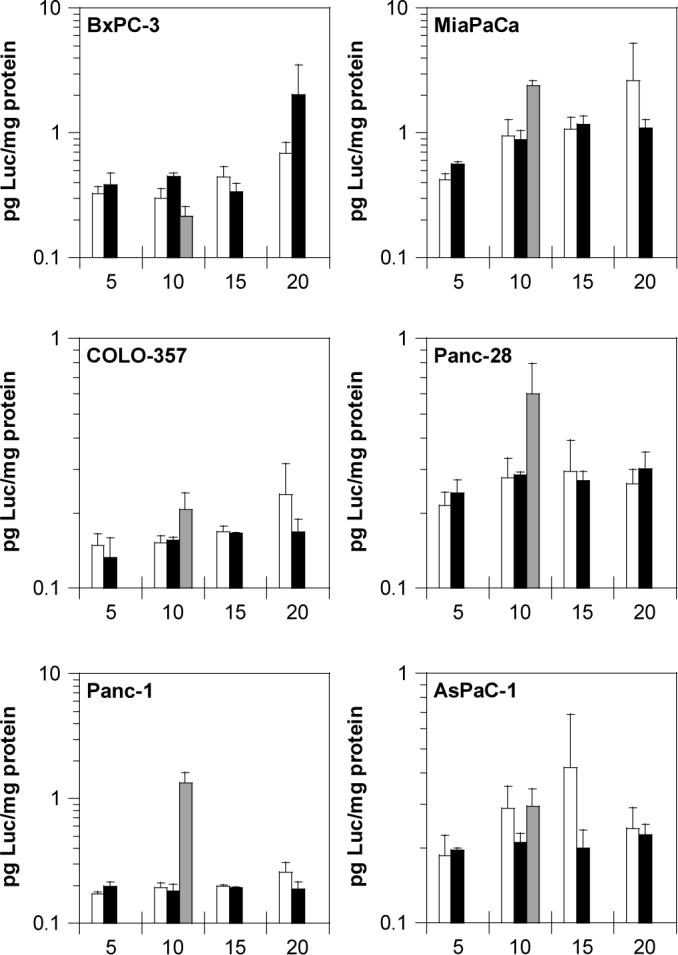
Transfection activity in a panel of human pancreatic cancer cell lines. Luciferase activity mediated by PDMAEMA1 (white bars), rPDMAEMA1 (black bars), and PEI (gray bars) polyplexes prepared at N:P 5, 10, 15, and 20 was measured after 24 h. Results are shown as mean pg of luciferase/mg of cellular protein ± S.D. (n =3)
3.4. Cytotoxicity
A promising gene delivery vector must be capable of mediating sufficient levels of gene expression without compromising on the viability of the host cells. Published reports suggest that reducible polycations based on polypeptides show only minimal cytotoxicity in the concentration range relevant for gene delivery [6, 9]. Light transmission micrographs of B16F10 cells in Figure 6 clearly document the effects of toxicity of the PDMAEMA polyplexes. Panels a, c and b, d show cells transfected with PDMAEMA and rPDMAEMA polyplexes at the two N:P ratios. Non-reducible polyplexes caused extensive levels of toxicity as evidenced by signs of growth inhibition and presence of rounded cells indicating cell stress. On careful observation, cellular debris can be visualized in panels a and c which is a typical feature of necrotic cell death, confirming published mechanisms of toxicity mediated by PDMAEMA [20]. On the contrary, cells transfected with reducible polyplexes remained healthy and appeared similar to control untreated cells (Figure 6e).
Figure 6.
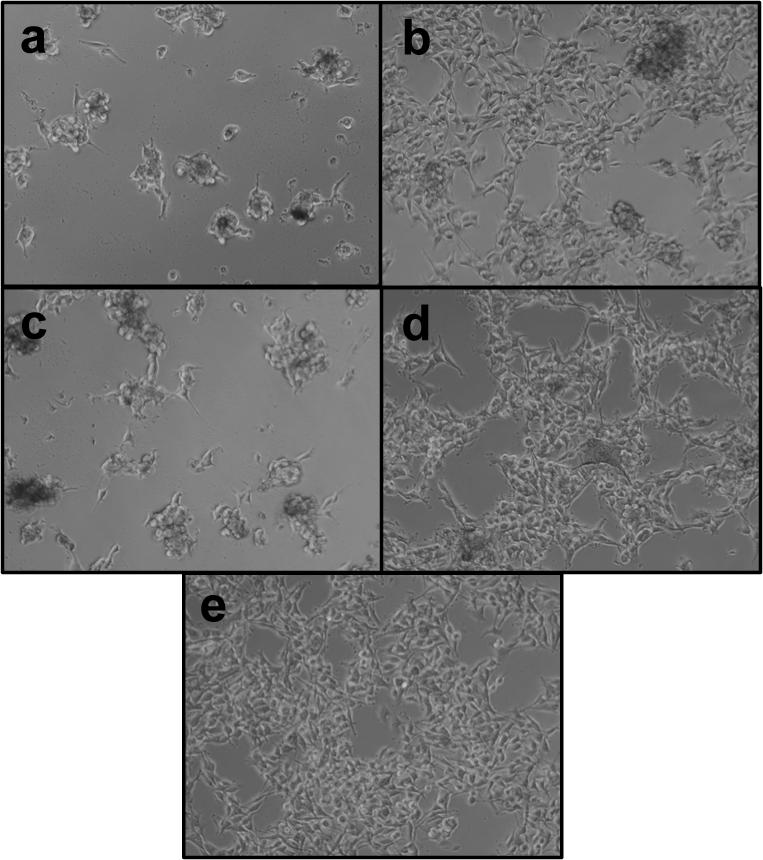
Cytotoxicity of rPDMAEMA1/DNA and PDMAEMA1/DNA polyplexes post transfection of B16F10 cells. Light transmission micrographs of the cells were taken 24 h after transfection. (a) PDMAEMA2/DNA (N:P 20), (b) rPDMAEMA2/DNA (N:P 20), (c) PDMAEMA2/DNA (N:P 30), (d) rPDMAEMA2/DNA (N:P 30), and (e) untreated cells.
In addition to the above, cytotoxicity of free polycations is a crucial phenomenon that needs to assessed since DMAEMA-based polymers, like PEI, rely heavily on the presence of free polycations for mediating efficient gene transfer. Therefore, cytotoxicity of free polycations was assessed by MTS assay which exploits metabolic activity of cells as a measure of cell viability (Figure 7). Toxicity of DMAEMA-based polycations was assessed in human endothelial EA.hy926 and human pancreatic cancer MiaPaCa cells. Cytotoxicity was measured after 16 hour incubation of the cells with the respective polycations at concentrations 10, 20, and 50 μg/mL. Overall, the data indicate that both reducible polycations imparted decreased levels of cytotoxicity compared to the non-reducible controls. The decreased cytotoxicity is believed to be a direct consequence of the intracellular degradation of the rPDMAEMA resulting in smaller fragments which leads to reduced binding affinity of the polycations with cellular membranes, essential macromolecules and reduced interference with cellular signal transduction pathways. In human endothelial cells, the IC50 value of PDMAEMA was less than 20 μg/mL, whereas rPDMAEMA did not mediate equivalent levels of toxicity even at a concentration as high as 50 μg/mL (Figure 7a). rPDMAEMA was nearly 3-fold and 10-fold less toxic compared to PDMAEMA at 20 and 50 μg/mL, respectively. Interestingly, PDMAEMA showed a more favorable cytotoxicity profile in pancreatic cancer cells compared to the endothelial cells (Figure 7b). A clear consequence of the impact of molecular weight on cytotoxicity is evident in MiaPaCa at the lowest concentration used. While PDMAEMA1 (Mn = 13,000 g/mol) shows minimal toxicity, PDMAEMA2 (Mn = 26,000 g/mol) reduced cell viability by about 40%. At 20 μg/mL, both rPDMAEMA were nearly 2- to 4-fold less toxic compared to the non-reducible PDMAEMA. At the highest concentration tested, both PDMAEMA and rPDMAEMA mediated significant levels of cytotoxicity reducing the cell viability to less than ∼35%. Neither the molar mass of the two DMAEMA oligomers nor the molar mass of the resultant reducible polycations appear to have a significant effect on the observed cytotoxicity except for the highest concentration in MiaPaCa cells where the rPDMAEMA based on a shorter oligomer shows lower cytotoxicity.
Figure 7.
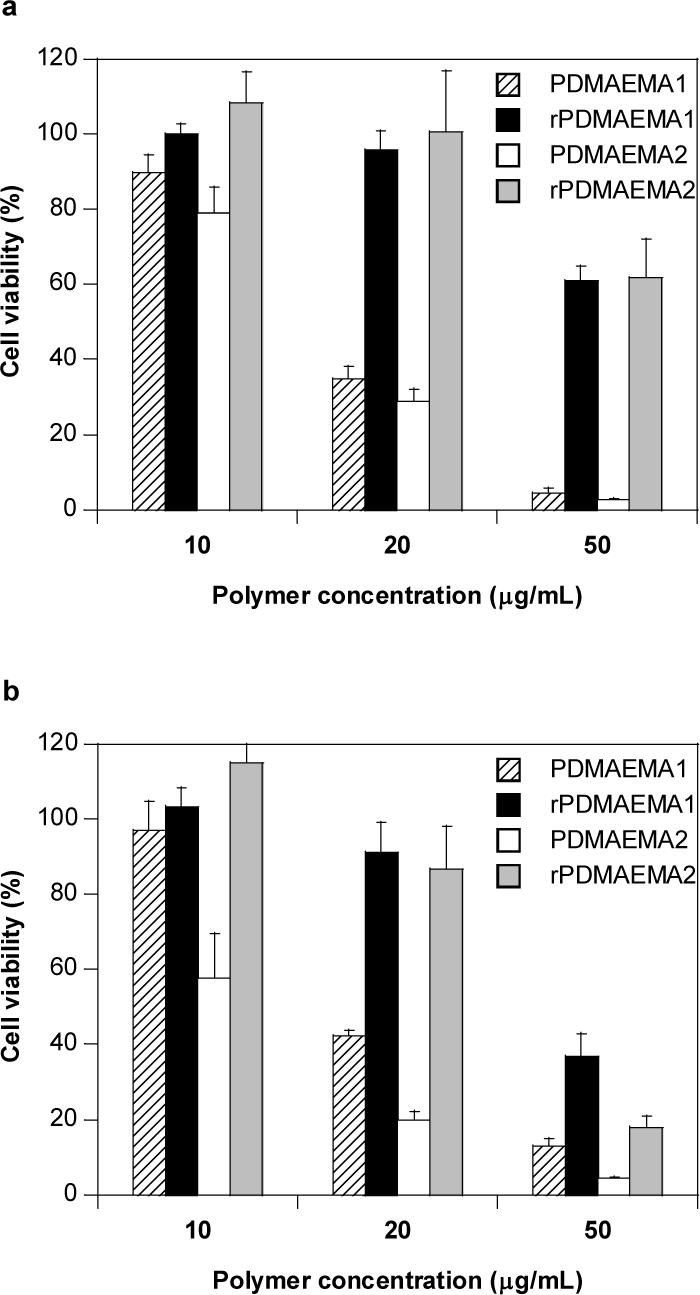
Cytotoxicity of rPDMAEMA and PDMAEMA polycations in (a) human endothelial EA.hy926 cells and (b) human pancreatic adenocarcinoma MiaPaCa cells. Metabolic activity was measured by MTS assay following 16 h incubation of the cells with the polycations. (mean ± S.D., n = 4)
4. CONCLUSIONS
Reducible PDMAEMA was successfully synthesized by an oxidation of telechelic bisthiol DMAEMA oligomers prepared by living free radical RAFT polymerization. The synthesized rPDMAEMA and its DNA polyplexes show similar physical properties and transfection activity to non-reducible control PDMAEMA, while exhibiting significantly reduced cytotoxicity in a range of cell lines. Overall, the synthetic approach reported here should be useful for synthesis of other reducible polymethacrylates, thus improving their prospects in gene delivery by, among others, offering the possibility to decrease toxicity and improving in vivo properties by allowing easier elimination from the organism due to an easy biodegradation to oligomer fragments. Finally, this approach also should be applicable to other non-ionic and anionic methacrylates and to an easy synthesis of hybrid reducible materials of polymethacrylates and a variety of dithiol-containing building blocks of organic or peptide origin.
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health (CA 109711).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Soundara Manickam D, Oupicky D. Polyplex gene delivery modulated by redox potential gradients. J. Drug Target. 2006;14(8):519–526. doi: 10.1080/10611860600834409. [DOI] [PubMed] [Google Scholar]
- 2.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, Kataoka K. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J. Am. Chem. Soc. 2004;126(8):2355–2361. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 3.McKenzie DL, Kwok KY, Rice KG. A potent new class of reductively activated peptide gene delivery agents. J. Biol. Chem. 2000;275(14):9970–9977. doi: 10.1074/jbc.275.14.9970. [DOI] [PubMed] [Google Scholar]
- 4.Read ML, Bremner KH, Oupicky D, Green NK, Searle PF, Seymour LW. Vectors based on reducible polycations facilitate intracellular release of nucleic acids. J. Gene Med. 2003;5(3):232–245. doi: 10.1002/jgm.331. [DOI] [PubMed] [Google Scholar]
- 5.Oishi M, Hayama T, Akiyama Y, Takae S, Harada A, Yamasaki Y, Nagatsugi F, Sasaki S, Nagasaki Y, Kataoka K. Supramolecular assemblies for the cytoplasmic delivery of antisense oligodeoxynucleotide: polyion complex (PIC) micelles based on poly(ethylene glycol)-SS-oligodeoxynucleotide conjugate. Biomacromolecules. 2005;6(5):2449–2454. doi: 10.1021/bm050370l. [DOI] [PubMed] [Google Scholar]
- 6.Soundara Manickam D, Oupicky D. Multiblock reducible copolypeptides containing histidine-rich and nuclear localization sequences for gene delivery. Bioconjug. Chem. 2006;17(6):1395–1403. doi: 10.1021/bc060104k. [DOI] [PubMed] [Google Scholar]
- 7.Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33(9):e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CP, Kim JS, Steenblock E, Liu D, Rice KG. Gene transfer with poly-melittin peptides. Bioconjug. Chem. 2006;17(4):1057–1062. doi: 10.1021/bc060028l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soundara Manickam D, Bisht HS, Wan L, Mao G, Oupicky D. Influence of TAT-peptide polymerization on properties and transfection activity of TAT/DNA polyplexes. J. Control. Release. 2005;102(1):293–306. doi: 10.1016/j.jconrel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Mo H, Koo H, Park JY, Cho MY, Jin GW, Park JS. Visualization of the degradation of a disulfide polymer, linear poly(ethylenimine sulfide), for gene delivery. Bioconjug. Chem. 2007;18(1):13–18. doi: 10.1021/bc060113t. [DOI] [PubMed] [Google Scholar]
- 11.Gosselin MA, Guo WJ, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug. Chem. 2001;12(6):989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 12.Kloeckner J, Wagner E, Ogris M. Degradable gene carriers based on oligomerized polyamines. Eur. J. Pharm. Sci. 2006;29(5):414–425. doi: 10.1016/j.ejps.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JF. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjug. Chem. 2007;18(1):138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 14.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, Engbersen JF, Feijen J. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjug. Chem. 2006;17(5):1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 15.Hoon Jeong J, Christensen LV, Yockman JW, Zhong Z, Engbersen JF, Jong Kim W, Feijen J, Wan Kim S. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28(10):1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Funhoff AM, van Nostrum CF, Lok MC, Kruijtzer JA, Crommelin DJ, Hennink WE. Cationic polymethacrylates with covalently linked membrane destabilizing peptides as gene delivery vectors. J. Control. Release. 2005;101(13):233–246. doi: 10.1016/j.jconrel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Van De Wetering P, Cherng JY, Talsma H, Crommelin DJA, Hennink WE. 2-(dimethylamino)ethyl methacrylate based (co)polymers as gene transfer agents. J. Control. Release. 1998;53:145–153. doi: 10.1016/s0168-3659(97)00248-4. [DOI] [PubMed] [Google Scholar]
- 18.Van De Wetering P, Cherng JY, Talsma H, Hennink WE. Relation between transfection efficiency and cytotoxicity of poly(2-(dimethylamino)ethyl methacrylate)/plasmid complexes. J. Control. Release. 1997;49:59–69. [Google Scholar]
- 19.Reschel T, Konak C, Oupicky D, Seymour LW, Ulbrich K. Physical properties and in vitro transfection efficiency of gene delivery vectors based on complexes of DNA with synthetic polycations. J. Control. Release. 2002;81(12):201–217. doi: 10.1016/s0168-3659(02)00045-7. [DOI] [PubMed] [Google Scholar]
- 20.Jones RA, Poniris MH, Wilson MR. pDMAEMA is internalised by endocytosis but does not physically disrupt endosomes. J. Control. Release. 2004;96(3):379–391. doi: 10.1016/j.jconrel.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Patton DL, Mullings M, Fulghum T, Advincula RC. A facile synthesis route to thiol-functionalized alpha,w-telechelic polymers via reversible addition fragmentation chain transfer polymerization. Macromolecules. 2005;38(20):8597–8602. [Google Scholar]
- 22.Mitsukami Y, Donovan MS, Lowe AB, McCormick CL. Water-soluble polymers. 81. Direct synthesis of hydrophilic styrenic-based homopolymers and block copolymers in aqueous solution via RAFT. Macromolecules. 2001;34(7):2248–2256. [Google Scholar]
- 23.Lai E, van Zanten JH. Monitoring DNA/poly-L-lysine polyplex formation with time-resolved multiangle laser light scattering. Biophys. J. 2001;80(2):864–873. doi: 10.1016/S0006-3495(01)76065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oupicky D, Reschel T, Konak C, Oupicka L. Temperature controlled behavior of self-assembly gene delivery vectors based on complexes of DNA with poly(L-lysine)-graft-poly(N-isopropylacrylamide) Macromolecules. 2003;36(18):6863–6872. [Google Scholar]
- 25.Fischer D, Dautzenberg H, Kunath K, Kissel T. Poly(diallyldimethylammonium chlorides) and their N-methyl-N-vinylacetamide copolymer-based DNA-polyplexes: role of molecular weight and charge density in complex formation, stability, and in vitro activity. Int. J. Pharm. 2004;280(12):253–269. doi: 10.1016/j.ijpharm.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Verbaan FJ, Oussoren C, van Dam IM, Takakura Y, Hashida M, Crommelin DJA, Hennink WE, Storm G. The fate of poly(2-dimethyl amino ethyl)methacrylate-based polyplexes after intravenous administration. Int. J. Pharm. 2001;214(12):99–101. doi: 10.1016/s0378-5173(00)00642-6. [DOI] [PubMed] [Google Scholar]
- 27.De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm. Res. 2000;17(2):113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 28.Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: Strategy for triggered intracellular activation of DNA delivery vectors. J. Am. Chem. Soc. 2002;124(1):8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 29.McCormick CL, Lowe AB. Aqueous RAFT Polymerization: Recent Developments in Synthesis of Functional Water-Soluble (Co)polymers with Controlled Structures. Acc. Chem. Res. 2004;37(5):312–325. doi: 10.1021/ar0302484. [DOI] [PubMed] [Google Scholar]
- 30.Moad G, Chong YK, Postma A, Rizzardo E, Thang SH. Advances in RAFT polymerization: the synthesis of polymers with defined end-groups. Polymer. 2005;46(19):8458–8468. [Google Scholar]
- 31.Goh YK, Monteiro MJ. Novel approach to tailoring molecular weight distribution and structure with a difunctional RAFT agent. Macromolecules. 2006;39(15):4966–4974. [Google Scholar]
- 32.Qiu XP, Winnik FM. Facile and efficient one-pot transformation of RAFT polymer end groups via a mild aminolysis/Michael addition sequence. Macromol. Rapid Commun. 2006;27(19):1648–1653. [Google Scholar]
- 33.Whittaker MR, Goh YK, Gemici H, Legge TM, Perrier S, Monteiro MJ. Synthesis of monocyclic and linear polystyrene using the reversible coupling/cleavage of thiol/disulfide groups. Macromolecules. 2006;39(26):9028–9034. [Google Scholar]
- 34.Oupicky D, Konak C, Ulbrich K, Wolfert MA, Seymour LW. DNA delivery systems based on complexes of DNA with synthetic polycations and their copolymers. J. Control. Release. 2000;65(12):149–171. doi: 10.1016/s0168-3659(99)00249-7. [DOI] [PubMed] [Google Scholar]
- 35.Dautzenberg H, Gao YB, Hahn M. Formation, structure, and temperature behavior of polyelectrolyte complexes between ionically modified thermosensitive polymers. Langmuir. 2000;16(23):9070–9081. [Google Scholar]
- 36.Blacklock J, Handa H, Soundara Manickam D, Mao G, Mukhopadhyay A, Oupicky D. Disassembly of layer-by-layer films of plasmid DNA and reducible TAT polypeptide. Biomaterials. 2007;28(1):117–124. doi: 10.1016/j.biomaterials.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 37.Gabrielson NP, Pack DW. Acetylation of polyethylenimine enhances gene delivery via weakened polymer/DNA interactions. Biomacromolecules. 2006;7(8):2427–2435. doi: 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]
- 38.Funhoff AM, van Nostrum CF, Koning GA, Schuurmans-Nieuwenbroek NM, Crommelin DJ, Hennink WE. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules. 2004;5(1):32–39. doi: 10.1021/bm034041+. [DOI] [PubMed] [Google Scholar]
- 39.Sonawane ND, Szoka FC, Jr., Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 40.von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E, Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Molec. Ther. 2006;14(5):745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Molec. Ther. 2005;12(3):468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 42.Rejman J, Conese M, Hoekstra D. Gene transfer by means of lipo- and polyplexes: role of clathrin and caveolae-mediated endocytosis. J. Liposome Res. 2006;16(3):237–247. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]