Abstract
Background
Emerging molecular, animal model and epidemiologic evidence suggests that Shiga-toxigenic Escherichia coli O157:H7 (STEC O157) isolates vary in their capacity to cause human infection and disease. The translocated intimin receptor (tir) and intimin (eae) are virulence factors and bacterial receptor-ligand proteins responsible for tight STEC O157 adherence to intestinal epithelial cells. They represent logical genomic targets to investigate the role of sequence variation in STEC O157 pathogenesis and molecular epidemiology. The purposes of this study were (1) to identify tir and eae polymorphisms in diverse STEC O157 isolates derived from clinically ill humans and healthy cattle (the dominant zoonotic reservoir) and (2) to test any observed tir and eae polymorphisms for association with human (vs bovine) isolate source.
Results
Five polymorphisms were identified in a 1,627-bp segment of tir. Alleles of two tir polymorphisms, tir 255 T>A and repeat region 1-repeat unit 3 (RR1-RU3, presence or absence) had dissimilar distributions among human and bovine isolates. More than 99% of 108 human isolates possessed the tir 255 T>A T allele and lacked RR1-RU3. In contrast, the tir 255 T>A T allele and RR1-RU3 absence were found in 55% and 57%, respectively, of 77 bovine isolates. Both polymorphisms associated strongly with isolate source (p < 0.0001), but not by pulsed field gel electrophoresis type or by stx1 and stx2 status (as determined by PCR). Two eae polymorphisms were identified in a 2,755-bp segment of 44 human and bovine isolates; 42 isolates had identical eae sequences. The eae polymorphisms did not associate with isolate source.
Conclusion
Polymorphisms in tir but not eae predict the propensity of STEC O157 isolates to cause human clinical disease. The over-representation of the tir 255 T>A T allele in human-derived isolates vs the tir 255 T>A A allele suggests that these isolates have a higher propensity to cause disease. The high frequency of bovine isolates with the A allele suggests a possible bovine ecological niche for this STEC O157 subset.
Background
Shiga-toxigenic Escherichia coli O157:H7 (STEC O157) is the major STEC serotype associated with human infection in the U.S. [1]. Cattle are the predominant North American reservoir of this zoonotic pathogen [2,3] and contact with infected livestock and ingestion of contaminated meat are frequent routes of human infection [4-7]. Other sources of STEC O157 infection are contaminated fruits, vegetables and water [8-10] and person-to-person contact [11,12]. From 1982–2002, there were 350 STEC O157 outbreaks reported in the U.S., resulting in 8,500 clinical cases, 1,500 hospitalizations and 40 deaths [13]. Human STEC O157 infections cause mild self-limiting diarrhea to severe disease including hemorrhagic colitis and hemolytic uremic syndrome (HUS) [14,15]. HUS, due to STEC O157 infection, is the leading cause of renal failure for children under the age of five years [1].
As with many infectious disease agents, STEC O157 strains appear to vary in their capacity to cause human infection and disease. For example, in the gnotobiotic pig challenge model, STEC O157 strains differ in both the clinical course they provoke and the histopathological lesions they induce [16]. Epidemiologic surveillance data in the U.S. also supports the idea of inter-strain variation in STEC O157 virulence. The annual U.S. incidence of clinical STEC O157 infections is estimated at 1.1 per 100,000 persons [1]. However, pooled data from five North American serological surveys found 11% of 2,251 healthy children and adults (11,000 per 100,000 persons) with serologic evidence of E. coli O157 exposure and/or subclinical infection [17-21]. On a smaller scale, the investigation of a recent STEC O157 outbreak linked with visiting an agricultural fair suggested that all STEC O157 are not equivalent in terms of their public health risk [22]. At least 25 people out of over 170,000 fair visitors who attended over a two-week period became ill with an STEC O157 isolate that shared the same pulse-field gel electrophoresis (PFGE) pattern. The outbreak investigation revealed that the fairground environment was heavily contaminated with multiple STEC O157 isolates with eight different PFGE patterns, including the outbreak strain. The presumed high human STEC O157 exposure but low human clinical disease incidence, suggested by both the surveillance and outbreak data, could be partially explained if only a subset of STEC O157 isolates present in the bovine (or other) zoonotic reservoirs were pathogenic to humans. Identifying markers for virulent strains as well as understanding the mechanisms responsible for disparities in virulence may provide new insights into the epidemiology and control of STEC O157 infections in both human and animal reservoirs.
An important step in the pathogenesis of human infection with STEC O157 is colonization of the lower gastrointestinal (GI) tract. STEC O157 have a number of virulence and putative virulence factors which aid in this colonization including the locus of enterocyte effacement (LEE), production of Shiga toxin 2, flagellin, OmpA, Lpf and ToxB [14,23-28]. The interaction of two LEE-encoded genes, tir and eae, is responsible for the tight bacterial adherence to host epithelial cells characteristic of STEC O157 infections. The eae-encoded ligand protein intimin is located on the bacterial outer membrane. The intimin receptor protein Tir is translocated into the epithelial cell by type III secretion and integrated in the host cell membrane [29,30]. Given the role of intimin and Tir in STEC O157 pathogenesis and the well documented role of cattle as a zoonotic reservoir, the purpose of this study was to characterize sequence variation in STEC O157 eae and tir genes and to evaluate whether it associates with human or bovine host origin.
Methods
Bacterial strains
For sequence discovery, 22 diverse STEC O157 isolates were assembled that varied by source, either human clinical (n = 9) or bovine (n = 13) (Table 1). A further 101 epidemiologically unrelated human clinical and 64 bovine isolates were included to estimate tir polymorphism allele frequencies. Each isolate was characterized by ELISA using anti-O157 and H7 monoclonal antibodies and multiplex PCR for stx1, stx2, eae, hlyA, rfbO157 and fliCH7 [31-34]. For the purpose of this study, isolates were defined as STEC O157 if they were E. coli O157 antigen positive by ELISA, rfbEO157 and fliCH7-positive by PCR, and stx1 and/or stx2 positive by PCR.
Table 1.
STEC O157 strains used for sequence polymorphism discovery in the tir gene and a representative strain from each tir genotype
| Strain | H antigen | Collection | Source | Reference | Discovery panel | tir genotypea |
| N245 | H7 | USMARC | Bovine | This paper | N | 1 |
| SS NE 1040 | H7 | USMARC | Bovine | [49] | N | 2 |
| CO 50-1 | H7 | USMARC | Bovine | [49] | Y | 3 |
| NE 972-1 | H7 | USMARC | Bovine | [49] | Y | 3 |
| KS 368-1 | H7 | USMARC | Bovine | [49] | Y | 4 |
| KS 546-2 | H7 | USMARC | Bovine | [49] | Y | 4 |
| NE 1370-3 | H7 | USMARC | Bovine | [49] | Y | 4 |
| TX 265-1 | H7 | USMARC | Bovine | [49] | Y | 4 |
| TX 376-2 | H7 | USMARC | Bovine | [49] | Y | 4 |
| TX 723-1 | H7 | USMARC | Bovine | [49] | Y | 4 |
| CO 147-1 | H7 | USMARC | Bovine | [49] | Y | 4 |
| NE 1127-1 | H7 | USMARC | Bovine | [49] | Y | 5 |
| TW07587 | H7 | EHEC 1–3 | Human | [50] | Y | 5 |
| 2 | NM | Acheson | Human | b | Y | 5 |
| USDA81 | H7 | CDC | Human | This paper | N | 6 |
| CO 713-5 | H7 | USMARC | Bovine | [49] | Y | 7 |
| MARC 611 | H7 | USMARC | Bovine | [49] | Y | 7 |
| NE 1270-2 | H7 | USMARC | Bovine | [49] | Y | 7 |
| 31277 | H7 | IDPH | Human | c | Y | 7 |
| 1 | H7 | Mandrell | Human | d | Y | 7 |
| 35150 | H7 | ATCC | Human | ATCC | Y | 7 |
| 43889 | H7 | ATCC | Human | ATCC | Y | 7 |
| 43890 | H7 | ATCC | Human | ATCC | Y | 7 |
| Sakai | H7 | RMID | Human | [45] | Y | 7 |
| TW04863 | H7 | EHEC 1–2 | Human | [50] | Y | 7 |
| N4009-114-4 | H7 | APHIS | Bovine | This paper | N | 8 |
| TW06555 | H7 | EHEC 1–6 | Human | [50] | N | 9 |
| TW00116 | H7 | EHEC 1–4 | Human | [50] | N | 10 |
a As described in Figure 1B.
b Dr. David Acheson, DHHS, FDA, College Park, MD.
c Enteric Lab, Illinois Department of Public Health.
d Dr. Robert Mandrell, USDA, ARS, Albany, CA.
PCR amplification, DNA sequencing and analysis
A 2,755-kb segment of the eae gene and 1,627-kb segment of the tir gene were amplified and sequenced using primers listed in Table 2. The amplification reactions contained 0.5 ng of DNA, 0.75 uM of each primer, 200 uM of each dNTP, 1.5 mM MgCl2 and 1 U of Platinum Taq DNA polymerase (Invitrogen Corporation, Carlsbad, CA) in a 55 ul reaction. PCR amplifications were performed using a PTC-200 (MJ Research, Waterton, MA) at the following conditions: 1 min at 95°C followed by 30 sec at 96°C, 30 sec at 52°C and 2 min at 72°C for 35 cycles and finally 72°C for 7 min for 1 cycle.
Table 2.
Oligonucleotides (5'-3') used in this study
| Oligonucleotide | Sequence | Orientationa | Application |
| tir:1U19 | atg cct att ggt aat ctt g | Sense | tir 5' amplification sequencing |
| tir:72U17 | aca aac cga cgg tgc ag | Sense | tir sequencing |
| tir:473L17 | atg ccc agc acc acc ac | Antisense | tir sequencing |
| tir:521U17 | aag ccc gcc aaa gga ta | Sense | tir RR1 sequencing |
| tir:967L17 | tct tcg cct gct gct tt | Antisense | tir RR1 sequencing |
| tir:1046U17 | cta aac gcc agg agg ag | Sense | tir RR2, 3, & 4 sequencing |
| tir:1572L17 | ggc gta atc cac cac tt | Antisense | tir RR2, 3, & 4 sequencing |
| tir:1638L16 | cgc tgg tgg gtt att c | Antisense | tir 3' amplification sequencing |
| eae:16U16 | tgt tat acc cgg acc c | Sense | eae 5' amplification sequencing |
| eae:135U19 | taa att ggg ttc gga ttc a | Sense | eae sequencing |
| eae:382L17 | aca aga ggt gcc gaa cc | Antisense | eae sequencing |
| eae:490U16 | aat tat gcg gca caa c | Sense | eae sequencing |
| eae:574L18 | gtt acc agc gat acc aag | Antisense | eae sequencing |
| eae:626U18 | att atg gaa cgg cag agg | Sense | eae sequencing |
| eae:886L19 | ttt tga aat agt ctc gcc a | Antisense | eae sequencing |
| eae:1090U17 | gat aag ctg cag tcg aa | Sense | eae sequencing |
| eae:1178L18 | ttt cat tac ccg tac cat | Antisense | eae sequencing |
| eae:1290U18 | atc agg cag ccg tta cga | Sense | eae sequencing |
| eae:1492L19 | ttc cgc tat gct gaa tct g | Antisense | eae sequencing |
| eae:1695U18 | gga taa gac ttc ggc taa | Sense | eae sequencing |
| eae:1732L18 | cgt cgc ggt ata agt aat | Antisense | eae sequencing |
| eae:1883U17 | cgc cag gac agg tcg tc | Sense | eae sequencing |
| eae:2135L17 | att tcc cgt ggt tgc tt | Antisense | eae sequencing |
| eae:2249U19 | ttg gta aca atg tca gag g | Sense | eae sequencing |
| eae:2358L13 | acc acc gct tgc t | Antisense | eae sequencing |
| eae:2360U17 | caa gcg gtg gtg atg gt | Sense | eae sequencing |
| eae:2699L17 | tcc aga acg ctg ctc ac | Antisense | eae sequencing |
| eae:2787L19 | tta ttc tac aca aac cgc a | Antisense | eae 3' amplification sequencing |
a With respect to tir and eae mRNA.
PCR products were purified and concentrated using the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA). DNA sequencing reactions were prepared using the ABI PRISM BigDye terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, CA) with slight modifications of the manufacturer's protocol to reduce the final volume to 10 ul. The sequencing reactions were cycled with a PTC-200 (MJ Research) at the following conditions: 1 min at 96°C followed by 30 sec at 96°C, 1 min at 50°C and 4 min at 60°C for 30 cycles. DNA sequences were determined with either an ABI PRISM 3700 DNA analyzer or an ABI PRISM 377 DNA sequencer (PE Applied Biosystems).
Nucleotide sequences were analyzed using SeqMan and alignments were constructed using Clustal X, both from the Lasergene software package (DNASTAR, Inc., Madison, WI). A consensus parsimony tree was generated in PHYLIP (version 3.65) from tir DNA sequences using the program PARS [35] and viewed in TreeView (version 1.6.6) [36].
Pulse field gel electrophoresis of isolates used for genotyping
PFGE was performed on all human and bovine derived E. coli O157:H7 isolates by using the PulseNet protocol and the restriction endonuclease XbaI [37]. Restriction fragment patterns were analyzed using Bionumerics version 4 (Applied Maths, Belgium).
Statistical analysis of tir variation and host association
The frequencies of each identified tir nucleotide or repeat polymorphism and tir genotype were compared between STEC O157 isolates of human and bovine origin. The data were analyzed as an unmatched case-control study by exact logistic regression using the LOGISTIC procedure of SAS 9.1 (SAS Institute, Inc., Cary, NC). The binary response variable (outcome) of interest was the probability of the strain being of bovine origin (case) vs human origin (control). Each tir polymorphism and genotype was converted into a categorical explanatory (predictor) variable, where each possible variant within a given polymorphism was coded separately. Genotype 10 and the most common variant for each polymorphism were used as reference conditions. The association of each tir polymorphism variant with the likelihood of being a case STEC O157 strain was examined by generating univariate exact odds ratios (OR) with exact 95% confidence intervals (CI) and corresponding p values. Stx profiles of isolates defined by PCR were also examined for association with human or bovine strain origin.
Results
Polymorphisms in STEC O157 tir and eae
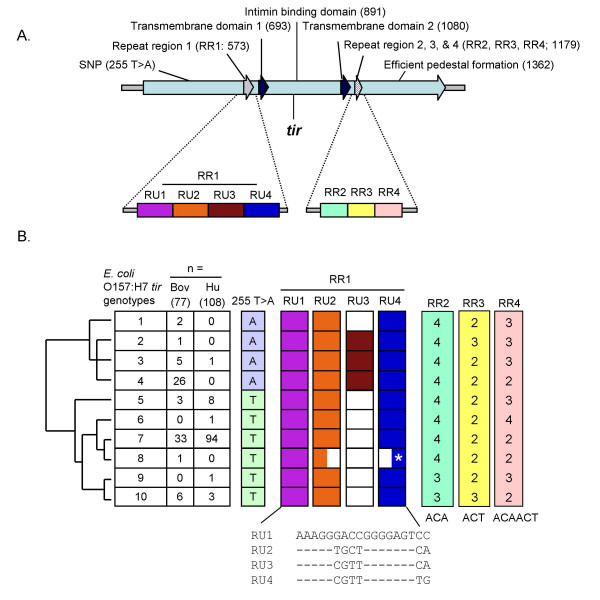
Five polymorphic loci were identified in a 1,627-kb segment of the STEC O157 tir gene [GenBank:DQ458771]. One was a single nucleotide polymorphism (SNP), 255 T>A (reference sequence is GenBank accession number BA000007 gene ECs4561). The minor allele of this non-synonymous polymorphism encodes aspartic acid while the other encodes glutamic acid. Four were repeat polymorphisms with the following properties: repeat region 1 (RR1, nucleotide position 573 in reference sequence BAB37984) containing up to four imperfect 18-bp repeat units (RU1-RU4), RR2 (nucleotide position 963) containing [ACA]n where n = 3 or 4, RR3 (nucleotide position 1,080) containing [ACT]n where n = 2 or 3 and RR4 (nucleotide position 1,179) containing [ACAACT]n where n = 2, 3 or 4 (Figure 1A). In one isolate, a chimeric repeat within RR1 was identified consisting of approximately the 5' half of RU2 and the 3' half of RU4 (Figure 1B).
Figure 1.
Physical maps and taxonomical relationships of the tir gene. (A) A physical map of the tir gene from STEC O157 (GenBank accession number BA000007 gene ECs4561). A 1.6-kb region of the tir gene was sequenced from 22 STEC O157 isolates and the polymorphisms mapped on the tir gene along with previously identified functional domains. (B) Consensus tree from seven equally parsimonious trees constructed in PHYLIP (version 3.65) [35] with the program PARS and viewed in TreeView (version 1.6.6) [36]. The tree was generated from seven variable sites (255 T>A, RR1-RU2, RR1-RU3, RR1-RU4, RR2, RR3 and RR4) and describes the taxonomical relationship of the tir nucleotide sequence with associated phenotypic and genotypic information. Ten genotypes resulting from one nucleotide polymorphism and four repeat region variations were identified from the 185 isolates. Colored boxes identify the different nucleotide variations and repeat units, while white boxes indicate missing repeat units. The sequences of the repeat regions are located below the colored boxes with the top line as the consensus sequence. A variant repeat unit (*) in repeat region 1 was identified that had the following sequence "AAAGGTGCTGGGGAGTTG".
The five polymorphic tir loci defined ten unique tir genotypes (Figure 1B). Two genotypes, 4 and 7, accounted for 83% (n = 185) of the isolates sequenced, while four genotypes were observed in only one isolate each (Figure 1B). A consensus parsimony tree generated from these genotypes defined two major clades (Figure 1B). Alleles of tir 255 T>A and RR1-RU3 were responsible for discrimination between these clades (Figure 1B). Alleles of these two polymorphisms are strongly correlated.
The sequence of a 2,755-bp region of the eae gene was identical in 42 of 44 STEC O157 isolates representing at least one isolate from each tir genotype [GenBank:EF540940]. Synonymous (eae 60 G>A reference sequence is GenBank accession number BA000007 gene ECs4559) and non-synonymous (eae 2414 C>T) SNPs were identified, each in isolates 2857\98 [GenBank:EF540939] and ATCC 43890 [GenBank:EF540941], respectively. The eae 2414 C>T results in a change from a threonine to an isoleucine (T805I). This isolate also contained tir genotype 7, the most common tir genotype.
Association of tir 255 T>A and RR1-RU3 alleles with host origin of STEC O157 isolates
Unmatched case-control analysis showed that only tir 255 T>A and RR1-RU3 (presence or absence) alleles were significantly associated with host origin. Specifically, isolates with tir 255 T>A A allele were 34.0 times more likely (5.7 to 1381.9 95% CI, p < 0.0001) to be of bovine than human origin. Similarly, isolates with RR1-RU3 present were 32.0 times more likely (5.3 to 1302.9 95% CI, p < 0.0001) to be of bovine than human origin. Because tir 255 T>A and RR1-RU3 alleles discriminate between genotypes 4 and 7, these genotypes also associate with host origin. Specifically, isolates with genotype 4 were 37.0 times more likely to be of bovine than human origin (6.6 to ∞ 95% CI, p < 0.0001), while those with genotype 7 were 2.9 times more likely to be of human than bovine origin (2.0 to 4.5 95% CI, p < 0.0001). Genotyping of 255 T>A on all study isolates reinforced this association, with 1 out of 108 human isolates (0.93%, 0.02 to 5.1 95% CI), as opposed to 34 out of 77 bovine isolates (44.2%, 32.8 to 55.9 95% CI), having an A at 255 T>A (p < 0.0001). Furthermore, stx1 and stx2 status (as determined by PCR) were statistically independent of bovine or human derived STEC O157 isolates (p = 0.12 and p = 0.37, respectively) and when all 185 STEC O157 isolates were compared by PFGE, there was no association of PFGE profiles with tir 255 T>A alleles.
Discussion
STEC O157 tir 255 T>A T allele is significantly overrepresented in human isolates relative to isolates with the A allele (99.1 vs. 0.9%, p < 0.0001). The reason for this host difference in 255 T>A allele frequency is unknown. It is possible that STEC O157 255 T>A A isolates are shed in cattle feces for shorter periods of time or in lower numbers than STEC O157 255 T>A T isolates, resulting in lower probability of product or environmental contamination and thus, lower human exposure and subsequent infection. Reduced human exposure could also occur if STEC O157 255 T>A A isolates are less able to survive on meat, on produce, in water or in other environmental sources of human infection. STEC O157 255 T>A A isolates maybe less virulent in humans so that they are less likely to cause clinical illness prompting fecal culture. Whatever the actual mechanism, the low 255 T>A A allele frequency in human isolates cannot be explained by a corresponding low frequency in the bovine reservoir.
Non-random distribution of E. coli O157:H7 subtypes among bovine and human isolates have been reported in previous investigations. Octamer-based genome scanning classified STEC O157 into two lineages (I and II), with human origin isolates biased towards lineage I [38]. Q-gene allelic variation (upstream of the prophage stx region), Shiga-toxin 2 production differences and Shiga toxin-encoding bacteriophage insertion site-defined genotypes also had biased distributions of isolates from bovine and human origin [39-41]. However, none of these previously described methods provided as clear a discrimination between human and bovine isolates as those described in this study. Furthermore, the presence of one or both stx1 and stx2 genes (as determined by PCR) was statistically independent of an isolate's tir 255 T>A allele or RU3 presence or absence. The high degree of discrimination provided by tir 255 T>A and the central role of Tir in human infection points towards a possible functional role, rather than solely as a marker, for this polymorphism.
Previous studies indicate a paucity of nucleotide polymorphisms in most STEC O157 genes [42-44]. The presence of five polymorphic loci with high minor allele frequency within tir, therefore, appears to be atypical in STEC O157. Furthermore, all five polymorphisms are non-synonymous. In contrast, only one low frequency synonymous polymorphism was found in eae, suggesting that these two loci are under different selective pressures. The association of tir 255 T>A T allele with human infection argues that host factors may impose some selection pressure on tir. The fact that the tir 255 T>A A minor allele frequency is over 30% in bovine isolates, where the frequency of minor alleles for most SNPs in STEC O157 is considerably less than that, also argues for some selection on this allele or another locus tightly linked to 255 T>A [43,44].
Limited information exists on complete tir gene sequence from STEC O157. Examination of the two published STEC O157 genomic sequences, EDL 933 and Sakai [45,46], showed that their tir genes were both genotype 7. Our sequencing of the tir gene from these two isolates confirmed this finding (data not shown for EDL 933). RR2, RR3 and RR4 together were previously used as a marker for high-resolution molecular typing of E. coli O157:H7 [47]. In the present study, the sequencing of a broad population of STEC O157 strains that included both human clinical and bovine reservoir isolates revealed additional informative tir polymorphisms, particularly the 255 T>A A allele and the presence of RR1-RU3.
Two polymorphic tir loci, tir 255 T>A and RR1-RU3, appear to have epidemiologic significance by their clear and strong association with isolate host source. Both loci are located near the amino terminus of Tir, a portion of the molecule that is normally located in the host cytosol during Tir-Intimin binding in host cell-bacterial adherence [48], in a region where no function has been described. However, these loci may have functional significance based on the biased distribution of their alleles in human derived isolates. One explanation for this could be variation in avidity, kinetics or tropism of adherence to epithelial cells, a major function of the Tir protein, from isolates with the tir 255 T>A A allele compared to isolates with the tir 255 T>A T allele. However, more investigation will be necessary to delineate the structure-function relationships of these tir polymorphisms.
Conclusion
Many host, bacterial and environmental factors impact whether or not infection results from human exposure to STEC O157. This study demonstrates that genomic polymorphisms in tir but not eae predict the likelihood that STEC O157 strains can cause human disease. Intriguing but unexplained findings include the host bias in tir allele frequency between human and bovine hosts. The over-representation of the tir 255 T>A T allele in human-derived isolates – vs the A allele proves its merit as a marker for virulence in humans. Also of interest is the high degree of tir sequence variation relative to that found in eae, even though the two proteins encoded by these genes interact together as receptor and ligand during adherence to host intestinal epithelial cells. Further research will be necessary to determine if the tir 255 T>A and RR1-RU3 polymorphisms are simply markers of strain virulence or are functional components of host-pathogen interactions.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JLB co-conceive the study and experimental design, carried out the DNA sequencing and editing of the tir and eae genes and drafted the manuscript. JEK provided isolates for the study, carried out the statistical analysis and helped edit the manuscript. MLC carried out the phylogenic analysis of the tir DNA sequence. LMD participated in the PFGE analysis of the isolates and helped edit the manuscript. MPH participated in DNA sequence analysis. WWL co-conceived the study and experimental design, carried out the initial sequencing of the tir and eae genes and helped draft and edit the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
We thank Drs. B. Swaminathan, T. Whittam, R. Mandrell, D. Acheson and T. Honda for providing isolates; Liz Ossian, Sandy Fryda-Bradley, Ron Mlejnek and Tammy Sorensen for technical assistance; and Joan Rosch and Sherry Kluver for secretarial assistance.
Contributor Information
James L Bono, Email: Jim.Bono@ars.usda.gov.
James E Keen, Email: Jim.Keen@ars.usda.gov.
Michael L Clawson, Email: Mike.Clawson@ars.usda.gov.
Lisa M Durso, Email: Lisa.Durso@ars.usda.gov.
Michael P Heaton, Email: Mike.Heaton@ars.usda.gov.
William W Laegreid, Email: laegreid@uiuc.edu.
References
- Corrigan JJ, Jr, Boineau FG. Hemolytic-uremic syndrome. Pediatr Rev. 2001;22:365–369. [PubMed] [Google Scholar]
- Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, Laegreid WW. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc Natl Acad Sci USA. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansheroff LJ, O'Brien AD. Escherichia coli O157:H7 in beef cattle presented for slaughter in the U.S.: higher prevalence rates than previously estimated [comment] Proc Natl Acad Sci USA. 2000;97:2959–2961. doi: 10.1073/pnas.97.7.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multistate outbreak of Escherichia coli O157:H7 infections associated with eating ground beef – United States, June–July 2002. MMWR Morb Mortal Wkly Rep. 2002;51:637–639. [PubMed] [Google Scholar]
- Outbreaks of Escherichia coli O157:H7 infections among children associated with farm visits – Pennsylvania and Washington, 2000. MMWR Morb Mortal Wkly Rep. 2001;50:293–297. [PubMed] [Google Scholar]
- Crump JA, Sulka AC, Langer AJ, Schaben C, Crielly AS, Gage R, Baysinger M, Moll M, Withers G, Toney DM, et al. An outbreak of Escherichia coli O157:H7 infections among visitors to a dairy farm. N Engl J Med. 2002;347:555–560. doi: 10.1056/NEJMoa020524. [DOI] [PubMed] [Google Scholar]
- Heuvelink AE, van Heerwaarden C, Zwartkruis-Nahuis JT, van Oosterom R, Edink K, van Duynhoven YT, de Boer E. Escherichia coli O157 infection associated with a petting zoo. Epidemiol Infect. 2002;129:295–302. doi: 10.1017/S095026880200732X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackers ML, Mahon BE, Leahy E, Goode B, Damrow T, Hayes PS, Bibb WF, Rice DH, Barrett TJ, Hutwagner L, et al. An outbreak of Escherichia coli O157:H7 infections associated with leaf lettuce consumption. J Infect Dis. 1998;177:1588–1593. doi: 10.1086/515323. [DOI] [PubMed] [Google Scholar]
- Outbreak of Escherichia coli O157:H7 and Campylobacter among attendees of the Washington County Fair-New York, 1999. MMWR Morb Mortal Wkly Rep. 1999;48:803–805. [PubMed] [Google Scholar]
- Hilborn ED, Mshar PA, Fiorentino TR, Dembek ZF, Barrett TJ, Howard RT, Cartter ML. An outbreak of Escherichia coli O157:H7 infections and haemolytic uraemic syndrome associated with consumption of unpasteurized apple cider. Epidemiol Infect. 2000;124:31–36. doi: 10.1017/S0950268899003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belongia EA, Osterholm MT, Soler JT, Ammend DA, Braun JE, MacDonald KL. Transmission of Escherichia coli O157:H7 infection in Minnesota child day-care facilities. JAMA. 1993;269:883–888. doi: 10.1001/jama.269.7.883. [DOI] [PubMed] [Google Scholar]
- Pavia AT, Nichols CR, Green DP, Tauxe RV, Mottice S, Greene KD, Wells JG, Siegler RL, Brewer ED, Hannon D, et al. Hemolytic-uremic syndrome during an outbreak of Escherichia coli O157:H7 infections in institutions for mentally retarded persons: clinical and epidemiologic observations. J Pediatr. 1990;116:544–551. doi: 10.1016/S0022-3476(05)81600-2. [DOI] [PubMed] [Google Scholar]
- Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis. 2005;11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton JC, Paton AW. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DR, Moxley RA, Francis DH. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv Exp Med Biol. 1997;412:53–58. doi: 10.1007/978-1-4899-1828-4_6. [DOI] [PubMed] [Google Scholar]
- Belongia EA, Chyou PH, Greenlee RT, Perez-Perez G, Bibb WF, DeVries EO. Diarrhea incidence and farm-related risk factors for Escherichia coli O157:H7 and Campylobacter jejuni antibodies among rural children. J Infect Dis. 2003;187:1460–1468. doi: 10.1086/374622. [DOI] [PubMed] [Google Scholar]
- Greatorex JS, Thorne GM. Humoral immune responses to Shiga-like toxins and Escherichia coli O157 lipopolysaccharide in hemolytic-uremic syndrome patients and healthy subjects. J Clin Microbiol. 1994;32:1172–1178. doi: 10.1128/jcm.32.5.1172-1178.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack JP, Jelacic S, Besser TE, Weinberger E, Kirk DJ, McKee GL, Harrison SM, Musgrave KJ, Miller G, Price TH, et al. Escherichia coli O157 exposure in Wyoming and Seattle: serologic evidence of rural risk. Emerg Infect Dis. 2003;9:1226–1231. doi: 10.3201/eid0910.020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond D, Johnson RP, Karmali MA, Petric M, Winkler M, Johnson S, Rahn K, Renwick S, Wilson J, Clarke RC, et al. Neutralizing antibodies to Escherichia coli Vero cytotoxin 1 and antibodies to O157 lipopolysaccharide in healthy farm family members and urban residents. J Clin Microbiol. 1996;34:2053–2057. doi: 10.1128/jcm.34.9.2053-2057.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JB, Clarke RC, Renwick SA, Rahn K, Johnson RP, Karmali MA, Lior H, Alves D, Gyles CL, Sandhu KS, et al. Vero cytotoxigenic Escherichia coli infection in dairy farm families. J Infect Dis. 1996;174:1021–1027. doi: 10.1093/infdis/174.5.1021. [DOI] [PubMed] [Google Scholar]
- Durso LM, Reynolds K, Bauer N, Jr, Keen JE. Shiga-toxigenic Escherichia coli O157:H7 infections among livestock exhibitors and visitors at a Texas County Fair. Vector Borne Zoonotic Dis. 2005;5:193–201. doi: 10.1089/vbz.2005.5.193. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Iimura M, Kaper JB, Torres AG, Kagnoff MF. Role of Shiga toxin versus H7 flagellin in enterohaemorrhagic Escherichia coli signalling of human colon epithelium in vivo. Cell Microbiol. 2006;8:869–879. doi: 10.1111/j.1462-5822.2005.00673.x. [DOI] [PubMed] [Google Scholar]
- Siegler RL, Obrig TG, Pysher TJ, Tesh VL, Denkers ND, Taylor FB. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic uremic syndrome. Pediatr Nephrol. 2003;18:92–96. doi: 10.1007/s00467-002-1035-7. [DOI] [PubMed] [Google Scholar]
- Tatsuno I, Horie M, Abe H, Miki T, Makino K, Shinagawa H, Taguchi H, Kamiya S, Hayashi T, Sasakawa C. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 2001;69:6660–6669. doi: 10.1128/IAI.69.11.6660-6669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, Kaper JB. Identification and characterization of lpfABCC'DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002;70:5416–5427. doi: 10.1128/IAI.70.10.5416-5427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Kaper JB. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect Immun. 2003;71:4985–4995. doi: 10.1128/IAI.71.9.4985-4995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/S0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- Gannon VP, D'Souza S, Graham T, King RK, Rahn K, Read S. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J Clin Microbiol. 1997;35:656–662. doi: 10.1128/jcm.35.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Keen JE, Westerman RB, Littledike ET, Kwang J. Monoclonal antibodies for detection of the H7 antigen of Escherichia coli. Appl Environ Microbiol. 1996;62:3325–3332. doi: 10.1128/aem.62.9.3325-3332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman RB, He Y, Keen JE, Littledike ET, Kwang J. Production and characterization of monoclonal antibodies specific for the lipopolysaccharide of Escherichia coli O157. J Clin Microbiol. 1997;35:679–684. doi: 10.1128/jcm.35.3.679-684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author:Department of Genome Sciences, Univerisity of Washington, Seattle; 2004. [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC Task Force PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7:382–389. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Nietfeldt J, Benson AK. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci USA. 1999;96:13288–13293. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Shaikh N, Holt NJ, Tarr PI, Konkel ME, Malik-Kale P, Walsh CW, Whittam TS, Bono JL. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl Environ Microbiol. 2007;73:671–679. doi: 10.1128/AEM.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune JT, Abedon ST, Takemura K, Christie NP, Sreevatsan S. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg Infect Dis. 2004;10:1482–1485. doi: 10.3201/eid1008.030784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JM, Wagner PL, Acheson DW, Waldor MK. Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl Environ Microbiol. 2003;69:1059–1066. doi: 10.1128/AEM.69.2.1059-1066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudva IT, Evans PS, Perna NT, Barrett TJ, Ausubel FM, Blattner FR, Calderwood SB. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J Bacteriol. 2002;184:1873–1879. doi: 10.1128/JB.184.7.1873-1879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller AC, McEllistrem MC, Stine OC, Morris JG, Jr, Boxrud DJ, Dixon B, Harrison LH. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J Clin Microbiol. 2003;41:675–679. doi: 10.1128/JCM.41.2.675-679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, Ribot EM, Fields PI, Whittam TS, Swaminathan B. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–767. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Keys C, Kemper S, Keim P. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J Appl Microbiol. 2005;98:928–940. doi: 10.1111/j.1365-2672.2004.02532.x. [DOI] [PubMed] [Google Scholar]
- Luo Y, Frey EA, Pfuetzner RA, Creagh AL, Knoechel DG, Haynes CA, Finlay BB, Strynadka NC. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature. 2000;405:1073–1077. doi: 10.1038/35016618. [DOI] [PubMed] [Google Scholar]
- Bono JL, Keen JE, Miller LC, Fox JM, Chitko-McKown CG, Heaton MP, Laegreid WW. Evaluation of a real-time PCR kit for detecting Escherichia coli O157 in bovine fecal samples. Appl Environ Microbiol. 2004;70:1855–1857. doi: 10.1128/AEM.70.3.1855-1857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam TS. Evolution of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. In: Kaper JB, O'Brien AD, editor. Escherichia coli O157:H7 and other Shiga toxin-producing E coli strains. Washington, D.C.: ASM Press; 1998. pp. 195–209. [Google Scholar]