Abstract
HIV infected patients harbor ~105–106 memory CD4 T-cells that contain fully integrated but transcriptionally silent HIV proviruses. While small in number, these latently infected cells form a drug-insensitive reservoir that importantly contributes to the life-long persistence of HIV despite highly effective antiviral therapy. In tissue culture, latent HIV proviruses can be activated when their cellular hosts are exposed to select proinflammatory cytokines or their T-cell receptors are ligated. However, due to a lack of potency and/or dose-limiting toxicity, attempts to purge virus from this latent reservoir in vivo with immune-activating agents including anti-CD3 antibodies and IL-2 have failed. A deeper understanding of the molecular underpinnings of HIV latency is clearly required, including determining whether viral latency is actively reinforced by transcriptional repressors, defining which inducible host transcription factors most effectively antagonize latency, and elucidating the role of chromatin in viral latency. Only through such an improved understanding will it be possible to identify combination therapies that might allow complete purging of the latent reservoir and realization of the difficult and elusive goal of complete eradication of HIV in infected patients.
Keywords: HIV, Latency, NF-κB, Tat
1. Introduction
A decade has now passed since the introduction of highly active antiretroviral therapy (HAART) comprised of combinations of active viral reverse transcriptase and protease inhibitors. These drugs have sharply altered the natural history of HIV infection allowing patients to live longer with a greatly improved quality of life. Despite initial hopes that HAART might be able to effect a cure after 2–3 years of intensive treatment [1], clinical studies have repeatedly revealed a prompt reemergence of virus and resumption of CD4+ T-cell depletion following drug withdrawal despite durable suppression of viremia [2, 3]. This viral recrudescence points to the existence of one or more drug-insensitive viral reservoirs that are capable of reseeding the infection [4]. Several reservoirs have been proposed, including cryptic low-level replication of virus in privileged anatomical compartments, long term retention of infectious virus on follicular dendritic cells, and continuous secretion of virus by long-lived infected cells [2, 5-8]. While low levels of persistent viral replication in various tissue compartments could respond to improved pharmaco-availability or intensification of anti-retroviral drug therapies, one reservoir will remain impervious to such improvements in therapy: a transcriptionally dormant form of HIV infection harbored in a state termed viral latency.
HIV latency was first documented in HIV-infected patients 1995 [9] and shortly thereafter in patients with successful HAART suppression of viremia [10]. T-cell cultures from infected patients were found to harbor a subset of cells that produce infectious virus only after stimulation with strong T-cell activators. Roughly 1 in 106 resting CD4+ T-cells harbor integrated and replication-competent but transcriptionally silent provirus, predicting a total patient load of <107 latently infected cells [11]. This latent reservoir is established within days following acute HIV infection, thus early application of antiretroviral therapy is not a viable strategy for dealing with HIV latency. Additionally, the latent reservoir is continuously repopulated throughout the course of active viral replication [12-14]. HAART administration results in a biphasic depletion of latently infected T-cells, with a period of rapid depletion for the first 3 months of therapy, followed by a more prolonged decay with a half-life of 6–44 months [14–18]. Based on the long half-life of these latent infected memory CD4 T cells, clearing the latent reservoir is predicted to require more than 60 years of antiviral drug therapy.
The durability of the pool of latently infected T-cells has prompted examination of the mechanisms underlying the establishment, maintenance, and resolution of HIV latency. Unfortunately, analysis of these events in vivo has been hampered by several factors. First, latently infected cells are rare. Second, enrichment of these cells is difficult as they lack expression of any viral markers, thus antibody-based purification strategies are ineffective [19]. Additionally, cells harboring defective integrated proviruses are found at frequencies 100–600-fold greater than replication competent latently infected cells in HAART-suppressed patients [11, 20]. The presence of this high background of defective viruses further confounds the analyses of fully infectious, but latent HIV provirus present in the blood of HAART-suppressed patients. As a consequence, most studies exploring the mechanistic basis of HIV latency have been conducted in cell-culture-based model systems. While these systems have limitations, they have revealed many aspects of the mechanisms underlying the establishment, maintenance, and resolution of HIV latency, and have pointed to new therapeutic methods to address viral persistence.
2. Establishing HIV Latency
Two general classes of HIV latency have been detected and can be segregated based on whether or not the virus has integrated into host chromosomes. These two forms are thus termed preintegration and postintegration viral latency. Preintegration latency is commonly observed following the fusion of HIV to resting, unactivated cells, (Figure 1) [21]. This latent state is characterized by incomplete reverse transcription of the viral genome as a consequence of the action of the low-molecular-mass form of APOBEC3G, which functions as a potent post-entry restriction factor for HIV [22]. In addition, the reduced nucleotide pools present in these cells contribute to inefficient reverse transcription [21, 23, 24]. Finally, the import of reverse transcribed HIV genomes into the nucleus is also restricted in resting T-cells as a result of reduced pools of ATP [22, 25]. However, preintegration latency does not appear to be as clinically important as postintegration latency due to its labile nature. The half-life of incompletely reverse-transcribed RNA genomes is roughly 1 day [26, 27]. As such, this latent form of HIV is lost rather rapidly, and cannot explain the long-term persistence of HIV in actively treated patients.
Figure 1.
T-cell activation status influences establishment of active, preintegration- or postintegration latency. (A) HIV infection of unactivated, resting T-cells results in labile preintegration latency with unintegrated, cytoplasmasmic provirus. (B) Infection of activated T-cells results in integrated provirus and active infection with efficient production of progeny virions. (C) Infection of cells in the process of resolving T-cell activation permits completion of reverse transcription and integration, but fails to support active expression of viral genes, i.e. postintegration latency.
The mechanisms promoting establishment of post-integration HIV latency are less clear than those driving preintegration latency. The majority of evidence indicates that HIV is unable to establish integration of the provirus into host DNA in circulating resting T-cells [21, 24, 25], suggesting that post-integration latency is established in activated T-cells. In this model, post-integration latency is established during the shut-off of T-cell activation. The observation that post-integration latency is commonly found in resting cells supports the notion that this form of infection may be established as cells retreat from an activated state. Studies of SCID-hu mice reconstituted with human thymus-liver explants indicate that HIV latency can be established during resolution of thymopoietic T-cell activation, before lymphocytes are fully developed [28]. In contrast, macaque studies indicate that hybrid HIV/simian immunodeficiency viruses (S/HIV) can establish postintegration latency in lymphocytes, but not in thymocytes [29]. The preponderance of postintegration latency in memory CD4 T-cells relative to naïve T-cells in infected patients suggests that T-cells infected during the resolution of a T-cell activating event represent the primary source of post-integration latency [11]. An alternative mechanism is suggested by studies of SIV indicating direct infection of resting T-cells in the vaginal mucosa of rhesus macaques [30]. It is unclear how the biology of macaque mucosal lymphocytes differs from their circulating human counterparts, but the possibility that a subset of resting lymphocytes is somehow permissive to infection cannot be dismissed. The predominance of latency within the memory T-cell compartment could be explained in this case by the selective expression of CCR5 on these cells, the preferred HIV coreceptor in early infection. Further studies will be required to determine the ultimate source of latently infected T-cells.
Although T-cell activation enhances expression of HIV, an activated T-cell millieu is not strictly required for HIV gene expression. For example, robust production of HIV virions occurs in 293T human kidney cells following infection with pseudotyped virions. Emerging evidence indicates that the site of proviral integration into the host genome plays a pivotal role in the establishment of postintegration HIV latency. HIV integrates into host chromosomal DNA in a non-random manner where preference is found for intronic regions of actively transcribed genes [31]. The site of proviral integration can strongly shape the level of basal viral gene expression, with ~100-fold differences in basal transcription observed between different integration loci [32]. Although initial reports suggested that integration into transcriptionally inactive regions of the genome is responsible for low basal levels of transcription [33], more thorough surveys show that latent HIV genomes are also commonly integrated into highly expressed host genes [34]. A study of proviral integration in resting CD4+ T-cells in HIV-infected patients on HAART similarly found >80% of integration events in host genes that are actively transcribed, although it is unclear whether these integration events are representative of defective integration events or true latent proviruses.
The finding that latent HIV proviruses can exist in actively transcribed genes is somewhat paradoxical in view of the notion that postintegration latency is a consequence of “transcriptional insufficiency.” Transcriptional interference might be in play. In the case of proviral integration with the same polarity as the surrounding host gene, “read-through” transcription from an upstream host promoter could displace key transcription factors from the HIV promoter, as observed in one HeLa cell model of HIV latency [35]. Alternatively, proviral integration in an opposing orientation to the surrounding gene might lead to collision of elongating polymerases from each promoter. Further, the placement of an active HIV promoter and host promoter in close proximity may result in silencing of both viral and host genes independent of direct polymerase displacement [34]. Thus, multiple mechanisms may contribute to the establishment of HIV latency.
3. Maintaining Post-Integration HIV Latency
HIV proviral DNA is organized into chromatin through interactions with octameric assemblies of cellular histone proteins, forming nucleosomes. Nucleosomes predictably occur in the 5′ long terminal repeat (LTR) of HIV and regulate basal transcriptional activity of the provirus [36]. Two nucleosomes, termed nuc-0 and nuc-1, encompass nt 40-200 and 452-596 respectively within the HIV LTR, and overlap binding sites for several transcription factors and the viral transcriptional initiation site [37, 38]. Several observations suggest that the status of these nucleosomes is a key modulator of HIV transcriptional activity. Nuc-1 is rapidly remodeled in response to T-cell activating stimuli, coincident with the activation of viral gene expression [38]. Similarly, induction of histone hyperacetylation, a histone modification associated with transcriptional activity (see [39] for review), by histone deacetylase (HDAC) inhibitors such as sodium butyrate, valproic acid, and trichostatin A induces remodeling of nuc-1 and transcriptional activation of HIV [40, 41]. The positive role of histone acetylation in HIV transcription is further supported by the observation that purified cellular acetyltransferases (HAT)s markedly stimulate transcriptional activity of in vitro chromatin-assembled HIV proviral DNA [42]. These findings have focused many studies on the identification of HDACs and HATs that regulate the acetylation of histones organizing the HIV LTR.
The histones comprising nuc-0 and nuc-1 in the HIV LTR are constitutively deacetylated in cellular models of HIV latency suggesting the presence and action of an HDAC. NF-κB p50 homodimers, LSF1, YY1, and thyroid hormone receptor have all been reported to recruit the cellular deacetylase HDAC1 to the HIV promoter through distinct binding sites with the LTR [43-45]. The deacetylated status of histones in the context of HDAC1 disfavors binding of core transcriptional machinery to the HIV LTR, including RNA polymerase II [45]. Displacement of HDAC1 from the HIV LTR or inhibition of its activity promotes effective RNA polymerase II binding thereby allowing for transcriptional initiation within the latent HIV LTR [45]. These observations suggested a new therapeutic strategy. Administration of the HDAC inhibitor valproic acid to HIV-infected patients on intensified antiretroviral therapy resulted in a 75% reduction in the frequency of latently infected cells [46]. While falling well short of complete elimination of the latent reservoir, this observed reduction underscores the contribution of HDAC-mediated processes to the maintenance of HIV latency.
Recent studies additionally point to a role for DNA CpG methylation in the establishment of certain forms of HIV latency. Methylated CpG sequences favor recruitment of cellular CPG-binding protein MBD2 and other regulatory proteins, providing an additional platform for the recruitment of HDACs [47]. Methylation of HIV DNA strongly suppresses viral gene expression [48, 49], and is the apparent basis of latency in the ACH-2 model of latency [50, 51]. However, this mechanism of latency does not appear to exist in all latently infected cell types, as evidenced by the rather weak induction of latent HIV expression in the J-Lat model of HIV-1 latency by CpG methylation inhibitor, 5-azacytidine [52]. Intriguingly, lentiviral infection enhances general DNA methylation of host genes through the upregulation of cellular DNA methyltransferase 1, suggesting elicitation of a potential innate protective cellular response [53, 54]. Further, long-term studies of HIV-based lentiviral vectors indicate that progressive DNA methylation underlies the eventual transcriptional shutdown that plagues this gene delivery method [55]. The contribution of CpG methylation to latency in vivo remains unknown.
4.1 Reactivating Latent HIV Proviruses – Initiation of Transcription
The activation of latent HIV proviruses can be induced by a wide range of T-cell stimuli, including T-cell receptor ligation by anti-CD3 antibodies, cytokines including IL-1β, IL-7, and TNF-α, and mitogens including PMA and prostratin [40, 56-61]. Each of these stimuli induce broad changes in T-cell activation status, and affect cellular and viral gene expression through both distinct and shared signaling pathways (Figure 2). Such signaling ultimately drives HIV transcription through the induction of activating cellular transcription factors, including NF-κB, NFAT, and AP-1, which bind to cognate enhancer sequences within the HIV LTR. The LTR contains several additional DNA binding domains that engage various cellular transcription factors, including Sp1, LEF-1, COUP-TF, YY1, Ets-1, and USF (reviewed in [62]). The Sp1 and κB binding sites are required for HIV-1 replication, whereas other sites enhance transcription but are nonetheless dispensable [63, 64]. The mechanism of Sp1 enhancement of HIV transcription is unclear; however, Sp1 association with TBP, TAF250, and TAF55 suggesting that Sp1 may direct the assembly of these key components of basal cellular transcriptional machinery on the LTR [65-67].
Figure 2.
Signaling networks linking T-cell activating stimuli to HIV-inducing transcription factors. NF-κB, NFAT, and AP-1 mediate activation of latent HIV in response to cytokines, TCR ligation, and mitogens.
NF-κB/Rel factors can promote both positive and negative effects on transcription depending on which Rel family member is bound to the duplicated κB enhancers present in the HIV LTR. In the J-Lat model of HIV latency, NF-κB p50 homodimers bind the HIV LTR in unstimulated cells, and promote recruitment of transcriptionally repressive HDAC1 [45, 68]. However, when T cells are activated, p50 homodimers are displaced by liberated cytoplasmic stores of transcriptionally activating NF-κB p50-RelA heterodimers, which in turn recruit the cellular histone acetyltransferase p300, driving localized histone acetylation and promoting transcriptional initiation [69, 70]. RelA additionally directs recruitment of the cellular RNA polymerase II kinase complexes including P-TEFb and TFIIH/CDK7 [70-72]. TFIIH/CDK7 and P-TEFb direct phosphorylation of serine-5 and serine-2 residues in the C-terminal domain of RNA polymerase II, respectively, modifications which promote promoter clearance and efficient transcriptional elongation. As a consequence of this modulation of chromatin and basal transcription factor recruitment, induction of NF-κB strongly enhances proviral transcriptional initiation rates [73]. These events appear to be important for the activation of HIV expression, as low levels of nuclear p50-RelA heterodimers are correlated with suppressed levels of HIV transcription [74]. Further, NF-κB inhibitors strongly restrict expression of HIV in response to T-cell activation [75]. The participation of other Rel-family proteins in HIV expression is less clear, but none appear to efficiently promote HIV expression [76]. c-Rel appears to function as a competitive inhibitor of RelA binding to the LTR and thus may inhibit HIV activation by displacing RelA [77]. Expanded analysis of the differences between these related transcription factors and their associated cofactors promises to provide important insights into the regulation of HIV latency.
T-cell receptor ligation additionally induces AP-1 and NFAT, which synergize with NF-κB to positively regulate HIV gene expression. NFAT enhances NF-κB-directed activation of HIV gene expression, but little is known about its action in the absence of active NF-κB [78]. Overexpression of NFATc in the transformed SupT1 T-cell line increases HIV replication, perhaps acting in synergy with NF-κB [79]. NFAT is a key regulator of IL-7 induced activation of latent HIV gene expression [61], and appears to be an important factor in the activation of latency in memory T cells in vivo [80]. The cofactors involved in NFAT-mediated transcriptional activation of HIV remain poorly defined, although reports of NFAT1 association with p300 and CBP suggest that, like NF-κB, members of the NFAT family may promote chromatin remodeling of the LTR [81]. Indeed, studies of host NFAT-responsive promoters indicate that NFAT binding induces extensive nucleosomal disruption, although this process is dependent on cooperative binding by AP-1 [82]. Suppression of AP-1 activation with upstream MAPK and MEK inhibitors suppresses, but does not eliminate TCR activation-induced expression of HIV [83]. Thus, though NFAT and AP-1 can positively regulate HIV gene expression, they are not required for activation of viral latency.
HIV transcription is further influenced by a number of non-inducible transcription factors. For example, LEF/TCF1-a binds the HIV LTR and promotes proviral expression by establishing a nucleosome-free region between Nuc-0 and Nuc-1 [84, 85]. This region may be further expanded by proximal binding of USF and Ets, proteins which cooperatively promote HIV transcription [86-88]. In contrast to the activating properties of these proteins, binding of the YY1 factor acts to repress HIV transcription in an LSF- and HDAC1-dependent manner [43, 89]. In view of the constitutive activity of these and other transcription factors, it is unclear how these interactions can be exploited to induce or permanently suppress expression of latent HIV proviruses.
4.2 Reactivating Latent HIV Proviruses- Stimulation of RNA Polymerase II Elongation
After successful initiation of transcription, expression of full-length HIV transcripts requires the concerted action of several cellular proteins to promote efficient elongation of the bound RNA Pol II complex. The absence or inactivity of one or more of these proteins, while generating short viral transcripts, fails to support effective viral replication. The HIV-transactivating protein, Tat, plays a central role in ensuring effective Pol II elongation. Tat binds to a stem loop structure, TAR, located in the 5′ region of all initiated HIV RNA transcripts. The binding of Tat to TAR promotes the recruitment of various cellular factors including kinases, methyltransferases, acetyltransferases and components of cellular mRNA capping machinery that contribute to Pol II elongation efficiency. Similarly, T-cell activation-induced binding of RelA to the HIV LTR is associated with enhanced transcriptional elongation, albeit with less efficiency than that induced by Tat [73]. This action of RelA or related factors is pivotal for the initial production of Tat.
Tat transcriptional activity is dependent on a direct interaction with P-TEFb, a cellular kinase comprised of CDK9 and CylinT1 (CycT1) heterodimers. P-TEFb phosphorylates serine-2 residues in the C-terminal domain (CTD) of the largest subunit of RNA polymerase II [90], a modification required for effective elongation by the bound polymerase complexes. P-TEFb additionally phosphorylates the negatively acting cellular transcriptional components NELF and DSIF, further promoting transcriptional activity [91]. Chemical inhibition of CDK9 with 5,6 dichloro-{beta}-D-ribofuranosylbenzimidazole (DRB) or genetic inhibition with CDK9 dominant negative mutants strongly impairs Tat-mediated activation of HIV gene expression [92, 93]. Limited expression of CycT1 in resting CD4 T cells also contributes to the restricted expression of HIV in some models of latency [94]. Indeed, CycT1 expression levels are effectively increased by several antagonists of HIV latency further suggesting that a lack of active P-TEFb contributes to HIV latency [95, 96].
P-TEFb kinase activity is constitutively restricted by its association with a small cellular RNA, 7SK, which serves as a scaffold for HEXIM1, a cellular protein with a C-terminal P-TEFb inhibitory domain [97]. T-cell activation promotes the release and activation of P-TEFb, driving strong expression of latent HIV [95]. Similarly, hexamethylene bisacetamide (HMBA), an inducer of HEXIM1 expression and P-TEFb release, enhances expression of HIV independently of both NF-κB and Tat [98]. This activity of HMBA has prompted hope that it might be useful as an antagonist of HIV latency provided it does not elicit unacceptable toxicity. Of note, HMBA also induces HEXIM1 expression, a response that likely forms a negative feedback loop for tight control of P-TEFb.
The transcriptionally active form of P-TEFb interacts with the cellular bromodomain protein BRD4, an interaction which can promote recruitment of the kinase complex to chromatin through interactions with acetylated histones [99, 100]. Surprisingly, while this interaction is important for basal expression of HIV, it appears dispensable for Tat-mediated transactivation. These findings highlight an important role for P-TEFb in HIV gene expression independent of its recruitment to TAR by Tat [99]. Indeed, RelA also assembles with P-TEFb and chemical inhibition of this kinase complex suppresses RelA-mediated transactivation of HIV, highlighting the common involvement of this kinase complex in both Tat and RelA action [70]. RelA-mediated recruitment of CDK9 to the HIV LTR promotes localized serine-2 phosphorylation of RNA polymerase II, however, this polymerase is subject to dephosphorylation and stalling [71]. In contrast, Tat appears to promote assembly of a mobile CDK9-RNA polymerase II complex which sustains serine-2 phosphorylation of RNA polymerase II as it transits downstream of the HIV TLR. These findings underscore the pivotal effects of P-TEFb on the HIV LTR involving both host and viral factors that ultimately shape the HIV transcriptional response.
Tat interacts with several additional cellular cofactors that influence transcriptional elongation. For example, Tat mediates recruitment of acetyltransferases p300 and P/CAF to the HIV LTR [101-103]. These interactions likely reinforce an acetylated, open chromatin environment favorable for continued rounds of transcriptional initiation. In addition to its role as a histone acetyltransferase, p300 acetylates Tat at K50, an activating modification which promotes release of P-TEFb, transfer of Tat to the elongating polymerase complex and subsequent interaction with the P/CAF acetyltransferase [104-106]. Tat is negatively regulated by arginine methylation via PRMT6, although it remains to be discerned precisely how this posttranslational modification impairs Tat action [107].
Increasing evidence suggests that transcription and pre-mRNA processing involving splicing, capping, and polyadenylation are intimately intertwined (see [108] for review). Tat may influence many of these processes. Recently, the interaction of P-TEFb with the cellular splicing factor SKIP was shown to be essential for Tat-mediated transactivation of HIV [109]. Indeed, Tat may influence splice site recognition through interaction with the ASF/SF-2 cellular splicing complex [110, 111]. Tat also interacts with cellular mRNA capping proteins Mce1 and Hcm1, which stimulate cotranslational capping and stabilization of nascent HIV transcripts, providing an additional mechanism to enhance HIV gene expression [112, 113].
Although Tat is classically recognized for its role as an enhancer of transcriptional elongation, it may also influence levels of transcriptional initiation. Tat can enhance HIV gene expression in the absence of HIV TAR, albeit with markedly less efficiency than in the presence of TAR [114]. Tat has been observed to direct partial assembly of the transcriptional preinitiation complex [115]. Studies of the chromatin-assembled HIV indicate that this effect of Tat is dependent on the Sp1 and NF-κB binding sites in the LTR [116]. Several studies suggest that Tat can activate expression via NF-κB, although no consensus has been reached in terms of mechanism. Tat may bind to κB DNA enhancer sequences and transactivate in the absence of NF-κB proteins [117]. Alternatively, Tat may stimulate acetylation of RelA/p50 complexes, enhancing their activity [118]. A further alternative is offered by observations of Tat-induced activation of upstream NF-κB kinases via a PKR-dependent mechanism [119, 120]. While not yet well understood, these effects of Tat on transcriptional initiation may contribute to activation of latent HIV proviruses.
Elongation of HIV transcripts is additionally dependent on the activity of cellular ATP-dependent chromatin remodeling enzymes in the SWI/SNF family, which promote remodeling of downstream nucleosomes, thus permitting transit of RNA polymerase II [121, 122]. As such, the absence or inactivity of SWI/SNF complexes may limit the generation of long HIV transcripts thereby promoting HIV latency. Mitogenic activation of T-cells induces concomitant recruitment of Jun-3, BRG-1 and ATF-3 subunits of SWI/SNF complexes to the HIV promoter [123]. Additionally, T-cell activation enhances the activity of ATP-dependent chromatin remodeling SWI/SNF enzymes through a poorly understood mechanism involving increased pools of inositol phosphate [124-126]. Further, Tat may promote SWI/SNF activity at the HIV LTR by directly recruiting the remodeling subunits Brm and INI1 [127, 128]. These findings raise the possibility that Tat enhances transcriptional elongation both by promoting RNA Pol II phosphorylation status and by stimulating chromatin remodeling.
4.3 Reactivating Latent HIV Proviruses- Role of Post-Transcriptional Events
Studies of HIV transcription using blood cells from infected patients suggest that events following viral transcription may also be important for HIV latency [129]. After proviral transcription is complete, fully spliced viral transcripts are rapidly exported out of the nucleus. These multiply spliced viral transcripts encoding the regulatory viral proteins tat, rev, and nef form the predominant cytoplasmic HIV transcripts. Conversely, expression of the viral Rev protein is required for efficient export of unspliced or singly spliced transcripts that encode the structural and enzymatic components of the virus as well as its genomic RNA (see [130] for review). Rev binds to an RNA stem-loop structure present in the Env region of viral transcripts termed the Rev response element (RRE). Rev recruits the RanGTP- dependent CRM1/exportin-1 protein that is essential for nuclear export of these incompletely spliced viral RNAs. No evidence has emerged implicating a modulation of RanGTP or CRM1 expression during T-cell activation thus it is unlikely that deficiencies in these factors provide a mechanistic basis for HIV latency. It is possible, however, that other unidentified cofactors in this export pathway are regulated during T-cell activation.
An new and interesting perspective on HIV RNA export has emerged with the recent observation that Tat and Rev transcripts are retained in the nucleus of resting CD4+ T-cells of infected patients treated with HAART [131]. This observation is in contrast to earlier studies that analyzed activated T-cell cultures. Of note, ectopic expression of the cellular RNA export protein PTB rescues export and expression of latent virus in infected patient blood and expression of PTB is induced by T-cell activation. These findings raise the possibility of a new mechanism regulating some forms of HIV latency.
5. Future Directions
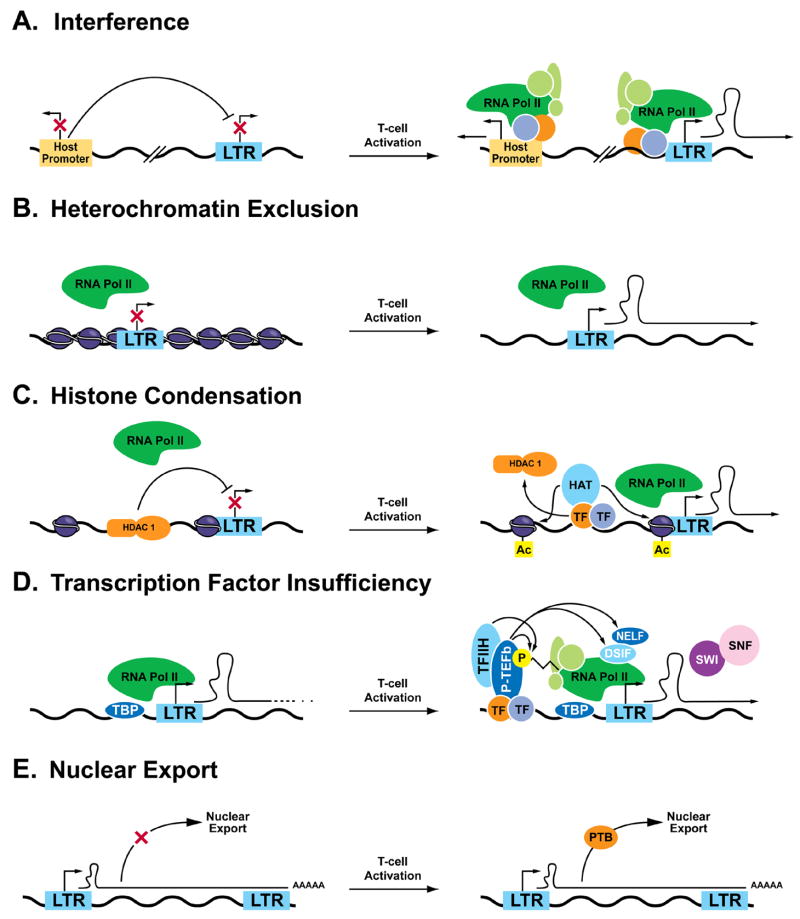
The study of HIV latency over the past decade has identified multiple mechanisms underlying this persistent and drug-insensitive form of viral infection. The site of proviral integration, chromatin status, active repression, limited access to positive transcription factors, absence of Tat required for RNA polymerase II elongation, and altered nuclear export machinery have all been implicated as important contributors to the restriction of latent HIV gene expression (Figure 3). T-cell activation can impact each of these stages of viral gene expression. Thus, it is unclear which of these blocks represent the major contributors to HIV latency as observed in vivo. Future studies delineating the contribution of each of these processes to in vivo latency will be key to future progress of this field. A first step in that direction will likely involve the development of superior in vitro systems based in primary blood [132] to sort out the predominant mechanisms underlying HIV latency. Additionally, investigation of the possibility of a class of latently infected hematopoetic stem cells should be pursued [133], as these cells may require wholly different strategies than their differentiated counterparts.
Figure 3.
Molecular mechanisms of postintegration HIV-1 latency. Multiple mechanisms underlying HIV-1 latency have been observed in various models, including transcriptional interference by proximal promoters of host genes (A), exclusion of basal transcription factors from the HIV promoter by heterochromatin (B), or HDAC-mediated chromatin condensation (C), insufficiency of transcriptional coactivators (D), or basal nuclear export factors (E).
An important consideration for the future development of viral purging strategies is the limitations of existing antiretroviral compounds. Although current therapeutic regimens are effective at inhibiting viral replication and the establishment of infection in new cells, they do not direct the elimination of existing infected cells. While in most cases viral infection results in cell death, observations in cell culture suggest that latently infected cells can be activated, produce viral genes, and survive to return to a renewed state of latency [78]. Thus, latently infected cells may display relative resistance to virus-induced cytopathic effects. Strategies to directly induce cell death in activated latently infected cells may be necessary to eliminate this pool. An immunotoxin therapy employing a viral-envelope targeted antibody for example fused to Pseudomonas exotoxin A offers one potential approach for actively depletion of these cells [134, 135].
Recent studies have uncovered multiple mechanisms underlying latency, but none have identified a “silver bullet” therapy to address each mechanism. While the early approaches involving immune-activating therapies might target all forms of latency, these approaches have thus far proven unsatisfactory due both to lack of potency and unacceptable toxiciy [56, 136-139]. It seems likely that a multi-pronged therapeutic approach will be required to confront this problem. Such combination therapy might include specific HDAC inhibitors, P-TEFb inducers, and mild T-cell activating agents. The recent successful partial depletion of latent HIV with the HDAC inhibitor valproic acid offers some hope that the elucidation of the molecular mechanisms of latency will point to new drugs for addressing the daunting problem of HIV latency. The P-TEFb-inducing activity of HMBA provides additional promise. Recent studies indicate that in addition to its ability to promote HIV expression, HMBA inhibits HIV replication by promoting downregulation of CD4 surface expression [140], thus this compound is an intriguing potential adjuvant for future HIV therapies. Phase II clinical trials suggest that HMBA can be administered safely to patients, thus the dual actions of this compound’s ability to promote HIV expression while inhibiting new infection might be useful in HIV-infected patients [141].
The further delineation of mechanisms surrounding HIV latency will likely identify yet additional agents that can attack the latent reservoir. Although the complete elimination of latent HIV will undoubtedly be a difficult task, an improved understanding of how HIV latency is formed and maintained offers hope that new therapies can be rationally developed. The identification and assessment of new anti-HIV latency adjuvants over the coming years will be an exciting and much anticipated chapter in HIV research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–91. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, Davey RT, Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins JI, et al. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med. 2000;6:757–61. doi: 10.1038/77481. [DOI] [PubMed] [Google Scholar]
- 3.Davey RT, Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96:15109–14. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrager LK, D’Souza MP. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. Jama. 1998;280:67–71. doi: 10.1001/jama.280.1.67. [DOI] [PubMed] [Google Scholar]
- 5.Di Mascio M, Dornadula G, Zhang H, Sullivan J, Xu Y, Kulkosky J, et al. In a subset of subjects on highly active antiretroviral therapy, human immunodeficiency virus type 1 RNA in plasma decays from 50 to <5 copies per milliliter, with a half-life of 6 months. J Virol. 2003;77:2271–5. doi: 10.1128/JVI.77.3.2271-2275.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharkey ME, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan JL, et al. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imamichi H, Crandall KA, Natarajan V, Jiang MK, Dewar RL, Berg S, et al. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J Infect Dis. 2001;183:36–50. doi: 10.1086/317641. [DOI] [PubMed] [Google Scholar]
- 8.Hlavacek WS, Stilianakis NI, Notermans DW, Danner SA, Perelson AS. Influence of follicular dendritic cells on decay of HIV during antiretroviral therapy. Proc Natl Acad Sci U S A. 2000;97:10966–71. doi: 10.1073/pnas.190065897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–90. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 10.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 11.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–8. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 12.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A. 1998;95:8869–73. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruff CT, Ray SC, Kwon P, Zinn R, Pendleton A, Hutton N, et al. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol. 2002;76:9481–92. doi: 10.1128/JVI.76.18.9481-9492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blankson JN, Finzi D, Pierson TC, Sabundayo BP, Chadwick K, Margolick JB, et al. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J Infect Dis. 2000;182:1636–42. doi: 10.1086/317615. [DOI] [PubMed] [Google Scholar]
- 15.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 16.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–7. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 17.Ramratnam B, Mittler JE, Zhang L, Boden D, Hurley A, Fang F, et al. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat Med. 2000;6:82–5. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, et al. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med. 1999;340:1605–13. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 19.Brooks DG, Zack JA. Effect of latent human immunodeficiency virus infection on cell surface phenotype. J Virol. 2002;76:1673–81. doi: 10.1128/JVI.76.4.1673-1681.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–5. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 21.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–22. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 22.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–14. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 23.Zack JA, Haislip AM, Krogstad P, Chen IS. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–25. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. Embo J. 1990;9:1551–60. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–7. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Zhang H, Siliciano JD, Siliciano RF. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol. 2005;79:2199–210. doi: 10.1128/JVI.79.4.2199-2210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol. 2002;76:8518–31. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA. Generation of HIV latency during thymopoiesis. Nat Med. 2001;7:459–64. doi: 10.1038/86531. [DOI] [PubMed] [Google Scholar]
- 29.Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, et al. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol. 2003;77:4938–49. doi: 10.1128/JVI.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286:1353–7. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 31.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–9. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 32.Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. Embo J. 2001;20:1726–38. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. Embo J. 2003;22:1868–77. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, et al. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol. 2005;79:6610–9. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greger IH, Demarchi F, Giacca M, Proudfoot NJ. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 1998;26:1294–301. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeang KT, Berkhout B, Dropulic B. Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J Biol Chem. 1993;268:24940–9. [PubMed] [Google Scholar]
- 37.Van Lint C, Ghysdael J, Paras P, Jr, Burny A, Verdin E. A transcriptional regulatory element is associated with a nuclease-hypersensitive site in the pol gene of human immunodeficiency virus type 1. J Virol. 1994;68:2632–48. doi: 10.1128/jvi.68.4.2632-2648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verdin E, Paras P, Jr, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. Embo J. 1993;12:3249–59. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 40.Laughlin MA, Zeichner S, Kolson D, Alwine JC, Seshamma T, Pomerantz RJ, et al. Sodium butyrate treatment of cells latently infected with HIV-1 results in the expression of unspliced viral RNA. Virology. 1993;196:496–505. doi: 10.1006/viro.1993.1505. [DOI] [PubMed] [Google Scholar]
- 41.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. Embo J. 1996;15:1112–20. [PMC free article] [PubMed] [Google Scholar]
- 42.Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci U S A. 1998;95:12924–9. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol. 2000;74:6790–9. doi: 10.1128/jvi.74.15.6790-6799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsia SC, Shi YB. Chromatin disruption and histone acetylation in regulation of the human immunodeficiency virus type 1 long terminal repeat by thyroid hormone receptor. Mol Cell Biol. 2002;22:4043–52. doi: 10.1128/MCB.22.12.4043-4052.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. Embo J. 2006;25:139–49. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366:549–55. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 48.Schulze-Forster K, Gotz F, Wagner H, Kroger H, Simon D. Transcription of HIV1 is inhibited by DNA methylation. Biochem Biophys Res Commun. 1990;168:141–7. doi: 10.1016/0006-291x(90)91685-l. [DOI] [PubMed] [Google Scholar]
- 49.Bednarik DP, Cook JA, Pitha PM. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. Embo J. 1990;9:1157–64. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishida T, Hamano A, Koiwa T, Watanabe T. 5′ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology. 2006;3:69. doi: 10.1186/1742-4690-3-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Brien MC, Ueno T, Jahan N, Zajac-Kaye M, Mitsuya H. HIV-1 expression induced by anti-cancer agents in latently HIV-1-infected ACH2 cells. Biochem Biophys Res Commun. 1995;207:903–9. doi: 10.1006/bbrc.1995.1271. [DOI] [PubMed] [Google Scholar]
- 52.Pion M, Jordan A, Biancotto A, Dequiedt F, Gondois-Rey F, Rondeau S, et al. Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation. J Virol. 2003;77:4025–32. doi: 10.1128/JVI.77.7.4025-4032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang JY, Mikovits JA, Bagni R, Petrow-Sadowski CL, Ruscetti FW. Infection of lymphoid cells by integration-defective human immunodeficiency virus type 1 increases de novo methylation. J Virol. 2001;75:9753–61. doi: 10.1128/JVI.75.20.9753-9761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikovits JA, Young HA, Vertino P, Issa JP, Pitha PM, Turcoski-Corrales S, et al. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol Cell Biol. 1998;18:5166–77. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofmann A, Kessler B, Ewerling S, Kabermann A, Brem G, Wolf E, et al. Epigenetic regulation of lentiviral transgene vectors in a large animal model. Mol Ther. 2006;13:59–66. doi: 10.1016/j.ymthe.2005.07.685. [DOI] [PubMed] [Google Scholar]
- 56.Prins JM, Jurriaans S, van Praag RM, Blaak H, van Rij R, Schellekens PT, et al. Immuno-activation with anti-CD3 and recombinant human IL-2 in HIV-1-infected patients on potent antiretroviral therapy. Aids. 1999;13:2405–10. doi: 10.1097/00002030-199912030-00012. [DOI] [PubMed] [Google Scholar]
- 57.Popik W, Pitha PM. Role of tumor necrosis factor alpha in activation and replication of the tat-defective human immunodeficiency virus type 1. J Virol. 1993;67:1094–9. doi: 10.1128/jvi.67.2.1094-1099.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulkosky J, Culnan DM, Roman J, Dornadula G, Schnell M, Boyd MR, et al. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood. 2001;98:3006–15. doi: 10.1182/blood.v98.10.3006. [DOI] [PubMed] [Google Scholar]
- 59.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–2. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 60.Poli G, Kinter AL, Fauci AS. Interleukin 1 induces expression of the human immunodeficiency virus alone and in synergy with interleukin 6 in chronically infected U1 cells: inhibition of inductive effects by the interleukin 1 receptor antagonist. Proc Natl Acad Sci U S A. 1994;91:108–12. doi: 10.1073/pnas.91.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Managlia EZ, Landay A, Al-Harthi L. Interleukin-7 induces HIV replication in primary naive T cells through a nuclear factor of activated T cell (NFAT)-dependent pathway. Virology. 2006;350:443–52. doi: 10.1016/j.virol.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 62.Rohr O, Marban C, Aunis D, Schaeffer E. Regulation of HIV-1 gene transcription: from lymphocytes to microglial cells. J Leukoc Biol. 2003;74:736–49. doi: 10.1189/jlb.0403180. [DOI] [PubMed] [Google Scholar]
- 63.Sune C, Garcia-Blanco MA. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995;69:6572–6. doi: 10.1128/jvi.69.10.6572-6576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM, Nabel GJ. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. Embo J. 1993;12:3551–8. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emili A, Greenblatt J, Ingles CJ. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol Cell Biol. 1994;14:1582–93. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–6. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 67.Shao Z, Ruppert S, Robbins PD. The retinoblastoma-susceptibility gene product binds directly to the human TATA-binding protein-associated factor TAFII250. Proc Natl Acad Sci U S A. 1995;92:3115–9. doi: 10.1073/pnas.92.8.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montano MA, Kripke K, Norina CD, Achacoso P, Herzenberg LA, Roy AL, et al. NF-kappa B homodimer binding within the HIV-1 initiator region and interactions with TFII-I. Proc Natl Acad Sci U S A. 1996;93:12376–81. doi: 10.1073/pnas.93.22.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci U S A. 1997;94:2927–32. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–37. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 71.Williams SA, Kwon H, Chen LF, Greene WC. Sustained Induction of NF-kB is Required for Efficient Expression of Latent HIV-1. J Virol. 2007 doi: 10.1128/JVI.02074-06. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, et al. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. Embo J. 2006;25:3596–604. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.West MJ, Lowe AD, Karn J. Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J Virol. 2001;75:8524–37. doi: 10.1128/JVI.75.18.8524-8537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–3. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 75.Logeat F, Israel N, Ten R, Blank V, Le Bail O, Kourilsky P, et al. Inhibition of transcription factors belonging to the rel/NF-kappa B family by a transdominant negative mutant. Embo J. 1991;10:1827–32. doi: 10.1002/j.1460-2075.1991.tb07708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J, Perkins ND, Schmid RM, Nabel GJ. Specific NF-kappa B subunits act in concert with Tat to stimulate human immunodeficiency virus type 1 transcription. J Virol. 1992;66:3883–7. doi: 10.1128/jvi.66.6.3883-3887.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doerre S, Sista P, Sun SC, Ballard DW, Greene WC. The c-rel protooncogene product represses NF-kappa B p65-mediated transcriptional activation of the long terminal repeat of type 1 human immunodeficiency virus. Proc Natl Acad Sci U S A. 1993;90:1023–7. doi: 10.1073/pnas.90.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams SA, Chen LF, Kwon H, Fenard D, Bisgrove D, Verdin E, et al. Prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem. 2004;279:42008–17. doi: 10.1074/jbc.M402124200. [DOI] [PubMed] [Google Scholar]
- 79.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–44. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 80.Robichaud GA, Barbeau B, Fortin JF, Rothstein DM, Tremblay MJ. Nuclear factor of activated T cells is a driving force for preferential productive HIV-1 infection of CD45RO-expressing CD4+ T cells. J Biol Chem. 2002;277:23733–41. doi: 10.1074/jbc.M201563200. [DOI] [PubMed] [Google Scholar]
- 81.Garcia-Rodriguez C, Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP) J Exp Med. 1998;187:2031–6. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson BV, Bert AG, Ryan GR, Condina A, Cockerill PN. Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol Cell Biol. 2004;24:7914–30. doi: 10.1128/MCB.24.18.7914-7930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang X, Chen Y, Gabuzda D. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappaB. J Biol Chem. 1999;274:27981–8. doi: 10.1074/jbc.274.39.27981. [DOI] [PubMed] [Google Scholar]
- 84.Steger DJ, Workman JL. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. Embo J. 1997;16:2463–72. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, et al. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 86.Sieweke MH, Tekotte H, Jarosch U, Graf T. Cooperative interaction of ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. Embo J. 1998;17:1728–39. doi: 10.1093/emboj/17.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Du H, Roy AL, Roeder RG. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the Ad-ML promoters. Embo J. 1993;12:501–11. doi: 10.1002/j.1460-2075.1993.tb05682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seth A, Hodge DR, Thompson DM, Robinson L, Panayiotakis A, Watson DK, et al. ETS family proteins activate transcription from HIV-1 long terminal repeat. AIDS Res Hum Retroviruses. 1993;9:1017–23. doi: 10.1089/aid.1993.9.1017. [DOI] [PubMed] [Google Scholar]
- 89.Margolis DM, Somasundaran M, Green MR. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–10. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parada CA, Roeder RG. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–8. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 91.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24:787–95. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. Embo J. 1991;10:4189–96. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fujinaga K, Irwin D, Geyer M, Peterlin BM. Optimized chimeras between kinase-inactive mutant Cdk9 and truncated cyclin T1 proteins efficiently inhibit Tat transactivation and human immunodeficiency virus gene expression. J Virol. 2002;76:10873–81. doi: 10.1128/JVI.76.21.10873-10881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liou LY, Herrmann CH, Rice AP. Transient induction of cyclin T1 during human macrophage differentiation regulates human immunodeficiency virus type 1 Tat transactivation function. J Virol. 2002;76:10579–87. doi: 10.1128/JVI.76.21.10579-10587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sung TL, Rice AP. Effects of prostratin on Cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology. 2006;3:66. doi: 10.1186/1742-4690-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ghose R, Liou LY, Herrmann CH, Rice AP. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4(+) T lymphocytes by combination of cytokines. J Virol. 2001;75:11336–43. doi: 10.1128/JVI.75.23.11336-11343.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–82. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 98.Antoni BA, Rabson AB, Kinter A, Bodkin M, Poli G. NF-kappa B-dependent and -independent pathways of HIV activation in a chronically infected T cell line. Virology. 1994;202:684–94. doi: 10.1006/viro.1994.1390. [DOI] [PubMed] [Google Scholar]
- 99.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 100.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–34. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 101.Marzio G, Tyagi M, Gutierrez MI, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A. 1998;95:13519–24. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, et al. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- 103.Hottiger MO, Nabel GJ. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–6. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bres V, Tagami H, Peloponese JM, Loret E, Jeang KT, Nakatani Y, et al. Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. Embo J. 2002;21:6811–9. doi: 10.1093/emboj/cdf669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ott M, Dorr A, Hetzer-Egger C, Kaehlcke K, Schnolzer M, Henklein P, et al. Tat acetylation: a regulatory switch between early and late phases in HIV transcription elongation. Novartis Found Symp. 2004;259:182–93. discussion 93-6, 223–5. [PubMed] [Google Scholar]
- 106.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, et al. HIV-1 tat transcriptional activity is regulated by acetylation. Embo J. 1999;18:6106–18. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, Wainberg MA, et al. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol. 2005;79:124–31. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. Rna. 2004;10:1489–98. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bres V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19:1211–26. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Berro R, Kehn K, de la Fuente C, Pumfery A, Adair R, Wade J, et al. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J Virol. 2006;80:3189–204. doi: 10.1128/JVI.80.7.3189-3204.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bohne J, Krausslich HG. Mutation of the major 5′ splice site renders a CMV-driven HIV-1 proviral clone Tat-dependent: connections between transcription and splicing. FEBS Lett. 2004;563:113–8. doi: 10.1016/S0014-5793(04)00277-7. [DOI] [PubMed] [Google Scholar]
- 112.Chiu YL, Coronel E, Ho CK, Shuman S, Rana TM. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J Biol Chem. 2001;276:12959–66. doi: 10.1074/jbc.M007901200. [DOI] [PubMed] [Google Scholar]
- 113.Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell. 2002;10:585–97. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 114.Taylor JP, Pomerantz R, Bagasra O, Chowdhury M, Rappaport J, Khalili K, et al. TAR-independent transactivation by Tat in cells derived from the CNS: a novel mechanism of HIV-1 gene regulation. Embo J. 1992;11:3395–403. doi: 10.1002/j.1460-2075.1992.tb05418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Raha T, Cheng SW, Green MR. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 2005;3:e44. doi: 10.1371/journal.pbio.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.El Kharroubi A, Piras G, Zensen R, Martin MA. Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol Cell Biol. 1998;18:2535–44. doi: 10.1128/mcb.18.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dandekar DH, Ganesh KN, Mitra D. HIV-1 Tat directly binds to NFkappaB enhancer sequence: role in viral and cellular gene expression. Nucleic Acids Res. 2004;32:1270–8. doi: 10.1093/nar/gkh289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Furia B, Deng L, Wu K, Baylor S, Kehn K, Li H, et al. Enhancement of nuclear factor-kappa B acetylation by coactivator p300 and HIV-1 Tat proteins. J Biol Chem. 2002;277:4973–80. doi: 10.1074/jbc.M107848200. [DOI] [PubMed] [Google Scholar]
- 119.Demarchi F, Gutierrez MI, Giacca M. Human immunodeficiency virus type 1 tat protein activates transcription factor NF-kappaB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR. J Virol. 1999;73:7080–6. doi: 10.1128/jvi.73.8.7080-7086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Demarchi F, d’Adda di Fagagna F, Falaschi A, Giacca M. Activation of transcription factor NF-kappaB by the Tat protein of human immunodeficiency virus type 1. J Virol. 1996;70:4427–37. doi: 10.1128/jvi.70.7.4427-4437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 122.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–10. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 123.Henderson A, Holloway A, Reeves R, Tremethick DJ. Recruitment of SWI/SNF to the human immunodeficiency virus type 1 promoter. Mol Cell Biol. 2004;24:389–97. doi: 10.1128/MCB.24.1.389-397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 125.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–4. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 126.Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–6. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, et al. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. Embo J. 2006;25:1690–9. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, Ott M, et al. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem. 2006;281:19960–8. doi: 10.1074/jbc.M603336200. [DOI] [PubMed] [Google Scholar]
- 129.Lassen KG, Bailey JR, Siliciano RF. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J Virol. 2004;78:9105–14. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–24. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 131.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sahu GK, Lee K, Ji J, Braciale V, Baron S, Cloyd MW. A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T-lymphocytes. Virology. 2006;355:127–37. doi: 10.1016/j.virol.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 133.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–57. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brooks DG, Hamer DH, Arlen PA, Gao L, Bristol G, Kitchen CM, et al. Molecular characterization, reactivation, and depletion of latent HIV. Immunity. 2003;19:413–23. doi: 10.1016/s1074-7613(03)00236-x. [DOI] [PubMed] [Google Scholar]
- 135.Bera TK, Kennedy PE, Berger EA, Barbas CF, 3rd, Pastan I. Specific killing of HIV-infected lymphocytes by a recombinant immunotoxin directed against the HIV-1 envelope glycoprotein. Mol Med. 1998;4:384–91. [PMC free article] [PubMed] [Google Scholar]
- 136.Chun TW, Engel D, Mizell SB, Hallahan CW, Fischette M, Park S, et al. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat Med. 1999;5:651–5. doi: 10.1038/9498. [DOI] [PubMed] [Google Scholar]
- 137.Dybul M, Hidalgo B, Chun TW, Belson M, Migueles SA, Justement JS, et al. Pilot study of the effects of intermittent interleukin-2 on human immunodeficiency virus (HIV)-specific immune responses in patients treated during recently acquired HIV infection. J Infect Dis. 2002;185:61–8. doi: 10.1086/338123. [DOI] [PubMed] [Google Scholar]
- 138.Stellbrink HJ, Hufert FT, Tenner-Racz K, Lauer J, Schneider C, Albrecht H, et al. Kinetics of productive and latent HIV infection in lymphatic tissue and peripheral blood during triple-drug combination therapy with or without additional interleukin-2. Antivir Ther. 1998;3:209–14. [PubMed] [Google Scholar]
- 139.Lafeuillade A, Poggi C, Chadapaud S, Hittinger G, Chouraqui M, Pisapia M, et al. Pilot study of a combination of highly active antiretroviral therapy and cytokines to induce HIV-1 remission. J Acquir Immune Defic Syndr. 2001;26:44–55. doi: 10.1097/00126334-200101010-00006. [DOI] [PubMed] [Google Scholar]
- 140.Klichko V, Archin N, Kaur R, Lehrman G, Margolis D. Hexamethylbisacetamide remodels the human immunodeficiency virus type 1 (HIV-1) promoter and induces Tat-independent HIV-1 expression but blunts cell activation. J Virol. 2006;80:4570–9. doi: 10.1128/JVI.80.9.4570-4579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Andreeff M, Stone R, Michaeli J, Young CW, Tong WP, Sogoloff H, et al. Hexamethylene bisacetamide in myelodysplastic syndrome and acute myelogenous leukemia: a phase II clinical trial with a differentiation-inducing agent. Blood. 1992;80:2604–9. [PubMed] [Google Scholar]