Abstract
A family of microtubule (MT)-binding proteins, Orbit/multiple asters/cytoplasmic linker protein–associated protein, has emerged as an important player during mitosis, but their functional mechanisms are poorly understood. In this study, we used meiotic egg extracts to gain insight into the role of the Xenopus laevis homologue Xorbit in spindle assembly and function. Xorbit immunodepletion or its inhibition by a dominant-negative fragment resulted in chromosome alignment defects and aberrant MT structures, including monopolar and small spindles. Xorbit-depleted extracts failed to nucleate MTs around chromatin-coated beads, indicating its essential requirement for spindle assembly in the absence of centrosomes and kinetochores. Xorbit's MT stabilizing effect was most apparent during anaphase, when spindle MTs depolymerized rapidly upon Xorbit inhibition. Biochemical interaction between a COOH-terminal Xorbit fragment and the kinetochore-associated kinesin centromeric protein E may contribute to Xorbit's role in chromosome congression. We propose that Xorbit tethers dynamic MT plus ends to kinetochores and chromatin, providing a stabilizing activity that is crucial for spindle assembly and chromosome segregation.
Introduction
Spindle formation relies on intricate spatial and temporal control of microtubule (MT) dynamics and coordinated organization by motor proteins (for review see Gadde and Heald, 2004). Mitotic chromosomes play an active role in this process by stabilizing MTs in their vicinity and by forming attachments at their kinetochores that facilitate their metaphase alignment and anaphase segregation. However, the molecular mechanisms linking dynamic MTs to chromosomes are poorly understood.
Stabilization of MTs by mitotic chromosomes is most apparent and essential in systems that lack MT nucleation centers (centrosomes), but increasing evidence suggests that this is a conserved process operating in many cell types (Heald et al., 1996; Khodjakov et al., 2000; Megraw et al., 2001; Maiato et al., 2004; Rebollo et al., 2004). Using meiotic Xenopus laevis egg extracts is a useful way to study this phenomenon, as chromatin-coated beads are sufficient to induce spindle assembly in the absence of centrosomes and kinetochores (Heald et al., 1996). Dynamic MTs generated by chromatin are organized by MT-based motor proteins, which may contribute to chromatin–spindle interactions (Walczak et al., 1998).
A fundamentally different kind of MT connection occurs at the kinetochore, where plus ends of a MT bundle form a stable yet dynamic attachment capable of coupling MT depolymerization to chromosome movement. A variety of kinetochore-associated proteins have been implicated in this process, including dynein, kinesin 13 (mitotic centromere-associated kinesin [MCAK]/XKCM1), the chromosomal passenger complex, and kinesin 7 (centromeric protein E [CENP-E]). However, it is poorly understood how the kinetochore–MT interface mediates chromosome movements and which factors are involved.
A class of MT-associated proteins that concentrate at MT plus ends has emerged as a potential key player in chromosome–MT interactions during mitosis. These plus end–tracking proteins or +Tips, such as the cytoplasmic linker protein 170 (CLIP-170) and adenomatous polyposis coli (APC), localize to kinetochores during mitosis and have been suggested to participate in MT–kinetochore attachments (Dujardin et al., 1998; Fodde et al., 2001; Kaplan et al., 2001; Green et al., 2005). CLIP-associated proteins (CLASPs) have also been identified and have been shown to associate with kinetochores independently of MTs. Mutant analysis and RNA interference of the Drosophila melanogaster version, multiple asters/Orbit, revealed that it is required for chromosome alignment, kinetochore–MT attachment, and maintenance of spindle bipolarity (Inoue et al., 2000; Lemos et al., 2000; Maiato et al., 2002). Intriguingly, a study using photobleaching and microsurgery suggested that CLASP is involved in MT polymerization at plus ends essential for MT poleward flux (Maiato et al., 2005). Further evidence supporting a role for CLASP in mitosis results from studies in human cells and Caenorhabditis elegans embryos (Maiato et al., 2003; Cheeseman et al., 2005), but the molecular mechanisms behind CLASP protein function remain unclear.
Results and discussion
Xorbit is required for chromosome alignment and proper spindle formation
To investigate the role of CLASP in spindle assembly and chromosome segregation in egg extracts, we cloned the Xenopus homologue Xorbit (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200508180/DC1). Consistent with Orbit/CLASP localization in Drosophila and mammalian cells, Xorbit associates with spindle MTs, spindle poles, and kinetochores during metaphase in Xenopus egg extracts and shifts to the central spindle in late anaphase (Fig. S2; Inoue et al., 2000; Maiato et al., 2002, 2003).
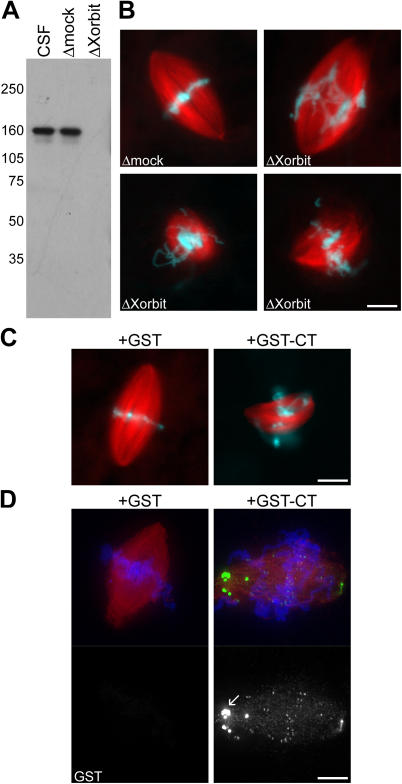
To assess the mitotic processes for which Xorbit is required, α-Xorbit antibody was used to quantitatively (>98%) deplete the protein from extracts arrested in metaphase of meiosis II (cytostatic factor–arrested [CSF] extract; Fig. 1 A). Spindle assembly reactions were performed by cycling CSF extract containing sperm nuclei through interphase to allow DNA and centrosome replication and then cycling back into metaphase (Desai et al., 1999). Although mock-depleted extracts yielded predominantly bipolar spindles with chromosomes congressed at the metaphase plate, Xorbit-depleted extracts generated aberrant spindles with severe chromosome alignment defects (Fig. 1 B). The average spindle length after Xorbit depletion was significantly shorter than controls (19.6 ± 3.8 μm and 31.8 ± 4.2 μm, respectively; n = 100; three experiments), and ∼20% of all structures were monopolar. We conclude that Xorbit depletion causes a metaphase phenotype similar to CLASP inhibition in other organisms (Inoue et al., 2000; Lemos et al., 2000; Maiato et al., 2002).
Figure 1.
Xorbit inhibition results in aberrant spindle structures and chromosome alignment defects. (A) Western blot of CSF Xenopus extract and extracts depleted using IgG (Δmock) or Xorbit antibodies (ΔXorbit) probed with α-Xorbit antibody. (B) Metaphase spindles assembled in mock- and Xorbit-depleted extracts. All three ΔXorbit images display chromosome alignment defects, and the two bottom spindles are also smaller in size. (C) GST or GST-CT were added at the start of mitosis, and metaphase spindles were examined. (D) Immunofluorescence with α-GST, shown in green in merged images. Arrow indicates aggregates of added protein. MTs are red and DNA is blue. Bars, 10 μm.
The COOH-terminal (CT) fragment of Xorbit causes dominant-negative effects similar to depletion
Previous domain analysis of human CLASP revealed a highly conserved CT domain capable of localizing to kinetochores (Fig. S1; Maiato et al., 2003). To potentially distinguish among Xorbit's different mitotic functions, we expressed the CT domain fused to GST (GST-CT) and tested its effects on spindle formation. In contrast to the GST control, spindles that formed in the presence of GST-CT displayed severe chromosome alignment defects, and many spindles appeared shorter in length (Fig. 1 C), which is a phenotype similar to Xorbit depletion. By immunofluorescence with an α-GST antibody, Xorbit GST-CT localized to spindle MTs and concentrated at kinetochores and spindle poles similar to endogenous Xorbit (Fig. 1 D). These results provide further evidence that the mitotic defects observed upon Xorbit depletion are specific. Because GST-CT did not prevent the association of endogenous Xorbit with chromosomes by biochemical analysis (not depicted), we speculate that it affects Xorbit's molecular interactions.
Xorbit is specifically required for MT stabilization around mitotic chromatin
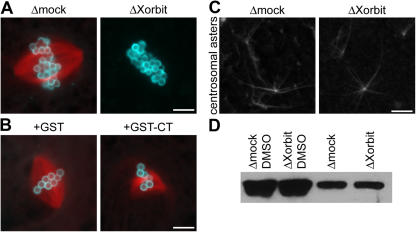
In addition to chromosome alignment defects indicative of a role at the kinetochore, the small spindle phenotype observed upon Xorbit inhibition suggested that Xorbit might also influence nonkinetochore MTs. To examine Xorbit's role in chromatin-induced MT stabilization, spindles were assembled around DNA-coated beads (Heald et al., 1996). In mock-depleted extracts, MT stabilization and organization generated bipolar spindles, whereas in Xorbit-depleted extracts, this process was severely inhibited (Fig. 2 A). No MT polymerization was observed at ∼98% of all bead clusters (n = 300; three experiments), suggesting that Xorbit plays a crucial role in chromatin-induced MT formation. Interestingly, GST-CT addition did not cause such a severe phenotype but predominantly resulted in small, distorted spindles (∼60% of all bead spindles; n = 300; three experiments; Fig. 2 B). This differs from the quite similar effect of Xorbit depletion and GST-CT addition on sperm spindle assembly, suggesting that endogenous Xorbit can still perform part of its function in the presence of GST-CT and that this function is redundant with that of centrosomes and/or kinetochores. One intriguing possibility is that Xorbit also acts at MT minus ends through nucleation/stabilization in addition to stabilizing dynamic plus ends. Consistent with a potential role at MT minus ends, Xorbit localizes to the poles of both bead and sperm spindles (Fig. S2 and not depicted).
Figure 2.
Xorbit is required for MT stabilization by mitotic chromatin. (A) Spindles assembled around chromatin-coated beads in mock- and Xorbit-depleted extracts. (B) GST or GST-CT were added at the start of mitosis, and metaphase chromatin bead spindles were examined. MTs are red and DNA is blue. (C) Centrosome asters were assembled in mock- and Xorbit-depleted extracts and were fixed after 10 min. Bars, 10 μm. (D) MTs polymerized after a 10-min incubation in the presence or absence of DMSO in mock- and Xorbit-depleted extracts were pelleted and analyzed by immunoblotting with α-tubulin antibody.
To determine whether Xorbit plays a general role in regulating MT dynamics, MT asters were generated in CSF extracts by adding either centrosomes as nucleation sites or DMSO, a MT-stabilizing agent that induces aster formation. No effect on aster assembly was observed by fluorescence microscopy upon Xorbit depletion in either assay (Fig. 2 C and not depicted). To confirm these observations more quantitatively, DMSO asters were pelleted and MT polymer levels were evaluated by α-tubulin immunoblotting, which revealed no obvious difference between mock- and Xorbit-depleted extracts (Fig. 2 D). These data suggest that Xorbit does not exert a global effect on MT dynamics but plays a role specifically in chromatin-driven MT assembly.
Xorbit plays a crucial role in stabilizing MTs during anaphase
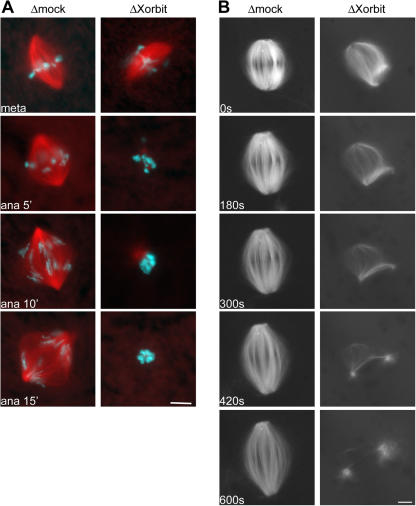
Previous studies and our characterization of Xorbit depletion establish an important function for Xorbit/CLASP in spindle assembly, but its function in later stages of mitosis has not been closely examined (Inoue et al., 2000; Lemos et al., 2000; Maiato et al., 2002, 2003). Therefore, we investigated the effects of Xorbit depletion on chromosome segregation and spindle dynamics during anaphase by adding calcium or an activated version of calcium/calmodulin-dependent kinase II to mock- or Xorbit-depleted cycled spindles arrested in metaphase. In control extracts, sister chromatids separated and segregated to opposite spindle poles, and kinetochore MTs shortened and spindle poles separated. Surprisingly, in Xorbit-depleted extracts, spindle MTs depolymerized within a few minutes of anaphase induction (Fig. 3 A). Time-lapse fluorescence microscopy further illustrated the dramatic spindle MT disassembly at anaphase onset in the absence of Xorbit (Fig. 3 B and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200508180/DC1).
Figure 3.
Xorbit is essential for MT stabilization during anaphase. Chromosome segregation in mock- and Xorbit-depleted extracts. (A) Once metaphase spindles assembled (top), anaphase was induced, and samples were fixed after 5, 10, and 15 min. MTs are red and DNA is blue. (B) Time-lapse fluorescence microscopy of X-rhodamine–labeled MTs in mock- and Xorbit-depleted spindles during anaphase. Time is in seconds after anaphase induction. Bars, 10 μm.
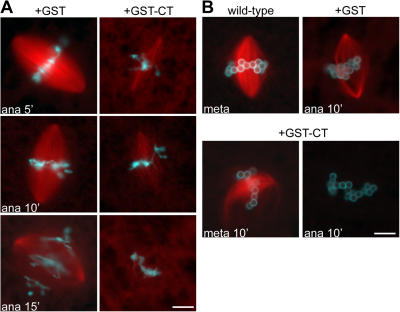
To determine whether the MT depolymerization was a consequence of aberrant metaphase spindle assembly or the result of a specific requirement for Xorbit during anaphase, we added the GST-CT construct to wild-type metaphase spindles and concomitantly triggered anaphase. Although the metaphase spindles were robust and normal in size, their MTs rapidly depolymerized upon anaphase onset when Xorbit was inhibited (Fig. 4 A). In contrast, if GST-CT was added without triggering anaphase, spindles persisted and gradually became small and distorted, as in Fig. 1 C (not depicted). To examine whether the depolymerization resulted from defects in kinetochore–MT interactions or because all spindle MTs require Xorbit activity to persist during anaphase, we compared the effects of GST-CT on chromatin bead spindles that were arrested in metaphase or induced to enter anaphase. Whereas GST-CT addition caused spindle distortion in metaphase, it caused complete MT depolymerization within 10 min of anaphase onset (Fig. 4 B). We conclude that Xorbit plays an essential role in MT stabilization during anaphase.
Figure 4.
Xorbit is required to maintain kinetochore and nonkinetochore MTs at anaphase onset. (A) Wild-type metaphase spindles were assembled, and GST or GST-CT was added at the same time anaphase was induced. Spindles were fixed 5, 10, and 15 min later. (B) Wild-type spindles were assembled around chromatin-coated beads, and GST or GST-CT was added during metaphase arrest (bottom left) or at the same time anaphase was induced (right), and spindles were fixed 10 min later. MTs are red and DNA is blue. Bars, 10 μm.
At the kinetochore, CLASP has been proposed to promote tubulin subunit addition at kinetochore fibers, driving poleward MT flux (Maiato et al., 2005), and our results are consistent with such a role for Xorbit/CLASP during metaphase. In anaphase, however, polymerization at the plus end stops and kinetochore MTs shorten, partly because of flux/minus end disassembly and partly because of plus end depolymerization (Maddox et al., 2003). Our results expand the role of Xorbit as a plus end–stabilizing factor that protects both kinetochore and nonkinetochore MTs from uncontrolled disassembly while attached chromosomes segregate.
The CT domain of Xorbit interacts with CLIP-170 and the kinetochore-associated motor CENP-E
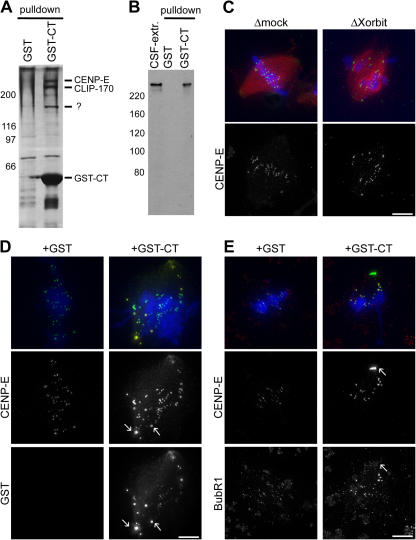
To investigate potential binding partners of Xorbit that could account for its mitotic phenotypes, we performed pull-down experiments with GST-CT and GST as a control in Xenopus laevis and Xenopus tropicalis extracts (Fig. 5 A). Mass spectrometry analysis revealed a Xenopus homologue of CLIP-170 as a specific GST-CT–interacting 250-kD protein. CLASP is known to interact with CLIP-170, which is preferentially associated with MT plus ends and is implicated in vesicle transport, but so far this interaction has only been reported in interphase (Akhmanova et al., 2001). Another high molecular mass protein that specifically pulled down with GST-CT was identified as kinesin 7 or CENP-E, a kinetochore-associated motor protein required for chromosome alignment (Schaar et al., 1997; Wood et al., 1997; McEwen et al., 2001). We confirmed this interaction with an α–CENP-E antibody, which recognized a specific band above 300 kD in CSF extracts and GST-CT pull-downs (Fig. 5 B). CENP-E did not coimmunoprecipitate with Xorbit antibodies, nor were its levels altered in Xorbit-depleted extracts (unpublished data), but it is likely that the COOH-terminal antibody disrupted the interaction between Xorbit and CENP-E. Furthermore, CENP-E localization to kinetochores in Xorbit-depleted extracts was not impaired (Fig. 5 C), suggesting that chromosome alignment defects in the absence of Xorbit reflect a functional interaction between the two proteins through the Xorbit CT domain. In the presence of GST-CT, CENP-E levels at kinetochores appeared slightly elevated, and aggregates of the two proteins often formed near spindle poles. These foci did not contain another kinetochore component, BubR1 (Fig. 5, D and E; arrows), indicating that the aggregates were not kinetochores and further supporting a biochemical interaction between CENP-E and GST-CT.
Figure 5.
Xorbit interacts with kinetochore kinesin 7 (CENP-E). (A) Silver-stained gel of GST and GST-CT pull-downs performed in Xenopus tropicalis extracts. GST-CT–interacting proteins analyzed by mass spectrometry are indicated on the right. The band at ∼160 kD could not be identified. (B) Western blot of CSF extract, GST, and GST-CT pull-downs probed with α–CENP-E antibody. (C) Metaphase spindles in mock- and Xorbit-depleted extracts stained for CENP-E. (D and E) Duplicated metaphase chromosomes formed in the presence of GST and GST-CT stained for CENP-E (D and E) and GST (D) or BubR1 (E). In merged images, CENP-E is shown in green, DNA in blue, and either MTs (C), GST-CT (D), or BubR1 (E) in red. Arrows indicate aggregates of GST-CT and CENP-E. Bars, 10 μm.
CENP-E is believed to somehow link the leading kinetochore to shrinking MT plus ends independent of ATP hydrolysis and to move the trailing kinetochore toward the growing MT plus end during antipoleward movement (Lombillo et al., 1995; Wood et al., 1997; Yao et al., 1997; Putkey et al., 2002). We propose that Xorbit supports CENP-E in coupling MT dynamics to chromosome movement either by a direct tethering activity or through its association with CENP-E. It is possible that Xorbit also functions through other factors during congression such as cytoplasmic dynein, because Xorbit interacts with CLIP-170, a dynein regulatory factor that has also been implicated in chromosome alignment (Fig. 5 A; Dujardin et al., 1998). Thus, Xorbit could function by physically linking chromosomes to dynamic plus ends during congression or by targeting/regulating necessary motor activities.
In summary, Xorbit plays an essential role in multiple aspects of spindle MT dynamics in Xenopus egg extracts. This differentiates Xorbit/CLASP from other +Tips that have been examined to date for roles in spindle assembly and function. For example, although APC is implicated in chromosome missegregation (Fodde et al., 2001; Kaplan et al., 2001; Green et al., 2005), the depletion of APC from egg extracts caused subtle defects in spindle morphology, except under conditions in which kinesin 13/XMCAK was codepleted (Banks and Heald, 2004; Dikovskaya et al., 2004). In contrast, EB1 influences global MT dynamics by promoting plus end polymerization (Tirnauer et al., 2002). Our study highlights Xorbit/CLASP as a critical factor linking chromosome segregation to MT dynamics during cell division.
Materials and methods
Cloning, expression, and purification of fusion proteins and antibody generation
Full-length Xorbit was generated by PCR from a cDNA library (gift from P. Budde, Cell Press, Boston, MA) with primer sequences derived from Xenopus EST clones homologous to human CLASP. GST-CT encoding the 282 CT amino acids fused to GST was expressed in Escherichia coli, purified by glutathione Sepharose 4B chromatography, and dialyzed into XB (10 mM Hepes, pH 7.7, 1 mM MgCl2, 0.1 mM CaCl2, 100 mM KCl, and 50 mM sucrose). α-Xorbit antibody was generated against amino acids 799–1,457 fused to GST (Covance Research Products) and was affinity purified.
Preparation of Xenopus egg extracts and in vitro assays
CSF extract was prepared, and spindle assembly reactions with replicated sperm chromosomes were performed as previously described (Desai et al., 1999). Extracts were driven into anaphase with calcium solution (4 mM CaCl2, 100 mM KCl, and 1 mM MgCl2) or an activated version of CamKII (CamKII plasmid was a gift from M. Doree, Centre National de la Recherche Scientifique, France) expressed in reticulocyte lysate.
Centrosome/DMSO aster formation and spindle assembly around chromatin-coated beads were assayed as previously described (Wignall and Heald, 2001). For MT pelleting assays, DMSO aster reactions were diluted with 30% glycerol in BRB80 (80 mM K-Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) and spun down through a cushion of 60% glycerol/BRB80. Pellets were analyzed by immunoblotting with a monoclonal α-tubulin antibody (1:5,000; E7; Developmental Studies Hybridoma Bank) using standard techniques.
Xorbit immunodepletion and inhibition by GST-CT
10 μg α-Xorbit antibody or random rabbit IgG (control; Sigma-Aldrich) was coupled to 50 μl protein A–Dynabeads (Dynal) and used to immunodeplete 150 μl CSF extract. Depletion was assessed by immunoblotting with 1 μg/ml α-Xorbit antibody. To inhibit Xorbit, GST-CT was added to the extract at a final concentration of 1.8 μM (approximately endogenous Xorbit levels) either upon entry into mitosis or at the onset of anaphase. As a control, 1.8 μM GST was added.
Immunofluorescence and microscopy
Spindle reactions were spun onto coverslips and fixed as previously described (Wignall and Heald, 2001). α-Xorbit and α–CENP-E antibody were used 1:3,000, α-BubR1 antibody was used at 1:5,000 (α–CENP-E and α-BubR1 antibodies were a gift from D. Cleveland, University of California, San Diego, La Jolla, CA) and α-GST antibody (gift from T.J. Maresca, University of California, Berkeley, Berkeley, CA) was used at 1:500. Images were collected with a fluorescence microscope (model BX51; Olympus) with a dry 40× NA 0.75 objective, a cooled CCD camera (model OrcaII; Hamamatsu), and MetaMorph software (Molecular Devices). Images in Figs. S2, 1 D, and 5 (C–E) were taken on an imaging station (DeltaVision; Applied Precision) equipped with an inverted microscope (model IX70; Olympus) with a 60× NA 1.35 oil immersion lens (Olympus) and a cooled CCD camera (model Photometrics; Roper Scientific). Images were taken with a Z-stack size of 0.2 μm and deconvolved, and stacks were projected into a single plane. Images were processed using Adobe Photoshop.
Pull-downs and mass spectrometry
100 μl of Xenopus laevis or tropicalis extract (a gift from M. Blower, University of California, Berkeley) incubated with 7 μM GST-CT was added to 60 μl glutathione Sepharose 4B slurry and rotated at 4°C. Extract was removed, and the slurry was washed once with XB, three times with XB + 100 mM KCl, once with XB, eluted with 25 μl 2× SDS sample buffer, and analyzed by immunoblotting with α–CENP-E antibody (1:5,000) or mass spectrometry.
For mass spectrometry analysis, protein bands were cut into small pieces, washed in 25 mM ammonium bicarbonate in 50% acetonitrile, reduced with DTT, alkylated with iodoacetamide, and digested with Sequencing Grade Modified Trypsin (Promega). Peptides were extracted with 5% trifluoroacetic acid (TFA)/50% acetonitrile and combined. Recovered peptides were purified with μ-C18 ZipTips (Millipore) and eluted. About 1 μl of the peptide mixture from each gel band was combined with an equal volume of matrix solution and allowed to dry on the MALDI target. The matrix solution used was a 10-mg/ml solution of α-cyano-4-hydroxycinnamic acid in 0.1% TFA/50% acetonitrile. Mass spectra were acquired on a MALDI-TOF mass spectrometer (Reflex III; Bruker). Proteins were identified by searching the SwissProt or NCBInr databases using MS-Fit, a program of ProteinProspector (University of California, San Francisco; http://prospector.ucsf.edu).
Online supplemental material
Fig. S1 shows a domain schematic and sequence alignment comparing Xorbit with human CLASP1 and 2 and a second Xenopus clone. Fig. S2 shows immunofluorescence localization of Xorbit in extract spindles in metaphase and anaphase. Videos show time-lapse fluorescence microscopy of spindle MTs after anaphase was induced in mock- (Video 1; total time of video is 20 min) or Xorbit-depleted (Video 2; total time is 12 min) extracts, performed using the wide-field Olympus microscope set-up described above. Frames were collected every 30 s and are displayed at a rate of 10 frames/s. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200508180/DC1.
Supplementary Material
Acknowledgments
Special thanks to Sharleen Zhou at the Howard Hughes Medical Institute Mass Spectrometry Facility (University of California, Berkeley) for assistance with mass spectrometry and protein identification. We acknowledge the gift of antibodies from D. Cleveland (α-BubR1 and α–CENP-E) and T.J. Maresca (α-GST) and the cDNA library from P. Budde. We would like to thank M. Blower for providing Xenopus tropicalis extract and for critical reading of the manuscript. Many thanks to Roshni Kasad for her contribution to the project as a rotation student. The authors are also extremely grateful to all members of the Heald, Weis, and Welch labs, past and present, for thoughtful discussions and support.
R. Heald is supported by the National Institutes of Health grant GM057839.
Abbreviations used in this paper: APC, adenomatous polyposis coli; CENP-E, centromeric protein E; CLASP, CLIP-associated protein; CLIP, cytoplasmic linker protein; CSF, cytostatic factor arrested; CT, COOH terminal; MT, microtubule.
References
- Akhmanova, A., C.C. Hoogenraad, K. Drabek, T. Stepanova, B. Dortland, T. Verkerk, W. Vermeulen, B.M. Burgering, C.I. De Zeeuw, F. Grosveld, and N. Galjart. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of MT dynamics in motile fibroblasts. Cell. 104:923–935. [DOI] [PubMed] [Google Scholar]
- Banks, J.D., and R. Heald. 2004. Adenomatous polyposis coli associates with the MT-destabilizing protein XMCAK. Curr. Biol. 14:2033–2038. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., I. MacLeod, J.R. Yates III, K. Oegema, and A. Desai. 2005. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr. Biol. 15:771–777. [DOI] [PubMed] [Google Scholar]
- Dikovskaya, D., I.P. Newton, and I.S. Nathke. 2004. The adenomatous polyposis coli protein is required for the formation of robust spindles formed in CSF Xenopus extracts. Mol. Biol. Cell. 15:2978–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., A. Murray, T.J. Mitchison, and C.E. Walzcak. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. [DOI] [PubMed] [Google Scholar]
- Dujardin, D., U.I. Wacker, A. Moreau, T.A. Schroer, J.E. Rickard, and J.R. De Mey. 1998. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 141:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde, R., R. Smits, and H. Clevers. 2001. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 1:55–67. [DOI] [PubMed] [Google Scholar]
- Gadde, S., and R. Heald. 2004. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14:R797–R805. [DOI] [PubMed] [Google Scholar]
- Green, R.A., R. Wollman, and K.B. Kaplan. 2005. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell. 16:4609–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R., R. Tournebize, T. Blank, R. Sandaltzopoulos, P. Becker, A. Hyman, and E. Karsenti. 1996. Self-organization of MTs into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 382:420–425. [DOI] [PubMed] [Google Scholar]
- Inoue, Y.H., M. do Carmo Avides, M. Shiraki, P. Deak, M. Yamaguchi, Y. Nishimoto, A. Matsukage, and D.M. Glover. 2000. Orbit, a novel MT-associated protein essential for mitosis in Drosophila melanogaster. J. Cell Biol. 149:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, K.B., A.A. Burds, J.R. Swedlow, S.S. Bekir, P.K. Sorger, and I.S. Nathke. 2001. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat. Cell Biol. 3:429–432. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., R.W. Cole, B.R. Oakley, and C.L. Rieder. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67. [DOI] [PubMed] [Google Scholar]
- Lemos, C.L., P. Sampaio, H. Maiato, M. Costa, L.V. Omel'yanchuk, V. Liberal, and C.E. Sunkel. 2000. Mast, a conserved MT-associated protein required for bipolar mitotic spindle organization. EMBO J. 19:3668–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo, V.A., C. Nislow, T.J. Yen, V.I. Gelfand, and J.R. McIntosh. 1995. Antibodies to the kinesin motor domain and CENP-E inhibit MT depolymerization-dependent motion of chromosomes in vitro. J. Cell Biol. 128:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P., A. Straight, P. Coughlin, T.J. Mitchison, and E.D. Salmon. 2003. Direct observation of MT dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., P. Sampaio, C.L. Lemos, J. Findlay, M. Carmena, W.C. Earnshaw, and C.E. Sunkel. 2002. MAST/Orbit has a role in MT-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J. Cell Biol. 157:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., E.A. Fairley, C.L. Rieder, J.R. Swedlow, C.E. Sunkel, and W.C. Earnshaw. 2003. Human CLASP1 is an outer kinetochore component that regulates spindle MT dynamics. Cell. 113:891–904. [DOI] [PubMed] [Google Scholar]
- Maiato, H., C.L. Rieder, and A. Khodjakov. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., A. Khodjakov, and C.L. Rieder. 2005. Drosophila CLASP is required for the incorporation of MT subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7:42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B.F., G.K. Chan, B. Zubrowski, M.S. Savoian, M.T. Sauer, and T.J. Yen. 2001. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 12:2776–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T.L., L.R. Kao, and T.C. Kaufman. 2001. Zygotic development without functional mitotic centrosomes. Curr. Biol. 11:116–120. [DOI] [PubMed] [Google Scholar]
- Putkey, F.R., T. Cramer, M.K. Morphew, A.D. Silk, R.S. Johnson, J.R. McIntosh, and D.W. Cleveland. 2002. Unstable kinetochore-MT capture and chromosomal instability following deletion of CENP-E. Dev. Cell. 3:351–365. [DOI] [PubMed] [Google Scholar]
- Rebollo, E., S. Llamazares, J. Reina, and C. Gonzalez. 2004. Contribution of noncentrosomal MTs to spindle assembly in Drosophila spermatocytes. PLoS Biol. 10.1371/journal.pbio.0020008. [DOI] [PMC free article] [PubMed]
- Schaar, B.T., G.K. Chan, P. Maddox, E.D. Salmon, and T.J. Yen. 1997. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139:1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J.S., S. Grego, E.D. Salmon, and T.J. Mitchison. 2002. EB1-MT interactions in Xenopus egg extracts: role of EB1 in MT stabilization and mechanisms of targeting to MTs. Mol. Biol. Cell. 13:3614–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., I. Vernos, T.J. Mitchison, E. Karsenti, and R. Heald. 1998. A model for the proposed roles of different MT-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8:903–913. [DOI] [PubMed] [Google Scholar]
- Wignall, S.M., and R. Heald. 2001. Methods for the study of centrosome-independent spindle assembly in Xenopus extracts. Methods Cell Biol. 67:241–256. [DOI] [PubMed] [Google Scholar]
- Wood, K.W., R. Sakowicz, L.S. Goldstein, and D.W. Cleveland. 1997. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 91:357–366. [DOI] [PubMed] [Google Scholar]
- Yao, X., K.L. Anderson, and D.W. Cleveland. 1997. The MT-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle MTs. J. Cell Biol. 139:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.