Abstract
The nuclear envelope defines the barrier between the nucleus and cytoplasm and features inner and outer membranes separated by a perinuclear space (PNS). The inner nuclear membrane contains specific integral proteins that include Sun1 and Sun2. Although the outer nuclear membrane (ONM) is continuous with the endoplasmic reticulum, it is nevertheless enriched in several integral membrane proteins, including nesprin 2 Giant (nesp2G), an 800-kD protein featuring an NH2-terminal actin-binding domain. A recent study (Padmakumar, V.C., T. Libotte, W. Lu, H. Zaim, S. Abraham, A.A. Noegel, J. Gotzmann, R. Foisner, and I. Karakesisoglou. 2005. J. Cell Sci. 118:3419–3430) has shown that localization of nesp2G to the ONM is dependent upon an interaction with Sun1. In this study, we confirm and extend these results by demonstrating that both Sun1 and Sun2 contribute to nesp2G localization. Codepletion of both of these proteins in HeLa cells leads to the loss of ONM-associated nesp2G, as does overexpression of the Sun1 lumenal domain. Both treatments result in the expansion of the PNS. These data, together with those of Padmakumar et al. (2005), support a model in which Sun proteins tether nesprins in the ONM via interactions spanning the PNS. In this way, Sun proteins and nesprins form a complex that links the nucleoskeleton and cytoskeleton (the LINC complex).
Introduction
The existence of distinct nuclear and cytoplasmic compartments is dependent upon the presence of a selective barrier called the nuclear envelope (NE). The NE consists of several structural elements (Burke and Stewart, 2002; Gruenbaum et al., 2005), the most prominent of which are the inner and outer nuclear membranes (INM and ONM, respectively). In most cells, these two membranes are separated by a regular gap of ∼50 nm, which is known as the perinuclear space (PNS). Periodic annular junctions between the two membranes form aqueous channels between the nucleus and the cytoplasm that accommodate nuclear pore complexes (NPCs) and, therefore, permit the movement of macromolecules across the NE.
In addition to its connections to the INM at the periphery of each NPC, the ONM also exhibits numerous continuities with the ER, to which it is functionally related. In this way, the INM, ONM, and ER form a single continuous membrane system. Similarly, the PNS represents a perinuclear extension of the ER lumen.
The final major structural feature of the NE is the nuclear lamina. This is a relatively thin (∼50 nm) protein meshwork associated with the nuclear face of the INM. The major components of the lamina are the A- and B-type lamins (Gerace et al., 1978). These are members of the larger intermediate filament family, and, like all intermediate filament proteins, they feature a central coiled-coil flanked by nonhelical head and tail domains (Gerace and Burke, 1988). The lamins are known to interact with components of the INM as well as with chromatin proteins. In this way, the lamina provides anchoring sites at the nuclear periphery for higher order chromatin domains.
In mammalian somatic cells, there are two major A-type lamins (lamins A and C) encoded by a single gene, Lmna (in mice). The B-type lamins, B1 and B2, are encoded by two separate genes (Hoger et al., 1988, 1990; Lin and Worman, 1993, 1995). Although B-type lamins are found in all cell types, the expression of A-type lamins is developmentally regulated (Stewart and Burke, 1987; Rober et al., 1989). Typically, A-type lamins are found in most adult cell types but are absent from those of early embryos. Mutations in the LMNA gene have been linked to a variety of human diseases (Burke and Stewart, 2002), many of which are associated with large-scale perturbations in nuclear organization. These observations have reinforced the view that the lamina is an important determinant of nuclear architecture and has an essential role in the maintenance of NE integrity.
Despite their numerous connections, the INM and ONM are biochemically distinct. Proteomic studies have revealed the existence of at least 50 integral membrane proteins that are enriched in NEs. Many of these appear to reside within the INM (Schirmer et al., 2003). Proteins become localized to the INM via a process of selective retention (Powell and Burke, 1990; Soullam and Worman, 1995; Ellenberg et al., 1997). In this scheme, membrane proteins synthesized on the peripheral ER or ONM may gain access to the INM by lateral diffusion via the continuities at the periphery of each NPC using an energy-dependent mechanism (Ohba et al., 2004). However, only those proteins that can interact with nuclear or INM/lamina components are retained and concentrated.
The recent identification of ONM-specific integral membrane proteins has raised some puzzling issues (Zhang et al., 2001; Zhen et al., 2002; Padmakumar et al., 2004). In particular, what prevents ONM proteins from simply drifting off into the peripheral ER? This question was originally addressed in Caenorhabditis elegans, where the localization of Anc-1, a very large type II ONM protein involved in actin-dependent nuclear positioning, was shown to be dependent upon Unc-84, an INM protein (Starr and Han, 2002). Localization of Unc-84 itself was found to be dependent upon the single C. elegans lamin (Lee et al., 2002). Based upon these findings, Starr and Han (2003) and Lee et al. (2002) proposed a novel model in which Unc-84 and Anc-1 would interact across the PNS via their respective lumenal domains. In this way, Unc-84 would act as a tether for Anc-1.
In mammalian cells, two giant (up to 1 MD) actin-binding proteins have been identified (variously termed NUANCE, nesprin 2 Giant [nesp2G], nesprin 1, enaptin, Syne 1, syne 2, and myne 1) as integral proteins of the ONM (Apel et al., 2000; Zhang et al., 2001; Mislow et al., 2002; Zhen et al., 2002; Padmakumar et al., 2004). These belong to a very large family of proteins encoded by the nesprin 1 and 2 genes (Zhang et al., 2001) and consist of a bewildering variety of alternatively spliced isoforms. The nesprins are related to Anc-1 as well as to a Drosophila melanogaster ONM protein, Klarsicht (Welte et al., 1998; Mosley-Bishop et al., 1999), in the possession of a 60–amino acid COOH-terminal KASH domain ( Klarsicht, Anc-1, and Syne homology). This domain comprises a single transmembrane anchor and a short segment of ∼40 residues that resides within the PNS.
One of the defining features of Unc-84 is a region of homology consisting of ∼200 amino acids with Sad1p, a Schizosaccharomyces pombe protein that is associated with the spindle pole body (Hagan and Yanagida, 1995). This region of homology is known as the SUN domain (Sad1p and UNc-84) and is believed to extend into the PNS. Mammalian cells also contain several SUN domain proteins. At least one of these, Sun2, has been shown to be an INM protein with the appropriate topology in which the SUN domain is localized in the PNS (Hodzic et al., 2004). It is tempting to speculate, based upon the model of Starr and Han (2003), that SUN domain proteins function as tethers for ONM-associated nesprins in mammalian cells.
Recently, Padmakumar et al. (2005) have shown that localization of nesp2G to the ONM is dependent upon an interaction with another mammalian SUN domain protein, Sun1. In this study, we provide evidence that Sun1 is inserted into the INM in such a way that its SUN domain, like that of Sun2, faces the PNS. In this way, we can conclusively demonstrate that Sun1 does indeed have the appropriate orientation, as assumed by Padmakumar et al. (2005), for its COOH-terminal domain to interact with the nesp2G KASH domain. The NH2-terminal region of both Sun1 and 2 face the nucleoplasm and interact with lamins. Surprisingly, our results indicate that Sun1 has a very strong preference for prelamin A. Sun1 is the only nuclear membrane protein described to date that exhibits such binding activity. This raises the distinct possibility that Sun1 may be involved in the targeting and assembly of newly synthesized lamin A. Finally, we demonstrate that Sun1 and 2 share some degree of functional redundancy and that both of these proteins cooperate in tethering nesp2G in the ONM. This tethering involves the establishment of molecular interactions that span the PNS and contributes to the remarkably regular spacing that is observed between the ONM and INM. Based upon our findings and upon those of Padmakumar et al. (2005), we are able to conclude that Sun1 and 2 function as key links in a molecular chain that connects the actin cytoskeleton via giant nesprin proteins to nuclear lamins and other components of the nuclear interior. We now refer to this assembly as the LINC complex (linker of nucleoskeleton and cytoskeleton).
Results
We have previously shown that Sun2 is an INM protein featuring an NH2-terminal nucleoplasmic domain and a significantly larger COOH-terminal domain that is localized within the PNS (Hodzic et al., 2004). This lumenal region of Sun2 contains a COOH-terminal SUN domain that is also found in C. elegans Unc-84. The SUN domain is found in at least two additional mammalian proteins (Fig. 1 A): Sun1 (GenBank/EMBL/DDBJ accession no. BC048156) and Sun3 (GenBank/EMBL/DDBJ accession no. BC026189).
Figure 1.
Sun1 is a ubiquitously expressed NE protein featuring a conserved COOH-terminal SUN domain. (A) ClustalW alignment of the COOH-terminal region of human Sun1–3 and C. elegans Unc-84 reveals the homology of the SUN domains. (B) Northern blot analysis of mRNA from multiple mouse tissues illustrates widespread expression of Sun1. A GAPDH probe was used as a loading control. (C) The NE localizations of Sun1 and 2 were determined by immunofluorescence microscopy using rabbit antibodies raised against recombinant Sun proteins. (D, left) HeLa cells transiently transfected with HA-tagged Sun1 confirm the predominant NE localization. HA-Sun1 was detected using an anti-HA monoclonal antibody. (D, right) Upon overexpression, HA-Sun1 is also detected in the ER. In each case, Sun protein localization is shown in red, whereas DNA, visualized using Höchst dye 33258, appears in blue.
Sun1 transcripts are present in a variety of tissues and cell types (Fig. 1 B). A comparison of Sun1 sequences in GenBank reveals that it exists in multiple, alternatively spliced isoforms. This conclusion is supported by Northern blot analysis, which reveals at least four or five discrete Sun1 transcripts in different tissues. We have not, however, surveyed these tissues for Sun1 isoforms. Immunofluorescence experiments using a polyclonal antibody raised against recombinant human Sun1 suggests that like Sun2, Sun1 is localized largely, if not exclusively, to the NE (Fig. 1 C). This is consistent with the appearance of Sun1 as well as Sun2 in a proteomic screen for NE-specific membrane proteins (Schirmer et al., 2003).
To further address the issue of Sun1 localization, we fused an HA epitope to the NH2 terminus of the largest isoform of mouse Sun1. After transfection into HeLa cells, the exogenous protein was found by immunofluorescence microscopy to be enriched at the NE (Fig. 1 D). At high expression levels, although still concentrated in the NE, HA-Sun1 began to appear in the peripheral ER (Fig. 1 D). Together, these observations confirm that Sun1 is a nuclear membrane protein. Similar experiments with HA-tagged Sun3 revealed a distribution that was more typical of an ER protein (not depicted). Furthermore, Northern blot analysis suggests that Sun3 is found primarily in the testis (not depicted). Consequently, all of our subsequent experiments focused on Sun1 and 2.
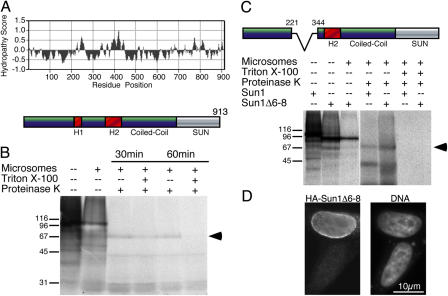
The primary structure of mouse Sun1 reveals no NH2-terminal signal sequence. However, two distinct hydrophobic domains, H1 and H2, are present (Fig. 2 A). H1 lies between residues 241 and 258, whereas the second and larger domain, H2, lies between residues 356 and 448. The presence of extended hydrophobic regions clearly raises questions concerning the topology of Sun1 within the nuclear membranes. An earlier study on Sun2 has shown that the SUN domain resides within the PNS (Hodzic et al., 2004). Is this also the case for Sun1? To address this, we used in vitro transcription/translation of Sun1 (tagged and untagged) both in the presence and absence of microsomes (Fig. 2 B). Digestion of Sun1 translation products with proteinase K revealed the existence of a 65–70-kD protected fragment in samples containing microsomes. Proteinase K digestion in the presence of Triton X-100 to permeabilize the microsomal membranes resulted in the complete loss of the protected fragment. Given the size of this protected fragment and the location of potential transmembrane domains, the most reasonable orientation for Sun1 would place its COOH terminus, including its SUN domain, within the microsome lumen. By extension, the SUN domain should thus reside within the PNS in vivo. This orientation is supported by additional experiments described below.
Figure 2.
Sun1 is a transmembrane protein with a lumenal COOH-terminal domain. (A) Hydropathy plot (Sweet and Eisenberg, 1983) of Sun1 reveals two hydrophobic domains (H1 and H2) upstream of the COOH-terminal coiled-coil and SUN domain. (B) When translated in vitro either in the presence or absence of microsomes, HA-tagged mouse Sun1, labeled with [35S]methionine/cysteine, appears as a ∼100-kD band, as revealed by SDS-PAGE. Subsequent proteinase K digestion of HA-Sun1 that had been translated in the presence of microsomes lead within 30–60 min to the quantitative loss of the full-length HA-Sun1 and the appearance of an ∼65–70-kD protected fragment (arrowhead). Inclusion of Triton X-100 in the digest to permeabilize the microsomes leads to the complete degradation of HA-Sun1 within 60 min. (C) An alternatively spliced isoform of Sun1 (Δ6–8) lacks the first hydrophobic domain (H1). When translated in vitro in either the presence or absence of microsomes, HA-Sun1Δ6–8 appears as a band that is predictably smaller than the full-length protein. However, both full-length HA-Sun1 and HA-Sun1Δ6–8 that were translated in the presence of microsomes and subjected to digestion with proteinase K yield identically sized protected fragments (arrowhead). Inclusion of Triton X-100 in the digestion reaction results in degradation of both proteins. (D) Immunofluorescence microscopy of HeLa cells transfected with HA-Sun1Δ6–8 reveals that the exogenous protein is localized at the NE. In this respect, HA-Sun1Δ6–8 is indistinguishable from full-length HA-Sun1. HA-Sun1Δ6–8 is detected with an anti-HA monoclonal antibody. The corresponding field labeled with Höchst dye to reveal cell nuclei is also shown.
To examine the roles of the two hydrophobic segments in Sun1 membrane anchoring, we took advantage of naturally occurring splice isoforms. Searches of GenBank reveal several mammalian Sun1 cDNAs lacking sequences within the NH2-terminal domain. Comparisons with genomic sequences indicate that at least one splice isoform of Sun1 lacks exons 6–8, corresponding to a deletion of residues 222–343 (Fig. 2 C). This particular Sun1 isoform is missing the first hydrophobic segment, H1. When this isoform (Sun1Δ6–8) was tagged with HA and transfected into HeLa cells, its localization at the NE was found to be indistinguishable from full-length Sun1 (Fig. 2 D). Translation of Sun1Δ6–8 in vitro in the presence of microsomes revealed a protease-resistant fragment identical in size with that derived from full-length Sun1 (Fig. 2 C). This finding presents us with two conclusions. First, it proves that the protected fragment must be derived from the COOH-terminal portion of the molecule. Second, if this first hydrophobic segment in Sun1 were to represent a transmembrane domain, its removal should logically alter the topology of the subsequent membrane-spanning sequences of Sun1 (i.e., H2) within the ER or nuclear membranes. This might potentially lead to the flipping of lumenal segments of newly synthesized Sun1 to the cytoplasmic aspect of the ER membrane or failure to insert H2 sequences into the membrane. Evidently, this does not happen in any detectable way. Therefore, our conclusion is that given this first hydrophobic sequence in Sun1 is dispensable with respect to membrane insertion, it does not function as an obligate membrane-spanning domain. Further secondary structure analyses of Sun1 using HMMTOP (Tusnady and Simon, 2001) indicate that the larger hydrophobic segment, H2, is capable of spanning the ER/nuclear membranes three times. Although this conformation for H2 still requires biochemical confirmation, it suggests that Sun1 is a multispanning protein in which the NH2-terminal domain faces the cytoplasm, whereas the COOH-terminal domain (including the SUN domain) is localized to the lumenal space.
To further examine Sun1 distribution and orientation in vivo, we prepared a form of mouse Sun1 that was tagged with an HA epitope at the NH2 terminus and a myc epitope at the COOH terminus (Fig. 3, HA-Sun1–myc). These analyses took advantage of the fact that low concentrations of digitonin can be used to selectively permeabilize the plasma membrane of cells while leaving the nuclear membranes and ER intact (Adam et al., 1990). For these experiments, HeLa cells expressing HA-Sun1–myc were fixed with formaldehyde and permeabilized on ice for 15 min with 0.003% digitonin. The cells were then labeled with either rabbit anti-myc or rabbit anti-HA. After PBS washes to remove unbound antibodies, the cells were refixed, permeabilized with Triton X-100, and further incubated with mouse antibodies against either HA or myc. In this way, the first tag could be assayed for accessibility at the nuclear surface, whereas the second tag could be used to define the localization and expression level of HA-Sun1–myc. As shown in Fig. 3, the myc tag was never visible at the nuclear surface (or at any other location) after digitonin permeabilization regardless of the expression level of HA-Sun1–myc. The HA tag was also undetectable at the nuclear surface after digitonin permeabilization of cells expressing low levels of HA-Sun1–myc. At high expression levels, however, the tag was detectable at the nuclear surface and was associated with a cytoplasmic reticular structure corresponding to the peripheral ER. Clearly, the NH2-terminal, but not the COOH-terminal, domain of ER-associated Sun1 is exposed to the cytoplasm. Altogether, these findings are consistent with the view that Sun1 is a component of the INM and that its COOH-terminal domain resides with the PNS.
Figure 3.
The SUN domain of Sun1 is located within the PNS, whereas the NH 2 -terminal domain is exposed to the nucleoplasm. To examine the orientation of Sun1, HA and myc epitope tags were placed at the NH2 and COOH termini, respectively. HeLa cells were transfected with the double-tagged construct and processed for immunofluorescence microscopy after 24 h. After fixation, the cells were permeabilized with 0.003% digitonin and incubated with an anti-epitope tag antibody (mouse monoclonal). Subsequently, the cells were refixed and permeabilized with Triton X-100 to expose the lumenal compartment to a second anti-epitope tag antibody (rabbit polyclonal). Neither epitope tag was significantly detectable after digitonin permeabilization in HeLa cells expressing low to moderate levels of HA-Sun1–myc. After Triton X-100 permeabilization, both myc and HA were readily detected at the NE. In HeLa cells with elevated expression of HA-Sun1–myc, the HA but not myc epitope tag was detected at the ER after digitonin permeabilization. Both myc and HA tags were identifiable after treatment with Triton X-100. These data indicate that the NH2- and COOH-terminal domains of Sun1 reside on opposite sides of the INM, with the COOH-terminal domain located in the PNS.
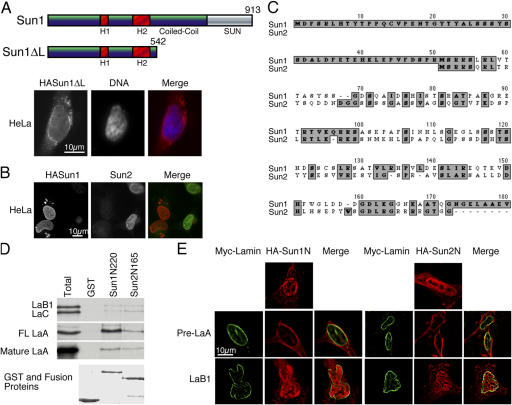
The aforementioned data, as well as our previously published study (Hodzic et al., 2004), indicate that the NH2-terminal domains of Sun1 and 2 are exposed to the nucleoplasm and, consequently, are accessible for interaction with nuclear components. Given the role that such interactions play in the appropriate targeting of INM proteins, it is not surprising that the lumenal domain of Sun1 is entirely dispensable with respect to Sun1 localization (Fig. 4 A). In this respect, Sun1 mimics Sun2 (Hodzic et al., 2004). Additional experiments suggest that Sun1 and 2 share overlapping interactions. Overexpression of HA-Sun1 in HeLa cells causes displacement of endogenous Sun2 from the NE (Fig. 4 B). However, the converse is not the case (not depicted). The implication is that Sun1 and 2 share a subset of interactions that are required for Sun2 localization. Sun1 retention must be dependent upon additional binding partners.
Figure 4.
Interactions between the nucleoplasmic domains of Sun1 and 2 with A-type lamins. (A) An HA-Sun1ΔL construct lacking the SUN domain and most of the lumenal coiled-coil localizes to the NE in a manner similar to the full-length protein. Anti-HA labeling is in red, whereas DNA, visualized with Höchst dye, is in blue. (B) Overexpression of HA-Sun1 leads to the loss of endogenous Sun2 in HeLa cells, suggesting that these two proteins share common binding partners. (C) A ClustalW alignment of the NH2-terminal sequences of mouse Sun1 and 2 identifies multiple clusters of homologous amino acids within a region of ∼120 residues. Sun1 exhibits a unique NH2-terminal extension of 50 amino acid residues. (D) The first 222 residues of Sun1 or the first 165 residues of Sun2 (this represents the entire nucleoplasmic domain of the latter) were fused to GST and, with GST alone (Coomassie stain, bottom), were used to pull down 35S-labeled, in vitro translated lamins B1, C, mature A, and full-length (FL) A (Total). Unlike GST alone, Sun1N220 and Sun2N165 pulled down lamins B1, C, and mature A at similar levels. However, Sun1N222 displayed a higher affinity for FL LaA than did Sun2N165. Evidently, Sun1 has a very strong preference for full-length (or pre) lamin A over mature lamin A (or full-length lamins C and B1). (E) An HA tag was inserted at the NH2 terminus of the same nucleoplasmic segments of both Sun1 and 2 (as described in D), and the tagged proteins were expressed in HeLa cells. Both of these exogenous proteins appear enriched in the nucleoplasm (top). As observed by deconvolution microscopy, cotransfection of the myc-tagged full-length lamin A (green) with HA-Sun1N222 or HA-Sun2N165 (red) resulted in the recruitment of both HA-Sun proteins to the NE (middle). Myc–lamin B1, in contrast, fails to produce such an effect. Evidently, the nucleoplasmic domains of Sun1 and 2 can interact with lamin A in vivo.
With what proteins might the Sun1 and 2 nucleoplasmic domains interact? Our initial thoughts were that the Sun proteins might associate with lamina components. To address this, we first adopted an in vitro approach to determine whether A- and B-type lamins could interact with the NH2-terminal domain of either protein. We prepared a GST fusion protein containing the first 165 amino acids of the Sun2 sequence (Sun2N165). This represents most of the Sun2 nucleoplasmic domain. A similar, although slightly larger (the first 220 amino acids), fusion protein was also prepared using Sun1 sequences that encompassed the bulk of the nonalternatively spliced region of the nucleoplasmic domain (Sun1N220). This region of Sun1 exhibits significant sequence similarity to the NH2-terminal domain of Sun2 over a region of ∼120 residues (Fig. 4 C), although Sun1 does display a unique 50-residue NH2-terminal extension. Both of these fusion proteins as well as a GST control were used in pull-down experiments using in vitro translated lamins as targets.
Four lamin species were used in these experiments (Fig. 4 D): lamin B1 (LaB1), lamin C (LaC), full-length lamin A (FL LaA), and mature lamin A. Full-length lamin A contains a CaaX motif and should be farnesylated in the reticulocyte lysate (Vorburger et al., 1989). However, it does not undergo detectable COOH-terminal proteolysis because the microsome-free in vitro translation mix lacks the appropriate processing enzymes. The mature lamin A cDNA contains a premature stop codon at position 647. In this way, it mimics processed (i.e., mature) lamin A. As shown in Fig. 4 D, GST-Sun2N165 bound all four lamin species, although the interaction with lamins B1 and C appeared barely more than the background observed with GST alone. Similarly, GST-Sun1N220 was also found to interact with all four lamin species. As was the case with Sun2N165, the interactions with lamins B1 and C were relatively weak. However, Sun1N220 showed a very strong preference for unprocessed (full-length lamin A) versus mature lamin A.
To determine whether these in vitro interactions might have any relevance in vivo, we prepared HA-tagged versions of both Sun1N220 and Sun2N165. Upon introduction into HeLa cells, both proteins accumulated within the nucleoplasm (Fig. 4 E), although a significant cytoplasmic pool was always present. We also prepared a form of lamin A (preLaA) containing an L647R mutation. This lamin A mutant is cleavage resistant and, therefore, retains its farnesylated COOH-terminal peptide. Cotransfection of HeLa cells with preLaA along with either HA-Sun1N220 or HA-Sun2N165 lead to a dramatic decline in the nucleoplasmic concentration of both Sun protein fragments coincident with recruitment to the nuclear periphery. Lamin B1, on the other hand, had no such effect. These results indicate that the Sun proteins do indeed have the capacity to interact with lamin A in vivo.
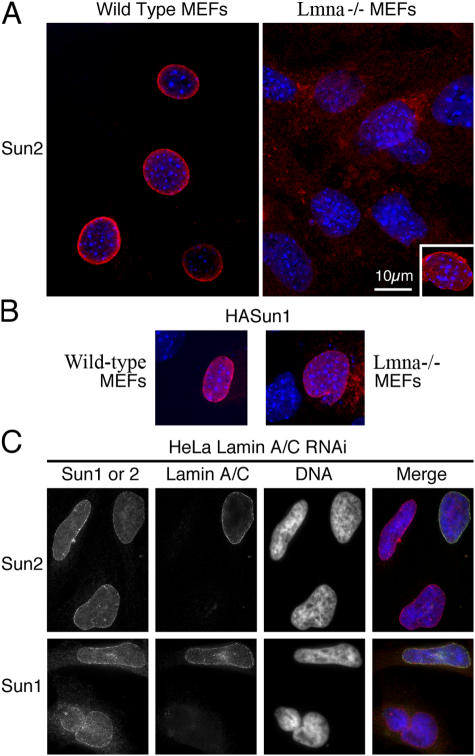
To determine whether this interaction with lamin A is required for Sun2 retention at the NE, we first performed immunofluorescence experiments on fibroblasts derived from both wild-type and Lmna-null mouse embryonic fibroblasts (MEFs). Although Sun2 was detected at the NE of all wild-type cells, in the majority of Lmna-null MEFs, Sun2 was dispersed throughout cytoplasmic membranes (Fig. 5 A). At first, these results do indeed implicate A-type lamins in the appropriate localization of Sun2 within the INM. However, there is clearly a minority of Lmna-null cells in which Sun2 is fully retained at the NE (Fig. 5 A, inset). Furthermore, loss of Sun2 from the INM is not reversed simply by introducing lamin A and/or C by transfection into Lmna-null MEFs (not depicted). Evidently, although A-type lamins could contribute to Sun2 localization, they are not the only determinants. This suggestion is reinforced by experiments in HeLa cells in which we eliminated A-type lamins by RNA interference (RNAi). After 48–72 h of RNAi treatment, lamin A/C was undetectable in many cells. However, the NE localization of Sun2 was barely affected (Fig. 5 C).
Figure 5.
A-type lamin independent retention of SUN domain proteins at the NE. (A) Endogenous Sun2 is frequently, but not always (inset), lost from the NE in Lmna-null MEFs. In wild-type MEFs, Sun2 is always found at the NE. (B) When HA-Sun1 was introduced by transfection into either wild-type or Lmna-null MEFs, it was always found to localize appropriately to the NE. (C) Depletion of HeLa cells of A-type lamins by RNAi had no significant effect on endogenous Sun1 or 2 localization. In the merged images, Sun protein localization is shown in red, A-type lamin localization (lamin A/C) is shown in green, and nuclei are revealed in blue using Höchst dye. The inference is that A-type lamins have, at best, a limited role in the retention of Sun2 at the NE and no significant role at all in the retention of Sun1.
In contrast to Sun2, we could find no evidence whatsoever for any lamin-dependent localization of Sun1. We took the approach of introducing HA-Sun1 into both wild-type and Lmna-null MEFs. In either case, exogenous Sun1 was always found at the nuclear periphery (Fig. 5 B). Similarly, in HeLa cells depleted of A-type lamins by RNAi, endogenous Sun1 always remained concentrated at the NE (Fig. 5 C). Regardless of A-type lamin content, we have never observed cells in which Sun1 is substantially mislocalized. Thus, although Sun proteins demonstrably interact with A-type lamins, this interaction is not required for their localization in the INM.
Studies in C. elegans have shown that the prototype SUN domain protein Unc-84 is required for the appropriate localization of Anc-1 in the ONM (Lee et al., 2002; Starr and Han, 2002). Giant nesprin family members are also known to localize to the ONM (Zhang et al., 2001; Zhen et al., 2002; Padmakumar et al., 2004) and, like Anc-1, feature a COOH-terminal KASH domain. Therefore, we examined the role that mammalian SUN domain proteins might play in the localization of one of these nesprin proteins (nesp2G; ∼800 kD).
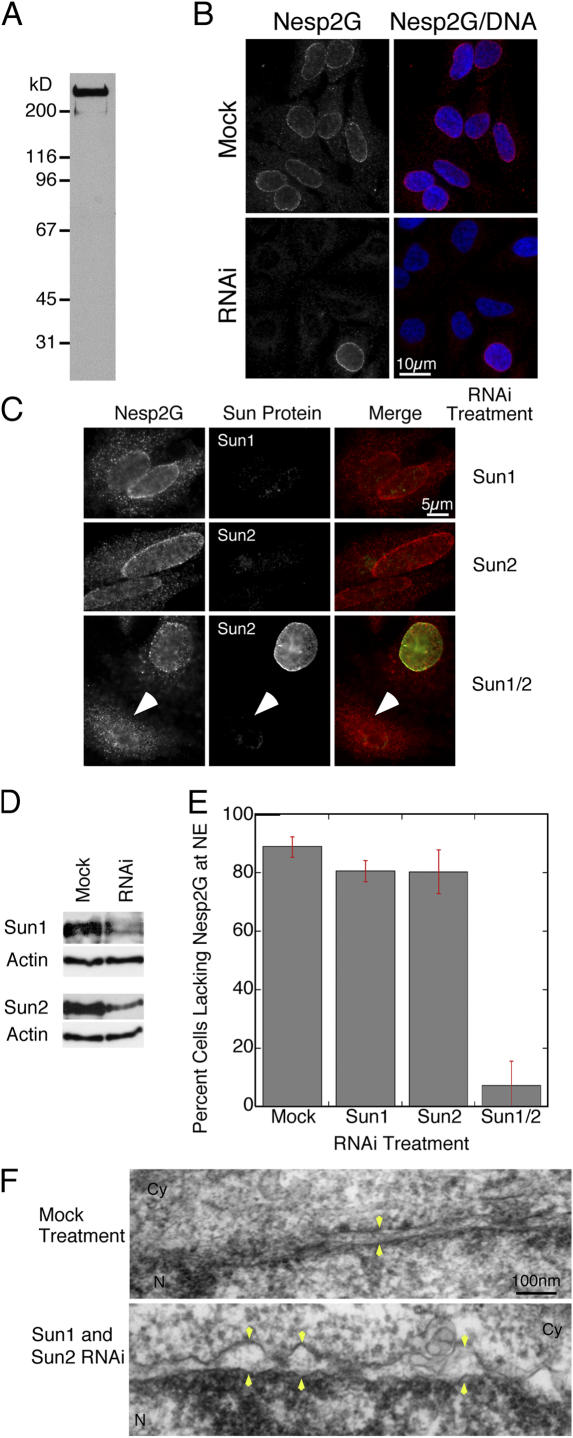
To accomplish this, we raised an antibody against the NH2-terminal actin-binding domain (ABD) of nesp2G. The affinity-purified antibody recognized a very large (>400 kD) protein on Western blots of HeLa cell lysates (Fig. 6 A). At longer exposure times, lower molecular mass bands appeared, possibly corresponding to smaller nesprin 2 isoforms (Zhang et al., 2001) or to degradation products. Immunofluorescence microscopy using digitonin permeabilization revealed that the anti-nesp2G antibody decorated the cytoplasmic surface of the NE (Fig. 6 B, mock). This labeling pattern was abolished by RNAi treatment of cells using nesprin 2–specific SmartPool oligonucleotides. Clearly, our antibody recognizes a very large ONM-associated nesprin 2 isoform, which is almost certainly nesp2G. Permeabilization of non–RNAi-treated cells with Triton X-100 yielded additional intranuclear labeling (unpublished data). This confirms a previous study suggesting that smaller nesprin 2 variants reside within the nucleus (Zhang et al., 2005). For all of our subsequent experiments, we used digitonin permeabilization to ensure that we were looking exclusively at nesp2G that was localized in the ONM.
Figure 6.
The retention of nesp2G at the ONM requires the expression of SUN domain proteins. (A) Western blot of a HeLa lysate fractionated by SDS-PAGE and probed with an affinity-purified antibody raised against the ABD of human nesp2G identifies a very large (>400 kD) protein. (B) Immunofluorescence microscopy of digitonin-permeabilized HeLa cells using the anti-ABD antibody reveals labeling of the cytoplasmic face of the NE. Depletion of nesprin 2 (all splice isoforms) in HeLa cells by RNAi leads to a loss of ONM labeling in the majority of cells (bottom). These data are consistent with the recognition of nesp2G by the anti-ABD antibody. (C) The reduction of either Sun1 or 2 levels by RNAi had only a marginal effect on nesp2G localization. However, the combined depletion of both Sun1 and 2 induced a dramatic loss of nesp2G from the ONM (arrowheads). Overall, we observed an ∼80% decline in the number of cells exhibiting NE-associated nesp2G (E). In total, 100–200 cells were scored for each category in three separate experiments. Error bars represent SEM. (D) Western blot analysis reveals that both Sun1 and 2 RNAi treatments lead to a substantial decline in Sun1 and 2 protein levels. The same blots were probed with an anti-actin antibody to confirm equal loading. (F) Thin section EM of cells subjected to the double (Sun1 and 2) RNAi treatment revealed frequent expansion of the PNS and increased separation of the INM and ONM (arrowheads). No such effect was observed in mock-treated cells. The nuclear interior (N) and cytoplasm (Cy) is indicated in each panel.
To explore the roles of Sun1 and 2 in nesp2G localization, we first adopted an RNAi-based approach. When we depleted HeLa cells of either Sun1 or 2 (Fig. 6, C and D), we could find little effect on the localization of nesp2G at the ONM (Fig. 6, C and E). However, codepletion of Sun1 and 2 led to the elimination of nesp2G from the ONM (Fig. 6, C and E). Quantitative analysis indicated an 80% reduction in the number of cells with detectable NE-associated nesp2G after codepletion of Sun1 and 2 versus mock RNAi treatment (Fig. 6 E). Ultrastructural analyses revealed changes in NE morphology in cells codepleted of both Sun proteins. Mock-treated cells displayed the usual uniform spacing between the ONM and INM of ∼50 nm. In the double RNAi-treated cells, however, the ONM was clearly dilated with obvious expansion of the PNS to 100 nm or more (Fig. 6 F).
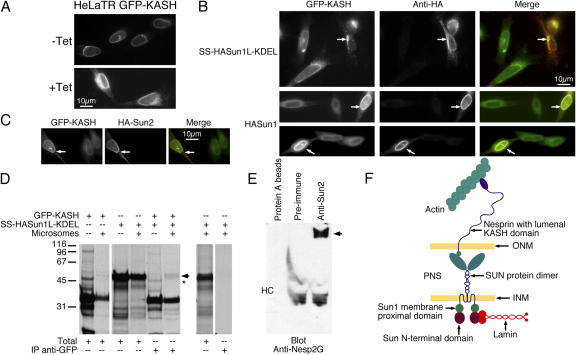
If SUN and KASH domain proteins form a molecular link across the PNS (Starr and Han, 2003), it should be possible to use a dominant-negative approach to break this linkage. In this strategy, we used almost the entire lumenal domain of Sun1 tagged at its NH2 terminus with HA (HA-Sun1L), which we introduced in soluble form into the lumen of the ER and PNS (Fig. 7 A). To accomplish this, we fused the signal sequence and signal peptidase cleavage site of human serum albumin onto the NH2 terminus of HA-Sun1L to yield SS–HA-Sun1L. To prevent its secretion, we fused a KDEL tetrapeptide to the COOH terminus of SS–HA-Sun1L, forming SS–HA-Sun1L–KDEL (Fig. 7 A). Synthesis of this chimeric protein in vitro in the presence of microsomes yielded a protein product of the appropriate size, which was resistant to digestion by proteinase K (Fig. 7 B). Clearly, the signal sequence directs HA-Sun1L to the microsome lumen. The shift up in molecular weight of latent HA-Sun1L–KDEL is likely a result of NH2-linked glycosylation (Sun1 has two potential glycosylation sites in its lumenal domain). Upon transfection into HeLa cells, HA-Sun1L–KDEL was found to accumulate intracellularly within the peripheral ER and PNS. Examination of the distribution of nesp2G in transfected cells revealed that it was completely eliminated from the ONM (Fig. 7 C). EM analysis of these cells exposed clear dilation of the ONM and expansion of the PNS (Fig. 7 D). This phenotype is indistinguishable from that associated with Sun1/2 codepletion by RNAi (Fig. 6 F).
Figure 7.
A soluble form of the Sun1 lumenal domain causes a loss of nesp2G from the ONM. (A) A signal sequence (SS), HA tag, and KDEL motif were added to the NH2 and COOH termini, respectively, of the Sun1 lumenal domain (SS–HA-Sun1L–KDEL). (B) SS–HA-Sun1L–KDEL, when translated in vitro in the presence of microsomes, is completely resistant to digestion by proteinase K. If Triton X-100 is included in the digestion reaction to permeabilize the microsomes, the ∼50-kD SS–HA-Sun1L–KDEL translation product (arrow) is completely degraded. These data demonstrate that the signal sequence is fully functional in directing HA-Sun1L–KDEL to the microsomal lumen. (C) When introduced by transfection into HeLa cells, the SS–HA-Sun1L–KDEL localizes both to the peripheral ER and to the PNS, which is revealed by immunolabeling with an anti-HA monoclonal antibody. Cells expressing SS–HA-Sun1L–KDEL (red in merged images) exhibit a very obvious loss of nesp2G (green in merged images) from the ONM. (D) Thin section EM of HeLa cells expressing SS–HA-Sun1L–KDEL (Transf.) revealed increased separation between the INM and ONM and expansion of the PNS (arrowheads). This effect was not observed in nontransfected cells (NT). The effects of SS–HA-Sun1L–KDEL expression were identical to those observed after codepletion of Sun1 and 2 by RNAi (Fig. 6).
In further experiments, we took advantage of a cDNA encoding a chimeric protein in which GFP is fused to the NH2 terminus of the nesprin 2 KASH domain (Zhang et al., 2001). This fusion protein localizes to the NE with the GFP exposed to the cytoplasm/nucleoplasm. The KASH domain is integrated into the NE, with its 40-residue COOH-terminal domain residing within the PNS. We prepared a HeLa cell line harboring GFP-KASH under the control of a tetracycline-inducible promoter (HeLaTR GFP-KASH). In the absence of tetracycline, GFP-KASH is present at low levels and localizes exclusively to the NE (Fig. 8 A, −Tet). After tetracycline induction, large amounts of GFP-KASH may be observed in both the NE and peripheral ER (Fig. 8 A, +Tet). Introduction of SS–HA-Sun1–KDEL into these cells leads to the complete loss of NE-associated GFP-KASH. Indeed, all of the GFP-KASH, regardless of expression level, appears to be recruited into vesicular structures, potentially as a prelude to degradation (Fig. 8 B, top; arrows). Conversely, when full-length Sun1 is introduced into tetracycline-induced cells, it leads to the recruitment of GFP-KASH from the peripheral ER to the NE (Fig. 8 B, middle and bottom; arrows). This is exactly what one would predict if Sun proteins function as tethers for KASH domain proteins. A similar effect was also observed when HA-Sun2 was introduced into the tetracycline-induced cells (Fig. 8 C), with GFP-KASH recruited to and stabilized at the NE. Altogether, these results can only be interpreted in terms of lumenal interactions between SUN domain and KASH domain proteins. Furthermore, they suggest that both Sun1 and 2 can interact in vivo with the nesprin 2 KASH domain, which is consistent with our RNAi results indicating that both SUN domain proteins contribute to nesp2G anchoring.
Figure 8.
The nesp2G KASH domain interacts with the Sun1 lumenal domain. (A) Fluorescence microscopy of HeLa cells expressing a tetracycline-inducible GFP-KASH fusion protein (HeLaTR GFP-KASH). In the absence of tetracycline (−Tet), GFP-KASH is expressed at low levels and is localized exclusively to the NE. After induction with tetracycline for 24 h (+Tet), GFP-KASH is found throughout the peripheral ER as well as the NE. Expression levels of GFP-KASH within the cell population is extremely uniform both before and after induction. (B) Transfection of SS–HA-Sun1–KDEL into the HeLaTR GFP-KASH cells after tetracycline induction resulted in the complete loss of GFP-KASH from the NE. This was accompanied by the formation of cytoplasmic aggregates (arrows, top). Conversely, introduction of full-length HA-Sun1 into these cells resulted in the recruitment of GFP-KASH to the NE (arrows, middle and bottom). (C) HA-Sun2 was found to have a similar effect (arrows). In the merged images in B and C, GFP-KASH is presented in green, whereas HA-Sun is shown in red. (D) To identify an in vitro interaction between KASH and SUN domains, GFP-KASH, SS–HA-Sun1–KDEL, or both proteins were translated in reticulocyte lysate containing [35S]methionine/cysteine in either the presence or absence of microsomes (Totals). Anti-GFP immunoprecipitation of a fraction of each sample revealed the pull-down of SS–HA-Sun1–KDEL by GFP-KASH when both proteins were cotranslated in the presence of microsomes (arrow). A slightly faster migrating band (asterisk) was detected in the absence of microsomes. However, this band originates in the GFP-KASH translation and is unrelated to HA-Sun1L–KDEL. Molecular masses are indicated in kD. (E) Immunoprecipitation of HeLa cell lysates with anti-Sun2 antibodies coprecipitates a very large anti-nesp2G immunoreactive protein (arrow). HC indicates the position of immunoglobulin heavy chains. These data suggest a significant interaction between KASH and Sun proteins that must involve their lumenal domains. These findings allow us to propose a model for the LINC complex (F) in which nuclear components, including lamins, bind to the INM SUN domain proteins. They, in turn, bind to the KASH domain of the actin-associated giant nesprins on the ONM. Thus, the LINC complex establishes a physical connection between the nucleoskeleton and the cytoskeleton.
To further define Sun1/2–KASH interactions, we set out to identify Sun–KASH complexes both in vivo and in vitro. For the former, we performed immunoprecipitation analyses of Sun2. As shown in Fig. 8 E, a high molecular weight protein recognized by our antibody against nesp2G was found to coimmunoprecipitate with Sun2 from HeLa cell lysates. As a complement to these experiments, we synthesized SS–HA-Sun1–KDEL and GFP-KASH in vitro. Interactions between these proteins were then analyzed by immunoprecipitation using antibodies against GFP. As revealed in Fig. 8 D, only when the SUN and KASH domain proteins were cosynthesized in vitro in the presence of microsomes could we detect HA-Sun1–KDEL in immunoprecipitates performed with the anti-GFP antibody. Together, all of these data provide strong evidence for the interaction, either direct or indirect, between Sun1/2 and nesp2G. Such an interaction spanning the PNS provides an obvious mechanism for the Sun1/2-dependent tethering of nesp2G in the ONM.
Discussion
We have shown that Sun1 is an INM protein with an NH2-terminal nucleoplasmic domain of ∼350 amino acids and a larger COOH-terminal domain of ∼500 amino acids, including the SUN domain, that resides in the PNS. In this way, the topology of Sun1 matches that of another INM protein, Sun2, to which it is related. Based upon structural predictions, it is likely that Sun1 possesses three closely spaced transmembrane domains between residues 356 and 448. A separate hydrophobic region, H1, that is situated closer to the NH2 terminus does not appear to function as a membrane anchor. This conclusion is based upon the behavior of naturally occurring splice isoforms that lack this hydrophobic sequence. A third mammalian Sun protein, Sun3, is also an integral membrane protein with a lumenal COOH-terminal SUN domain and a relatively small cytoplasmic NH2-terminal domain (unpublished data). In this way, Sun3 conforms to the general topological organization of other Sun family members.
The organization of the lumenal domains of Sun1 and 2 bears some comment. The membrane proximal sequences of both proteins are predicted to form a coiled-coil. The implication is that these proteins may form homodimers. Given the number of residues within the Sun1 coiled-coil region, this could potentially project ∼25–30 nm into the PNS and would terminate in a pair of globular SUN domains. The coiled-coil domain of Sun2 is of a similar size. In both cases, the overall conformation of the Sun protein lumenal domain would be that of a flower on a stalk, which could potentially bridge the gap between the lumenal faces of the INM and ONM.
The exact mechanism by which Sun1 and 2 are localized to the INM has yet to be resolved, although it is likely to involve the type of selective retention that has been observed for other INM proteins. What is clear is that the lumenal domain of both proteins is entirely dispensable for appropriate localization. This is exactly the reverse of what is observed for nesprin proteins (including nesp2G) of the ONM, where the lumenal and transmembrane domains (comprising the KASH domain) are essential for their retention at the nuclear periphery (Zhang et al., 2001).
On the nucleoplasmic side of the INM, the Sun1 and 2 NH2-terminal domains contain regions of similarity within the first ∼200 amino acid residues. This common NH2-terminal region interacts, to a greater or lesser extent, with A-type lamins. In the case of Sun2, there is some evidence that A-type lamins might contribute to Sun2 localization in the INM. However, whether this requires a direct interaction with A-type lamins is less clear. Certainly, the concentration of Sun2 in the INM is A-type lamin independent in some cells. Furthermore, even in Lmna-null MEFs in which Sun2 is mislocalized, the mere reintroduction of A-type lamins fails to recruit Sun2 to the INM, at least within a period of ∼24 h (unpublished data). It seems more likely to us that A-type lamins may have indirect effects on Sun2, perhaps by altering the accessibility of chromatin proteins with which Sun2 might interact.
In the case of Sun1, there is no evidence that lamins play any role in its localization to the INM. However, Sun1 displays an extremely robust interaction with prelamin A. Newly synthesized lamin A undergoes extensive COOH-terminal processing (Sinensky et al., 1994). This involves farnesylation of the COOH-terminal CaaX (single letter code where C is cysteine, a is any amino acid with an aliphatic side chain, and X is any amino acid) motif followed by endoproteolysis to remove the aaX residues and carboxy methylation of the farnesyl cysteine. Once incorporated into the nuclear lamina, a second cleavage event after Y646 yields mature lamin A (Weber et al., 1989). This cleavage of prelamin A at Y646 abolishes any strong interaction with Sun1. Because prelamin A exists only transiently in normal cells, it seems unlikely that its interaction with Sun1 could contribute to Sun1 localization. In our opinion, it is more likely that Sun1 might function in the organization of newly synthesized lamin A within the nuclear lamina. This suggestion is currently under investigation.
Starr and Han (2002) have shown that the C. elegans SUN domain protein Unc-84 is required for the localization of Anc-1 in the ONM. They have proposed a model in which the lumenal domain of Unc-84, which itself is retained in the INM through interactions with the single C. elegans lamin, forms a complex with the lumenal KASH domain of Anc-1. In this way, Unc-84 and Anc-1 would provide links in a molecular chain that spans the PNS and connects the actin cytoskeleton to the nuclear lamina. Because similar SUN and KASH domain molecules are widely represented in the animal kingdom, we attempted to determine whether the Unc-84/Anc-1 paradigm was applicable in mammalian systems. We used a combination of RNAi and a dominant-negative form of Sun1 to test this model. We found that both Sun1 and 2 contribute to the tethering of nesp2G in the ONM. Elimination of either Sun protein by RNAi had little or no effect on nesp2G localization in HeLa cells. However, codepletion of Sun1 and 2 leads to the loss of nesp2G from the ONM. This was accompanied by separation of the ONM and INM, leading to expansion of the PNS. The implication here is that links between the Sun proteins in the INM and KASH proteins in the ONM help to maintain the remarkably regular spacing of the nuclear membranes. This view was reinforced by the findings that overexpression of a soluble form of the lumenal domain of Sun1 (SS–HA-Sun1L–KDEL) induced essentially the same phenotype: loss of nesp2G from the ONM and expansion of the PNS.
In a complementary series of experiments, SS–HA-Sun1L–KDEL expression was also found to lead to the loss of GFP-KASH from the NEs of HeLa cells. This effect can only be accounted for by perturbation of lumenal interactions. Conversely, overexpression of full-length Sun1 (or Sun2) leads to the recruitment of GFP-KASH to the NE. All of these results are predictable on the basis of SUN domain proteins functioning as translumenal tethers for KASH domain proteins.
Our final experiments demonstrated the existence of SUN–KASH complexes. Immunoprecipitates of Sun2 from HeLa extracts were found to contain nesp2G. Similarly, in vitro translation of SS–HA-Sun1L–KDEL and GFP-KASH leads to the formation of HA-Sun1L–KDEL–GFP-KASH complexes provided that microsomes were present in the translation mix. While this manuscript was in preparation, Padmakumar et al. (2005) published a study that demonstrated a similar interaction between SUN domain and KASH domain proteins. Their results suggest, however, that rather than interacting with the SUN domain itself, the KASH domain actually bound to a region of the polypeptide chain that is proximal to the SUN domain. This region is present in our Sun1L-based dominant-negative mutant.
All of our data suggest that the two Sun proteins are the key to the appropriate localization of nesp2G in the ONM. In contrast to previously published findings (Libotte et al., 2005), we could find no evidence of a role for A-type lamins. This is not surprising given that in our HeLa cells, the localization of Sun1 and 2 appear relatively insensitive to A-type lamin expression (or depletion). Padmakumar et al. (2005) reached exactly the same conclusion with respect to Sun1. However, in Lmna-null MEFs, Sun2 is frequently lost from the NE. Given that expression levels of Sun1 appear to vary somewhat between different tissues, it is conceivable that in at least some cell types, nesp2G localization to the ONM might be sensitive to A-type lamin expression.
Altogether, our findings and those of Padmakumar et al. (2005) are entirely consistent with the model proposed by Starr and Han (2003) in which SUN and KASH domain proteins form a link across the PNS (Fig. 8 F). In addition, in C. elegans, a similar mechanism may well operate in the tethering of Zyg-12, a NE protein that is required for dynein-mediated centrosome positioning (Malone et al., 2003). As well as tethering ONM proteins, our data would suggest that SUN–KASH linkages further contribute to the structural integrity of the NE in maintaining the precise separation of the two nuclear membranes. Furthermore, given that the giant nesprins are actin-binding proteins, the SUN–KASH links provide direct molecular connections between the actin cytoskeleton and the nuclear interior. We now refer to this molecular chain as the LINC complex.
Many studies have documented mechanical coupling between the nucleus and the cytoplasm. Maniotis et al. (1997) used microneedle-mediated deformation of the cytoplasm of cultured cells to demonstrate mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm. More recently, Lammerding et al. (2004) were able to show that fibroblasts derived from Lmna-null mouse embryos have impaired mechanically activated gene transcription. In related studies, Broers et al. (2004) have shown that these same cells exhibited reduced mechanical stiffness and perturbations in the organization of the cytoskeleton. The existence of the LINC complex provides a basis for these various observations in that it may integrate the nucleus into a protein matrix that includes the cytoskeleton, extracellular matrix, and cell–cell adhesion complexes. This mechanical link not only provides structural continuity within and between cells, but it also allows for a direct physical signaling pathway from the cell surface to the nucleus, potentially facilitating rapid and regionalized gene regulation.
Materials and methods
Cell culture
HeLa cells and MEFs, both Lmna +/+ and Lmna −/− (Sullivan et al., 1999), were maintained in 7.5% CO2 and at 37°C in DME (GIBCO BRL) plus 10% FBS (Hyclone), 10% penicillin/streptomycin (GIBCO BRL), and 2 mM glutamine.
Antibodies
The following antibodies were used in this study: the monoclonal antibody against lamins A and C (XB10) has been described previously (Raharjo et al., 2001). The monoclonal antibodies 9E10 and 12CA5 against the myc and HA epitope tags were obtained from the American Type Culture Collection and Covance, respectively. Rabbit antibodies against the same epitopes were obtained from AbCam. Rabbit antibodies against Sun1 and 2 were raised against GST fusion proteins as previously described (Hodzic et al., 2004). The chicken antibody against the ABD of nesp2G was raised against an ABD-GST fusion protein by Aves Labs, Inc. Affinity purification of the IgY was performed in two stages. In the first step, an affinity column was prepared consisting of GST cross-linked to glutathione agarose (Sigma-Aldrich) using 40 mM dimethyl pimelimidate in 0.2 M borate buffer, pH 9.0, for 1 h at 4°C. 5 ml IgY solution was passed over this column (1-ml bed volume), and the flow through was collected. This flow through was applied to a second 1-ml column prepared from ABD-GST that was also cross-linked to glutathione agarose. Antibody bound to the column was eluted at pH 2.8 in 0.2 M glycine-HCl. The antibody eluate was neutralized with 3 M Tris, pH 8.8, and stored at 4°C with 1 mM sodium azide. Secondary antibodies conjugated with AlexaFluor dyes were obtained from Invitrogen. Peroxidase-conjugated secondary antibodies were obtained from Biosource International.
Immunofluorescence microscopy
Cells were grown on glass coverslips and fixed in 3% formaldehyde (prepared in PBS from PFA powder) for 10 min followed by a 5-min permeabilization with 0.2% Triton X-100. The cells were then labeled with the appropriate antibodies plus the DNA-specific Höchst dye 33258. For experiments involving selective permeabilization, the cells were first fixed in 3% formaldehyde. This was followed by permeabilization in 0.003% digitonin in PBS on ice for 15 min (Adam et al., 1990). The cells were then labeled with appropriate primary and secondary antibodies. For certain double-label experiments, a single primary antibody was applied after the digitonin permeabilization. After removal of unbound antibody with three PBS washes, the cells were refixed for 5 min (in 3% formaldehyde) and subjected to a further permeabilization step in 0.2% Triton X-100. The second primary antibody was then applied followed by appropriate secondary antibodies. Specimens were observed using a microscope (model DMRB; Leica). Images were collected using a CDC camera (CoolSNAP HQ; Photometrics) linked to a Macintosh G4 computer running IPLab Spectrum software (Scanalytics).
EM
Cells grown in 35-mm petri dishes were fixed in 3% glutaraldehyde and 0.2% tannic acid in 200 mM sodium cacodylate buffer for 1 h at room temperature. Postfixation was performed in 2% OsO4 for 20 min. The cells were dehydrated in ethanol, lifted from the culture dish using propylene oxide, and infiltrated with Polybed 812 resin. Polymerization was performed at 60°C for 24 h. Silver-gray sections were cut using an ultramicrotome (Leica) equipped with a diamond knife. The sections were stained with uranyl acetate and lead citrate and examined in an electron microscope (model 7000; Hitachi).
Short inhibitory RNA (siRNA) methods
HeLa cells were depleted of Sun1, Sun2, nesprin 2, and lamins using appropriate SmartPool oligonucleotide duplexes (Dharmacon). Cells were exposed to each siRNA in the presence of OligofectAMINE (Invitrogen) precisely as recommended by the manufacturer. Cells were subjected to siRNA treatment for periods up to four days. However, most of our analyses were performed after 2–3-d treatments.
Immunoblotting and gel electrophoresis
Cells (siRNA- or mock-treated) grown in 35-mm tissue culture dishes were washed once in PBS and lysed in a buffer containing 50 mM Tris-HCl, pH 7.4, 500 mM NaCl, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF, and 1:1,000 CLAP (10 mg/ml in DMSO each of chymostatin, leupeptin, antipain, and pepstatin). The lysate was centrifuged for 5 min in an Eppendorf centrifuge at 4°C. Proteins in the supernatant were precipitated by the addition of TCA to a final concentration of 10%. The precipitate was washed with ethanol/ether and solubilized in SDS-PAGE sample buffer. Protein samples were fractionated on polyacrylamide gels (7.5, 10, or 4–15% gradient as appropriate; Laemmli, 1970) and transferred onto nitrocellulose filters (usually BA85; Schleicher and Schuell) using a semidry blotting apparatus manufactured by Hoeffer Scientific Instruments Inc. Filters were blocked and labeled with primary antibodies and peroxidase-conjugated secondary antibodies exactly as previously described (Burnette, 1981). Blots were developed using ECL (GE Healthcare) and exposed to X-OMAT film (Kodak) for appropriate periods of time.
Immunoprecipitations
For Sun2/nesp2G coimmunoprecipitations, three subconfluent 35-mm dishes of HeLa cells were each extracted with 1 ml each of PBS containing 0.1% Triton X-100, 1:1,000 CLAP, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, and 1 mM sodium vanadate. The dishes were rocked at 4°C for 15 min, and the cell lysates were centrifuged for 10 min at maximum speed and at 4°C in a microcentrifuge. The pellets were then reextracted in a total volume of 3 ml radioimmunoprecipitation assay buffer for 15 min, also at 4°C. After centrifugation at 4°C for 10 min, the supernatants were pooled and divided into 500-μl aliquots. Each aliquot was then incubated for 14 h at 4°C with appropriate combinations of immune and preimmune sera and protein A–Sepharose beads. At the end of this period, the beads were collected by brief centrifugation and washed three times in PBS containing 0.1% Triton X-100 and once in PBS alone. Finally, the beads were suspended in SDS-PAGE sample buffer, heated to 95°C for 5 min, and analyzed by SDS-PAGE and Western blotting.
All other immunoprecipitations were performed in TNX (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 0.5% Triton X-100) containing 1:1,000 CLAP. Incubation with antibodies and protein A–Sepharose beads was for 1 h at 4°C with continuous gentle mixing. At the end of this period, the beads were collected by brief centrifugation and were washed three times in TNX and once in 50 mM Tris-HCl, pH 7.4. Finally, the beads were processed for electrophoresis as described above.
In vitro translations
In vitro translations were performed in 25-μl reaction volumes using the TNT T7 coupled transcription translation system (Promega). Each translation reaction contained 20 μl TNT master mix and was programmed with 1 μl plasmid DNA at a concentration of 0.1 μg/μl. Labeling of translation products was accomplished by the inclusion of 10 μCi 35S Translabel (MP Biolabs). Where appropriate, up to 3 μl of canine pancreatic microsomes (Promega) was also included in each reaction. Translation reactions were assembled on ice before incubation at 30°C for 90 min. At the end of this period, translation mixes were further processed for in vitro binding studies (see In vitro pull-down…proteins) or were subjected to digestion with proteinase K in order to define Sun protein topology. Proteinase K digestions were performed on ice for periods of up to 1 h. Each digestion mix (10-μl total volume) contained 5 μl of the complete in vitro translation reaction, 1 μl proteinase K (from a 1-mg/ml stock solution), 1 μl of 10× compensation buffer (containing 0.5 M sucrose, 50 mM Tris-HCl, pH 7.6, and 200 mM potassium acetate), and, where appropriate, 1 μl of a 10% solution of Triton X-100. Termination of digestion was accomplished by the addition of 100 μl of 10% TCA to precipitate the proteins. Precipitates were washed in ethanol/ether, air dried, and dissolved in 25 μl SDS-PAGE sample buffer by incubation at 37°C.
Plasmids
A mouse Sun1 cDNA (IMAGE clone ID 5321879) was obtained from Invitrogen. To generate Sun1 tagged at the NH2 terminus with an HA epitope, Sun1 cDNA flanked by 5′Sal1 and 3′Afl2 restriction sites was amplified by PCR using primers 5′-GAACGTCGACTTTTCTCGGCTGCACACGTACACC-3′ and 5′-CTGGCTTAAGCTACTGGATGGGCTCTCCG-3′. The PCR product was digested with Sal1 and Afl2 and inserted downstream of an HA tag sequence in the vector pCDNA3.1(−). This vector was prepared from pcDNA3.1(−) containing HA–lamin A (Raharjo et al., 2001) by digestion with Xho1 and Afl2. The resulting plasmid was pcDNA3.1(−)HA-Sun1.
pcDNA3.1(−)HA-Sun1 was used as a template for generating further Sun1 constructs. The Sun1 splice isoform Δ6–8 was created by inverse PCR using primers 5′-CGTGGTTTGAGAGTCCTGTCTCTGG-3′ and 5′-GACCTCTTGGTTCAAGCACTGCGAAGG-3′ to amplify the entire plasmid outside of the deleted region. The PCR product was digested with the Klenow fragment of DNA polymerase and was circularized by ligation. A double epitope-tagged Sun1 construct, HA-Sun1–myc, was made by PCR using primers 5′-GCATCTGAAGACCAGCTGAG-3′ and 5′-TAAACTTAAGCTAGAGATCCTCTTCTGAGATGAGTTTTTGTTCCTCAGCCTGGATGGGCTCTCCGTGGACTCG-3′ to insert a 3′ myc tag followed by an AflII restriction site. The fragment was then cloned back into the HindIII–AflII site of the original template.
To prepare a form of EGFP-KASH that could be translated in vitro, the BamHI–Nhe1 digested fragment of pEGFP-KASH was subcloned into the BamHI–Nhe1 site of pCDNA3.1(−) to yield pcDNA3.1EGFP-KASH. EGFP-KASH was also inserted into the tetracycline-inducible expression vector pcDNA4TO. This was accomplished by amplifying EGFP-KASH by PCR using the primers 5′-TAAACTTAAGCACCATGGTGAGCAAGGGCGAGGAGC-3′ and 5′-TAAAGCGGCCGCCTATGTGGGGGGTGGCCCATTGGTGTACC-3′. The 1,003-bp product was cut with Afl2 and Not1 and was ligated into similarly cut pcDNA4TO.
The soluble Sun1 lumenal domain construct SS–HA-Sun1L–KDEL, which was targeted to the ER and PNS, was prepared in three stages. The first step involved ligation of a double-stranded oligonucleotide encoding the entire NH2-terminal signal sequence of human serum albumin, which was ligated into pcDNA3.1(−) between Nhe1 and Apa1 sites to yield pcDNA3.1SS. In the second intermediate step, the 5′ end of HA–lamin A was amplified by PCR using the pair of primers 5′-AATTGGGCCCGCTTACCCTTACGATGTACCG-3′ and 5′-ATATCTTAAGCAGCGCATCCGCCAGCCGGCTC-3′. The 787-bp PCR product was ligated downstream of the signal sequence in pcDNA3.1SS between the Apa1 and Afl2 sites to yield pcDNA3.1SS-HALaA5′. For the final step, the lamin A sequences were excised using Xho1 and Afl2. To prepare the Sun1 lumenal domain sequence incorporating a KDEL motif, PCR was performed using mouse Sun1 cDNA as a template and using the primers 5′-AGAGGGTCGACGATTCCAAGGGCATGCATAG-3′ and 5′-CTGGCTTAAGCTACAACTCATCTTTCTGGATGGGCTCTCCGTGGAC-3′. The resultant 1,403-bp product was cut with Sal1 and Afl2 and was ligated into the Xho1–Afl2 cut vector. The resulting plasmid was pcDNA3.1SS–HA-Sun1L–KDEL. All enzymes were obtained from New England Biolabs, Inc.
Transfections
Plasmids were introduced into HeLa cells using the Polyfect reagent as described by the manufacturer (QIAGEN). Transfections were normally performed in six-well plates. In brief, 1.5 μg plasmid DNA was combined with 100 μl of serum-free medium and 12 μl Polyfect. After a 10-min room temperature incubation, this mixture was combined with 600 μl of complete medium. The entire volume was then placed on the cells with an additional 1.5 ml of complete medium. The cells were then returned to the tissue culture incubator for 12–24 h. At the end of this period, the cells were processed as appropriate.
Preparation of GST fusion proteins
GST-Sun1 and Sun2 fusion proteins were prepared using the plasmids pGEX-4T3Sun1N220 and pGEX-4T3Sun2NP. These plasmids were created by amplifying 5′ sequences of Sun1 and 2 by PCR. The Sun1 PCR product encoded the first 222 amino acids of the NH2-terminal domain, whereas the Sun2 sequences encoded the bulk of NH2-terminal domain of 165 amino acids. These PCR products were inserted into pGEX4T3 (GE Healthcare) between BamHI and EcoRI sites. The primers used for Sun1 were 5′-CGCGGATCCGACTTTTCTCGGCTGCAC-3′ and 5′-CCGGAATTCTTAGCGTGGTTTGAGAGTCCT-3′, whereas the Sun2 primer pair consisted of 5′-CGCGGATCCTCCCCGAAGAAGCCAGCGCCTCACG-3′ and 5′-CCGGAATTCTTAGGAGCCCGCCCGTGAGACGGC-3′. A single colony of Bl-21 cells transformed with either plasmid was grown overnight in 10 ml Luria-Bertani containing 100 μg/ml ampicillin and was induced with 0.1 mM IPTG for 4 h at 37°C. The cells were harvested by 15-min centrifugation at 4°C and at 3,200 g in an Eppendorf table-top centrifuge. The bacterial pellets were resuspended by trituration in 1 ml of lysis buffer consisting of STE (150 mM NaCl, 10 mM Tris, pH 8.0, and 1 mM EDTA) containing 5 mM DTT and 0.25% sarkosyl (N-laurylsarcosine). The suspension was sonicated to achieve maximum cell breakage and was centrifuged at maximum speed in a microcentrifuge for 10 min at 4°C. The supernatant was then transferred to a fresh microcentrifuge tube containing 30 μl of a 50% suspension in PBS of swollen glutathione agarose beads. The cleared bacterial lysate and beads were then incubated with continuous mixing at 4°C for 1 h. At the end of this period, the beads were washed three to five times with ice-cold STE and twice with ice-cold binding buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 0.1% Triton X-100, and 1 mM DTT).
In vitro pull-down with GST fusion proteins
1.5 μg of plasmid DNA (pcDNA3.1HA–lamin A, –mature lamin A, –lamin B1, and –lamin C) was included in 25 μl TNT-coupled transcription/translation mixes containing 10 μCi 35S Translabel and incubated at 30°C for 90 min. 1 ul of each reaction was retained for the analysis of total translation products, whereas the remainder was incubated with 10 ul GST–agarose beads in 600 μl of binding buffer (containing 10 μg/ml chymostatin, leupeptin, antipain, pepstatin, and 1 mM PMSF) for 30 min at room temperature with constant mixing. After a low speed centrifugation at 800 g, the supernatant was split into three tubes containing 5 ul GST–agarose, GST-Sun1N220, or GST-Sun2NP beads. The suspensions were then incubated for 45 min at room temperature with constant mixing. Finally, the beads were washed three to five times with binding buffer containing 1 mM DTT. After the final wash, the binding buffer was replaced with 20 μl SDS-PAGE sample buffer. The samples were subsequently fractionated by SDS-PAGE. The gels were stained with Coomassie blue R-250, impregnated with Amplify (GE Healthcare), dried, and exposed to X-OMAT film (Kodak).
Northern blot analysis
Double-stranded DNA probes consisting of the 5′ (650 bp) and 3′ (860 bp) fragments of mSun1 and the full-length human Sun3 (1,050 bp) were generated by PCR. Incorporation of [32P]dCTP into the PCR products was accomplished by random priming using the Rediprime II Random Primer Labeling System (GE Healthcare) using 15 ng (in 45 μl Tris-EDTA buffer) denatured DNA. A mouse multiple tissue Northern blot (BLOT-2; Sigma-Aldrich) containing 2 ug polyA+ (per lane) RNA isolated from 10 different mouse organs (brain, heart, liver, kidney, spleen, testis, lung, thymus, placenta, and skeletal muscle tissues of BALB/c mice) was hybridized independently with each of the prepared probes, including one against glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1.4 ng/ml in 6 ml PerfectHyb Plus hybridization buffer) for 17 h (Sigma-Aldrich). Between each hybridization, the blot was stripped of the probe according to the manufacturer's instructions.
Acknowledgments
We would like to acknowledge Manfred Lohka, Colin Stewart, and Phyllis Hansen for valuable discussions.
This work was supported by grants from the National Institutes of Health to B. Burke and P.D. Stahl. D. Hodzic was supported by a fellowship from the Muscular Dystrophy Association.
Abbreviations used in this paper: ABD, actin-binding domain; INM, inner nuclear membrane; MEF, mouse embryonic fibroblast; NE, nuclear envelope; NPC, nuclear pore complex; ONM, outer nuclear membrane; PNS, perinuclear space; RNAi, RNA interference; siRNA, short inhibitory RNA.
References
- Adam, S.A., R.E. Sterne-Marre, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, E.D., R.M. Lewis, R.M. Grady, and J.R. Sanes. 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275:31986–31995. [DOI] [PubMed] [Google Scholar]
- Broers, J.L., E.A. Peeters, H.J. Kuijpers, J. Endert, C.V. Bouten, C.W. Oomens, F.P. Baaijens, and F.C. Ramaekers. 2004. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 13:2567–2580. [DOI] [PubMed] [Google Scholar]
- Burke, B., and C.L. Stewart. 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3:575–585. [DOI] [PubMed] [Google Scholar]
- Burnette, W.N. 1981. ‘Western blotting’: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195–203. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., E.D. Siggia, J.E. Moreira, C.L. Smith, J.F. Presley, H.J. Worman, and J. Lippincott-Schwartz. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace, L., and B. Burke. 1988. Functional organization of the nuclear envelope. Annu. Rev. Cell Biol. 4:335–374. [DOI] [PubMed] [Google Scholar]
- Gerace, L., A. Blum, and G. Blobel. 1978. Immunocytochemical localization of the major polypeptides of the nuclear complex-lamina fraction: interphase and mitotic distribution. J. Cell Biol. 79:546–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., A. Margalit, R.D. Goldman, D.K. Shumaker, and K.L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31. [DOI] [PubMed] [Google Scholar]
- Hagan, I., and M. Yanagida. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodzic, D.M., D.B. Yeater, L. Bengtsson, H. Otto, and P.D. Stahl. 2004. Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 279:25805–25812. [DOI] [PubMed] [Google Scholar]
- Hoger, T.H., G. Krohne, and W.W. Franke. 1988. Amino acid sequence and molecular characterization of murine lamin B as deduced from cDNA clones. Eur. J. Cell Biol. 47:283–290. [PubMed] [Google Scholar]
- Hoger, T.H., K. Zatloukal, I. Waizenegger, and G. Krohne. 1990. Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma. 99:379–390. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lammerding, J., P.C. Schulze, T. Takahashi, S. Kozlov, T. Sullivan, R.D. Kamm, C.L. Stewart, and R.T. Lee. 2004. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.K., D. Starr, M. Cohen, J. Liu, M. Han, K.L. Wilson, and Y. Gruenbaum. 2002. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol. Biol. Cell. 13:892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libotte, T., H. Zaim, S. Abraham, V.C. Padmakumar, M. Schneider, W. Lu, M. Munck, C. Hutchison, M. Wehnert, B. Fahrenkrog, et al. 2005. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol. Biol. Cell. 16:3411–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F., and H.J. Worman. 1993. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 268:16321–16326. [PubMed] [Google Scholar]
- Lin, F., and H.J. Worman. 1995. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 27:230–236. [DOI] [PubMed] [Google Scholar]
- Malone, C.J., L. Misner, N. Le Bot, M.C. Tsai, J.M. Campbell, J. Ahringer, and J.G. White. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836. [DOI] [PubMed] [Google Scholar]
- Maniotis, A.J., C.S. Chen, and D.E. Ingber. 1997. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 94:849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mislow, J.M., M.S. Kim, D.B. Davis, and E.M. McNally. 2002. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J. Cell Sci. 115:61–70. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop, K.L., Q. Li, L. Patterson, and J.A. Fischer. 1999. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 9:1211–1220. [DOI] [PubMed] [Google Scholar]
- Ohba, T., E.C. Schirmer, T. Nishimoto, and L. Gerace. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar, V.C., S. Abraham, S. Braune, A.A. Noegel, B. Tunggal, I. Karakesisoglou, and E. Korenbaum. 2004. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 295:330–339. [DOI] [PubMed] [Google Scholar]
- Padmakumar, V.C., T. Libotte, W. Lu, H. Zaim, S. Abraham, A.A. Noegel, J. Gotzmann, R. Foisner, and I. Karakesisoglou. 2005. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118:3419–3430. [DOI] [PubMed] [Google Scholar]
- Powell, L., and B. Burke. 1990. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the association of p55 with the nuclear lamina. J. Cell Biol. 111:2225–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raharjo, W.H., P. Enarson, T. Sullivan, C.L. Stewart, and B. Burke. 2001. Nuclear envelope defects associated with LMNA mutations causing dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. J. Cell Sci. 114:447–457. [DOI] [PubMed] [Google Scholar]
- Rober, R.-A., K. Weber, and M. Osborn. 1989. Differential timing of lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 105:365–378. [DOI] [PubMed] [Google Scholar]
- Schirmer, E.C., L. Florens, T. Guan, J.R. Yates III, and L. Gerace. 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 301:1380–1382. [DOI] [PubMed] [Google Scholar]
- Sinensky, M., K. Fantle, M. Trujillo, T. McLain, A. Kupfer, and M. Dalton. 1994. The processing pathway of prelamin A. J. Cell Sci. 107:61–67. [DOI] [PubMed] [Google Scholar]
- Soullam, B., and H.J. Worman. 1995. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 130:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D.A., and M. Han. 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 298:406–409. [DOI] [PubMed] [Google Scholar]
- Starr, D.A., and M. Han. 2003. ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 116:211–216. [DOI] [PubMed] [Google Scholar]
- Stewart, C., and B. Burke. 1987. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 51:383–392. [DOI] [PubMed] [Google Scholar]
- Sullivan, T., D. Escalante-Alcalde, H. Bhatt, M. Anver, N. Bhat, K. Nagashima, C.L. Stewart, and B. Burke. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147:913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet, R.M., and D. Eisenberg. 1983. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J. Mol. Biol. 171:479–488. [DOI] [PubMed] [Google Scholar]
- Tusnady, G.E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics. 17:849–850. [DOI] [PubMed] [Google Scholar]
- Vorburger, K., G.T. Kitten, and E.A. Nigg. 1989. Modification of nuclear lamin proteins by a mevalonic acid derivative occurs in reticulocyte lysates and requires the cysteine residue of the C-terminal CXXM motif. EMBO J. 8:4007–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, K., U. Plessmann, and P. Traub. 1989. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 257:411–414. [DOI] [PubMed] [Google Scholar]
- Welte, M.A., S.P. Gross, M. Postner, S.M. Block, and E.F. Wieschaus. 1998. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 92:547–557. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., J.N. Skepper, F. Yang, J.D. Davies, L. Hegyi, R.G. Roberts, P.L. Weissberg, J.A. Ellis, and C.M. Shanahan. 2001. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114:4485–4498. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., C.D. Ragnauth, J.N. Skepper, N.F. Worth, D.T. Warren, R.G. Roberts, P.L. Weissberg, J.A. Ellis, and C.M. Shanahan. 2005. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 118:673–687. [DOI] [PubMed] [Google Scholar]
- Zhen, Y.Y., T. Libotte, M. Munck, A.A. Noegel, and E. Korenbaum. 2002. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115:3207–3222. [DOI] [PubMed] [Google Scholar]