Abstract
The organization of genes within the nucleus may influence transcription. We have analyzed the nuclear positioning of the coordinately regulated α- and β-globin genes and show that the gene-dense chromatin surrounding the human α-globin genes is frequently decondensed, independent of transcription. Against this background, we show the frequent juxtaposition of active α- and β-globin genes and of homologous α-globin loci that occurs at nuclear speckles and correlates with transcription. However, we did not see increased colocalization of signals, which would be expected with direct physical interaction. The same degree of proximity does not occur between human β-globin genes or between murine globin genes, which are more constrained to their chromosome territories. Our findings suggest that the distribution of globin genes within erythroblast nuclei is the result of a self-organizing process, involving transcriptional status, diffusional ability of chromatin, and physical interactions with nuclear proteins, rather than a directed form of higher-order control.
Introduction
Regulated gene expression is an essential feature of tissue-specific differentiation involving transcription factor binding and an interplay of chromatin modifications. It has been proposed that the position of genes within the nucleus may be another important facet of the regulation of differential gene expression, where certain areas of the nucleus are repressive and others support or enhance transcription (for reviews see Kosak and Groudine, 2004; Misteli, 2004). There are also compelling arguments against any requirement for genes to be at a specific location within the nucleus for transcription to proceed. Phosphorylated RNA polymerase II and global nascent RNA can be visualized throughout the entire volume of the nucleus, with no evidence that the periphery, apart from the nuclear rim, is a repressive environment to transcription or that the interior of the nucleus is enriched in transcription sites (Iborra, 2002; Sadoni and Zink, 2004). The whole nucleoplasm appears to be accessible to protein complexes (Misteli, 2001; Verschure et al., 2003), and even mitotic chromatin has been shown to be accessible to transcription factors and chromatin proteins (Chen et al., 2005).
To address whether nuclear organization has a functional role, we have chosen to follow the behavior of the coregulated α- and β-globin genes in primary human erythroblasts during terminal erythroid differentiation (Wickramasinghe, 1975). Transcription from these genes is regulated in a tissue-specific manner, so that balanced amounts of protein from both genes are produced over the course of several days, as erythroblasts undergo differentiation. This balance is achieved despite the fact that the α- and β-globin genes lie on separate chromosomes and in very different chromatin contexts. The α-globin genes are subtelomeric on human chromosome (HSA)16 in a gene-rich, GC-rich domain of constitutively open chromatin (Vyas et al., 1992) that was early replicating in all of the cell types examined (Smith and Higgs, 1999). In contrast, the β-globin genes are located on HSA11, in an AT-rich region of late-replicating, tissue-restricted genes, and become early replicating and sensitive to DNase1 digestion only in erythroblasts (Epner et al., 1988; Higgs et al., 1998; Reik et al., 1998). The β-globin genes associate with pericentromeric heterochromatin in cycling lymphocytes, where they are not expressed, but sit apart from heterochromatin in proerythroblasts, whereas the α-globin genes never associate with heterochromatin, irrespective of cell type or transcriptional status (K.E. Brown et al., 2001). Furthermore, the two genes occupy different regions of the nucleus; HSA16 is most frequently located in the middle and inner zone in lymphoblastoid cells, whereas HSA11 is more commonly found at the nuclear periphery (Boyle et al., 2001). The α-globin genes are also located outside their chromosome territory in lymphoblasts far more frequently than the β-globin genes (Mahy et al., 2002). The α- and β-globin genes, therefore, represent very different characteristics when silent. If positional or organizational changes are necessary for the regulated and balanced expression of these genes, then they should be apparent during the course of erythroid differentiation, when the globin genes become highly expressed.
In this study, we have monitored the changes in positioning and expression of the α- and β-globin genes at successive stages of terminal erythroid differentiation. We have characterized precisely when the globin genes are transcribed during differentiation and compared these findings with their positions. When actively transcribing, the human α- and β-globin genes are frequently located very close together, but not colocalized, at splicing factor–enriched nuclear speckles. This proximity of globin genes is not observed to the same degree in the mouse.
Results
Sorting enriched populations of human erythroblasts at distinct stages of differentiation
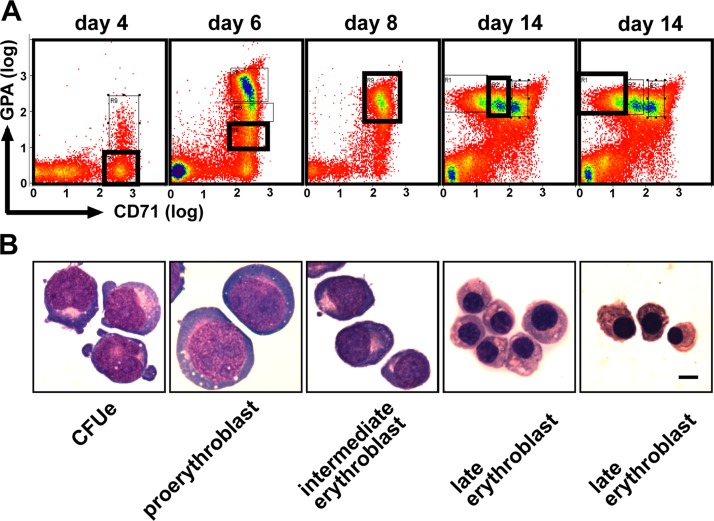
We wanted to characterize organization within the nuclei of primary differentiating erythroblasts, which fully recapitulate terminal erythroid differentiation, rather than using long-established and karyotypically abnormal cell lines. To obtain reproducible populations of erythroblasts, we induced erythroid differentiation in primary mononuclear cells and then sorted erythroblasts at different stages of differentiation based on cell surface markers. Mononuclear cells from buffy coat were cultured through a two-phase liquid culture system (Fibach et al., 1989) that induces circulating erythroid burst-forming unit cells to expand and terminally differentiate to erythrocytes. Cells were harvested for sorting at 4, 6, 8, and 14 d after the addition of erythropoietin in the second phase of culture. Cells were sorted for CD36 (glycoprotein IV receptor; days 4 and 6 only), CD71 (transferrin receptor), and glycophorin A (GPA).
The appropriate gates were selected to provide populations of early erythroid colony-forming unit (CFUe)–enriched day 4, proerythroblast day 6, intermediate day 8, and late day 14 stages of differentiation (Fig. 1 A ). The gates chosen were based on those described by Socolovsky et al. (2001) for mouse erythroblasts. The morphology of cells within each population showed restricted variation and were greatly enriched and distinct compared with unsorted cells (Fig. 1 B). CFUes and proerythroblasts were the only fractions that were difficult to distinguish based on morphology alone. Gated fractions from days 4 and 6 were plated out for culture of CFUes. This indicated that the human CFUe day 4 population is as enriched for CFUes (86%; unpublished data) as we are able to prepare from mouse erythroblasts (>80%; Anguita et al., 2004).
Figure 1.
Cell sorting for human primary erythroblasts at defined stages of differentiation. (A) Flow sorting profiles of erythroblast cultures at successive days after the addition of erythropoietin, as indicated. Cells were sorted based on GPA and CD71 staining from the bold black rectangular gate in each profile. (B) Representative MGG-stained cells from each gated fraction. Bar, 5 μm.
Maximal expression of the globin genes occurs in intermediate erythroblasts
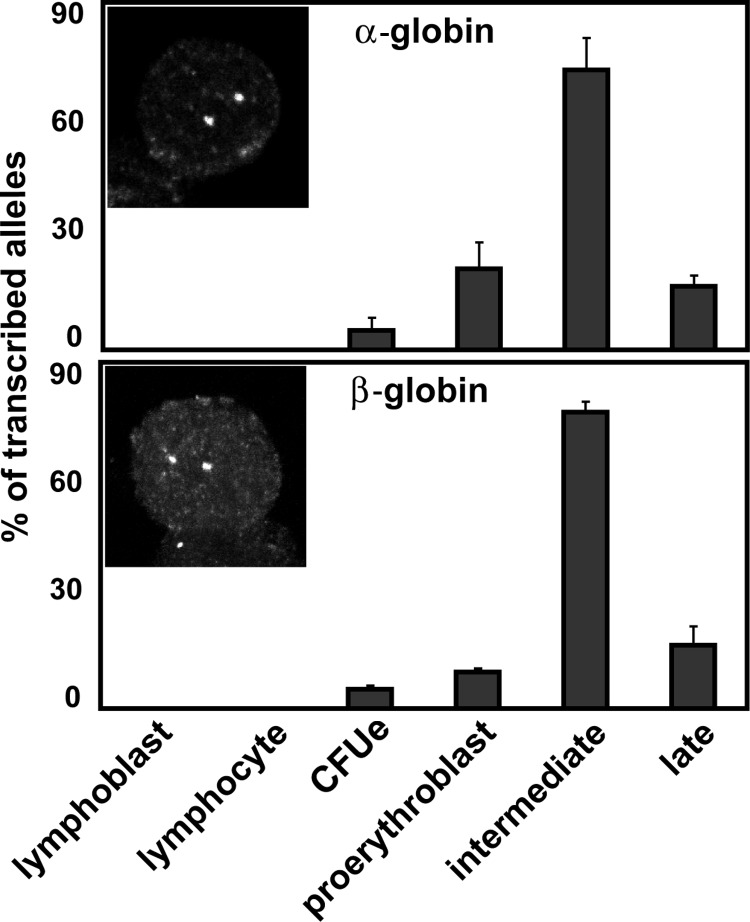
To evaluate nuclear organization in relation to transcription in these primary human cells, we needed to establish the pattern of transcription from the globin genes during the course of erythroblast differentiation. We analyzed the percentage of transcribed loci for α- and β-globin using RNA FISH in all successive fractions of differentiating erythroblasts (n = 187–372), together with lymphoblasts (n = 100) and activated T lymphocytes (n = 100; Fig. 2 ). Transcription from both α- and β-globin genes peaked in intermediate erythroblasts, with nascent transcript signals scored for 73% of α-globin loci and 77% of β-globin loci. The percentage of signals declined in subsequent fractions, to almost nothing in late erythroblasts. For both α- and β-globin, there are significant differences in the proportion of transcribing loci between intermediate erythroblasts and all other cell populations (P < 0.02). We also undertook three-dimensional (3D) DNA FISH on cells from the same cultures of erythroblasts. We were able to detect 91% of α-globin genes by DNA FISH, whereas we only detected nascent transcript at 3% of the possible sites in late erythroblasts. This suggests that probes and antibodies used in detection are not hindered in accessing sites within very condensed nuclei and, therefore, that our scores for the percentage of RNA FISH signals observed are an accurate reflection of the percentage of transcribing sites.
Figure 2.
Maximal expression of globin genes occurs in intermediate erythroblasts. Globin gene transcription in nonerythroid cells and sorted successive stages of differentiating erythroblasts. Error bars represent one standard deviation, calculated from two to five hybridizations. Representative nuclei show RNA FISH detection of nascent transcripts from the α- and β-globin genes.
We conclude that the α- and β-globin genes are both transcribed most highly in intermediate erythroblasts, rather than in proerythroblasts, where there is the most global transcription during erythroid differentiation (unpublished data). We also noted that 10% of all β-globin RNA signals sat at the nuclear periphery, indicating that the periphery is not a repressive environment for the β-globin genes. These data provided us with a clear framework to examine any changes in position or association of the globin genes in relation to transcriptional status.
The position of the globin genes in relation to their chromosome territories does not correlate with transcriptional status
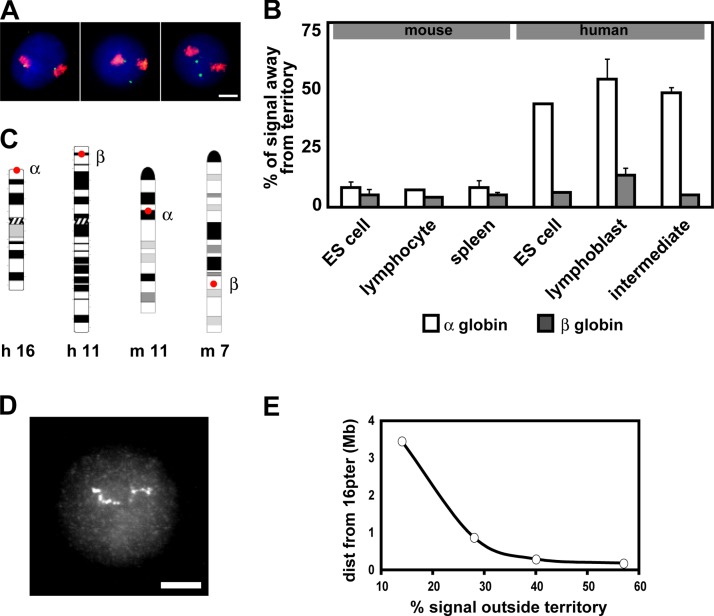
The human α- and β-globin gene loci are located in very different chromosomal contexts (Higgs et al., 1998), and we already know that they exhibit different positions in respect to their chromosome territories when silent (Mahy et al., 2002). We examined the location of the human globin genes in relation to their chromosome territories in human primary intermediate erythroblasts, where the globin genes are maximally expressed, and in human lymphoblasts and embryonic stem (ES) cells, where the globin genes are transcriptionally silent. We hybridized probes for the α- and β-globin genes, together with whole chromosome paints for HSA16 and HSA11, respectively, and scored the proportion of FISH signals sitting away from their territory (Fig. 3, A and B ). The cells analyzed were fixed in methanol-acetic acid rather than in PFA, as this compares with previous studies involving other loci (Volpi et al., 2000; Mahy et al., 2002). We found that 52% (n = 500) of human α-globin genes sit away from their HSA16 territory in lymphoblasts, which concurs with an earlier study (Mahy et al., 2002). We found a similar frequency in ES cells (42%; n = 100) and also in intermediate erythroblasts where globin genes are active (45%; n = 200). The human β-globin genes did not localize away from their HSA11 territories with high frequency in any of the cell types examined (5 to 13%; n = 100–500). Therefore, although the human α- and β-globin genes show significantly different positioning in respect to their chromosome territories (P < 0.02), we did not find any evidence that this correlates with their transcriptional status.
Figure 3.
Human α-globin genes extend out of the chromosome territory irrespective of transcriptional status. (A and B) DNA FISH signals for α- and β-globin genes were scored for position in relation to their chromosome territory, delineated by whole chromosome FISH paint in erythroid and nonerythroid cell types. (A) Three representative lymphoblast nuclei are depicted with both α-globin gene signals sitting at or in the HSA16 territory, one sitting at and one located away from the territory, and both located away from their territories, respectively. Bar,5 μm. (B) The percentage of α-globin (white bars) and β-globin (gray bars) gene FISH signals located away from their chromosome territory in different mouse and human cell types. Error bars represent one standard deviation, calculated from two to five hybridizations. (C) Ideograms of HSA16, HSA11, MMU11, and MMU7, showing the locations of the α- and β-globin genes. (D) DNA FISH signals for a pool of 24 cosmids, from a 2-Mb contig within 16p13.3, which was hybridized to a lymphoblast nucleus. The contig can be seen extending across half the diameter of the nucleus. Bar, 5 μm. (E) Positioning of DNA FISH signals for four cosmids from 16p13.3, in respect to HSA16 territory. Cosmids are respectively centered within the linear genome at 3.5 Mb and 874, 292, and 181 kb away from the HSA16 short-arm telomere.
In addition, we know that the chromosomal contexts of the human and mouse α-globin genes are very differen The human α-globin genes are located very close (162 kb) to the HSA16 short-arm telomere, whereas in the mouse, the α-globin genes sit close to the HSA11 centromere in a region of lower gene density (Fig. 3 C). Therefore, we also looked at the location of the murine globin genes in respect to the main body of their chromosomes in anemic mouse spleen, where the globin genes are active in the majority of cells, and in murine ES cells and primary activated lymphocytes, where they are silent (Fig. 3 B). None of the three cell types, ES cells (n = 350), lymphocytes (n = 100), and spleen (n = 150), had a high frequency of α-globin sitting away from the murine chromosome (MMU)11 territory (8, 7, and 8%, respectively). In addition, the mouse β-globin genes did not often localize away from their MMU7 territories (4–5%; n = 100–350). As in human nuclei, we found no correlation of transcriptional status with the positions of the murine globin genes, in respect to their chromosome territories. We did find that the α-globin genes rarely localize away from their chromosome territory in the mouse, as opposed to human cells, where they are frequently found outside their territory (P < 0.01 for human intermediates, compared with mouse spleen). In contrast, the β-globin genes, which are not subtelomeric in human or mouse (Fig. 3 C), rarely localized outside their territories in any cell type.
The position of the human α-globin genes very close to the HSA16 telomere may mean that they are less frequently packaged into the main body of the chromosome. We hybridized a pool of 24 cosmids from a 2-Mb contig around the human α-globin genes and found that this region can extend to 3.5 μm, which can be half the diameter of a lymphoblast nucleus (Fig. 3 D).We hybridized four selected cosmids from the contig, together with an HSA16 paint, to lymphoblast nuclei (Fig. 3 E) and found that probes for sites 874, 292, and 181 kb away from the telomere showed progressively more frequent positions outside the HSA16 territory (28, 40, and 57%, respectively; n = 400 for each). This suggests that the human α-globin genes are not on a loop of chromatin protruding out of the territory, but rather are on a terminal piece of chromatin that frequently extends away from the body of the chromosome.
α-Globin genes frequently associate with each other and with β-globin genes in transcribing cells
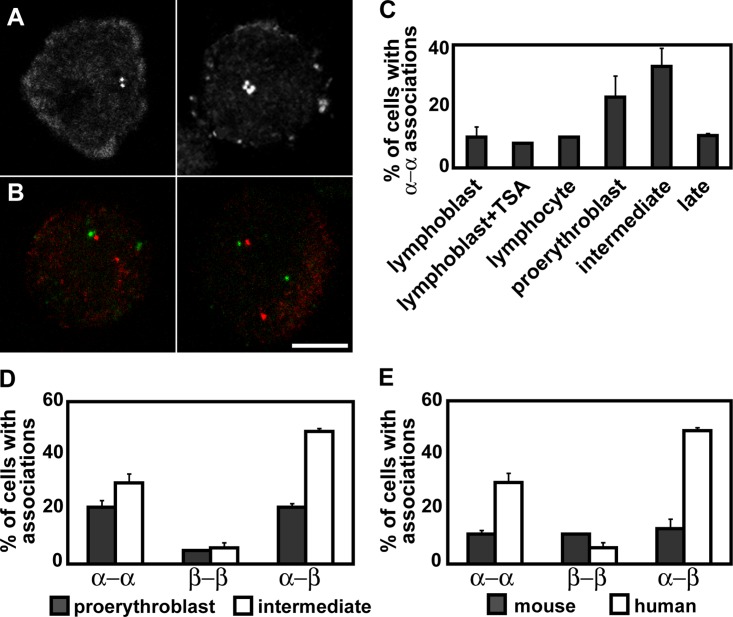
We next examined the positioning of homologous and cotranscribed α- and β-globin genes relative to each other, during terminal erythroid differentiation. All cells were scored for whether signals were separate, physically juxtaposed, or actually colocalized with overlapping fluorescent signals. Measurements between signals scored as juxtaposed did not exceed 1 μm, with a mean distance of 0.48 μm and a standard deviation of 0.19 μm. Signals separated by >1 μm were not scored as juxtaposed. There was an average incidence of 3% colocalization of homologous α-globin signals and of α- and β-globin signals, whereas homologous β-globin FISH signals were hardly ever colocalized. A degree of random colocalization can occur between probes in interphase nuclei (Dewald et al., 1993), and there were no significant differences in the degree of colocalization in different cell types or in line with transcription. Therefore, all percentages given in this section represent a pool of juxtaposed and colocalized scores. We have described juxtaposed and colocalized signals as “associating”—this term does not imply a physical interaction, merely a physical proximity.
First, we looked at association between the two homologous signals of the α- and β-globin genes. In nonexpressing lymphoblasts (n = 500) and lymphocytes (n = 100), the association between globin homologues in 3D FISH preparations was a little higher for α–α associations, at 10% of nuclei scored, than for β− associations, at 3 and 4%, respectively. In erythroblasts (n = 200–700), the level of β−
associations, at 3 and 4%, respectively. In erythroblasts (n = 200–700), the level of β− associations remained unchanged, but α–α associations increased and peaked in intermediate erythroblasts at 33% (n = 700; Fig. 4, A and C
). The level of α–α associations in intermediate erythroblasts was significantly different (P < 0.005) than those scored in lymphoid and other erythroid cell populations. We also treated lymphoblasts with trichostatin A (TSA), which inhibits deacetylation and induces transcription from the α-globin genes (>10-fold increase; Garrick, D., personal communication). There was no change in the frequency of α–α associations in treated cells (8%; n = 100; Fig. 4 C), suggesting that such association is not an essential requirement for transcription to proceed. The increased association cannot be caused by changes in nuclear volume during differentiation for two reasons. First, intermediate erythroblasts have a nuclear volume similar to that of lymphoblasts (unpublished data) but very different levels of association of the α-globin genes. Second, the association levels of the β-globin genes do not change significantly in the different cell types scored.
associations remained unchanged, but α–α associations increased and peaked in intermediate erythroblasts at 33% (n = 700; Fig. 4, A and C
). The level of α–α associations in intermediate erythroblasts was significantly different (P < 0.005) than those scored in lymphoid and other erythroid cell populations. We also treated lymphoblasts with trichostatin A (TSA), which inhibits deacetylation and induces transcription from the α-globin genes (>10-fold increase; Garrick, D., personal communication). There was no change in the frequency of α–α associations in treated cells (8%; n = 100; Fig. 4 C), suggesting that such association is not an essential requirement for transcription to proceed. The increased association cannot be caused by changes in nuclear volume during differentiation for two reasons. First, intermediate erythroblasts have a nuclear volume similar to that of lymphoblasts (unpublished data) but very different levels of association of the α-globin genes. Second, the association levels of the β-globin genes do not change significantly in the different cell types scored.
Figure 4.
Interphase association of α- and β-globin genes in primary intermediate erythroblast nuclei. (A) DNA FISH showing association of homologous α-globin genes. (right) Replicated genes. (B) RNA FISH showing α-globin (red) and β-globin (green) gene transcripts associating. Bar, 5 μm. (C) Proportion of cells where the homologous α-globin DNA FISH signals are associating in human lymphoid and erythroid cell types. Lymphoblasts were untreated or treated with TSA. (D) Proportion of cells with RNA FISH signals for all four globin gene transcripts, where the homologous α-globin signals, homologous β-globin signals, or α- and β-globin signals are associating, in human proerythroblast and intermediate erythroblast nuclei. (E) Proportion of cells with RNA FISH signals as in D, in mouse and human intermediate erythroblast nuclei. Error bars represent one standard deviation, calculated from two to fourteen hybridizations.
We also looked at the association of the cotranscribed α- and β-globin genes. We looked specifically at RNA FISH preparations, where we knew that all signals represented a gene in the process of transcription, and we used unsorted cells for this analysis, to minimize any potential disruption to nuclear organization. 7 d after the addition of erythropoietin, an erythroid culture contains both proerythroblasts and intermediate erythroblasts. These two stages of differentiation can be distinguished visually on the basis of size and the amount of cytoplasmic RNA FISH signal because intermediate erythroblasts accumulate globin mRNA in the cytoplasm, which can be detected with probes covering exons of the α- or β-globin genes. We scored the percentage of association of homologous loci in cells with two clear α- or β-globin transcript signals and the percentage of α- and β-globin association in cells with all four signals present. The α–α associations rose from 20.5% (n = 400) in proerythroblasts to 30% (n = 600) in intermediate erythroblasts, whereas β−β associations hardly changed, at 5% (n = 300) and 6% (n = 400), respectively. We found that α- and β-globin genes were associating in 21% (n = 100) of proerythroblast nuclei, where all four genes were active, increasing to 49% (n = 250) in intermediate erythroblasts (Fig. 4, B and D), where a few nuclei were also observed with three (two α and one β) globin loci associating. The increase in the number of associating globin loci mirrors the increase in the percentage of transcribing loci.
The fact that the degree of association was greater in intermediate erythroblasts than in proerythroblasts in cells where all four globin loci have a nascent transcript signal (Fig. 4 D;P < 0.0025 for both α−α and α−β associations) implies that association is more likely to occur when globin transcription is most frequent, as we have demonstrated for the intermediate stage of erythroid differentiation.
To confirm that this association was specific to the globin genes, we asked whether α-globin would associate with other chromosome regions. We chose the subtelomeric probe for HSA2 short arm to cohybridize, via DNA FISH, with α-globin to lymphoblasts and intermediate erythroblasts because this would occupy a more peripheral location in the nucleus, like the β-globin genes. We found no difference in association of α-globin and the 2p subtelomere in the two cell types (13%; n = 100 for each).
We went on to look at the association between globin genes in the mouse, given the different chromatin context of the murine α-globin genes, and found a very different picture than in human cells. By analysis of RNA FISH signals in mouse intermediate erythroblasts, we found that the percentage of α−α associations was 11% (n = 200) and of β−β associations was also 11% (n = 200). Association between α- and β-globin genes was equal to 13% of cells with all four globin loci transcribing (n = 244). Therefore, the high degree of association we have identified between active globin genes in human erythroblasts does not occur in the mouse (Fig. 4 E; P < 0.05, comparing human and mouse α−α and α−β associations). There were significantly more β−β associations scored in mouse than in human erythroid cells (P < 0.02).
Globin genes contact nuclear speckles when transcribing
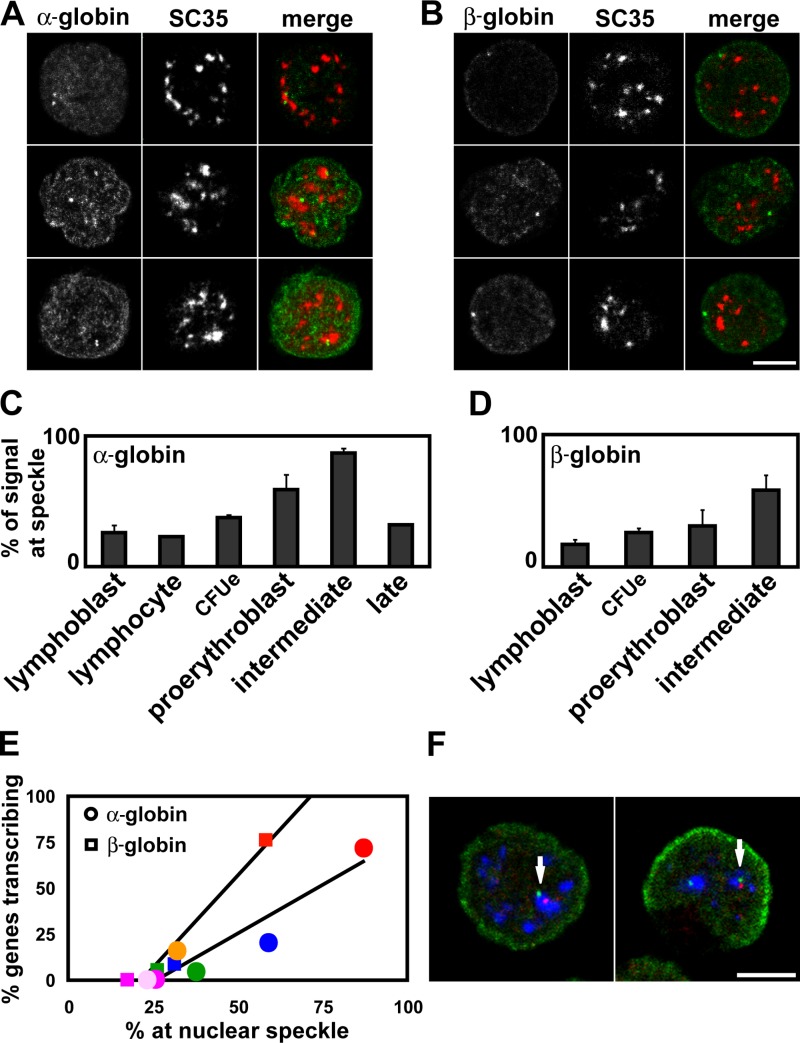
After identifying the association between globin gene loci, we explored whether this was occurring within any specific nuclear subcompartment. Proteins recognizing different nuclear bodies and globin gene DNA were detected by immuno-FISH to different cell types, and the number of gene signals directly contacting or sitting away from the nuclear body were recorded (n = 100–300 cells for each hybridization). We found no indication that the globin genes were associating in erythroid cells at PML bodies, detected with PML antibody, or at nucleoli, detected with upstream binding factor antibody or a nucleolar-organizer region DNA probe (unpublished data). We did find that the globin genes had increased contact with SC35-positive nuclear speckles (Spector, 1993). The frequency of contact between α-globin genes and speckles was 26% in lymphoblasts and 23% in lymphocytes. This level of contact may be caused, in part, by the transcriptional activity of housekeeping genes in the region surrounding the α-globin genes. It was higher in early erythroblasts, and peaked in the intermediate erythroblast population at 87% before falling again in the late erythroblast population (Fig. 5, A and C ). There was a significant difference between the proportion of α-globin loci contacting nuclear speckles in lymphoblasts and in erythroid intermediates (P < 0.02). We established that this was not attributable to differences in the size of the SC35 compartment by comparing the relative volume occupied by the SC35 signal in lymphoblast and intermediate nuclei (lymphoblasts, 0.064 ± 0.009; intermediates, 0.061 ± 0.008). Fewer β-globin than α-globin genes contacted SC35-positive nuclear speckles in lymphoblasts, but the percentage was greater in erythroid cells, peaking at 58% in intermediate erythroblasts (Fig. 5, B and D). We found that there was a good correlation between the percentage of genes transcribing in each erythroid population and the percentage of genes contacting nuclear speckles for both α-globin (R2 = 0.89) and β-globin (R2 = 0.95; Fig. 5 E). The number of genes located at speckles clearly increased as the amount of transcription from the globin genes increased, although the degree of this contact was more marked for the α-globin genes. We looked at the contact of actual transcripts with speckles by RNA immuno-FISH, which does not involve a denaturing step, so that the 3D architecture of the cell is well preserved. We found that 100% (n = 81) of α-globin transcripts were located at speckles in intermediate erythroblasts. However, this association with large nuclear speckles cannot be essential for transcription in general because only 67% (n = 52) of β-globin transcripts were located at speckles in the same experiments.
Figure 5.
Location of α- and β-globin genes in respect to SC35-enriched nuclear speckles. (A) Representative intermediate erythroblast nuclei for immuno-FISH detection of α-globin genes with SC35 protein. Both α-globin signals contacting the same speckle (top), the α-globin signals contacting different speckles (middle), and one replicated α-globin signal at a speckle (bottom). (B) Representative intermediate erythroblast nuclei for immuno-FISH detection of β-globin genes with SC35 protein. (top) One β-globin signal contacting a small speckle in the outer shell of the nucleus; (middle and bottom) the β-globin signals sitting away from any speckles. Bar, 5 μm. (C and D) Proportion of α-globin (C) and β-globin (D) FISH signals contacting a nuclear speckle in different cell types. Error bars represent one standard deviation, calculated from two to three hybridizations. (E) Proportion of globin genes transcribing (Fig. 2) plotted against their location at nuclear speckles. α-globin (circles) and β-globin (squares) FISH signals were scored in lymphoblasts (pink), lymphocytes (pale pink), CFUes (green), proerythroblasts (blue), intermediate eryth-roblasts (red), and late erythroblasts (orange). (F) Immuno-FISH showing α-globin (red) and β-globin (green) genes associating at the same nuclear speckle (arrow). Bar, 5 μm.
We have shown that the globin genes associate with nuclear speckles, and with each other, when transcribing. It seemed likely that these associations were related, thus, we undertook three-color immuno-FISH hybridizations, where we detected the α- and β-globin genes together with the SC35 splicing factor speckles. We were able to demonstrate that the association between α-globin genes, and between α- and β-globin genes, occurred at nuclear speckles in intermediate erythroblasts (100%; n = 43; Fig. 5 F); of these, 82% of associating α-globin genes and 78% of associating α- and β-globin genes were contacting the same nuclear speckle. Thus, the human globin genes exhibit a remarkable degree of association at the same speckle when transcribing.
Discussion
Tissue-specific gene expression is influenced by the availability of transcription factors and the epigenetic modulation of higher order chromatin structure. Chromatin at the globin gene loci is modified in committed erythroid progenitors, but the genes transcribe at high levels several cell cycles later, in intermediate erythroblasts. Our aim has been to understand what contribution the positioning of the globin genes within the nucleus may make to their transcriptional regulation during terminal erythroid differentiation. By comparing results in human and murine cells, we have shown that the positioning of the globin genes is not determined simply by their transcriptional status, but is also dependent on the underlying chromosomal context.
Globin genes and chromatin decondensation
Gene-dense arrays of chromatin have been reported to unfold out of the chromosome territory more frequently when genes within the arrays are being transcribed (Volpi et al., 2000;Williams et al., 2002; Mahy et al., 2002; Chambeyron and Bickmore, 2004a). We have not found this to be the case for either of the globin gene clusters because approximately half of α-globin genes sit outside the HSA16 territory, irrespective of their transcriptional status, whereas the β-globin genes sit most frequently within the HSA11 territory, both in cells where they are silent and in cells where they are highly expressed. This most likely reflects the chromatin environments where the α- and β-globin genes reside. The human α-globin genes are located in a very gene-dense region of open chromatin that is especially rich in housekeeping genes (Vyas et al., 1992; Daniels et al., 2001; Flint et al., 2001), and which might be expected to be physically decondensed even in the absence of globin expression (Gilbert et al., 2004). Indeed, this region has previously been shown in human lymphoblasts to localize the furthest away from a chromosome territory, out of 17 sites on four different chromosomes (Mahy et al., 2002). Furthermore, the α-globin genes lie <200 kb away from the telomere, and we have shown that this region extends in a linear fashion out of the chromosome territory. We see a different picture for α-globin in the mouse, where the genes are located toward the centromere of MMU11. In mouse ES cells, lymphocytes, and spleen, the α-globin region most frequently localizes within the territory, irrespective of transcriptional status. When we compare the two genomes, there are 102 genes in the 2 Mb surrounding the human α-globin locus, whereas in the mouse, although there is the same gene density in the immediate syntenic region (138 kb), beyond that there are only 21 genes in total across a 2-Mb region. We infer that the subtelomeric location, together with surrounding gene density, is a critical factor in determining the extension of the human α-globin genes away from the chromosome territory. This may not be a feature common to all subtelomeric genes because not all subtelomeres locate away from their territories in the same way (Mahy et al., 2002), and other subtelomeres do not have the same gene density (Daniels et al., 2001).
In contrast to α-globin, the β-globin genes are embedded in a region of tissue-restricted genes that has previously been shown to localize within the HSA11 territory in lymphoblasts (Mahy et al., 2002). We found that human β-globin is most frequently localized within the territory in lymphoid and ES cells and that this does not change with transcription in erythroblasts. A previous study found that β-globin did loop out of the chromosome before transcription (Ragoczy et al., 2003), although this involved the behavior of human and mouse genes in an inducible mouse erytholeukemia background, rather than in human primary erythroblasts. Certainly, there is evidence that individual genes do not always loop out from their territories when active (Scheuermann et al., 2004), and can even transcribe from within a domain of closed chromatin fibers (Gilbert et al., 2004). When we compare the number of genes in a 2-Mb region around both human globin genes, there is little difference in the actual gene density (α, 102; β, 77), only in the number of housekeeping genes. Therefore, in agreement with a previous study (Mahy et al., 2002), transcriptional activity across a region must also feature in the positioning of genes, in respect to their territories.
It is remarkable that the α- and β-globin genes behave so differently, despite the need for coordinated regulation, and that the α-globin genes behave so differently in human and mouse cells. Our findings are a specific example of a global relationship between chromatin fiber structure and nuclear organization (Gilbert et al., 2004) and suggest that extension of a gene away from the chromosome territory is dependent on a combination of properties of the surrounding chromatin, such as gene density, chromatin fiber structure, transcriptional activity across the region, and location on the chromosome, rather than a directed requirement to access an environment conducive to transcription.
Interchromosomal association of globin genes
The most striking finding of this study is that homologous α-globin genes, and α- and β-globin genes, frequently associate when transcribing. Some colocalization of transcribing α- and β-globin genes (7%) has also recently been documented in the mouse (Osborne et al., 2004). There are examples where homologous genes and cotranscribed genes show associations in the nucleus. The ribosomal RNA genes come together in nucleoli to create specialized functional environments, and a similar clustering of transfer RNA genes has been reported in yeast (Thompson et al., 2003). Recently, it has been recognized that eukaryotic genes that are coexpressed or that have products involved in the same protein complexes or metabolic pathway are often clustered in the same region of chromosome. It has been suggested that this clustering may facilitate expression by creating a microenvironment enriched in the appropriate factors or promoting association with nuclear structures such as SC35 domains (Hurst et al., 2004). Such a model has been extended to consider the possibility that homologous genes and other cotranscribed regions may participate in expression neighborhoods (Kosak and Groudine, 2004; Chambeyron and Bickmore, 2004b). This is precisely the situation we have identified for the α- and β-globin genes during erythroid differentiation; however, we have also been able to show that association between globin genes at nuclear speckles is not obligatory for transcription. Furthermore, the fact that associations between active globin genes increase from proerythroblasts to intermediate erythroblasts, as transcription becomes more frequent, would suggest that they are a consequence of transcription rather than a requirement for transcriptional regulation.
The phenomena of trans-homologue enhancer–promoter interactions and pairing-sensitive repression have been reported in Drosophila melanogaster (Bantignies et al., 2003; Ronshaugen and Levine, 2004). Could the associations we observe mean that the globin genes are physically interacting? We did not see a high frequency of signals actually overlapping, and the distances separating associating signals (mean 480 nm) did not suggest to us that the genes are sharing transcription factories, which are only 50 nm in diameter, as detected by bromouridine triphosphate incorporation (Iborra, 2002). Two recent papers also report associations between different genes. Osborne et al. (2004) found via FISH that transcribing genes on the same chromosome could colocalize in mouse nuclei at foci of RNA pol II. However, the distances between these signals are in the same range (mean, ∼900 nm) as the associations between globin genes we describe and greater than transcription factories. It is possible that both observations result from the same processes in the nucleus. Another paper (Spilianakis et al., 2005) reports that alternatively expressed cytokine genes on two different chromosomes colocalize before transcription from either locus and suggest that this may be a general phenomenon where coordinate or alternative regulation is important. We have carefully followed transcription from the globin genes at successive stages of differentiation, and we have found no evidence for increased association between globin genes before transcription.
The α-globin genes associate with each other much more frequently than do the β-globin genes. This is most likely a reflection of a much greater freedom of movement for the α-globin genes—they are frequently extended out of the chromosome territory and must be able to diffuse through a larger subvolume of the nucleus than the β-globin genes. Importantly, when we looked at the positioning of active globin genes in the mouse, we did not find the same levels of association as in human cells, just as the α-globin genes are more constrained to their chromosome territory in the mouse. Therefore, association between active globin genes may be a result of transcription, but the degree to which this occurs is dependent on the chromatin context in which the genes are embedded.
Association of globin genes with the splicing factor compartment
There have been several reports of genes associating with different nuclear compartments when active and inactive (Brown et al., 1997; Jolly et al., 1999; Francastel et al., 2001; Kosak et al., 2002; Nielsen et al., 2002; Shopland et al., 2003; Moen et al., 2004; Wang et al., 2004; Zink et al., 2004), and this has led to the proposition that the position of a gene or gene cluster within the nucleus may serve to enhance or suppress transcription (Verschure et al., 2003; Misteli, 2004). We have examined the nuclear locations of the highly expressed globin genes during terminal erythroid differentiation and have identified an association between α- and β-globin genes and aggregations of splicing factors in nuclear speckles. This association is erythroid-specific and correlates with changes in gene expression during erythroid differentiation.
Pre-mRNA splicing factors are distributed diffusely throughout the nucleoplasm, but are also enriched by 5–10-fold in 20–50 nuclear speckles (Spector, 1993; Lamond and Spector, 2003). Several papers record the association of active genes and gene regions with nuclear speckles (Nielsen et al., 2002; Shopland et al., 2003; Moen et al., 2004), although not all genes behave this way. Speckles re-form after each cell division (Prasanth et al., 2003) in what is currently thought to be a macro-molecule-driven self-assembly (Hancock, 2004) caused by protein–protein and protein–RNA interactions, where speckles may nucleate randomly, or at sites that have already begun transcribing (Lamond and Spector, 2003). Despite the fact that the α- and β-globin genes are both highly expressed in the same cells, we find that the α-globin genes contact the speckles more frequently than β-globin genes. It is possible that only transient contact is required; however, our observations may be a result of the relative positions and diffusional ability of the globin genes within the nucleus. Speckles nucleate in the interchromatin space and occur less frequently around the nuclear periphery. Our studies have shown that active β-globin genes are frequently found in the peripheral zone of the nucleus and are also more constrained to the chromosome territory than the α-globin genes. Crucially, for our understanding of this association, we show that about one-third of transcribing β-globin genes do not contact the speckles. These genes must be using diffuse splicing factors for their prespliceosomal complexes. Therefore, the tight association between transcribing genes and nuclear speckles, which is virtually 100% for the α-globin genes, cannot be generally obligatory for transcription or RNA processing, despite a close correlation with levels of transcription. In support of this assumption, it has been shown that the disassembly of the interchromatin granule compartment does not abolish transcription itself (Sacco-Bubulya and Spector, 2002).
Speckles are positionally stable within the nucleus, and it has been suggested that they act as “road blocks” to diffusing genes (Parada and Misteli, 2002). However, this would not explain the frequent association of globin genes at the same speckle. The globin genes are expressed very frequently in intermediate erythroblasts, much more so than housekeeping genes (Lockhart and Winzeler, 2000; unpublished data). The nucleation of large concentrations of splicing factors into speckles at sites of transcription may actually pull transcribing genes, particularly more mobile genes such as α-globin, into an association. There is the possibility that there may be subsets of speckles with different constituents; however, other coregulated genes that have been examined do not appear to associate at speckles (Nielsen et al., 2002; Shopland et al., 2003). This may reflect a variety of factors: their frequency of transcription, their positioning within the nucleus and on the chromosome, and gene density, gene activity, and modifications of the surrounding chromatin. This does not exclude the possibility that association of cotranscribed genes at speckles may indeed enhance the levels of ongoing transcription by enrichment in the local environment of essential factors as they dynamically interact with the chromatin. There may even be an increased potential for interchromosomal interactions between enhancers and promoters. Currently, we have little idea of how promiscuous transcriptional activation may be.
In summary, the associations we have identified appear to be directly related to transcription from the globin genes. Yet, we have found no evidence that they are essential for transcription to proceed because we show that globin genes can transcribe when free of any association with other globin genes or nuclear speckles and that these associations are less frequent in the mouse. By comparing the behavior of the coregulated α- and β-globin genes, we suggest that their frequent associations with each other at nuclear speckles are a result of a combination of factors present in the nucleus, involving chromatin diffusion, transcriptional status, and the formation of new speckles. Current thinking suggests that nuclear location and association of genes may be playing an important role in the regulation of gene expression. This study highlights the fact that considerable care is required in the interpretation of such data, which may be influenced by aspects of genome structure and chromatin environment that play little or no role in the regulation of gene expression.
Materials and methods
Cell culture
Primary human differentiating erythroblasts were obtained using a two-phase liquid culture system, as previously described (Fibach et al., 1989). In brief, mononuclear cells were obtained from peripheral blood buffy coat by centrifugation on a gradient of Ficoll-Hypaque and seeded at 2.5 × 106 cells/ml in α-minimal essential medium with 10% fetal calf serum, 1 μg/ml cyclosporin A, and 10% conditioned medium from cultures of the 5637 bladder carcinoma cell line for growth factors. The cells were cultured at 37°C for 6–7 d (phase I), and the nonadherent cells were washed and reseeded in fresh medium with 30% fetal calf serum, 1% deionized BSA, 1 U/ml of human recombinant erythropoietin, 10-5 M β mercaptoethanol, 10-6 mol/l dexamethasone, 0.3 mg/ml holotransferrin, and 10 ng/ml of human stem cell factor. Cells were cultured (phase II) for a further 4 (CFUe), 6 (proerythroblast), 8 (intermediate erythroblast), or 14 d (late erythroblast). Progression of the cells through terminal erythroid differentiation was monitored by May-Grunwald Giemsa (MGG)–stained cytospins. Concerns that the sorting process would disrupt organization within nuclei were addressed by returning aliquots of sorted cells to culture, where their ability to proceed to terminal erythroid differentiation in a normal manner was confirmed. Lymphoblastoid cell lines were established from normal individuals and cultured in RPMI with 10% fetal calf serum. Early passages were used. TSA (Sigma-Aldrich), when used, was added at 1 μM for the last 8 h of culture. Human activated T lymphocytes were obtained from whole blood by culture for 3 d in RPMI/10% fetal calf serum in the presence of 2% phytohemagglutinin M (GIBCO BRL) and 20 U/ml interleukin-2. Mouse-activated B lymphocytes were obtained from a suspension of spleen cells cultured in the same manner as mononuclear cells, but with 50 ng/ml lipopolysaccharide as mitogen. ES cells were cultured as described (Smith, 1991; Thomson et al., 1998) and harvested by standard cytogenetic techniques for two-dimensional FISH. ES cells were provided by A. Smith (mouse; The Institute for Stem Cell Research, The University of Edinburgh, Edinburgh, Scotland) and P. Andrews (human; University of Sheffield, Sheffield, England). Suspensions of mouse intermediate erythroblasts were obtained from adult mouse spleen treated with 1-acetyl-2-phenylhydrazine for 4 and 5 d (Spivak et al., 1973), under license from the Oxford University Local Ethical Review Process.
Flow cytometry
Cells were sorted on a fluorescence-activated cell sorter (MoFlo; DakoCytomation) to purify populations of cells at specific stages of differentiation, as described previously (Socolovsky et al., 2001), but based on human cell surface markers CD36, CD71, and GPA. The antibodies used were anti–CD36-PE (BD Biosciences; days 4 and 6), anti–CD71-FITC (DakoCytomation), anti–GPA-APC (BD Biosciences; days 4 and 6), and anti–GPA-RPE (DakoCytomation; days 8 and 14).
Probes
Probes used for RNA FISH were either pools of hapten-labeled oligonucleotides (Eurogentec) designed from within the human and mouse α- or β-globin gene introns (sequences provided by P. Fraser, The Babraham Institute, Cambridge, England), nick-translated plasmids covering the human α-globin gene pRA.03 (a cloned 1.3-kb PstI fragment), or the human β-globin gene pN1β7 (a cloned 1.8-kb XbaI fragment). The globin DNA FISH probes used were cosmid HSGG1 (Daniels et al., 2001) or BAC RP11-344L6 (AC007604) for human α-globin; cosmid 202 (a gift from B. Morley, Imperial College School of Medicine, London, England) or BAC pBeloBac11 (Kaufman et al., 1999) for human β-globin; BAC 14567 (Incyte) for mouse α-globin; and BAC 357K20 (Research Genetics) for mouse β-globin. Additional human FISH probes used were a subtelomeric clone for HSA2p (J. Brown et al., 2001), cosmids 439A6, 419C1, and GG1 from a 2-MB contig of HSA16p13.3 (Daniels et al., 2001), cosmid 330H2 (a gift from N. Doggett, Los Alamos National Laboratory, Los Alamos, NM), BAC clone dJ1174A5 for NOR (courtesy of M. Rocchi [Universita' di Bari, Bari, Italy] and P. Finelli [University of Milan, Milan, Italy]), and whole chromosome paints for HSA11 and HSA16 (Cambio-Biosys).
Nick translation labeling of probes
2 μg of BAC, cosmid, or plasmid DNA were labeled, as previously described (Smith and Higgs, 1999).
RNA FISH
RNA FISH was performed essentially as described previously (Gribnau et al., 1998), but using 0.02% pepsin. DNP-labeled oligonucleotides were detected with rat anti-DNP (MONOSAN), donkey anti–rat Cy3, and goat anti–horse Cy3 (Jackson ImmunoResearch Laboratories); digoxigenin (DIG)-labeled probes were detected with antibody layers of sheep anti-DIG FITC (Roche) and rabbit anti–sheep FITC (Vector Laboratories); biotinylated probes were detected with Avidin-Cy3.5 (GE Healthcare). Cells were mounted in Vectashield (Vector Laboratories) with 1 μg/ml DAPI counterstain. For RNA FISH analysis of sorted cell populations, sorted fractions were put back into culture for 6 h to ensure reestablishment of transcription patterns. In test cultures, this time period proved to be an adequate interval for recovery of transcription, but short enough to avoid further differentiation.
Two-dimensional DNA FISH
Methanol-acetic acid–fixed cells were prepared for hybridization with 100 μg/ml RNase in 2× SSC at 37°C for 30 min, washed in 2×SSC, and dehydrated. They were then denatured in 70% formamide/2× SSC solution at 70°C for 2 min and dehydrated.
Cosmid and BAC probes labeled with DIG were combined with Cot-1 DNA and denatured in hybridization mix (50% [vol/vol] formamide, 10% [wt/vol] dextran sulfate, 1% [vol/vol], Tween 20, and 2× SSC, pH 7.0) at 75°C for 5 min, and then preannealed at 37°C for 20 min. Biotinylated whole human chromosome paints (Cambio, Ltd.) were prepared according to the manufacturer. For each slide, 100 ng of cosmid or BAC probe in 5 μl was combined with 10 μl denatured chromosome paint and applied to the slide. Slides were hybridized overnight at 42°C. After hybridization, slides were washed four times in 50% formamide/2× SSC at 45°C for 3 min, four times in 2× SSC at 45°C for 3 min, and four times in 0.1 × SSC at 60°C for 3 min. The slides were blocked in 3% BSA/4× SSC at RT for 30 min, after which DIG-labeled and biotinylated probes were detected and mounted as in the previous section.
3D DNA FISH
FISH was also performed on cells where the 3D structure was preserved by PFA fixation. Cells were washed and allowed to settle on poly-l-lysine–treated coverslips for 10 min. The coverslips were fixed in 4% PFA for 15 min and permeabilized in 0.2% Triton X-100 in PBS for 12 min, at RT. RNA was removed with 100 μg/ml RNase in 2×SSC at 37°C for 1 h, and cells were denatured in 3.5 N HCl for 10 min at RT and neutralized in ice-cold PBS. Probes with Cot-1 DNA were denatured in hybridization mix as the previous section at 95°C for 10 min, preannealed at 37°C for 20 min, and hybridized to the denatured cells at 42°C for 48 h. Cells were washed twice in 2× SSC at 37°C for 30 min, once in 1× SSC at RT for 30 min, and blocked in 3% BSA in 4× SSC at RT for 30 min. DIG-labeled probes were detected with antibody layers of sheep anti-DIG FITC (Roche), followed by rabbit anti-sheep FITC (Vector Laboratories), both at RT for 30 min. Biotin-labeled probes were detected with antibody layers of avidin Cy3.5 (GE Healthcare) and biotinylated anti-avidin (Vector Laboratories). Coverslips were washed between layers in SSCT (4× SSC with 0.05% Tween 20), rinsed in PBS, and mounted in Vectashield with 1 μg/ml DAPI or 200 nM TOPRO3 (Invitrogen) counterstain.
Immunofluorescence
Cells were washed and allowed to settle on poly-l-lysine–treated coverslips for 10 min. Cells were fixed in 2% PFA for 15 min and permeabilized in 0.2% Triton X-100 in PBS for 12 min at RT. Nonspecific sites were blocked using 5% normal goat serum/5% fetal calf serum in PBS at RT for 30 min. Antibodies were prepared in blocking solution at the following concentrations: rabbit anti-PML (sc-5621; Santa Cruz Biotechnologies, Inc.) 1:100; rabbit anti–upstream binding factor (serum 6'2; a gift from M.Valdivia, Universidad de Cádiz, Cádiz, Spain), 1:1,000; mouse anti-SC35 (Sigma-Aldrich) 1:500. The secondary antibodies used were goat anti–mouse Cy5 (Jackson ImmunoResearch Laboratories) and horse anti–mouse Texas red (Vector Laboratories). Coverslips were mounted in Vectashield with DAPI, as in the previous section.
Immuno-FISH
Immunofluorescence was first performed essentially as described in the previous section, with the exception that cells were fixed in 4% PFA and permeabilized in 1% Triton X-100. After immunofluorescence, cells were postfixed in 4% PFA for 15 min at RT, followed by RNA removal by RNase in 100 μg/ml 2× SSC (Sigma-Aldrich) at 37°C for 1 h. Cells were denatured in 3.5 N HCl at RT for 20 min and neutralized with three brief washes in ice-cold PBS. Probes were prepared as described in the 3D DNA FISH section. Hybridization was performed overnight at 37°C. Washes and detection of DNA probes were performed as described in the 3D DNA FISH section.
Immuno RNA FISH
Immuno RNA FISH was adapted from Chaumeil et al. (2004). Cells were fixed in 3.7% PFA in PBS at 37°C for 10 min, permeabilized in 0.5% Triton X-100 in PBS on ice for 6 min, and blocked in 1% BSA at RT for 15 min. Primary and secondary antibodies were prepared in blocking solution, as described in the Immunofluorescence section. After protein detection, the slides were postfixed in 3.7% PFA in PBS at RT for 10 min. Slides were washed in PBS, rinsed in 2× SSC, and hybridized with probes, as described in RNA FISH. Detection of probes is also as described in the RNA FISH section.
Image analysis
MGG-stained cytospins were imaged on a microscope (BX60; Olympus) with a Q Imaging camera and OpenLab (Improvision) software. The chromosome territory images were taken on a BX60 microscope with a PSI MacProbe v4.3 image analysis package (Applied Imaging) and a Sensys charge-coupled device camera (Photometrics). All other fluorescent preparations were analyzed with a confocal microscope (Radiance 2000; Bio-Rad Laboratories) system on a microscope (BX51; Olympus). All images were taken through a 100× objective (UplanF1; Olympus), numerical aperture 1.3, using Lasersharp software (Bio-Rad Laboratories). Z stack images were acquired at 0.2-μm intervals. Contrast-stretch and gamma adjustments were made using Photoshop (Adobe) only for display in figures. Distance measurements between associating FISH signals used LaserPix software (Bio-Rad Laboratories). Estimations of nuclear volume were made by applying the volume formula for an oblate ellipsoid to maximal radii obtained from confocal Z stacks of nuclei. Volume density of the speckles was calculated using stacks of images that cover the entire volume of the nuclei. In each plane, we measured the area covered by the speckles and the area of the nuclei. The volume density of the speckles was calculated as the sum of speckle areas divided by the sum of the nuclear areas for each nucleus. This was repeated for 10 lymphoblast and 10 intermediate erythroblast nuclei.
Acknowledgments
We thank Sue Butler, Jackie Sloane-Stanley, and Kathryn Robson for provision of cells and Jim Hughes for advice on oligonucleotide probes and bioinformatics analysis. We also thank Bernie Morley for a β-globin cosmid.
This work was supported by the Medical Research Council.
Abbreviations used in this paper: 3D, three dimensional; CFUe, erythroid colony-forming unit; DIG, digoxigenin; ES, embryonic stem; GPA, glycophorin A; MGG, May-Grunwald Giemsa; TSA, trichostatin A.
References
- Anguita, E., J. Hughes, C. Heyworth, G.A. Blobel, W.G. Wood, and D.R. Higgs. 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies, F., C. Grimaud, S. Lavrov, M. Gabut, and G. Cavalli. 2003. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 17:2406–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, S., S. Gilchrist, J. Bridger, N. Mahy, J. Ellis, and W. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. [DOI] [PubMed] [Google Scholar]
- Brown, J., K. Saracoglu, S. Uhrig, M. Speicher, R. Eils, and L. Kearney. 2001. Subtelomeric chromosome rearrangements are detected using an innovative 12-color FISH assay (M-TEL). Nat. Med. 7:497–501. [DOI] [PubMed] [Google Scholar]
- Brown, K., S. Guest, S. Smale, K. Hahm, M. Merkenschlager, and A. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854. [DOI] [PubMed] [Google Scholar]
- Brown, K.E., S. Amoils, J.M. Horn, V.J. Buckle, D.R. Higgs, M. Merkenschlager, and A.G. Fisher. 2001. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602–606. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., and W.A. Bickmore. 2004. a. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron, S., and W.A. Bickmore. 2004. b. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 16:256–262. [DOI] [PubMed] [Google Scholar]
- Chaumeil, J., I. Okamoto, and E. Heard. 2004. X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 376:405–419. [DOI] [PubMed] [Google Scholar]
- Chen, D., M. Dundr, C. Wang, A. Leung, A. Lamond, T. Misteli, and S. Huang. 2005. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, R., J. Peden, C. Lloyd, S. Horsley, K. Clark, C. Tufarelli, L. Kearney, V. Buckle, N. Doggett, J. Flint, and D. Higgs. 2001. Sequence, structure and pathology of the fully annotated terminal 2 Mb of the short arm of human chromosome 16. Hum. Mol. Genet. 10:339–352. [DOI] [PubMed] [Google Scholar]
- Dewald, G., C. Schad, E. Christensen, A. Tiede, A. Zinsmeister, J. Spurbeck, S. Thibodeau, and S. Jalal. 1993. The application of fluorescent in situ hybridization to detect Mbcr/abl fusion in variant Ph chromosomes in CML and ALL. Cancer Genet. Cytogenet. 71:7–14. [DOI] [PubMed] [Google Scholar]
- Epner, E., W. Forrester, and M. Groudine. 1988. Asynchronous DNA replication within the human beta-globin gene locus. Proc. Natl. Acad. Sci. USA. 85:8081–8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibach, E., D. Manor, A. Oppenheim, and E.A. Rachmilewitz. 1989. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood. 73:100–103. [PubMed] [Google Scholar]
- Flint, J., C. Tufarelli, J. Peden, K. Clark, R.J. Daniels, R. Hardison, W. Miller, S. Philipsen, K.C. Tan-Un, T. McMorrow, et al. 2001. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum. Mol. Genet. 10:371–382. [DOI] [PubMed] [Google Scholar]
- Francastel, C., W. Magis, and M. Groudine. 2001. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc. Natl. Acad. Sci. USA. 98:12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., S. Boyle, H. Fiegler, K. Woodfine, N.P. Carter, and W.A. Bickmore. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 118:555–566. [DOI] [PubMed] [Google Scholar]
- Gribnau, J., E. de Boer, T. Trimborn, M. Wijgerde, E. Milot, F. Grosveld, and P. Fraser. 1998. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 17:6020–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, R. 2004. Internal organisation of the nucleus: assembly of compartments by macromolecular crowding and the nuclear matrix model. Biol. Cell. 96:595–601. [DOI] [PubMed] [Google Scholar]
- Higgs, D., J. Sharpe, and W. Wood. 1998. Understanding alpha globin gene expression: a step towards effective gene therapy. Semin. Hematol. 35:93–104. [PubMed] [Google Scholar]
- Hurst, L., C. Pal, and M. Lercher. 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5:299–310. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J. 2002. The path that RNA takes from the nucleus to the cytoplasm: a trip with some surprises. Histochem. Cell Biol. 118:95–103. [DOI] [PubMed] [Google Scholar]
- Jolly, C., C. Vourc'h, M. Robert-Nicoud, and R.I. Morimoto. 1999. Intron-independent association of splicing factors with active genes. J. Cell Biol. 145:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, R.M., C.T. Pham, and T.J. Ley. 1999. Transgenic analysis of a 100-kb human beta-globin cluster-containing DNA fragment propagated as a bacterial artificial chromosome. Blood. 94:3178–3184. [PubMed] [Google Scholar]
- Kosak, S.T., and M. Groudine. 2004. Form follows function: the genomic organization of cellular differentiation. Genes Dev. 18:1371–1384. [DOI] [PubMed] [Google Scholar]
- Kosak, S.T., J.A. Skok, K.L. Medina, R. Riblet, M.M. Le Beau, A.G. Fisher, and H. Singh. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and D.L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605–612. [DOI] [PubMed] [Google Scholar]
- Lockhart, D., and E. Winzeler. 2000. Genomics, gene expression and DNA arrays. Nature. 405:827–836. [DOI] [PubMed] [Google Scholar]
- Mahy, N.L., P.E. Perry, and W.A. Bickmore. 2002. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847. [DOI] [PubMed] [Google Scholar]
- Misteli, T. 2004. Spatial positioning; a new dimension in genome function. Cell. 119:153–156. [DOI] [PubMed] [Google Scholar]
- Moen, P.T., Jr., C.V. Johnson, M. Byron, L.S. Shopland, I.L. de la Serna, A.N. Imbalzano, and J.B. Lawrence. 2004. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell. 15:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J.A., L.D. Hudson, and R.C. Armstrong. 2002. Nuclear organization in differentiating oligodendrocytes. J. Cell Sci. 115:4071–4079. [DOI] [PubMed] [Google Scholar]
- Osborne, C., L. Chakalova, K. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071. [DOI] [PubMed] [Google Scholar]
- Parada, L., and T. Misteli. 2002. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 12:425–432. [DOI] [PubMed] [Google Scholar]
- Prasanth, K., P. Sacco-Bubulya, S. Prasanth, and D. Spector. 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 14:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy, T., A. Telling, T. Sawado, M. Groudine, and S.T. Kosak. 2003. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 11:513–525. [DOI] [PubMed] [Google Scholar]
- Reik, A., A. Telling, G. Zitnik, D. Cimbora, E. Epner, and M. Groudine. 1998. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol. 18:5992–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen, M., and M. Levine. 2004. Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev. Cell. 7:925–932. [DOI] [PubMed] [Google Scholar]
- Sacco-Bubulya, P., and D.L. Spector. 2002. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156:425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni, N., and D. Zink. 2004. Nascent RNA synthesis in the context of chromatin architecture. Chromosome Res. 12:439–451. [DOI] [PubMed] [Google Scholar]
- Scheuermann, M.O., J. Tajbakhsh, A. Kurz, K. Saracoglu, R. Eils, and P. Lichter. 2004. Topology of genes and nontranscribed sequences in human interphase nuclei. Exp. Cell Res. 301:266–279. [DOI] [PubMed] [Google Scholar]
- Shopland, L.S., C.V. Johnson, M. Byron, J. McNeil, and J.B. Lawrence. 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. 1991. Culture and differentiation of embryonic stem cells. J. Tissue Cult. Methods. 13:89–94. [Google Scholar]
- Smith, Z.E., and D.R. Higgs. 1999. The pattern of replication at a human telomeric region (16p13.3): its relationship to chromosome structure and gene expression. Hum. Mol. Genet. 8:1373–1386. [DOI] [PubMed] [Google Scholar]
- Socolovsky, M., H. Nam, M. Fleming, V. Haase, C. Brugnara, and H. Lodish. 2001. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 98:3261–3273. [DOI] [PubMed] [Google Scholar]
- Spector, D. 1993. Nuclear organization of pre-mRNA processing. Curr. Opin. Cell Biol. 5:442–447. [DOI] [PubMed] [Google Scholar]
- Spilianakis, C., M. Lalioti, T. Town, G. Lee, and R. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature. 435:637–645. [DOI] [PubMed] [Google Scholar]
- Spivak, J., D. Toretti, and H. Dickerman. 1973. Effect of phenylhydrazine-induced hemolytic anemia on nuclear RNA polymerase activity of the mouse spleen. Blood. 42:257–266. [PubMed] [Google Scholar]
- Thompson, M., R.A. Haeusler, P.D. Good, and D.R. Engelke. 2003. Nucleolar clustering of dispersed tRNA genes. Science. 302:1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, J., J. Itskovitz-Eldor, S. Shapiro, M. Waknitz, J. Swiergiel, V. Marshall, and J. Jones. 1998. Embryonic stem cell lines derived from human blastocysts. Science. 282:1145–1147. [DOI] [PubMed] [Google Scholar]
- Verschure, P., I. van der Kraan, E. Manders, D. Hoogstraten, A. Houtsmuller, and R. van Driel. 2003. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 4:861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi, E.V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R.D. Campbell, M. Goldsworthy, S.H. Powis, J. Ragoussis, et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576. [DOI] [PubMed] [Google Scholar]
- Vyas, P., M. Vickers, D. Simmons, H. Ayyub, C. Craddock, and D. Higgs. 1992. Cis-acting sequences regulating expression of the human alpha-globin cluster lie within constitutively open chromatin. Cell. 69:781–793. [DOI] [PubMed] [Google Scholar]
- Wang, J., C. Shiels, P. Sasieni, P.J. Wu, S.A. Islam, P.S. Freemont, and D. Sheer. 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe, S. 1975. Erythropoiesis. In Human Bone Marrow. Blackwell Scientific Publications, Oxford. 162–232.
- Williams, R.R., S. Broad, D. Sheer, and J. Ragoussis. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163–175. [DOI] [PubMed] [Google Scholar]
- Zink, D., M. Amaral, A. Englmann, S. Lang, L. Clarke, C. Rudolph, F. Alt, K. Luther, C. Braz, N. Sadoni, et al. 2004. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 166:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]