Abstract
Regulated phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF2α) by the endoplasmic reticulum (ER) stress-activated protein kinase PERK modulates protein synthesis and couples the production of ER client proteins with the organelle's capacity to fold and process them. PERK activation by ER stress is known to involve transautophosphorylation, which decorates its unusually long kinase insert loop with multiple phosphoserine and phosphothreonine residues. We report that PERK activation and phosphorylation selectively enhance its affinity for the nonphosphorylated eIF2 complex. This switch correlates with a marked change to the protease sensitivity pattern, which is indicative of a major conformational change in the PERK kinase domain upon activation. Although it is dispensable for catalytic activity, PERK's kinase insert loop is required for substrate binding and for eIF2α phosphorylation in vivo. Our findings suggest a novel mechanism for eIF2 recruitment by activated PERK and for unidirectional substrate flow in the phosphorylation reaction.
Introduction
The early steps in biogenesis of most secreted and membrane-bound proteins take place in association with the ER (Baumann and Walz, 2001). A dedicated machinery of chaperones and enzymes located within the organelle's lumen and on its membrane accomplish this important task (Fewell et al., 2001). Homeostasis in the ER is maintained by several signal transduction pathways that couple the load of newly synthesized ER client proteins to the organelle's capacity to cope with that load. These signal transduction pathways are collectively referred to as the ER unfolded protein response (UPRer), and they are activated by ER stress, a departure from equilibrium between client proteins, and the organelle's capacity to cope with their load (Patil and Walter, 2001; Kaufman, 2002).
Three distinct ER-associated proteins are known to initiate signaling in the UPRer. Two of these, IRE1 and ATF6, are concerned exclusively with activating genes whose products enhance the secretory machinery's capacity to cope with client proteins (Mori, 2000; Patil and Walter, 2001; Kaufman, 2002). The third upstream component of the UPRer, PERK, is uniquely responsible for coupling stress on the luminal side of the ER to rates of protein synthesis on the cytoplasmic side (Harding et al., 1999, 2000b). In the absence of PERK, unregulated protein synthesis overwhelms the capacity of the ER's luminal machinery, leading to unmitigated ER stress, protein misfolding, and cellular dysfunction and death (Harding et al., 2000b, 2001). Cells with physiologically high levels of ER client protein flux are especially dependent on the PERK-mediated arm of the UPRer, and its inactivation adversely affects the function of organs with high secretory rates (Harding et al., 2001; Zhang et al., 2002).
PERK is a type I ER membrane protein consisting of a stress-sensing luminal domain connected by a transmembrane segment to a cytoplasmic effector domain. The latter consists of an unusually large protein kinase domain with a single known substrate, serine 51 of the α subunit of eukaryotic translation initiation factor 2 (eIF2α; Harding et al., 1999; Ron and Harding, 2000; Ron, 2002). The phosphorylation of eIF2 converts it to an inhibitor of the GTP exchange factor eIF2B and, thereby, attenuates the recycling of eIF2 and the rate of translation initiation (Hinnebusch, 2000). In addition to this mechanism for regulating ER client protein load, PERK-dependent eIF2α phosphorylation also promotes the transcriptional activation of a large number of UPRer target genes (Harding et al., 2000a, 2003). This occurs through translational up-regulation of the transcription factor ATF4 (Harding et al., 2000a; Lu et al., 2004a; Vattem and Wek, 2004) and other less well-defined mechanisms (Deng et al., 2004). Thus, PERK-mediated eIF2α phosphorylation has a pervasive role in both the translational and transcriptional components of the UPRer (Ron, 2002).
In the compensated ER, PERK's stress-sensing luminal domain is maintained in a repressive complex with chaperones. As ER stress develops and the balance between chaperones and unfolded client proteins is disturbed, PERK's luminal domain is released from its repressive complex with chaperones. This leads to oligomerization of PERK in the plane of the ER membrane and promotes the transautophosphorylation of a large number of residues in PERK's kinase domain (Bertolotti et al., 2000). Several of these residues lie in PERK's activation loop, and their phosphorylation is likely to play a part in the conventional mechanism by which the catalytic activity of protein kinases is activated. However, other phosphorylated residues are located on PERK's unusually large kinase insert loop (Ma et al., 2001). The latter is a feature conserved in PERK homologues from diverse species; however, the importance of the kinase insert loop to PERK function has not been previously explored. In this study, we report that PERK activation and transautophosphorylation selectively recruit its substrate, the nonphosphorylated eIF2 complex, to the kinase and that this mechanism of substrate recruitment, which requires the large kinase insert loop, is indispensable for eIF2α phosphorylation in vivo.
Results
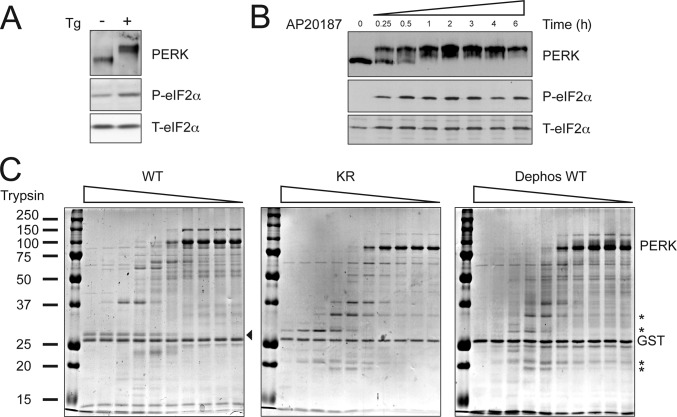
PERK activation is associated with extensive transautophosphorylation, which leads to marked retardation in the protein's mobility on SDS-PAGE (Harding et al., 1999; Bertolotti et al., 2000). These activation-dependent changes in PERK mobility were observed both in endogenous PERK, which were activated by treating cells with the ER stress–inducing agent thapsigargin, and in an artificial PERK derivative, Fv2E-PERK, in which the protein's COOH-terminal kinase domain has been fused to a drug-dependent oligomerization domain (Lu et al., 2004b). Treatment of Fv2E-PERK–expressing cells with the oligomerization-inducing ligand, the otherwise inert drug AP20187, elicited a similar marked shift in the kinase's mobility (Fig. 1, A and B ).
Figure 1.
Activation-dependent conformational changes correlate with eIF2 recruitment by PERK. (A) Immunoblot of immunopurified PERK, phosphorylated eIF2α (P-eIF2α), and total eIF2α (T-eIF2α) from mouse fibroblasts exposed to 400 nM thapsigargin, an ER stress–inducing agent, for 1 h. (B) Immunoblots of FV2E-PERK, P-eIF2α, and T-eIF2α from Fv2E-PERK–expressing CHO cells exposed to 10 nM AP20187, the activating ligand, for the indicated period of time. (C) Coomassie-stained SDS-PAGE of purified bacterially expressed wild-type PERK kinase domain (WT), K618R mutant (KR), and purified wild-type PERK kinase domain that had been dephosphorylated in vitro with λ-phage phosphatase (Dephos WT) followed by partial tryptic digest. Tryptic fragments specific to the inactive conformation are indicated by asterisks. A fragment specific to the active conformation is indicated by an arrowhead. Small amounts of contaminating trypsin stable GST were also released from the glutathione–Sepharose beads affinity matrix after cleavage at the TEV site and are visible on the gel (GST).
A similar activity-dependent shift in PERK mobility was observed in a bacterially expressed fusion protein of PERK's kinase domain and GST (GST-PERK) because of constitutive dimerization through the GST portion. After cleavage of the GST moiety, the wild-type active PERK kinase domain migrated on an SDS-PAGE above the 100-kD marker with an apparent molecular mass of 105 kD, whereas a point mutation (lysine 618 to arginine) predicted to disrupt the kinases' ability to coordinate the γ phosphate of ATP migrated as a 85-kD protein (Fig. 1 C). The role of phosphorylation in affecting these marked differences in protein mobility was demonstrated by the observation that dephosphorylation of wild-type PERK in vitro reduced its mobility to that of the inactive mutant (Fig. 1 C).
These phosphorylation-dependent changes in mobility on SDS-PAGE were also reflected in the protease sensitivity profile of the purified bacterially expressed proteins. Both the inactivating point mutation and the dephosphorylated PERK kinase had a similar protease sensitivity profile consisting of several protease-resistant fragments (Fig. 1 C, asterisks) that were not observed in the active wild-type protein. Overall, the active kinase was more protease sensitive, which is consistent with the known tendency of active kinases to assume a more open conformation that promotes ATP's access to the active site (Huse and Kuriyan, 2002). These observations suggested that PERK autophosphorylation leads to marked conformational shifts.
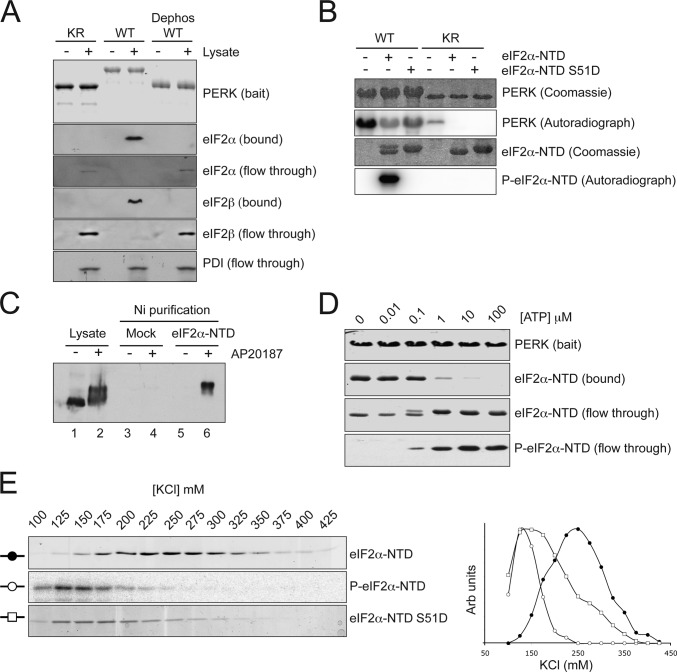
To examine the possible impact of these conformational shifts in the kinase on substrate recruitment, we incubated cell lysates with immobilized, purified, bacterially expressed wild-type, K618R mutant and in vitro dephosphorylated wild-type GST-PERK and measured the recruitment of eIF2. Wild-type GST-PERK strongly bound the eIF2 complex in the lysate, as reflected by the large amount of eIF2α and eIF2β immunoreactivity that remained associated with the resin after extensive washes. In contrast, the mutant PERK and the dephosphorylated wild-type PERK had no measurable interaction with eIF2, which was recovered in the flow-through of the binding reaction (Fig. 2 A ).
Figure 2.
Preferential binding of unphosphorylated eIF2α to activated PERK. (A) Immunoblots of bound and soluble proteins from cell lysate that had been incubated with the indicated GST-PERK proteins immobilized on a glutathione–Sepharose bead matrix. The bait proteins were stained with Coomassie (top). (B) Autoradiograph and corresponding Coomassie-stained gel of 32P-labeled proteins after incubation of bacterially expressed wild-type and S51D mutant eIF2α-NTD with wild-type or K618R mutant GST-PERK and γ-[32P]ATP. (C) Immunoblot of FV2E-PERK from untreated cells and cells treated with 10 nM AP20187 for 1 h. Lanes 1 and 2 are the lysate input, and lanes 3–6 are the material bound to Ni-NTA agarose beads that had not (mock) or had been coated with 6His-tagged eIF2α-NTD. (D) Coomassie-stained proteins (top three panels) and antiphospho-eIF2α blot (bottom) from bound and soluble fractions of an experiment in which purified eIF2α-NTD was added to immobilized GST-PERK in the presence of the indicated concentration of ATP. Note the selective elution of the phosphorylated product from the immobilized kinase. (E) Coomassie-stained SDS-PAGE (top and bottom) and autoradiograph of the top panel of the proteins eluted at the indicated salt concentrations from activated GST-PERK bound to a glutathione–Sepharose bead affinity matrix. The matrix of the top and middle panels was loaded with a mixture of unphosphorylated eIF2α-NTD and tracer amounts of radiolabeled phosphorylated P–eIF2α-NTD, whereas the bottom panel was loaded with purified eIF2α-NTD bearing a phosphomimetic mutation S51D. A graphical representation of the same data is also presented.
A nuclear magnetic resonance–based structural analysis reveals that eIF2α is a bipartite protein with globular COOH- and NH2-terminal domains (NTDs) connected by a flexible linker. The phosphorylated residue serine 51 is found on a relatively flexible loop extending from the globular core of the NTD (Ito et al., 2004). To further characterize the activity-dependent interaction between PERK and its substrate, we sought to determine whether it could be reconstituted with the NTD of eIF2α (eIF2α-NTD). First, we established that eIF2α-NTD is a substrate for PERK; wild-type GST-PERK readily phosphorylated eIF2α-NTD in vitro but failed to phosphorylate a point mutant in which serine 51 was replaced by aspartic acid (Fig. 2 B). In the absence of substrate, 32P is incorporated into the wild-type kinase through transautophosphorylation. Interestingly, both eIF2α-NTD and the nonphosphorylatable S51D mutant inhibit this transautophosphorylation, likely through the masking of the relevant sites on PERK. Next, purified bacterially expressed eIF2α-NTD tagged on its COOH terminus with six histidines was immobilized on a Ni–nitrilotriacetic acid (NTA) affinity column. Lysates of untreated and AP20187-treated Fv2E-PERK–expressing cells were passed over this affinity resin, and the binding of Fv2E-PERK was measured by immunoblotting. Only the activated low-mobility PERK bound to the eIF2α-NTD affinity matrix (Fig. 2 C). These observations indicate that the NTD of eIF2α specifically interacts with the activated PERK kinase domain.
To examine the impact of eIF2α phosphorylation on its interaction with the kinase, we combined bacterially expressed eIF2α-NTD with immobilized active GST-PERK in the presence of various concentrations of ATP. We then separated the bound complex from the free proteins, which were then resolved by SDS-PAGE. The amount of eIF2α-NTD that associated with PERK was markedly diminished by the addition of ATP, and the protein emerging in the flow-through had a lower mobility and reacted with a monoclonal antibody that selectively detects phosphorylated eIF2α (Fig. 2 D).
To further characterize these apparent differences in avidity of active PERK for its substrate (nonphosphorylated eIF2α) and its product (phosphorylated eIF2α), we mixed tracer amounts of 32P-phosphorylated eIF2α-NTD with the nonphosphorylated protein, bound the mixture to immobilized GST-PERK, and challenged the complex with increasing concentrations of salt. The nonphosphorylated eIF2α, which was detected by staining, eluted from the resin at a peak of 225 mM KCl, whereas the phosphorylated eIF2α-NTD, which was detected by autoradiography of the same gel, eluted at a lower salt concentration (peak of 125 mM KCl). The phosphorylation mimetic mutation serine 51 to aspartate also destabilized the complex with PERK, eluting at a similar low salt concentration in a parallel experiment (Fig. 2 E). These observations indicate that active PERK preferentially recruits its substrate, the nonphosphorylated eIF2α, and that the substrate, once phosphorylated, loses its attraction for the kinase.
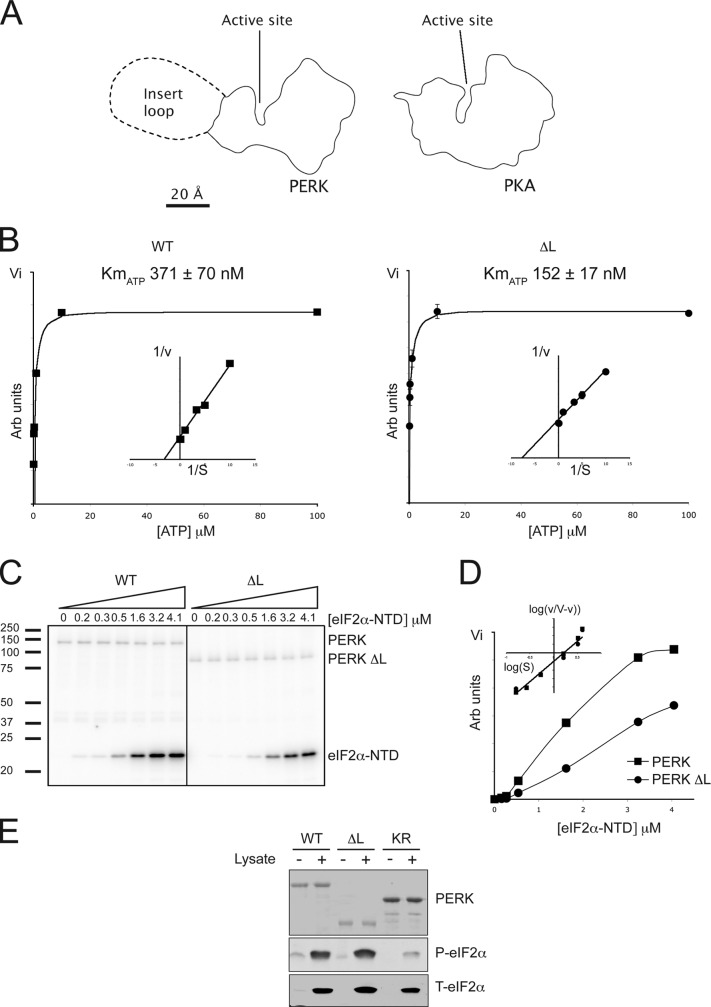
Most serine and threonine residues phosphorylated in active PERK are found on the kinase's large insert loop (Ma et al., 2001). A model of PERK (based on the crystalographically determined structure of the related kinase GCN2; Padyana et al., 2005) predicts that this large kinase insert loop, which is modeled as a single globular domain in scale with the PERK kinase, will extend from the surface of the NH2-terminal lobe at a distance from the catalytic site and, thus, is likely to be dispensable for catalytic activity (Fig. 3 A ). To test this notion, we deleted the large kinase insert loop of PERK and compared the catalytic activity of the wild-type and Δ-loop versions of bacterially expressed GST-PERK. First, we measured the catalytic activity in terms of the incorporation of radiolabeled phosphate into the kinase itself in a transautophosphorylation reaction. The reaction exhibited typical Michaelis-Menten kinetics with a measured Km for ATP of 317 ± 70 nM for the wild-type kinase and a lower Km of 152 ± 17 nM for the Δ-loop mutant (Fig. 3 B). The kinetics of eIF2α-NTD phosphorylation by both the wild-type and Δ-loop generated nonlinear Lineweaver-Burke plots, indicating non–Michaelis-Menten kinetics. Hill plot analysis yielded Hill coefficients of 2.5 for both enzymes, indicating positive cooperation, which, furthermore, is consistent with an apparent dimeric or possibly multimeric configuration of the active PERK kinase domain as revealed by gel filtration (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200508099/DC1). The calculated K0.5 for eIF2α-NTD of both enzymes was also similar (Fig. 3, C and D; and Table I ) and agreed with similar measurements conducted on the related eIF2α kinases PKR and HRI (Mellor and Proud, 1991; Dey et al., 2005). Furthermore, like the wild-type protein, GST-PERK Δ-loop was able to phosphorylate the intact eIF2 complex when added to cell lysates (Fig. 3 E). Based on these observations, we conclude that PERK Δ-loop is uncompromised in its catalytic activity.
Figure 3.
Deletion of PERK's kinase insert loop does not abolish the in vitro kinase activity. (A) The outline of protein kinase A and PERK's cytosolic domain based on a crystal structure (PKA) or a model (PERK). Proteins are drawn to scale, and PERK's large kinase insert loop is modeled as a compact globular structure. (B) ATP dependence of autokinase activity of wild-type PERK and PERK lacking the kinase insert loop (ΔL). Lineweaver-Burk plot inset; representative experiment shown. (C) Autoradiograph of eIF2α-NTD phosphorylation reaction performed with the indicated kinase in the presence of varying concentrations of substrate. (D) Graphical representation of P–eIF2α-NTD levels from C. Hill plot inset; representative experiment shown. (E) Immunoblots of phosphorylated and total eIF2α from reactions in which reticulocyte lysate had been incubated with the indicated PERK preparations; the latter are revealed by Coomassie staining (top).
Table I. Kinetic coefficients of wild-type and Δ-loop PERK enzymes.
| Autokinase |
eIF2α directed |
|||
|---|---|---|---|---|
| Kinase | KmATP | KmATP | K0.5eIF2α-NTD | Hill coefficient |
| (nM) | (nM) | (μM) | ||
| PERK | 371 ± 70 | 305 ± 50 | 1.38 ± 0.07 | 2.53 ± 0.09 |
| PERK Δ-loop | 152 ± 17 | 185 ± 34 | 1.44 ± 0.18 | 2.45 ± 0.10 |
The KmATP of wild-type PERK and PERK Δ-loop's autokinase activity was measured by filter-binding assay. The KmATP and K0.5eIF2α-NTD of wild-type PERK and PERK Δ-loop–mediated eIF2α-NTD phosphorylation was measured by SDS-PAGE autoradiography as shown in Fig. 3 C (n = 3; mean ± SEM).
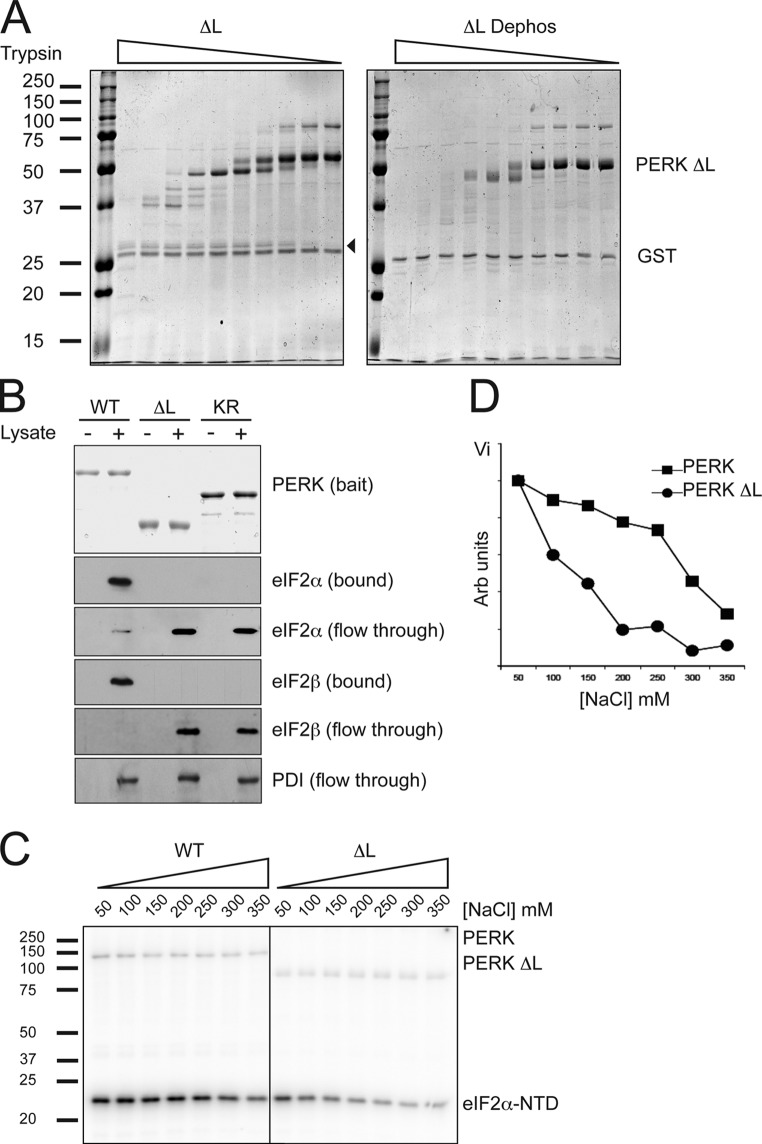
To determine whether deletion of the kinase insert loop impacted the activation-dependent conformational shifts in the kinase domain, we compared the protease digestion profiles of purified bacterially expressed GST-PERK Δ-loop with that of the same protein after de-phosphorylation in vitro. Comparison of Figs. 1 C and 4 A shows that most of the protease-resistant fragments observed in the dephosphorylated wild-type full-length kinase domain and in the full-length inactive K618R mutant (Fig. 1 C) were missing in the dephosphorylated PERK Δ-loop (Fig. 4 A). The presence of a common 27-kD phosphorylation-dependent trypsin-resistant fragment suggests that the active wild-type and Δ-loop proteins share a common core.
Figure 4.
PERK's kinase insert loop is required for efficient recruitment of substrate. (A) Coomassie-stained partial tryptic digest of TEV-cleaved recombinant Δ-loop PERK and dephosphorylated Δ-loop PERK as in Fig. 1 C. A fragment specific to the active conformation is indicated by an arrowhead. (B) Immunoblots of bound and soluble proteins from cell lysate that had been incubated with the indicated GST-PERK proteins immobilized on a glutathione–Sepharose bead matrix. The bait proteins were stained with Coomassie (top). (C) Autoradiography of eIF2α-NTD phosphorylated by the indicated PERK kinase preparations at increasing salt concentrations. (D) Graphical representation of P–eIF2α-NTD levels from C.
Having established that the kinase insert loop contributes substantively to the activation-dependent changes in PERK's kinase domain, we sought to determine whether it also influences substrate recruitment. Therefore, we compared the binding of eIF2 in cell lysates to immobilized wild type and the Δ-loop mutant GST-PERK. Under identical conditions, the wild-type protein bound all of the eIF2 in the lysate, whereas the Δ-loop mutant was as ineffective as the catalytically dead K618R mutant GST-PERK (Fig. 4 B). These differences in avidity of the wild-type and mutant kinase were also reflected in the salt sensitivity of eIF2α-NTD phosphorylation. Whereas the phosphorylation of eIF2α-NTD by wild-type GST-PERK was maintained at salt concentrations of up to 250 mM, the GST-PERK Δ-loop's activity toward its substrate declined significantly between 50 and 150 mM of salt (Fig. 4, C and D). Salt had a similar negligible effect on the two kinases' catalytic activities, as reflected by the undiminished incorporation of label into the kinases themselves. We concluded that although dispensable to catalytic activity, PERK's large kinase insert loop contributes substantively to substrate recruitment in vitro.
A stable interaction between phosphorylated residues on PERK's kinase domain and its substrate predict that substrate binding would protect these residues from dephosphorylation in vitro. To test this hypothesis, we radiolabeled GST-PERK by autophosphorylation in the presence of γ-[32P]ATP and incubated the purified, radiolabeled protein with bacterially expressed λ-phage phosphatase in the absence or presence of wild-type eIF2α-NTD or in the presence of a mutant S51D substrate. PERK dephosphorylation was significantly more attenuated by the wild-type substrate than by the binding-defective mutant (Fig. 5 , compare A with B). Furthermore, an important fraction of the phosphorylated residues protected by the substrate are likely located on the kinase's insert loop, as the substrate had a very modest protective effect when applied to radiolabeled PERK that lacked the loop (Fig. 5 C). To test whether the recruitment of substrate through interaction with phosphorylated residues within the insert loop was independent of kinase activity, we mutated the serine and threonine residues of this domain in the catalytically inactive K618R GST-PERK to aspartates, mimicking their phosphorylation. We then repeated the in vitro pull-down experiments with eIF2α-NTD in the presence and absence of ATP (Fig. 5 D). Wild-type GST-PERK recruited its substrate, releasing phosphorylated product in the presence of ATP. The K618R mutation abolished this interaction, but recruitment could be restored by phosphomimetic mutations within the insert loop, which failed to restore kinase activity. These experiments support the interaction of substrate with phosphorylated residues within the insert loop of PERK.
Figure 5.
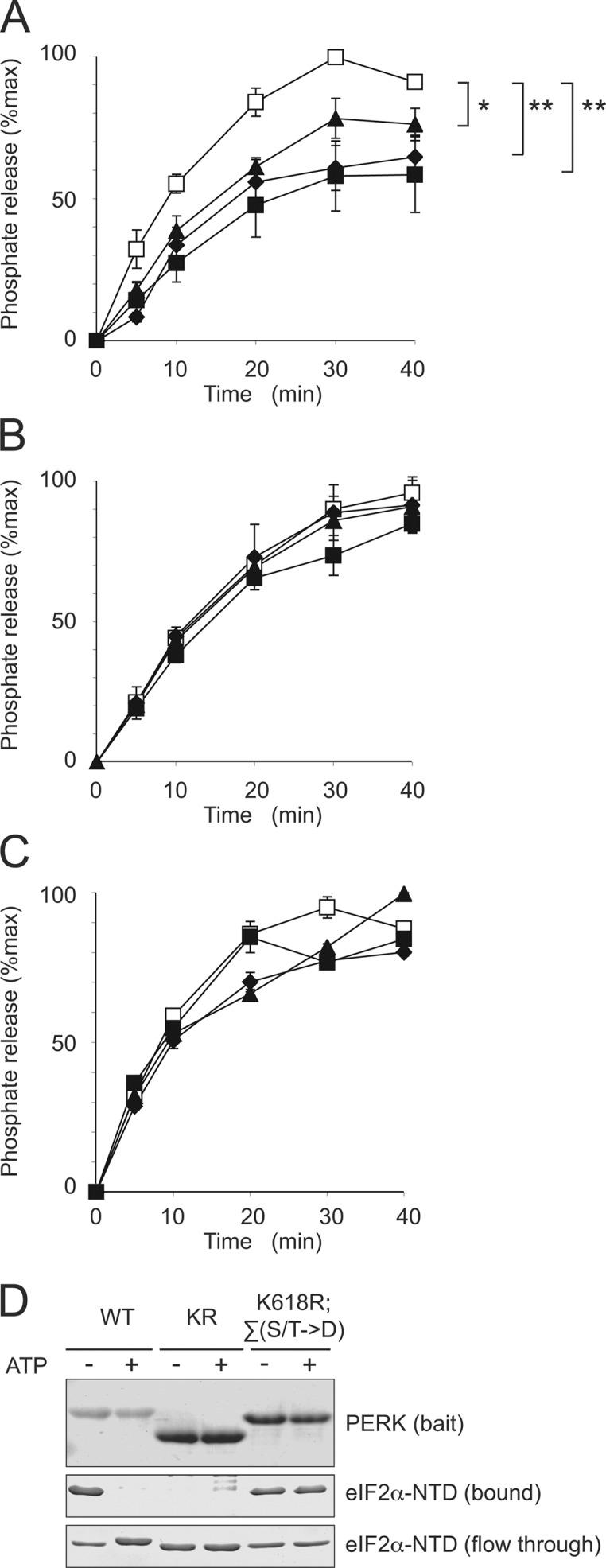
Phosphorylated residues on PERK interact with eIF2α. (A and B) Release of 32P from GST-PERK by λ-phage phosphatase in the absence (open square) and presence of eIF2α-NTD (A) or S51D eIF2α-NTD (B; 1.5 mg/ml, closed triangle; 3.75 mg/ml, closed diamond; 7.5 mg/ml, closed square). P values for the divergence of curves were determined by two-factor analysis of variance between groups: *, P < 0.05; **, P < 0.01. (C) Release of 32P from GST-PERK Δ-loop by λ-phage phosphatase in the absence (open square) and presence of eIF2α-NTD (1.5 mg/ml, closed triangle; 3.75 mg/ml, closed diamond; 7.5 mg/ml, closed square). (D) Coomassie-stained proteins from bound and soluble fractions of an experiment in which purified eIF2α-NTD was added to immobilized GST-PERK (WT), GST-PERKK618R (KR), and GST-PERKK618R;Σ(S/T→D) in which insert loop serine and threonine residues were mutated to aspartates (K618R;∑(S/T→D)) in the absence or presence of 10 μM ATP. Error bars represent SEM.
To determine whether the kinase insert loop's role in substrate recruitment in vitro is mirrored by its activity in vivo, we constructed oligomerization-activatable 6His-tagged versions of wild-type Fv2E-PERK, inactive K618R mutant, and Δ-loop mutant Fv2E-PERK and transduced these into mammalian cells. Both the wild-type and Δ-loop proteins were activated by adding AP20187 compound to the culture media, as reflected by their shift in mobility, whereas the K618R mutant was inactive as expected (Fig. 6 A ). Isolation of the kinase and associated cellular proteins by Ni-NTA affinity chromatography showed that eIF2 bound only to the activated wild-type kinase but not to the two mutants. Furthermore, levels of phosphorylated eIF2α increased selectively in the drug-treated cells transduced with the wild-type protein but not in cells transduced with the two mutants (Fig. 6 A). These observations indicate that in cells, activation-dependent substrate recruitment and eIF2α phosphorylation depend on PERK's kinase insert loop.
Figure 6.
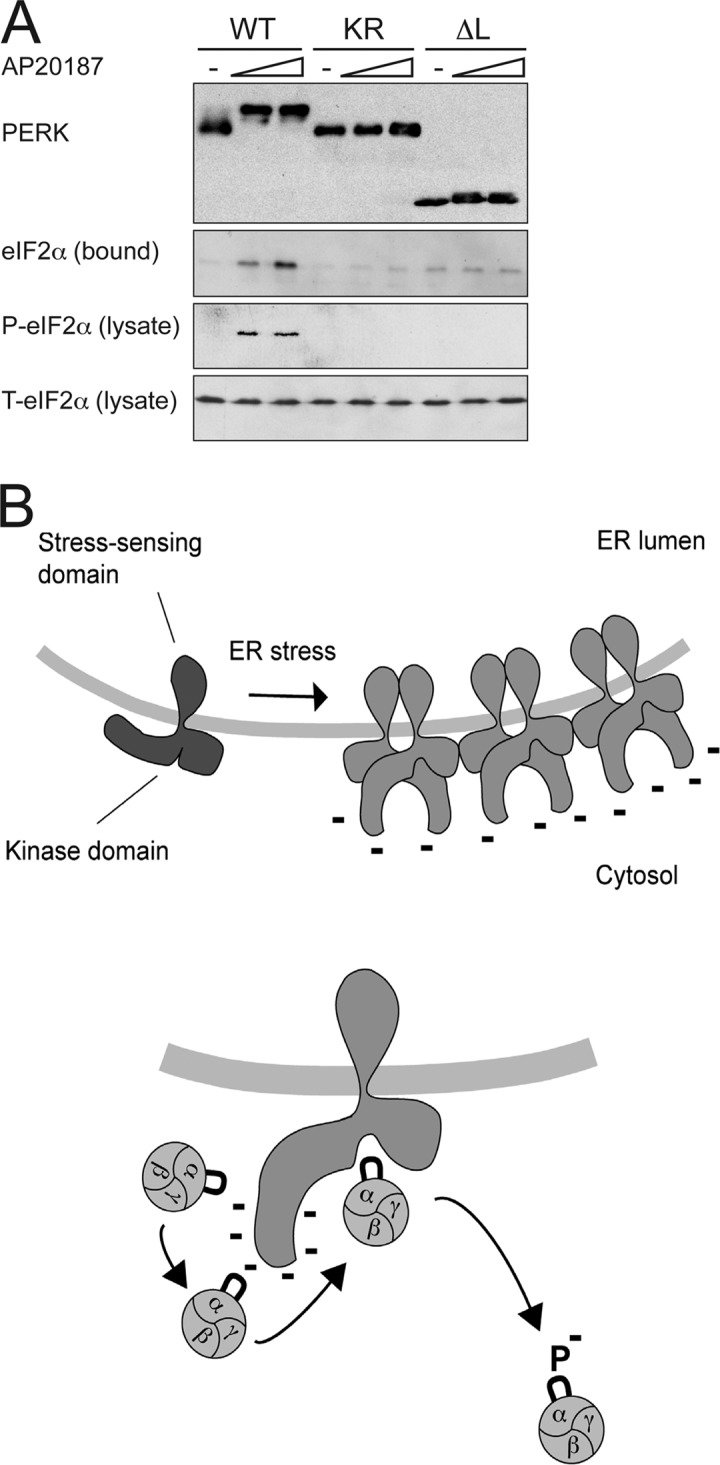
PERK's kinase insert loop is essential for in vivo recruitment and phosphorylation of eIF2α. (A) Anti-myc immunoblot of affinity-purified His6-FV2E-PERK-9E10, associated eIF2α, phosphorylated eIF2α (P-eIF2α), and total eIF2α (T-eIF2α) in the lysate of untreated and AP20187-treated (100 nM and 1 μM) 293T cells expressing the wild-type protein (WT), the K618R mutant (KR), and the Δ-loop version (ΔL). (B) Cartoon representing a hypothetical model of the interaction of activated PERK with its substrate. ER stress promotes oligomerization of PERK in the plane to the membrane, leading to transautophosphorylation. eIF2 complex is recruited to the highly phosphorylated insert loop before being passed on to the catalytic center. Phosphorylation of eIF2 (denoted as P) prevents further interaction with the insert loop and leads to product release.
Discussion
This study has uncovered a novel mechanism for substrate recruitment by the activated eIF2α kinase PERK (summarized in Fig. 6 B). In its nonphosphorylated inactive form, PERK has a low affinity for its substrate. Upon activation, PERK undergoes phosphorylation-dependent conformational changes that markedly increase its affinity for the substrate, the nonphosphorylated form of eIF2. This change in affinity depends on the unusually large kinase insert loop, which is dispensable for catalytic activity itself. Phosphorylation of the substrate results in a marked decline in its affinity for the kinase and likely promotes substrate dissociation. The phosphorylated eIF2 product is free to migrate away from the kinase and to interact with and inhibit the guanine nucleotide exchange factor, thereby attenuating protein synthesis.
The kinase insert loop, which connects strands 4 and 5 of the large β sheet in the NH2-terminal lobe of all known protein kinases, is highly variable in size (Hanks and Hunter, 1995). However, to date, no function has been specifically attributed to this portion of any known kinase. The tyrosine kinase domain of the platelet-derived growth factor receptor has a large insert loop that undergoes tyrosine phosphorylation upon receptor activation, but it connects structural elements in the COOH-terminal lobe (Kazlauskas and Cooper, 1989) that are distinct from those connected by PERK's loop. PERK has one of the largest kinase insert loops of all known serine/threonine protein kinases. Although the primary amino acid sequence of the kinase insert loop is poorly conserved among PERK homologues, its unusual size is a conserved feature and one that it also shares with the related eIF2α kinase GCN2. Phosphopeptide mapping indicates that several transautophosphorylation events on PERK take place on residues in the kinase insert loop (Ma et al., 2001). The presence of numerous phosphorylated residues on the loop in a context lacking sequence conservation and the marked salt sensitivity of the interaction between active PERK and eIF2α, which is further weakened when the latter is phosphorylated, suggests that eIF2 recruitment relies heavily on ionic interactions.
Our data are consistent with a simple model whereby autophosphorylation rearranges the structure of the loop exposing the phosphorylated residues to the solvent. The substrate serine 51 of eIF2α is exposed on a loop (residues 47–67) containing six arginines and a lysine (Ito et al., 2004). These exposed residues likely create a positively charged surface that may interact with the negatively charged phosphorylated residues on the PERK kinase insert loop, recruiting the substrate. Our experiments also indicate that more than one molecule of eIF2α-NTD is able to dock on activated PERK in vitro (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200508099/DC1), suggesting the existence of multiple substrate-binding sites on the activated enzyme. Phosphorylation of serine 51 would tend to reduce these charge interactions either by repelling the negatively charged residues on the phosphorylated kinase or by neutralizing intramolecular interactions with the basic residues on eIF2α's phosphorylation loop; either mechanism would account for the reduced affinity of the kinase for its phosphorylated substrate.
The nearly identical catalytic features of the wild-type and Δ-loop kinases (Fig. 3 and Table I) argue against a specific role for the loop in orienting the substrate for catalysis and suggest that the recruitment mechanism described here may rely on determinants in eIF2α that are dispensable for the phosphorylation reaction itself. In this regard, it is noteworthy that mutation of the three arginine residues immediately COOH-terminal to serine 51 on eIF2α's phosphorylation loop (RRR52–54→VKA) had only a modest inhibitory effect on the ability of the mutant protein to serve as a substrate for GCN2 in yeast or for PKR in vitro (Dey et al., 2005). Our findings are easier to reconcile with a sequential mode of action whereby the phosphorylated loop facilitates substrate recruitment to PERK and hands it off to the catalytic core of the enzyme. Phosphorylation of the substrate disfavors rebinding and, thereby, promotes unidirectional substrate flow.
This proposed mechanism of substrate recruitment helps explain how PERK, which is a relatively scarce enzyme, can nonetheless rapidly phosphorylate significant amounts (∼20%) of its abundant substrate eIF2α in ER stressed cells (Harding et al., 1999). Many signaling modules consist of a kinase and downstream effectors held together in a preformed complex; however, eIF2 has a catalytic role in translation, which likely precludes a persistent association with the kinase. Furthermore, recent evidence suggests that in yeast, eIF2B, the GTP exchange factor that is inhibited by eIF2α phosphorylation, is located in discrete cytosolic foci through which eIF2 must traffic (Campbell et al., 2005). These considerations favor a mechanism whereby the substrate is rapidly recruited to the kinase solely when the latter is activated and then is rapidly released upon phosphorylation.
Materials and methods
Expression plasmids
The expression plasmids PerkKD-pGEX4T-1 and PerkKD(KA)-pGEX4T-1 encoding GST fusion proteins of mouse PERK residues 537–1,114 wild type and the kinase-dead K618A mutant have been previously described (Harding et al., 1999). PerkKD(K618R mutant)-pGEX4T-1 has incorporated a K618R mutation in the PERK cDNA by PCR. PerkKD(ΔL)-pGEX4T-1 was generated by excising residues 643–886 of wild-type PERK. Tobacco etch virus (TEV)–cleavable GST fusion proteins were generated by transfer of the coding sequences of wild-type, K618R mutant, and Δ-loop PERK into pGV67 (a gift from B. Nolan, Yale, New Haven, CT). Mammalian expression vectors for 6His-tagged Fv2E-PERK wild type, K618R mutant, and Δ-loop were generated by transferring the coding sequence form Fv2E-Perk-pBABE (Lu et al., 2004b) into pcDNA1/Amp containing a 6His cassette. eIF2α-NTD encoding residues 1–185 of human eIF2α with three solubilizing mutations was bacterially expressed from codon optimized vector 2aOPTx3M(1–185)pET-30a(+) (Ito et al., 2004). eIF2α-NTD S51D was generated by transferring residues 4–185 from 2aOPTs51dx3M(4–302) (Ito et al., 2004) into pET-30a(+). λ-Phage phosphatase cDNA was generated by PCR from phage DNA and cloned into the expression vector pDUET (Novagen). Mutation of 57 serine and threonine residues within the insert loop to aspartic acid was performed by means of custom gene synthesis (GenScript Corp.).
Cell culture and transfection
Stable clones of CHO cells expressing Fv2E-PERK have been described previously (Lu et al., 2004a). 293T cells were transfected with His6-FV2E-PERK–expressing plasmids and, 48 h later, were treated with AP20187 (ARIAD). Lysates were prepared in 0.5% Triton X-100, 100 mM NaCl, 20 mM Tris, pH 7.4, 1 mM DTT, 1 mM PMSF, 4 μg/ml aprotinin, 2 μg/ml pepstatin A, 10 mM terasodium pyrophosphate, 15.5 mM β-glycerophosphate, and 100 mM NaF.
Limited tryptic proteolysis
Bacterially expressed GST fusion proteins were first cleaved with TEV protease to liberate the GST portion. Thereafter, they were incubated with serial dilutions of sequencing grade trypsin over the range 20 ng/ml to 5 μg/ml (1047841; Roche) for 30 min at room temperature in 100 mM Tris, pH 8.0.
Immunoblot and immunoprecipitation
Total eIF2α was detected with a mAb to human eIF2α (a gift of the late E. Henshaw), and phosphorylated eIF2α was detected with an epitope-specific antiserum (RG0001; Research Genetics). eIF2β was detected with a mAb (a gift from S. Kimball, Pennsylvania State University, University Park, PA). Protein disulfide isomerase was detected with a mAb (SPA891; StressGen Biotechnologies). PERK was immunoprecipitated and detected with polyclonal antisera as previously described (Bertolotti et al., 2000). Myc-epitope was detected with a mAb (9E10).
Kinetic analysis
Bacterially expressed GST-PERK proteins were incubated with Mg-ATP at the indicated concentrations and spiked 1:200 with γ-[32P]ATP (MP Biochemical, Inc.). Reactions were terminated by the addition of TCA, and insoluble protein was bound to glass microfiber filters for quantification by liquid scintillation counting. Data was fitted to Lineweaver-Burke double reciprocal plots. In experiments to determine the Km for ATP during eIF2α-NTD phosphorylation, eIF2α-NTD was at a concentration of 20 μM. In experiments to determine the K0.5 for eIF2α-NTD, the ATP concentration was at 100 μM.
Phosphatase protection assay
Bacterially expressed GST-PERK or GST-PERK Δ-loop was dephosphorylated by incubation with λ-phage phosphatase. Subsequent incubation with γ-[32P]ATP led to rephosphorylation and the incorporation of 32P; unincorporated label was removed by gel filtration and dialysis. Samples of labeled kinase were then incubated at 37°C in the presence of limiting concentrations of λ-phage phosphatase with or without substrate addition. Samples were taken at intervals, and protein was precipitated by TCA. Free, released 32P was measured by liquid scintillation counting.
Copurification assays
Unless specified otherwise, copurification experiments were performed in buffer containing 0.1% Triton X-100, 2 mM MgCl2, 20 mM Tris, pH 7.4, and NaCl 150 mM. Glutathione–Sepharose beads and Ni-NTA agarose beads were washed three times in the same buffer. When Ni-NTA beads were used, 10 mM imidazole was included during binding and 20 mM during washes. Binding took place at 4 or 30°C for 30 min.
In experiments requiring dephosphorylated PERK, bacterially expressed protein was purified on glutathione–Sepharose beads and incubated with bacterially expressed λ-phage phosphatase at 30°C for 1 h. Beads were washed extensively with 450 mM NaCl and phosphate buffer to remove contaminating phosphatase.
Online supplemental material
Fig. S1 shows that active wild-type PERK cytosolic domain migrates in oligomeric form during size exclusion chromatography, whereas an inactive mutant remains monomeric. Fig. S2 shows the saturability of eIF2α-NTD binding to PERK and suggests that its stoichiometry is not 1:1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200508099/DC1.
Supplementary Material
Acknowledgments
We thank Huiqing Zeng for carrying out preliminary experiments in this project; Gerhard Wagner (Harvard Medical School, Boston, MA), Assen Marintchev (Harvard Medical School, Boston, MA), and Steven Hubbard (New York University) for advice; Scot Kimball for the eIF2β antibody; Brad Nolan for pGV67 vector; and the ARIAD corporation for the FV2E dimerization system and AP20187 compound.
This work was supported by the National Institutes of Health grants ES08681 and DK47119 to D. Ron and a Wellcome Trust Fellowship to S.J. Marciniak.
S.J. Marciniak's present address is Cambridge Institute for Medical Research, Addenbrooke's Hospital, Cambridge CB2 2XY, UK.
Abbreviations used in this paper: eIF, eukaryotic translation initiation factor; NTA, nitrilotriacetic acid; NTD, NH2-terminal domain; UPRer, ER unfolded protein response.
References
- Baumann, O., and B. Walz. 2001. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205:149–214. [DOI] [PubMed] [Google Scholar]
- Bertolotti, A., Y. Zhang, L. Hendershot, H. Harding, and D. Ron. 2000. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat. Cell Biol. 2:326–332. [DOI] [PubMed] [Google Scholar]
- Campbell, S.G., N.P. Hoyle, and M.P. Ashe. 2005. Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translation control. J. Cell Biol. 170:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J., P.D. Lu, Y. Zhang, D. Scheuner, R.J. Kaufman, N. Sonenberg, H.P. Harding, and D. Ron. 2004. Translational repression mediates activation of Nuclear Factor kappa B by phosphorylated translation initiation factor 2. Mol. Cell. Biol. 24:10161–10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, M., B. Trieselmann, E.G. Locke, J. Lu, C. Cao, A.C. Dar, T. Krishnamoorthy, J. Dong, F. Sicheri, and T.E. Dever. 2005. PKR and GCN2 kinases and guanine nucleotide exchange factor eukaryotic translation initiation factor 2B (eIF2B) recognize overlapping surfaces on eIF2alpha. Mol. Cell. Biol. 25:3063–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell, S.W., K.J. Travers, J.S. Weissman, and J.L. Brodsky. 2001. The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35:149–191. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., and T. Hunter. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576–596. [PubMed] [Google Scholar]
- Harding, H., Y. Zhang, and D. Ron. 1999. Translation and protein folding are coupled by an endoplasmic reticulum resident kinase. Nature. 397:271–274. [DOI] [PubMed] [Google Scholar]
- Harding, H., I. Novoa, Y. Zhang, H. Zeng, R.C. Wek, M. Schapira, and D. Ron. 2000. a. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding, H., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. b. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904. [DOI] [PubMed] [Google Scholar]
- Harding, H., H. Zeng, Y. Zhang, R. Jungreis, P. Chung, H. Plesken, D. Sabatini, and D. Ron. 2001. Diabetes Mellitus and excocrine pancreatic dysfunction in Perk−/− mice reveals a role for translational control in survival of secretory cells. Mol. Cell. 7:1153–1163. [DOI] [PubMed] [Google Scholar]
- Harding, H., Y. Zhang, H. Zeng, I. Novoa, P. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In Translational Control of Gene Expression. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 185–243.
- Huse, M., and J. Kuriyan. 2002. The conformational plasticity of protein kinases. Cell. 109:275–282. [DOI] [PubMed] [Google Scholar]
- Ito, T., A. Marintchev, and G. Wagner. 2004. Solution structure of human initiation factor eIF2alpha reveals homology to the elongation factor eEF1B. Structure (Camb). 12:1693–1704. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 110:1389–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas, A., and J.A. Cooper. 1989. Autophosphorylation of the PDGF receptor in the kinase insert region regulates interactions with cell proteins. Cell. 58:1121–1133. [DOI] [PubMed] [Google Scholar]
- Lu, P.D., H.P. Harding, and D. Ron. 2004. a. Translation re-initiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P.D., C. Jousse, S.J. Marciniak, Y. Zhang, I. Novoa, D. Scheuner, R.J. Kaufman, D. Ron, and H.P. Harding. 2004. b. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y., Y. Lu, H. Zeng, D. Ron, W. Mo, and T.A. Neubert. 2001. Characterization of phosphopeptides from protein digests using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and nanoelectrospray quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 15:1693–1700. [DOI] [PubMed] [Google Scholar]
- Mellor, H., and C.G. Proud. 1991. A synthetic peptide substrate for initiation factor-2 kinases. Biochem. Biophys. Res. Commun. 178:430–437. [DOI] [PubMed] [Google Scholar]
- Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 101:451–454. [DOI] [PubMed] [Google Scholar]
- Padyana, A.K., H. Qiu, A. Roll-Mecak, A.G. Hinnebusch, and S.K. Burley. 2005. Structural basis for autoinhibition and mutational activation of eukaryotic initiation factor 2alpha protein kinase GCN2. J. Biol. Chem. 280:29289–29299. [DOI] [PubMed] [Google Scholar]
- Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349–355. [DOI] [PubMed] [Google Scholar]
- Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Invest. 110:1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, D., and H. Harding. 2000. PERK and translational control by stress in the endoplasmic reticulum. In Translational Control of Gene Expression. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 547–560.
- Vattem, K.M., and R.C. Wek. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., B. McGrath, S. Li, A. Frank, F. Zambito, J. Reinert, M. Gannon, K. Ma, K. McNaughton, and D.R. Cavener. 2002. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22:3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.