Abstract
Membrane fusion in the secretory pathway is mediated by SNAREs (located on the vesicle membrane [v-SNARE] and the target membrane [t-SNARE]). In all cases examined, t-SNARE function is provided as a three-helix bundle complex containing three ∼70–amino acid SNARE motifs. One SNARE motif is provided by a syntaxin family member (the t-SNARE heavy chain), and the other two helices are contributed by additional t-SNARE light chains. The syntaxin family is the most conformationally dynamic group of SNAREs and appears to be the major focus of SNARE regulation. An NH2-terminal region of plasma membrane syntaxins has been assigned as a negative regulatory element in vitro. This region is absolutely required for syntaxin function in vivo. We now show that the required function of the NH2-terminal regulatory domain (NRD) of the yeast plasma membrane syntaxin, Sso1p, can be circumvented when t-SNARE complex formation is made intramolecular. Our results suggest that the NRD is required for efficient t-SNARE complex formation and does not recruit necessary scaffolding factors.
Introduction
SNAREs are required for membrane fusion in the eukaryotic secretory pathway (Weber et al., 1998; Chen and Scheller, 2001; Ungar and Hughson, 2003). The concerted assembly of SNARE subunits is carefully regulated at many levels by intrinsic protein conformations and extrinsic regulatory proteins. Characterization of both the molecular properties and assembly of the SNARE complex is imperative to understand mechanistic details of membrane fusion.
SNARE complex assembly at the plasma membrane begins with a binary association between the syntaxin component (the t-SNARE heavy chain) and the SNAP25 homologue (t-SNARE light chains), resulting in a functional t-SNARE complex. In the case of the yeast plasma membrane homologues (Sso1p or Sso2p and Sec9p), the formation of this binary complex (three SNARE domains) is rate limiting for the overall process of SNARE complex assembly (Nicholson et al., 1998). Although the subunit composition of the yeast plasma membrane t-SNARE complex is clearly one Sso1p or Sso2p and one Sec9p (Nicholson et al., 1998; Fiebig et al., 1999), the stoichiometry of the neuronal counterpart is debated.
Increasing evidence suggests that four SNARE domains form a t-SNARE complex with two syntaxin1A proteins and one SNAP25 in vitro (Margittai et al., 2001; Kim et al., 2002; Zhang et al., 2002). The functional consequences of a four-stranded t-SNARE complex remain unclear because this species has yet to be demonstrated in vivo. However, most t-SNARE complexes that form on internal membranes use three different proteins to form a functional t-SNARE (Fukuda et al., 2000). In this case, one syntaxin family member serves as a t-SNARE heavy chain, and two nonsyntaxin proteins provide t-SNARE light chain function. The v-SNARE, imbedded in the vesicle membrane in vivo, associates with the t-SNARE complex to complete the ternary complex. In all known instances, a single, membrane-integral protein provides v-SNARE function. High resolution crystal structure determination of a stable proteolytic fragment of the neuronal ternary SNARE complex showed that the assembled ternary complex is a parallel ∼12-nm, four-stranded helical bundle with one helix contributed by syntaxin1A, one from vesicle-associated membrane protein, and two helices from SNAP25 (Sutton et al., 1998).
Syntaxins exhibit various conformations that are an intrinsic part of SNARE complex formation. Biophysical characterization of SNARE proteins in various free and complexed states has yielded important conformational information (Fernandez et al., 1998; Lerman et al., 2000; Misura et al., 2000; Munson et al., 2000). Free syntaxins are almost entirely α-helical, whereas SNAP25 and Sec9p as well as the v-SNAREs, VAMP2 (vesicle-associated membrane protein 2), and Snc1/2p are unstructured in solution (Rice et al., 1997; Fiebig et al., 1999; Lerman et al., 2000; Munson et al., 2000). Secondary structure is induced in t-SNARE light chains when they associate with the syntaxin component during t-SNARE complex formation. Similarly, α-helical structure is induced in the v-SNARE as it enters the ternary complex (Fasshauer et al., 1997a,b; Nicholson et al., 1998).
One of the first indications that the various conformational states of syntaxin1A are functionally important came from studies examining the interactions of the SNARE recycling machinery, SNAP and NSF, with syntaxin1A. Upon ATP hydrolysis, NSF promoted a conformational change in syntaxin1A (referred to as syntaxin* in Hanson et al., 1995) that made it refractory to further SNARE binding. The physical basis for this change is likely mediated through the binding of an NH2-terminal domain back onto a COOH-terminal segment, which prevents further protein–protein interactions (Calakos et al., 1994). Structural analysis has confirmed this association between the NH2 and COOH termini of syntaxins (Fiebig et al., 1999; Munson et al., 2000).
Although the conformational gymnastics of syntaxins are well documented, the precise in vivo role for the various states remains undetermined. All syntaxins appear to have a large NH2-terminal regulatory domain (NRD; also called the HABC domain; Teng et al., 2001). The NRD exhibits inhibitory functions in vitro. Binding studies have documented that the binary association between the syntaxin heavy chain and the t-SNARE light chains are adversely affected by the presence of this sequence (Nicholson et al., 1998; Margittai et al., 2003). The strength of the interaction between the NRD and the syntaxin core SNARE domain appears to be much stronger for yeast Sso1p than for neuronal syntaxin1A. Removal of the NRD in Sso1p results in a 3,000-fold increase in the SNAP25 homologous region of Sec9p (Sec9c) binding (Nicholson et al., 1998), whereas similar experiments with syntaxin1A and SNAP25 showed a much more modest sevenfold increase in t-SNARE complex formation in the absence of the syntaxin1A NRD (Margittai et al., 2003). Additionally, the NRD region of syntaxin 1A is completely dispensable for in vitro fusion (Parlati et al., 1999), yet the NRD of Sso1p is required for plasma membrane SNARE function in vivo for unknown reasons (Munson et al., 2000).
This study further characterizes the function of the Sso1p NRD in vivo using chimeric proteins that alter the NH2-terminal sequence of Sso1p. We found that replacing the NRD of Sso1p with the homologous sequence from the neuronal plasma membrane SNARE, syntaxin 1A, did not restore function. However, when the t-SNARE complex was made intramolecular by physically linking the SNAP25 homologous region of Sec9p to Sso1p without the NRD sequence, in vivo function was restored. Conformation of an intramolecular t-SNARE was demonstrated by a single point mutation in the Sec9c portion of the t-SNARE chimera (Q468R) that abrogated Sso1p function. These results suggest that the NRD is primarily involved in facilitating t-SNARE complex formation or preventing inappropriate associations with the H3 domain of Sso1p in vivo.
Results
The Sso1p NRD is necessary for in vivo function but not for in vitro fusion
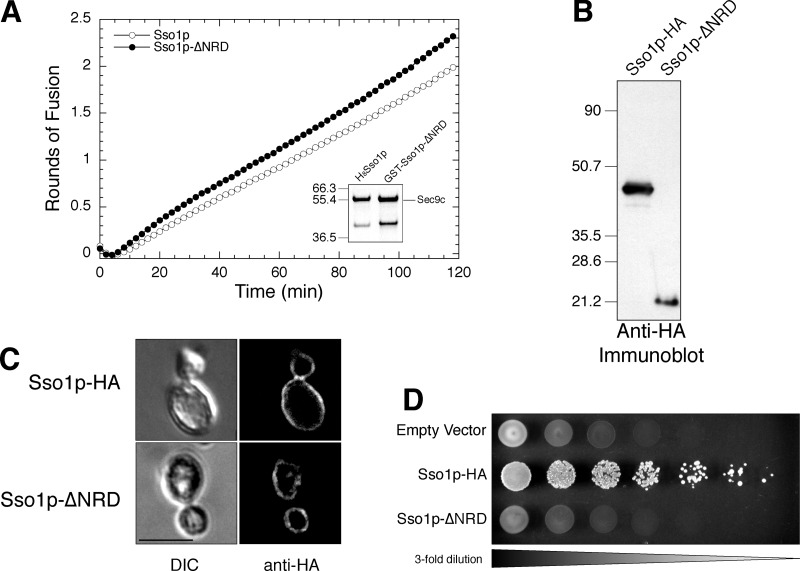
We began by confirming that, like syntaxin1A, the NRD of Sso1p was not required for in vitro fusion. Recombinant His8-Sso1p and GST-Sso1p-ΔNRD proteins were used to form t-SNARE complexes with GST-Sec9c in detergent, which were subsequently reconstituted into phosphatidylcholine/phosphatidylserine liposomes by detergent dilution and dialysis (Scott et al., 2003). Fig. 1 A shows that Sso1p lacking the NRD sequence (closed circles) fuses comparably to full-length Sso1p (open circles) with fluorescent Snc1p liposomes. Recent work with proteoliposomes produced by detergent-assisted insertion into preformed liposomes yielded a similar result (Chen et al., 2004).
Figure 1.
Sso1p-ΔNRD is fusion competent in vitro but does not function in vivo. (A) In vitro fusion. Kinetic fusion graph comparing wild-type Sso1p (open circles) with Sso1p-ΔNRD (filled circles), illustrating that both are fusion competent with Snc1p fluorescent donor liposomes. (Inset) Coomassie-stained SDS-PAGE gel of the two t-SNARE liposomes. (B) Relative protein expression in yeast cell extracts of JMY303 and JMY305 was determined by immunoblot analysis detecting an HA epitope with the monoclonal antibody 16B12 (Covance). (C) Plasma membrane localization. Differential interference contrast (DIC) images and indirect immunofluorescence images are shown for JMY303 (Sso1p-HA) and JMY305 (Sso1p-ΔNRD). Localization was determined by staining with an anti-HA antibody. Bar, 5 μm. (D) Growth on 5-FOA. Three-fold serial dilutions of JMY302 (empty vector), JMY303 (Sso1p-HA), and JMY305 (Sso1pΔNRD) were spotted onto synthetic complete media with 2% galactose containing 1 mg/ml 5-FOA and grown at 30°C for 72 h.
We also examined the function of Sso1p-ΔNRD in vivo using a haploid plasmid shuffle strain that contains a genomic deletion in both SSO1 and SSO2 genes (JMY128). The viability of this strain is maintained by a low-copy URA3 plasmid expressing Sso1p under the control of its endogenous promoter (pJM198). The Sso1p-ΔNRD plasmid, driven by the galactose-inducible GAL1-10 promoter, was transformed into the plasmid shuffle strain and maintained on glucose. Sso1p-ΔNRD expression was induced by shifting to growth on 2% galactose and was plated onto synthetic complete media lacking histidine with the drug 5-fluoroorotic acid (5-FOA). This drug is metabolized to a toxic intermediate in cells that express the URA3 gene product, thereby counterselecting for this marker. Because the wild-type Sso1p on the URA3 plasmid is required for viability, only coexpressing plasmids that produce a functional Sso1p will survive in the presence of 5-FOA. Fig. 1 D shows threefold serial dilutions of cells spotted on 5-FOA containing media. Wild-type Sso1p-HA driven by the GAL1-10 promoter was also included as a positive control. These data show that Sso1p-ΔNRD cannot support the required function of Sso1p in vivo, similar to previous results with different length NH2-terminal truncations (Munson et al., 2000). The validity of this negative result was bolstered by the conformation of Sso1p-ΔNRD protein expression (Fig. 1 B) and, more importantly, by appropriate localization to the yeast plasma membrane (Fig. 1 C).
What is the role of the NRD in vivo? Several possibilities can be envisioned. The NRD of Sso1p could interact with regulatory proteins that control fusion independently of t-SNARE complex formation, thereby serving as a scaffold for recruiting other proteins to the site of fusion. Alternatively, the NRD could serve as an intramolecular chaperone preventing inappropriate associations with the Sso1p SNARE “core” domain (also known as the H3 domain; Fig. 2 ). This function would also control access of Sec9p to the H3 region of Sso1p, thereby serving as a kinetic barrier regulating t-SNARE complex formation. We can begin to discriminate between these possibilities by modulating the ability of Sso1p and Sec9p to form t-SNARE complexes in vivo. If the NRD is regulating access by chaperoning the H3 domain, then increasing the local concentration of Sec9p, which is normally present at levels 5–10-fold less than Sso1p in wild-type yeast (see Fig. 7; Lehman et al., 1999), may allow yeast to survive without the NRD of Ssop. However, if the NRD is a scaffold, it will be required regardless of the extent of t-SNARE complex formation. An increase in overall t-SNARE complex formation could be achieved in a variety of ways, including an increase in global expression of soluble Sec9p by plasmid-based overexpression. However, this may not substantially increase Sec9p levels that have intimate contact with Sso1p or Sso2p. The ultimate means to achieve a stoichiometric level of Sec9p at the precise place where Ssop is located would be to covalently attach Sec9p to Sso1p, resulting in all three helices of the t-SNARE complex being expressed as a single protein. Such a chimeric protein would allow an intramolecular t-SNARE complex to form and remove any kinetic barrier.
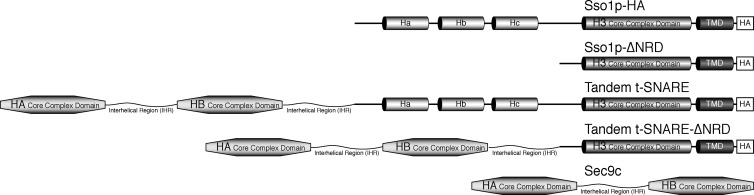
Figure 2.
Domain structure of t-SNARE proteins. Schematic representation of the domain structure of Sso1p, Sso1p-ΔNRD, Sec9c, and the chimeric tandem t-SNAREs. Sso1p-ΔNRD contains amino acids 179–290 of Sso1p. Sec9c is the SNAP25 homologous portion of Sec9p (amino acids 401–651). The tandem t-SNARE contains Sec9c and an additional copy of the Sec9p interhelical region (IHR; amino acids 499–588) covalently linked to the NH2 terminus of Sso1p. The additional Sec9p IHR allows sufficient conformational freedom to assume the necessary parallel orientation. The tandem t-SNARE–ΔNRD contains Sec9c, an additional copy of the Sec9p IHR (amino acids 499–588) linked to the NH2 terminus of Sso1p-ΔNRD. HA, helix A; HB, helix B; TMD, transmembrane domain.
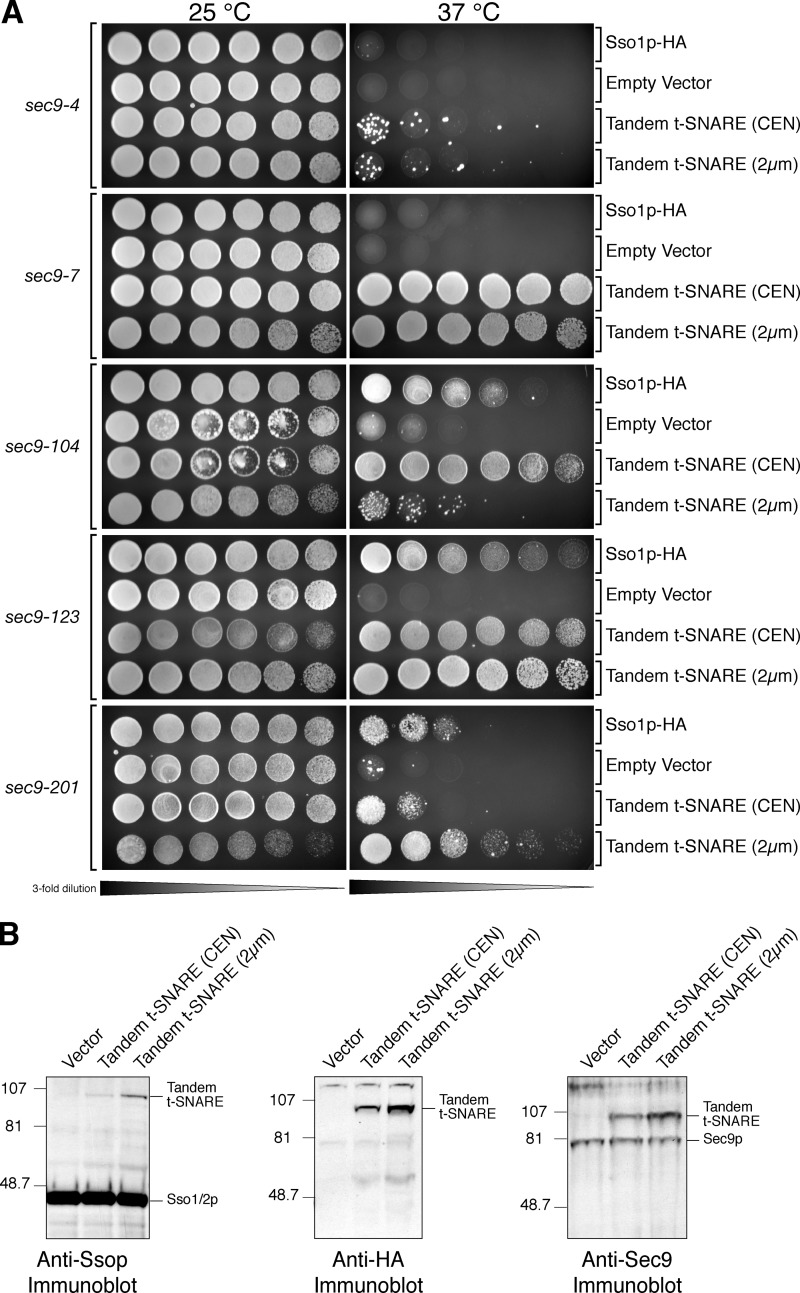
Figure 7.
The tandem t-SNARE provides Sec9p function. (A) Threefold serial dilutions of each indicated cell culture (sec9-4 [JMY335-338], sec9-7 [JMY339-342], sec9-104 [JMY343-346], sec9-123 [JMY347-350], and sec9-201 [JMY351-354]) were spotted onto plates containing synthetic complete media with 2% glucose. One plate was grown at permissive temperature (25°C), and one was grown at restrictive temperature (37°C) for ∼2.5 d. The tandem t-SNARE efficiently complements the sec9-7ts, sec9-104ts, sec9-123ts, and sec9-201ts alleles but minimally complements the sec9-4ts allele. (B) Tandem t-SNARE expression levels. Whole cell extracts of JMY363 (vector), JMY364 (tandem t-SNARE; CEN), and JMY365 (tandem t-SNARE; 2 μm) were prepared by glass bead lysis. The indicated whole cell extracts were resolved by SDS-PAGE on a 4–10% BisTris NuPAGE gel (left and middle) and blotted with anti-Ssop antisera (∼95 μg of total protein per lane, left) or anti-HA antisera (∼75 μg of total protein per lane, middle). Additionally, a 7% standard gel (∼110 μg of total protein per lane, right) was probed with anti-Sec9 antisera.
Creation of tandem t-SNAREs
To this end, we created a series of chimeric proteins termed tandem t-SNAREs that physically link the SNAP25 homologous region of Sec9p to Sso1p. Fig. 2 illustrates a schematic representation of these proteins. We appended Sec9c (amino acids 401–651) and an additional copy of the Sec9p interhelical region (IHR; amino acids 499–588) in between Sec9c and Sso1p to the NH2 terminus of full-length Sso1p (tandem t-SNARE). The additional Sec9p IHR was added to produce a chimera that contains sufficient conformational freedom to form the parallel three-helix bundle t-SNARE complex. Crystallographic analysis (Munson et al., 2000) determined that the NH2 terminal end of the H3 domain begins at residue 185. The next chimera removes the NRD and linker sequences, leaving the H3 domain, the juxtamembrane region, and the transmembrane domain (amino acids 179–290; tandem t-SNARE–ΔNRD).
These chimeric proteins were expressed under the control of the inducible GAL1-10 promoter from a high-copy number vector; however, expression levels were within about one- to twofold of episomal Sso1p in the haploid plasmid shuffle strain and endogenous Sso1p and Sso2p in a wild-type strain (Figs. 3 C , 4 C , and not depicted). Expression was examined by immunoblotting with an anti-Ssop and anti-Sec9p antibody (Figs. 3 C and 4 C; see Fig. 7 B). Localization was verified by indirect immunofluorescence microscopy using a COOH-terminal HA tag encoded by the parent vector (Figs. 3 B, 4 B, and 5 B ).
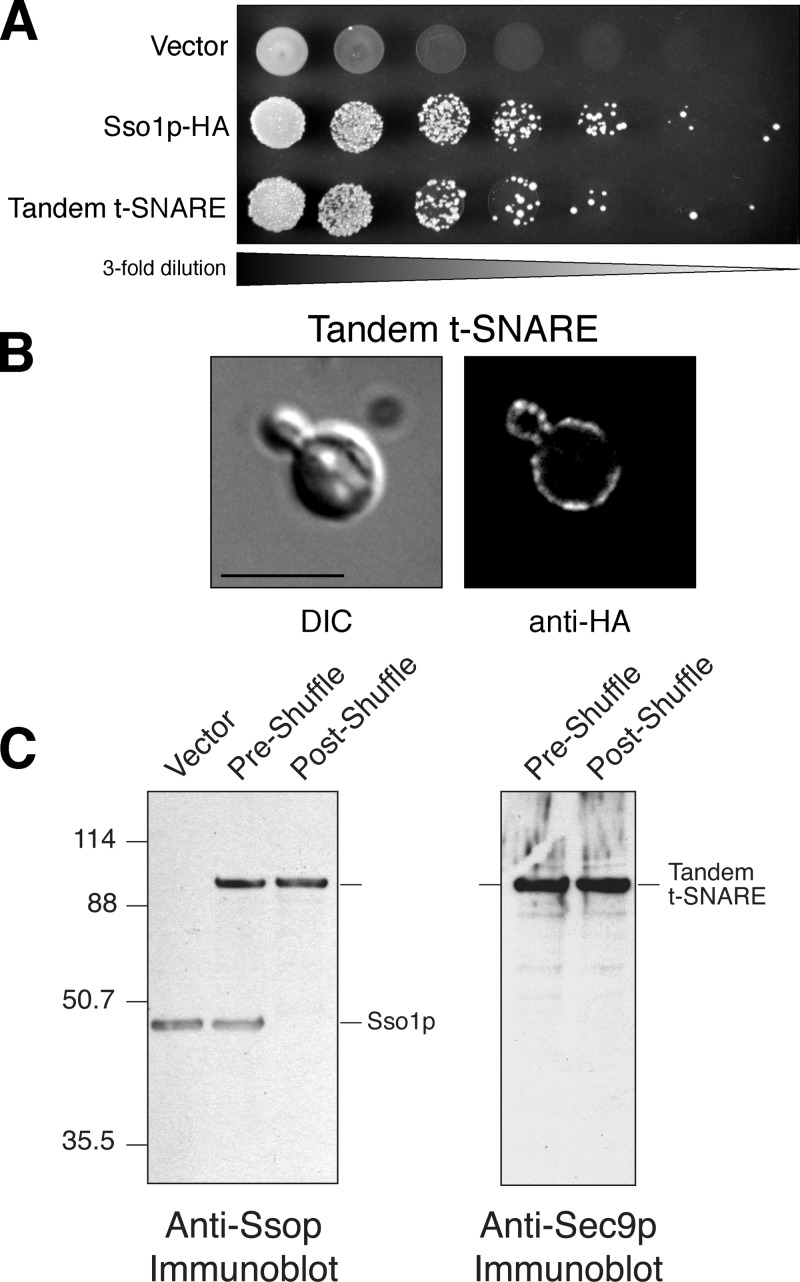
Figure 3.
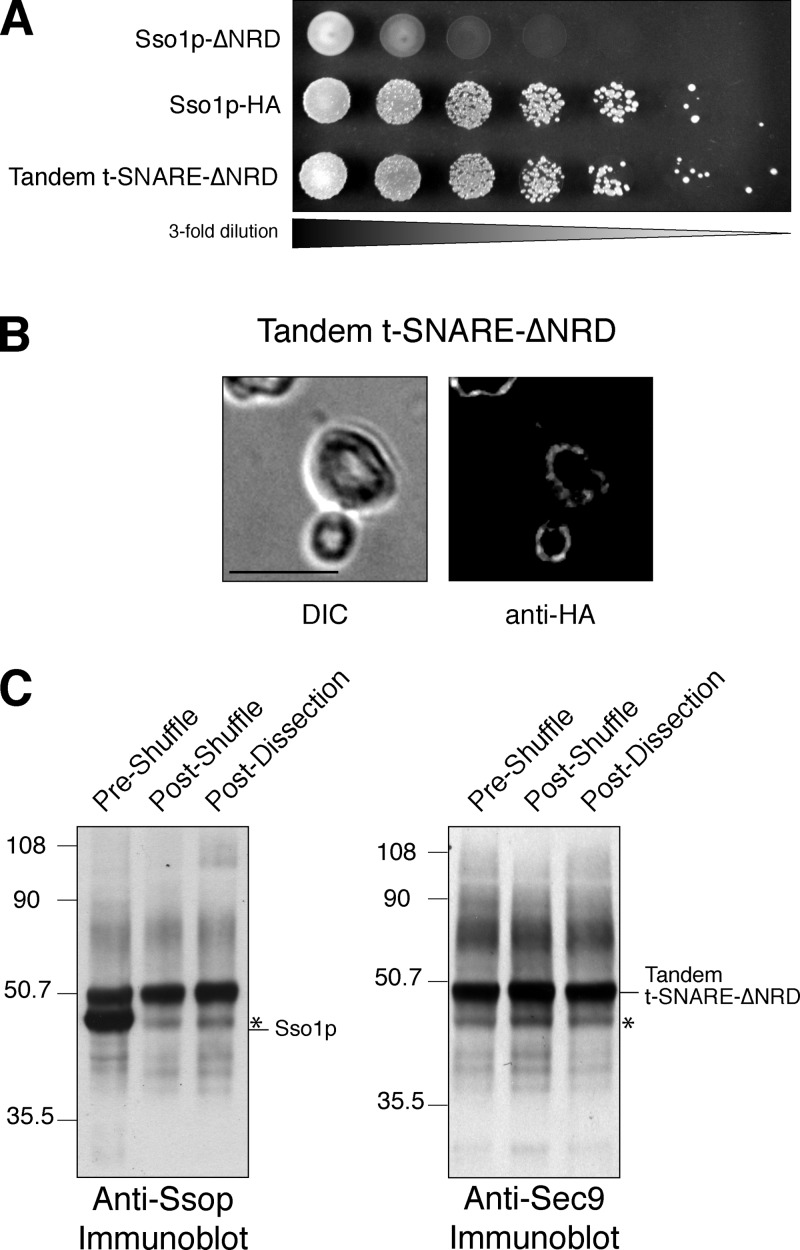
The tandem t-SNARE can serve as the sole source of Ssop. (A) Growth on 5-FOA. Threefold serial dilutions of JMY302 (empty vector), JMY303 (Sso1p-HA), and JMY306 (tandem t-SNARE) were spotted onto synthetic complete media with 2% galactose containing 1 mg/ml 5-FOA and grown at 30°C for 72 h. (B) Plasma membrane localization. Differential interference contrast (DIC) images and indirect immunofluorescence images are shown for JMY306. Localization was determined by staining with an anti-HA antibody. Bar, 5 μm. (C) Whole cell extracts of JMY302 (empty vector), JMY306 (preshuffle), and JMY307 (postshuffle) were resolved by SDS-PAGE on a 4–10% BisTris NuPAGE gel and blotted with anti-Ssop antisera (∼29 μg of total protein per lane, left) or anti-Sec9p antisera (∼118 μg of total protein per lane, right). Note that endogenous Sec9p is not visualized because it is not efficiently transferred in this gel system.
Figure 4.
The tandem t-SNARE–ΔNRD can function as the only source of Ssop. (A) Growth on 5-FOA. Threefold serial dilutions of JMY305 (Sso1 p-ΔNRD), JMY303 (Sso1p-HA), and JMY308 (tandem t-SNARE–ΔNRD) were spotted onto synthetic complete media with 2% galactose containing 1 mg/ml 5-FOA and grown at 30°C for 72 h. (B) Plasma membrane localization. Differential interference contrast (DIC) images and indirect immunofluorescence images are shown for JMY308. Localization was determined by staining with an anti-HA antibody. Bar, 5 μm. (C) Whole cell extracts of JMY308 (preshuffle), JMY309 (postshuffle), and JMY298 (postdissection) were resolved by SDS-PAGE on a 4–10% BisTris NuPAGE gel and blotted with anti-Ssop antisera (∼29 μg of total protein per lane, left) or anti-Sec9p antisera (∼115 μg of total protein per lane, right). The asterisk shows a tandem t-SNARE–ΔNRD proteolytic product that migrates at the same molecular weight as Ssop.
Figure 5.
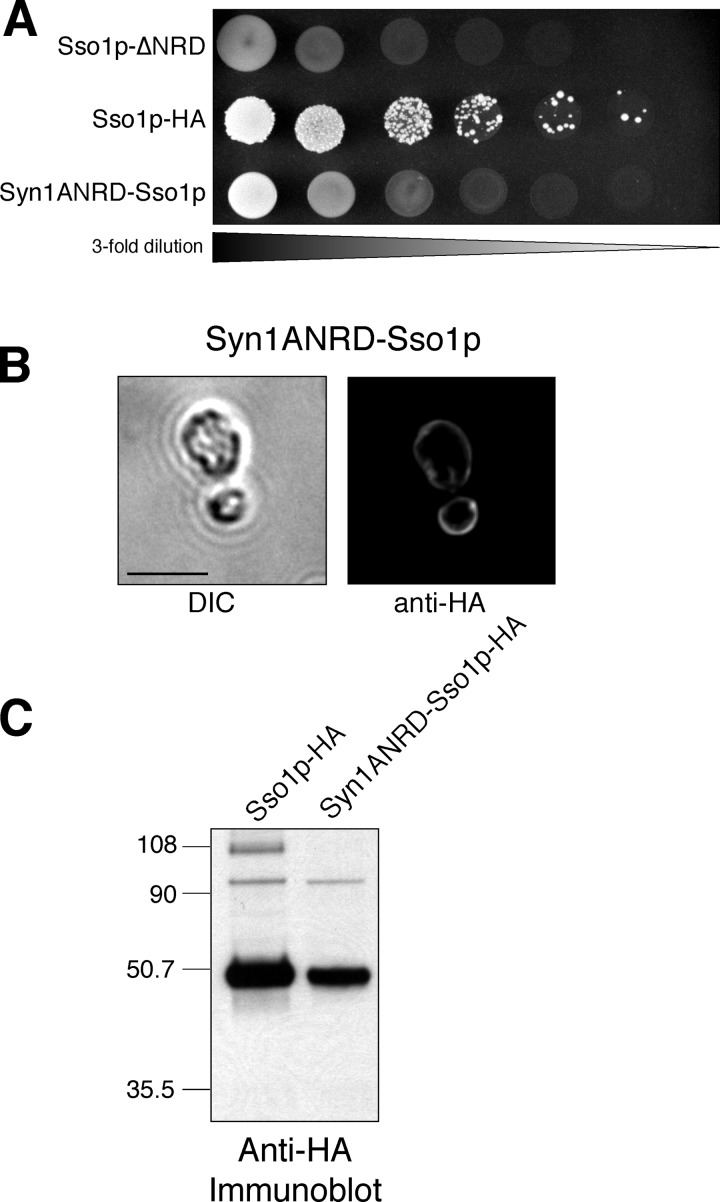
The syntaxin1A NRD cannot replace the Sso1p NRD in vivo. (A) Growth on 5-FOA. Threefold serial dilutions of JMY305 (Sso1p-ΔNRD), JMY303 (Sso1p-HA), and JMY310 (Syn1ANRD-Sso1p) were spotted onto synthetic complete media with 2% galactose containing 1 mg/ml 5-FOA and grown at 30°C for 72 h. (B) Plasma membrane localization. A differential interference contrast (DIC) image and an indirect immunofluorescence image are shown for JMY310. Localization was determined by staining with an anti-HA antibody. Bar, 5 μm. (C) Expression of the rSyn1A-Sso1p chimera. Whole cell extracts of JMY303 and JMY310 were prepared by glass bead lysis. Total protein (∼34 μg per lane) was resolved by SDS-PAGE on a 4–10% BisTris NuPAGE gel and probed with an anti-HA antibody.
The tandem t-SNARE can serve as the sole source of Sso1p
The functionality of the tandem t-SNARE plasmids was tested in the haploid plasmid shuffle strain (JMY128). Fig. 3 A shows threefold serial dilutions of cells spotted on 5-FOA containing media. Wild-type Sso1p was also included as a positive control. These data show that the full-length tandem t-SNARE is able to grow in the presence of 5-FOA nearly to the same degree as wild-type Sso1p. Cells carrying only the parental vector were unable to grow as expected. This result demonstrates that Sso1p function is not inhibited when Sec9c is physically linked to the NH2 terminus of Sso1p.
If the tandem t-SNARE is capable of providing sole Ssop function as our drug selection suggests, then the drug-selected strain, or “postshuffle” strain, should only contain the tandem t-SNARE plasmid under the control of the GAL1-10–inducible promoter as the sole source of Sso protein. Growth of this strain is completely dependent on galactose as a carbon source, suggesting that the tandem t-SNARE is the only source of functional Sso1p (unpublished data). To examine the expression levels of all sources of Sso1p, we made total yeast extracts from cells harvested before and after selection on 5-FOA. These extracts were probed with polyclonal antibodies directed against both isoforms of Ssop to detect the tandem t-SNARE and wild-type Sso1p. Fig. 3 C demonstrates that the tandem t-SNARE is the only source of Sso1p in the postshuffle strain and that it is only modestly expressed (2.2-fold above episomal Sso1p) in the plasmid shuffle strain. Proper protein localization to the plasma membrane was also confirmed by immunofluorescence microscopy (Fig. 3 B). We also measured the growth rate of the tandem t-SNARE strains before and after selection. The doubling time for cells expressing both wild-type Sso1p and the tandem t-SNARE were substantially longer than those with only wild-type Sso1p (257 vs. 497 min). Growth is restored to approximately normal rates (224 min for wild type vs. 237 min) when the tandem t-SNARE is the only source of Sso1p in the postshuffle strain. This suggests that the tandem t-SNARE behaves as a slightly dominant-negative protein when both forms of Sso1p are present and that the dominant-negative effect of the tandem t-SNARE is caused by an interaction with wild-type Sso1p. Evidence of the tandem t-SNARE dominant-negative effect can also be seen in the BY1 genetic background used for analysis of SEC9 function (see Fig. 7). In accordance with the near wild-type growth rate, the extent of secretion measured by external invertase or α factor was similar in cells expressing Sso1p-HA or the tandem t-SNARE (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200507138/DC1).
The NRD of Sso1p is dispensable in the context of a tandem t-SNARE
To examine the contribution of the NRD, we next analyzed the tandem t-SNARE lacking the NRD portion of Sso1p (tandem t-SNARE–ΔNRD) in a similar manner. Cells expressing the tandem t-SNARE–ΔNRD were selected on 5-FOA (Fig. 4 A) in parallel with a wild-type Sso1p control plasmid and the NRD-deleted Sso1p without the addition of Sec9c. Similar to the full-length tandem t-SNARE, yeast expressing both wild-type Sso1p and the tandem t-SNARE–ΔNRD were slow growing, indicating a dominant-negative phenotype (doubling time of ∼541 min). However, growth was restored to near normal rates after selection against the wild-type plasmid in the postshuffle strain (224 min for wild type vs. 241 min). Additionally, secretion of invertase in the postshuffle strains does not appear to be impaired (Fig. S1). These results suggest that the tandem t-SNARE–ΔNRD is fully functional, similar to the full-length tandem t-SNARE. Protein expression (Fig. 4 C) and localization (Fig. 4 B) were also confirmed. This experiment clearly demonstrates that the Sso1p NRD is no longer required for cell viability when Sec9c is covalently attached. In addition to counterselection on 5-FOA, the functionality of the tandem t-SNARE–ΔNRD was also examined by tetrad dissection (unpublished data). Although sporulation and germination are very poor (Herman and Rine, 1997; Deutschbauer et al., 2002; Jantti et al., 2002), haploid spores could be isolated that were disrupted for both chromosomal copies of Sso1p and Sso2p but contained the tandem t-SNARE–ΔNRD as the only source of Sso protein (Fig. 4 C).
The expression level of the tandem t-SNARE–ΔNRD was reduced relative to the full-length tandem t-SNARE. Although the tandem t-SNARE–ΔNRD is expressed from a 2-μm based multicopy plasmid under transcriptional control of the strong GAL1-10 promoter, the expression level of the chimeric protein is slightly less than (∼95%) what Sso1p expressed under the control of the SSO1 promoter from a centromeric plasmid (Fig. 4 C). The protein at approximately the same molecular weight as Sso1p in the postshuffle and postdissection extracts (Fig. 4 C, asterisk) is a proteolytic fragment derived from the tandem t-SNARE–ΔNRD detected by both the Ssop and Sec9c antibodies (Fig. 4 C).
The homologous syntaxin1A NRD cannot replace the Sso1p NRD in vivo
Next, we examined the possibility that any sequence appended to the NH2 terminus of ΔNRD-Sso1p might maintain Sso1p function. It is possible, although unlikely, that Sec9c is simply masking the Sso1p H3 domain and not forming a functional t-SNARE complex, and any sequence that interacts with the H3 domain will suffice. To test this, we added the NRD (amino acids 1–182) of the homologous neuronal syntaxin rat syntaxin1A. Although Sso1p and syntaxin1A are only 27% identical (40% similar) in amino acid sequence, both fold into a very comparable closed conformation. When the structure of Sso1p was compared with syntaxin1A in the syntaxin–n–Sec1 complex (a stabilized closed conformation), the root mean square deviation was only 1.1 Å for 111 Cα pairs (Munson et al., 2000). Although the syntaxin1A NRD is structurally similar to the endogenous Sso1p NRD, the rSyn1A-Sso1p chimera was unable to provide Ssop function even though it was well expressed and localized to the plasma membrane (Fig. 5).
A point mutation in the Sec9c portion of the tandem t-SNARE eliminates Sso1p function in vivo
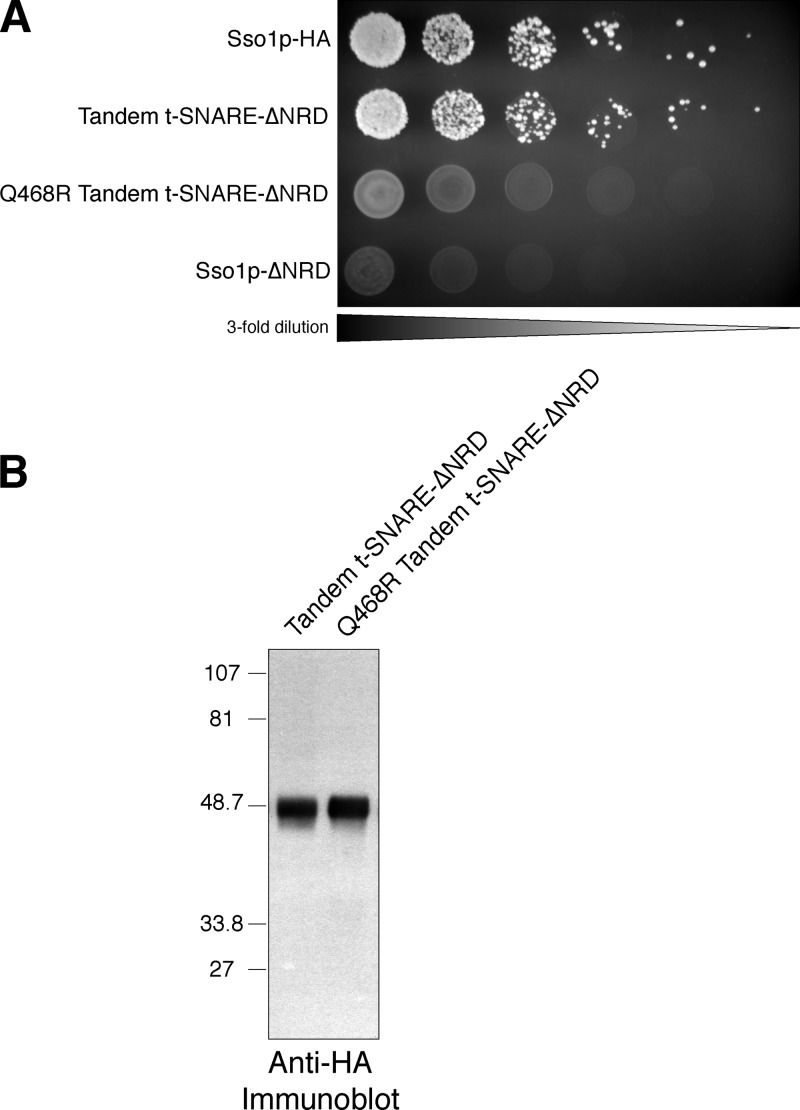
Although we are confident that the covalently attached Sec9c and Sso1p are working in tandem as an intramolecular t-SNARE complex, we sought to confirm this by creating a specific point mutation in Sec9c that should abolish SEC9 function. We mutated a conserved glutamine residue in helix A of Sec9c (Q468 in the full-length Sec9p) to an arginine. This residue is one of four residues, one from each SNARE core helix, that compose the so-called “zero layer” or ionic layer in the SNARE coil (Sutton et al., 1998). In the prototypical neuronal SNARE structure, as well as in the yeast plasma membrane counterpart, these four residues consist of three glutamines and one arginine (3Q:1R). One glutamine is contributed by the SNARE core domain of Sso1p, the second is from helix A of Sec9p, and the third is contributed by helix B of Sec9p. The single arginine in the ionic layer is provided by Snc1/2p. These residues have a defined role in SNARE complex formation and are thought to set the “register” of the SNARE domain coiled-coil. A previous study has shown that a single mutation of Q468R renders Sec9p nonfunctional in vivo and reduces SNARE complex assembly in vitro (Katz and Brennwald, 2000). We reasoned that the Q468R mutation in the context of the tandem t-SNARE–ΔNRD would also produce a nonfunctional t-SNARE complex by this very specific change. Fig. 6 shows that the Q468R tandem t-SNARE–ΔNRD is unable to provide Sso1p function. This experiment demonstrates that a single residue change in Sec9c, known to disrupt Sec9p function, abolishes the function of the attached Sso1p, strongly supporting our assertion that the Sec9c–Sso1p–ΔNRD chimera is functioning as an intramolecular t-SNARE complex.
Figure 6.
The Q468R point mutation in the Sec9c segment of the tandem t-SNARE abolishes Sso1p function. (A) Growth on 5-FOA. Threefold serial dilutions of JMY303 (Sso1p-HA), JMY308 (tandem t-SNARE–ΔNRD), JMY334 (Q468R tandem t-SNARE–ΔNRD), and JMY305 (Sso1p-ΔNRD) were spotted onto synthetic complete media with 2% galactose containing 1 mg/ml 5-FOA and grown at 30°C for 72 h. (B) Whole cell extracts of JMY308 (tandem t-SNARE–ΔNRD) and JMY334 (Q468R tandem t-SNARE–ΔNRD) were resolved by SDS-PAGE on a 4–10% BisTris NuPAGE gel and blotted with an anti-HA antisera (∼21 μg of total protein per lane).
The tandem t-SNARE provides Sec9p function
Our functional analysis of the tandem t-SNARE chimeras also included examining Sec9p function. The tandem t-SNARE, under transcriptional control of the SSO1 promoter, was transformed into a wild-type yeast strain as well as several temperature-sensitive SEC9 mutant alleles. These strains were spotted onto synthetic media containing glucose and grown at permissive temperature (25°C) or restrictive temperature (37°C). Fig. 7 shows that the tandem t-SNARE on both low- (CEN) and high- (2 μm) copy plasmids permits efficient growth of sec9-7, sec9-123, sec9-104, and sec9-201, confirming that tandem t-SNAREs also provide Sec9p function, likely by intragenic complementation. Interestingly, the high- and low-copy vectors have differential effects with the various sec9ts alleles. Higher expression of the tandem t-SNARE more efficiently complements sec9-123 and sec9-201, whereas the low-copy vector is more effective with sec9-7 and possibly sec9-104 (Fig. 7 A). The tandem t-SNARE also minimally complements the sec9-4 allele under both conditions (Fig. 7 A). However, the tandem t-SNARE does not function as the only source of SEC9, as it was unable to complement a SEC9-null allele (not depicted).
The expression level of the tandem t-SNAREs and the expression relative to total endogenous Sso1p and Sso2p as well as endogenous Sec9p is measured in Fig. 7 B. The centromeric tandem t-SNARE is ∼5% of endogenous Sso1/2p, whereas the 2-μm tandem t-SNARE is ∼13% (Fig. 7 B, left). The amount of tandem t-SNARE from the CEN vector is very similar to endogenous levels of Sec9p (Fig. 7 B, right). Growth of the sec9 ts alleles at the nonpermissive temperature is not likely a result of suppression by overexpression of the Sso1p component of the tandem t-SNARE because low levels of the protein are expressed compared with endogenous Sso1p. However, some suppression of sec9-104, sec9-123, and sec9-201 is observed when Sso1p alone is expressed from its endogenous promoter on a centromeric plasmid. Conversely, no growth at 37°C is seen in the sec9-4 and sec9-7 strains when Sso1p alone was expressed from its own promoter on a centromeric plasmid (Fig. 7) or overexpressed with the alcohol dehydrogenase promoter on a 2-μm vector (not depicted). We were not able to complement any of the sec9 ts alleles with the tandem t-SNARE lacking the NRD, most likely because of significantly lower expression of these proteins and the added complication of all tandem t-SNAREs behaving as dominant-negative mutants when wild-type Sso1/2p is present (Fig. 7 A and not depicted).
The tandem t-SNARE does not suppress other late-acting Sec mutants
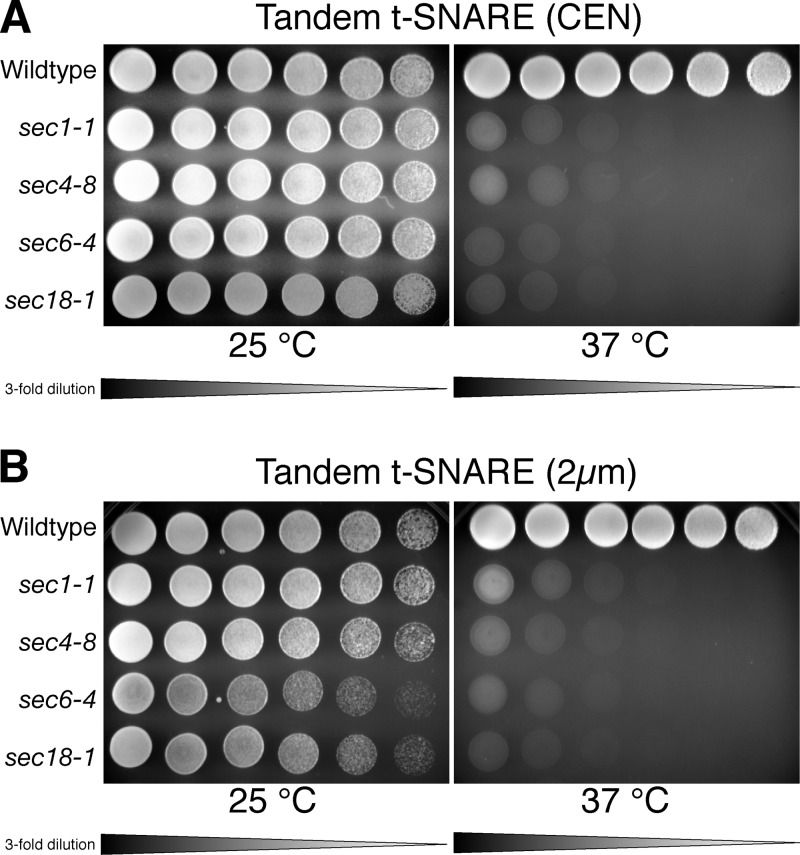
An intramolecular t-SNARE complex also affords us the opportunity to examine other late-acting sec mutants thought to play a role in regulated SNARE assembly. We transformed four sec mutants with the tandem t-SNARE in high (Fig. 8 A ) and low copy (Fig. 8 B). We examined the SNARE regulator Sec1p, the Rab-GTPase family member Sec4p, the exocyst component Sec6p, and the general SNARE chaperone Sec18p. The tandem t-SNARE was unable to suppress mutations in these genes, suggesting that they function in roles other than, or in addition to, t-SNARE complex formation.
Figure 8.
The tandem t-SNARE does not suppress other late-acting sec mutants. Threefold serial dilutions of each indicated cell culture were spotted onto plates containing synthetic complete media with 2% glucose. Three late-acting sec mutants were transformed with the tandem t-SNARE in low copy (A; sec1-1 [JMY355], sec4-8 [JMY356], and sec6-4 [JMY357]) or high copy (B; sec1-1 [JMY359], sec4-8 [JMY360], and sec6-4 [JMY361]) and grown at permissive (25°C) or restrictive temperature (37°C) for ∼2.5 d. The general SNARE chaperone Sec18 (sec18-1 [JMY358 and JMY362]) was used as a control. No suppression was observed for these mutants.
Discussion
In this study, we show that Sso1p lacking the NRD is fully functional for fusion in vitro (Fig. 1 A) but cannot provide Sso1p function in vivo (Figs. 1 D, 4 A, and 5 A), suggesting that the NRD is important for regulating SNARE complex formation. Replacement of the Sso1p NRD by the homologous sequence from the neuronal plasma membrane syntaxin, syntaxin1A, did not restore function (Fig. 5). However, we show that plasma membrane syntaxin function can be provided by a chimeric protein composed of Sso1p covalently linked to the t-SNARE light chains provided by Sec9c (Fig. 3). We also demonstrate that yeast can dispense with the NRD of Sso1p in the context of a tandem t-SNARE, illustrating that this otherwise essential portion of Sso1p is no longer required if the other t-SNARE component is physically connected (Fig. 4).
Much to our surprise, the tandem t-SNARE could not provide full Sec9p function. Although we were able to complement five sec9 ts alleles to some degree, we could not complement a SEC9 null with a tandem t-SNARE plasmid (Fig. 7 and not depicted). We suggest that restoration of growth by the tandem t-SNARE in the sec9 ts alleles does indicate that the tandem t-SNARE provides Sec9p function because the various sec9 ts mutations map to different sites of the protein. These observations suggest the intriguing possibility that Sec9p has roles in addition to exocytosis. The nature of the tandem t-SNARE chimera is such that we permanently affix Sec9c to the plasma membrane, preventing it from cycling through the cytoplasm. The mechanism of wild-type Sec9p membrane association is largely unknown, but is unlikely to be entirely through its interaction with Sso1/2p. If Sec9p provides another required function not localized to the plasma membrane, our chimera would not support this role. Recent results with the neuronal isoform of Sec9p, SNAP25, suggest that an additional function may be on internal membranes because SNAP25 was found to drive membrane fusion when complexed with syntaxin 13 on an endosomal membrane (Sun et al., 2003).
Another interpretation of our results is that Sec9c is simply replacing the chaperone function of the Sso1p NRD rather than functioning as an active participant in a t-SNARE complex. In this case, endogenous Sec9p would be required for viability. Although this formally remains a possibility as a result of our inability to cover a Sec9p-null allele, our intragenic complementation of multiple temperature-sensitive SEC9 alleles makes this possibility unlikely. Furthermore, this interpretation does not change our primary conclusion that the NRD cannot be required as a scaffold given that Sso1p without an NRD is still functional. Additionally, careful consideration of the amount of endogenous Sec9p and its localization relative to the tandem t-SNARE further limits the possibility that the physically attached Sec9c is chaperoning access of the Sso1p H3 domain for endogenous Sec9p. In our experiments examining Sso1p function, the tandem t-SNARE is expressed at >10 times the concentration of total endogenous Sec9p, which is present both in the cytoplasm and on membranes. For endogenous Sec9p to function with the truncated Sso1p in the context of the tandem t-SNARE, it must first attach to the plasma membrane if it is present in the soluble pool or, minimally, locate the Sso1p portion of the tandem t-SNARE in the plane of the bilayer. Next, it must be present in a sufficiently high concentration that it can displace, or minimally compete with, a covalently attached version of itself that is essentially at an infinite concentration because it is permanently attached. Such a concentration of Sec9p on the membrane in the vicinity of Sso1p seems unachievable. Additionally, we have shown that a single point mutation in the Sec9c portion of the tandem t-SNARE–ΔNRD eliminates Sso1p function (Fig. 6). In the context of the Q468R mutant, the interaction of the Sec9c portion of the tandem t-SNARE is likely reduced; however, this construct is nonfunctional in a situation where endogenous Sec9p should have greater access to the H3 domain of Sso1p. Finally, the addition of the homologous NRD from syntaxin 1A does not produce a functional chimera, a domain that has a reasonable possibility of providing a chaperone function for the Sso1p H3 domain.
We also found that both the full-length and NH2-terminally truncated tandem t-SNAREs were dominant interfering when a wild-type copy of Sso1p was present in the same cells. This result suggests that wild-type Sso1p may interact with the tandem t-SNARE. One possible explanation for this observation is that a four-helix complex composed of the core H3 domain of endogenous Sso1p and the three-helix bundle of the tandem t-SNARE form in the same membrane. This dead-ended cis-SNARE complex would likely be toxic and difficult for the SNARE recycling machinery Sec17p and Sec18p to resolve and recycle. Clearly, other interpretations are also possible.
Much of what we know about the in vivo function of the Sso1p NRD was derived from work that generated mutants of Sso1p inefficient at forming a “closed” conformation. Munson and Hughson (2002) found that targeted mutations “opened” the Sso1p structure and virtually eliminated the strong kinetic impediment to t-SNARE complex formation with Sec9c in vitro. They also determined that these open mutants could provide Sso1p function in vivo. These findings suggest that although deletion of the NH2 terminus was fatal, significantly changing the rate of t-SNARE complex formation was tolerated substantially. One interpretation was that additional machinery may use the Sso1p NH2 terminus as a scaffold for their function during fusion. On the surface, our results are seemingly at odds with this interpretation; however, a closer evaluation of the results with the open mutants suggests that this may not be the case. Although mutations in the noncore domain in Sso1p strongly affect its ability to form the closed conformation, it does not eliminate it. The observed in vitro enhancement of t-SNARE complex formation is large (1,100–1,300×), but not as large as removing the NH2 terminus entirely (∼3,000×). As Nicholson et al. (1998) noted, there is still a factor of three “inhibition” with the open mutants, and all of these measurements are kinetic effects in vitro. Additionally, Sso1p is in large excess (∼10×) relative to its partner Sec9 (Lehman et al., 1999). These results clearly make room for the possibility that the mutant Sso1p can still be closed, although not nearly as well. We would argue that the open mutants can still spend a significant portion of their lifetime in a closed conformation in vivo, allowing for the interpretation that the NRD is still influencing t-SNARE complex formation.
Our data strongly suggest that the NRD of Sso1p and, by extension, all plasma membrane syntaxins are involved in facilitating t-SNARE complex formation or preventing inappropriate associations with the H3 portion of Sso1p. However, our data effectively eliminate a scaffold function of the NRD to recruit other required components to the site of SNARE action.
Materials and methods
Reagents
All lipids were purchased from Avanti Polar Lipids, Inc.; detergents were obtained from Calbiochem (n-octyl β-glucopyranoside) and Fisher Scientific (Triton X-100); 5-FOA was purchased from Zymo Research; and G418 sulfate was obtained from Research Product International Corp. Bacterial and yeast media components, including yeast peptone dextrose (YPD), synthetic complete media, raffinose, and amino acid and nucleotide supplements, were obtained from Qbiogene; yeast nitrogen base was purchased from Difco; bacto agar was obtained from BD Biosciences; and the carbon sources glucose and galactose were purchased from Fisher Scientific. Restriction endonucleases were purchased from New England Biolabs, Inc. Tgo polymerase was obtained from Roche, and oligonucleotides were purchased from Integrated DNA Technologies. The monoclonal anti-HA 16B12 antibody was purchased from Covance, and secondary antibodies were purchased from the following companies: goat anti–mouse IgG HRP from Rockland Immunochemicals; goat anti–rabbit IgG Fc HRP from Pierce Chemical Co.; and AlexaFluor488 goat anti–mouse IgG from Invitrogen.
Strain construction
All strains made for this study are in the W303 background. JMY120 (Table I ) is a haploid strain with the SSO1 loci disrupted with the kanamycin resistance gene. The genetic disruption was produced by creating a kanMX2 fragment of 1,450 bp from pFA6 (Wach et al., 1994) using oligonucleotides 36 and 37 (Table II ). The PCR-amplified product was transformed into W3031A, plated onto YPD plates, and grown overnight at 30°C. The samples were then replica plated onto YPD plates with 200 μg/ml G-418 sulfate. Confirmation of sso1 deletions was performed by PCR analysis. H404 (obtained from H. Ronne, Swedish University of Agricultural Sciences, Uppsala, Sweden; Aalto et al., 1993) is a haploid strain with the SSO2 loci disrupted with the LEU2 gene. JMY123 is the diploid from the cross of JMY120 with H404. JMY128 was made by sporulating JMY123 transformed with pJM198 followed by tetrad dissection to obtain the double-deletion plasmid shuffle strain. JMY298 was made by sporulating JMY123 transformed with pJM311 followed by tetrad dissection to obtain the double-deletion strain with the tandem t-SNARE–ΔNRD.
Table I. Oligonucleotides.
| Oligo no. | Oligo name | Sequences |
|---|---|---|
| 36 | Sso1 kan | TTACAATTAAAAAAGGCAATTAAAAATAGAAACAAATCAAATGAGT TATAATAATGCTTGCCTCGTCCCCGCCGCGTCA |
| 37 | kan Sso1 | TTGATATACAAAAGGGGAGTTCGGATAGAATAGAAATATAGAAAAT AGTTGGAAGCGCACTTAACTTCGGATCTGGGCAG |
| 41 | Sso1-4 | GAGATATCCTCGAGACGCGTTTTGACAACAGCTGGG |
| 42 | Sso1-7 | GCGAATTCCTCGAGCTTAATGGACTTCCTGGAGGAGG |
| 43 | Sso1-8 | CGTCTAGAGCGGCCGCGAATGAGGTAGCATGTGAAAACG |
| 44 | Sso1-9 | GCGAATTCATGAATGCTAACAGACGTGG |
| 211 | Sso1-14 | GGTCCGGAAATGCTAACAGACGTGG |
| 216 | Sso1-15 | GGGAGCTCGCTTAATGGACTTCCTGGAGGAGG |
| 217 | Sso1-16 | CGTCTAGAGATTTGTTTCTATTTTTAATTGCC |
| 218 | Sso1-17 | GCCTCGAGTAATTCCAACTATTTTCTATATTTC |
| 219 | Sso1-18 | CCGGTACCGAGGTAGCATGTGAAAACGCGGGAG |
| 113 | Sec9c-1 | CGCCATGGAATTCATGCAGCGTGGTTACAAAAC |
| 179 | Sec9c-3 | TAGGATCCCCTCCGGAGATATCGATACGTATACCTGCCAGACGG |
| 180 | Sec9c-4 | CCATCGATCTGAACCGTTCTATCCTGGC |
| 181 | Sec9c-5 | TAGGATCCCCTCCGGAGTCATCTTCTTCATCGTTCTC |
| 301 | EcoRI-Syn1A-Sso1 | GCGAATTCATGAAGGACCGAACCCAGG |
| 302 | Syn1A-Sso1-ClaI | GCTCCGGAATCGATGATACCAGAGGCAAAGATG |
| 325 | Sec9c Q468Rf | GGGTATGCTGGGTCATCGATCTGAACAGCTGAAC |
| 326 | Sec9c Q468Rr | GTTCAGCTGTTCAGATCGATGACCCAGCATACCC |
Table II. Yeast strains.
| Strain name | Genotype | Source |
|---|---|---|
| W3031A | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11,15 can1-100 | R. Rothsteina |
| JMY120 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11,15 can1-100 sso1::kanMX2 | This study |
| H404 | MATα ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11,15 can1-100 sso2-d1::LEU2 | H. Ronne |
| JMY123 | MATa/MATα ade2-1/ade2-1 leu2-3,112/leu2-3,112 ura3-1/ura3-1 trp1-1/trp1-1 his3-11,15/his3-11, 15 can1-100/can1-100 SSO2/sso2-d1::LEU2 SSO1/sso1::kanMX2 | This study |
| JMY128 | MATα ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11,15 can1-100 sso1::kanMX2 sso2-d1::LEU2 [pJM198] | This study |
| JMY298 | MATα ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11,15 can1-100 sso1::kanMX2 sso2-d1::LEU2 [pJM311] | This study |
| JMY302 | JMY128 [pJM198], [pYX223] | This study |
| JMY303 | JMY128 [pJM198], [pJM290] | This study |
| JMY304 | JMY128 [pJM290] | This study |
| JMY305 | JMY128 [pJM198], [pJM222] | This study |
| JMY306 | JMY128 [pJM198], [pJM293] | This study |
| JMY307 | JMY128 [pJM293] | This study |
| JMY308 | JMY128 [pJM198], [pJM311] | This study |
| JMY309 | JMY128 [pJM311] | This study |
| JMY310 | JMY128 [pJM198], [pJM414] | This study |
| JMY334 | JMY128 [pJM198], [pJM429] | This study |
| JMY335 | BY41 [pJM198] | This study |
| JMY336 | BY41 [pRS426] | This study |
| JMY337 | BY41 [pJM334] | This study |
| JMY338 | BY41 [pJM427] | This study |
| JMY339 | BY70 [pJM198] | This study |
| JMY340 | BY70 [pRS426] | This study |
| JMY341 | BY70 [pJM334] | This study |
| JMY342 | BY70 [pJM427] | This study |
| JMY343 | BY392 [pJM198] | This study |
| JMY344 | BY392 [pRS426] | This study |
| JMY345 | BY392 [pJM334] | This study |
| JMY346 | BY392 [pJM427] | This study |
| JMY347 | BY200 [pJM198] | This study |
| JMY348 | BY200 [pRS426] | This study |
| JMY349 | BY200 [pJM334] | This study |
| JMY350 | BY200 [pJM427] | This study |
| JMY351 | BY445 [pJM198] | This study |
| JMY352 | BY445 [pRS426] | This study |
| JMY353 | BY445 [pJM334] | This study |
| JMY354 | BY445 [pJM427] | This study |
| JMY355 | BY29 [pJM334] | This study |
| JMY356 | BY33 [pJM334] | This study |
| JMY357 | BY37 [pJM334] | This study |
| JMY358 | RSY271 [pJM334] | This study |
| JMY359 | BY29 [pJM427] | This study |
| JMY360 | BY33 [pJM427] | This study |
| JMY361 | BY37 [pJM427] | This study |
| JMY362 | RSY271 [pJM427] | This study |
| JMY363 | BY1 [pRS246] | This study |
| JMY364 | BY1 [pJM334] | This study |
| JMY365 | BY1 [pJM427] | This study |
| BY1 | MATa ura3-52 | P. Brennwald |
| BY70 | MATa ura3-52 sec9-7 | P. Brennwald |
| BY41 | MATa ura3-52 sec9-4 | P. Brennwald |
| BY392 | MATa ura3-52 sec9-104 | P. Brennwald |
| BY200 | MATa ura3-52 sec9-123 | P. Brennwald |
| BY445 | MATa ura3-52 sec9-201 | P. Brennwald |
| BY33 | MATa ura3-52 sec4-8 | P. Brennwald |
| BY37 | MATa ura3-52 sec6-4 | P. Brennwald |
| BY29 | MATa ura3-52 sec1-1 | P. Brennwald |
| RSY271 | MATα ura3-52 his4-619 sec18-1 | R. Schekmanb |
Columbia University, New York, NY.
University of California, Berkeley, Berkeley, CA.
Plasmid construction
All plasmids were propagated in the E. coli strain DH5α, and standard DNA manipulation techniques were used. All PCR procedures were performed with Tgo polymerase. All other DNA modifying enzymes were obtained from New England Biolabs, Inc. pJM198 (Table III ) is a yeast expression vector coding for Sso1p under the transcriptional control of its own promoter. A 1,615-bp PCR fragment containing the SSO1 promoter, ORF, and terminator was generated from genomic DNA and the oligonucleotides 42 and 43. This fragment was cut with EcoRI and XbaI and ligated into pRS316 (Sikorski and Hieter, 1989) cut with the same enzymes. pJM222 is a yeast expression vector coding for the Sso1p-ΔNRD under the transcriptional control of the GAL1-10 promoter. A 356-bp fragment coding for amino acids 179–290 was generated by PCR using pJM88 (McNew et al., 2000) as template DNA and the oligonucleotides 41 and 44. This fragment was cut with EcoRI and MluI and ligated into pYX223 (Invitrogen) cut with the same enzymes. pJM290 was previously described (Scott et al., 2004). pJM291 is a yeast expression vector coding for the tandem t-SNARE under the transcriptional control of the GAL1-10 promoter. It was generated by PCR with oligonucleotides 113 and 179 using pJM89 as a template. The 794-bp PCR product was cut with EcoRI and BamHI, and the 794-bp fragment was ligated into pJM290 cut with the same enzymes. pJM293 is a yeast expression vector coding for the tandem t-SNARE with IHR under the transcriptional control of the GAL1-10 promoter. It was generated with PCR with oligonucleotides 180 and 181 and pJM89 as a template. The 294-bp fragment of the Sec9 IHR was digested with ClaI and BspEI and ligated into pJM291 cut with the same enzymes. pJM311 is a yeast expression vector coding for the tandem t-SNARE–ΔNRD under transcriptional control of the GAL1-10 promoter. It was generated with PCR using the oligonucleotides 211 and 41 with pJM88 as a template. The 358-bp PCR product was digested with BspEI and MluI and ligated into pJM293 cut with the same enzymes. pJM334 is a yeast expression vector coding for the tandem t-SNARE with IHR under the transcriptional control of the SSO1 promoter. The ∼1,977-bp SSO1 fragment was generated by digesting pJM293 with EcoRI and XhoI. The ∼496-bp fragment containing the 5′ untranslated region of SSO1 was PCR amplified from Saccharomyces cerevisiae genomic DNA as a template with oligonucleotides 216 and 217. The ∼310-bp fragment containing the 3′ untranslated region of SSO1 was PCR amplified from S. cerevisiae genomic DNA as a template with oligonucleotides 218 and 219. All of these fragments were assembled into pRS316, an URA-CEN plasmid (Sikorski and Hieter, 1989). pJM414 is a yeast expression vector coding for the rat syntaxin1A NRD-appended Sso1p-ΔNRD under transcriptional control of the GAL1-10 promoter. A 565-bp fragment coding for amino acids 1–178 of rat syntaxin1A was generated by PCR using pTW20 (Weber et al., 1998) as template DNA and oligonucleotides 301 and 302. This fragment was cut with EcoRI and ClaI and ligated into pJM311 cut with the same enzymes. pJM427 is a high-copy yeast expression vector coding for the tandem t-SNARE with IHR under the transcriptional control of the SSO1 promoter. The ∼2,783-bp fragment tandem t-SNARE with SSO1 promoter and terminator was generated by digesting pJM334 with KpnI and SacI and ligated into pRS426 cut with the same enzymes. pJM429 is a yeast expression vector coding for the tandem t-SNARE–ΔNRD with a Q468R point mutation in the Sec9p helix A and is under the transcriptional control of the GAL1-10 promoter. PCR sewing was used to make the specific point mutation. The first round of PCR used oligonucleotides 113 and 326 with pJM311 as the template to generate a 222-bp fragment and used oligonucleotides 41 and 325 with pJM311 as the template to generate a 1,177-bp fragment. The second round of PCR used oligonucleotides 113 and 41, with the two PCR products from the first round as template DNA to generate a ∼1,380-bp fragment. This fragment was cut with EcoRI and BspEI and ligated into pJM311 cut with the same enzymes. pJM367 (GST-Sso1p-ΔNRD) was a gift from Y.-K. Shin (Iowa State University, Aimes, IO) and has been previously described (Chen et al., 2004).
Table III. SNARE constructs.
| Plasmid | Chimera | Vector | Origin | Marker | Promoter |
|---|---|---|---|---|---|
| pJM88 | His8-Sso1p | pET24 | ColE1 | KAN | T7 |
| pJM90 | Snc2p-His6 | pET28a | ColE1 | KAN | T7 |
| BB442 | GST-Sec9c | pGEX-2T | ColE1 | AMP | Ptac |
| pJM367 | GST-Sso1p-ΔNRD | pGEX-KG | ColE1 | AMP | Ptac |
| pJM198 | Sso1p (no tag) | pRS316 | CEN6 | URA3 | SSO1 |
| pJM222 | ΔNRD-Sso1p-HA | pYX223 | 2μ | HIS3 | GAL1-10 |
| pJM290 | Sso1p-HA | pYX223 | 2μ | HIS3 | GAL1-10 |
| pJM291 | Tandem t-SNARE without IHR | pYX223 | 2μ | HIS3 | GAL1-10 |
| pJM293 | Tandem t-SNARE | pYX223 | 2μ | HIS3 | GAL1-10 |
| pJM311 | Tandem t-SNARE–ΔNRD | pYX223 | 2μ | HIS3 | GAL1-10 |
| pJM334 | Tandem t-SNARE | pRS316 | CEN6 | URA3 | SSO1 |
| pJM414 | Syn1ANRD-ΔNRD-Sso1p | pYX223 | 2μ | HIS3 | GAL1-10 |
| pJM427 | Tandem t-SNARE | pRS426 | 2μ | URA3 | SSO1 |
| pJM429 | [Q468R] Tandem t-SNARE–ΔNRD | pYX223 | 2μ | HIS3 | GAL1-10 |
Cell fixation and antibody staining
W3031A transformed with the indicated plasmids were grown at 30°C in synthetic complete media and analyzed by indirect immunofluorescence microscopy by standard methods (Burke et al., 2000) with an anti- HA mAb (16B12; 1:1,000; Covance) followed by fluorescent secondary antibodies (AlexaFluor488 goat anti–mouse IgG (Invitrogen) at 1:1,000. The cells were mounted on slides using the ProLong antifade kit (Invitrogen).
Microscopy
Fluorescent images were taken and analyzed with a microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) and a plan-Neofluar 100× NA 1.3 oil immersion objective (Carl Zeiss MicroImaging, Inc.) using filter sets for fluorescein (FITC, excitation 480 nm, emission 535 nm, and dichroic Q505LP; Chroma Technology Corp.). Images were captured at room temperature using a digital camera (CoolSNAP HQ; Roper Scientific) and MetaMorph Imaging software (version 6.1; Universal Imaging Corp.). The images were deconvolved using the no-neighbors algorithm and digitally magnified before assembly with Adobe Photoshop version 8.0. Several different isolates of the strain were examined to confirm the reproducibility of the observed localization of the indicated Sso1p.
5-FOA counterselection
The transformed JMY128 shuffle strain was grown in synthetic complete media minus histidine and uracil with 2% galactose at 30°C for 2 d followed by a subsequent back dilution and another overnight growth. 10-OD600 cells were spun down and resuspended in 1 ml sterile water. A threefold serial dilution with 20-μl spots were plated onto synthetic complete minus histidine with 2% galactose and 1 g/liter 5-FOA plates and incubated for 3 d at 30°C.
Western blotting of whole cell extracts
Total cell extracts were made by glass bead lysis of TCA-killed cells. The amount of total cell extract indicated in the figure legends was resolved by SDS-PAGE and probed with anti-HA or anti-Ssop antibodies. Primary antibodies were at 1:1,000 (anti-HA and anti-Ssop) or 1:2,000 (anti-Sec9p) dilutions. The HRP-conjugated secondary antibodies were at a 1:10,000 dilution. Immunoblots were developed using ECL detection (GE Healthcare).
sec ts complementation analysis
BY1, BY41 (sec9-4), BY70 (sec9-7), BY392 (sec9-104), BY200 (sec9-123), BY445 (sec9-201), BY29 (sec1-1), BY33 (sec4-8), BY37 (sec6-4) (obtained from P. Brennwald, University of North Carolina, Chapel Hill, NC), and RSY271 (sec18-1) (Novick et al., 1980) were transformed with pJM334, pJM407, and pJM427. sec9-104, -123, and -201 are unpublished sec9ts alleles. Clustered charge-to-alanine mutagenesis (Bennett et al., 1991; Gibbs and Zoller, 1991) was used to generate sec9-104, which contains two mutations, K434A and K437A, and sec9-123, which contains E591A and E593A. The third allele, sec9-201, was made by random mutagenesis. The strains were grown in synthetic complete media minus uracil with 2% glucose at 25°C overnight. 5-OD600 cells were spun down and resuspended in 1 ml sterile water. Threefold serial dilutions with 20-μl spots were plated onto synthetic complete minus uracil plates with 2% glucose. The plates were grown at 25 or 37°C for ∼2.5 d.
Protein production
GST-Sso1p-ΔNRD (185–290; C266A) was expressed in 12 liters of super broth media and induced with 1 mM IPTG at 30°C for 4 h. Protein was purified by GST affinity chromatography as described previously (McNew et al., 2000) except that cells were lysed in 1% Triton X-100, and the protein was eluted with 1% n-octyl β-glucopyranoside. His8-Sso1p (pJM88), Snc1p-His6 (pJM90), and GST-Sec9c (BB442) were expressed and purified as described previously (McNew et al., 2000).
Reconstitution into liposomes and fusion assays
All proteoliposomes were formed and used standard fusion assays as previously described (Scott et al., 2003).
Antibody production
The polyclonal anti-Sec9c antibodies (RC62 and RC63) were generated by Cocalico Biologicals, Inc. in rabbits immunized with recombinant Sec9c. Initial injections consisted of 200 μg/rabbit followed by 100 μg/rabbit boost injections. Antisera at a 1:2,000 dilution in TBS with 1% Tween-20 (TBS-T) was used for the detection of Sec9p in conjunction with a 1:10,000 dilution in TBS-T of secondary antibody goat anti–rabbit HRP. RC62 antisera was used throughout this study. The polyclonal anti-Ssop was previously described (Sogaard et al., 1994).
Online supplemental material
Fig. S1 shows the secretion in strains expressing Sso1p or tandem t-SNAREs. Secretion is quantitatively measured by analyzing externalized invertase activity and is qualitatively examined by mating factor secretion with a halo assay. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200507138/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Kirilee Wilson, Dr. Pat Brennwald, Dr. Tom Melia, Blair Doneske, and Travis Rodkey for their thoughtful comments on this manuscript as well as Neel Parishak (Rice University, Houston, TX) for a preliminary Sso1-ΔNRD expression construct and Song Liu for help with control 5-FOA experiments. We also thank Dr. Pat Brennwald for the sects strains, Hans Ronne for the sso2 deletion strain, and Dr. Yeon-Kyun Shin for the GST-Sso1 p-ΔNRD plasmid.
This work was supported by grants from The National Science Foundation (IBN-02126051) and The Robert A. Welch Foundation (C-1517) to J.A. McNew, a National Science Foundation Integrative Graduate Education and Research Traineeship Program grant (DGE-0114264) to J.S. Van Komen, and the Houston Live Stock Show and Rodeo to J.S. Van Komen and B.L. Scott.
Abbreviations used in this paper: IHR, interhelical region; NRD, NH2-terminal regulatory domain; 5-FOA, 5-fluoorotic acid.
References
- Aalto, M.K., H. Ronne, and S. Keranen. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12:4095–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, W.F., N.F. Paoni, B.A. Keyt, D. Botstein, A.J. Jones, L. Presta, F.M. Wurm, and M.J. Zoller. 1991. High resolution analysis of functional determinants on human tissue-type plasminogen activator. J. Biol. Chem. 266:5191–5201. [PubMed] [Google Scholar]
- Burke, D., D. Dawson, and T. Stearns. 2000. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. xvii, 205 pp.
- Calakos, N., M.K. Bennett, K.E. Peterson, and R.H. Scheller. 1994. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 263:1146–1149. [DOI] [PubMed] [Google Scholar]
- Chen, Y.A., and R.H. Scheller. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98–106. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Y. Xu, F. Zhang, and Y.K. Shin. 2004. Constitutive versus regulated SNARE assembly: a structural basis. EMBO J. 23:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer, A.M., R.M. Williams, A.M. Chu, and R.W. Davis. 2002. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 99:15530–15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer, D., D. Bruns, B. Shen, R. Jahn, and A.T. Brunger. 1997. a. A structural change occurs upon binding of syntaxin to SNAP-25. J. Biol. Chem. 272:4582–4590. [DOI] [PubMed] [Google Scholar]
- Fasshauer, D., H. Otto, W.K. Eliason, R. Jahn, and A.T. Brunger. 1997. b. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem. 272:28036–28041. [DOI] [PubMed] [Google Scholar]
- Fernandez, I., J. Ubach, I. Dulubova, X. Zhang, T.C. Sudhof, and J. Rizo. 1998. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 94:841–849. [DOI] [PubMed] [Google Scholar]
- Fiebig, K.M., L.M. Rice, E. Pollock, and A.T. Brunger. 1999. Folding intermediates of SNARE complex assembly. Nat. Struct. Biol. 6:117–123. [DOI] [PubMed] [Google Scholar]
- Fukuda, R., J.A. McNew, T. Weber, F. Parlati, T. Engel, W. Nickel, J.E. Rothman, and T.H. Sollner. 2000. Functional architecture of an intracellular membrane t-SNARE. Nature. 407:198–202. [DOI] [PubMed] [Google Scholar]
- Gibbs, C.S., and M.J. Zoller. 1991. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J. Biol. Chem. 266:8923–8931. [PubMed] [Google Scholar]
- Hanson, P.I., H. Otto, N. Barton, and R. Jahn. 1995. The N-ethylmaleimide-sensitive fusion protein and α-SNAP induce a conformational change in syntaxin. J. Biol. Chem. 270:16955–16961. [DOI] [PubMed] [Google Scholar]
- Herman, P.K., and J. Rine. 1997. Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 16:6171–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantti, J., M.K. Aalto, M. Oyen, L. Sundqvist, S. Keranen, and H. Ronne. 2002. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 115:409–420. [DOI] [PubMed] [Google Scholar]
- Katz, L., and P. Brennwald. 2000. Testing the 3Q:1R “rule”: mutational analysis of the ionic “zero” layer in the yeast exocytic SNARE complex reveals no requirement for arginine. Mol. Biol. Cell. 11:3849–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.S., D.H. Kweon, and Y.K. Shin. 2002. Membrane topologies of neuronal SNARE folding intermediates. Biochemistry. 41:10928–10933. [DOI] [PubMed] [Google Scholar]
- Lehman, K., G. Rossi, J.E. Adamo, and P. Brennwald. 1999. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J. Cell Biol. 146:125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman, J.C., J. Robblee, R. Fairman, and F.M. Hughson. 2000. Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry. 39:8470–8479. [DOI] [PubMed] [Google Scholar]
- Margittai, M., D. Fasshauer, S. Pabst, R. Jahn, and R. Langen. 2001. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J. Biol. Chem. 276:13169–13177. [DOI] [PubMed] [Google Scholar]
- Margittai, M., J. Widengren, E. Schweinberger, G.F. Schroder, S. Felekyan, E. Haustein, M. Konig, D. Fasshauer, H. Grubmuller, R. Jahn, and C.A. Seidel. 2003. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proc. Natl. Acad. Sci. USA. 100:15516–15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew, J.A., F. Parlati, R. Fukuda, R.J. Johnston, K. Paz, F. Paumet, T.H. Sollner, and J.E. Rothman. 2000. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 407:153–159. [DOI] [PubMed] [Google Scholar]
- Misura, K.M., R.H. Scheller, and W.I. Weis. 2000. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 404:355–362. [DOI] [PubMed] [Google Scholar]
- Munson, M., and F.M. Hughson. 2002. Conformational regulation of SNARE assembly and disassembly in vivo. J. Biol. Chem. 277:9375–9381. [DOI] [PubMed] [Google Scholar]
- Munson, M., X. Chen, A.E. Cocina, S.M. Schultz, and F.M. Hughson. 2000. Interactions within the yeast t-SNARE sso1p that control SNARE complex assembly. Nat. Struct. Biol. 7:894–902. [DOI] [PubMed] [Google Scholar]
- Nicholson, K.L., M. Munson, R.B. Miller, T.J. Filip, R. Fairman, and F.M. Hughson. 1998. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat. Struct. Biol. 5:793–802. [DOI] [PubMed] [Google Scholar]
- Novick, P., C. Field, and R. Schekman. 1980. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 21:205–215. [DOI] [PubMed] [Google Scholar]
- Parlati, F., T. Weber, J.A. McNew, B. Westermann, T.H. Sollner, and J.E. Rothman. 1999. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc. Natl. Acad. Sci. USA. 96:12565–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, L.M., P. Brennwald, and A.T. Brunger. 1997. Formation of a yeast SNARE complex is accompanied by significant structural changes. FEBS Lett. 415:49–55. [DOI] [PubMed] [Google Scholar]
- Scott, B.L., J.S. Van Komen, S. Liu, T. Weber, T.J. Melia, and J.A. McNew. 2003. Liposome fusion assay to monitor intracellular membrane fusion machines. Methods Enzymol. 372:274–300. [DOI] [PubMed] [Google Scholar]
- Scott, B.L., J.S. Van Komen, H. Irshad, S. Liu, K.A. Wilson, and J.A. McNew. 2004. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J. Cell Biol. 167:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard, M., K. Tani, R.R. Ye, S. Geromanos, P. Tempst, T. Kirchhausen, J.E. Rothman, and T. Sollner. 1994. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 78:937–948. [DOI] [PubMed] [Google Scholar]
- Sun, W., Q. Yan, T.A. Vida, and A.J. Bean. 2003. Hrs regulates early endosome fusion by inhibiting formation of an endosomal SNARE complex. J. Cell Biol. 162:125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, R.B., D. Fasshauer, R. Jahn, and A.T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 395:347–353. [DOI] [PubMed] [Google Scholar]
- Teng, F.Y., Y. Wang, and B.L. Tang. 2001. The syntaxins. Genome Biol. 2:REVIEWS3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar, D., and F.M. Hughson. 2003. SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19:493–517. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10:1793–1808. [DOI] [PubMed] [Google Scholar]
- Weber, T., B.V. Zemelman, J.A. McNew, B. Westermann, M. Gmachl, F. Parlati, T.H. Söllner, and J.E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell. 92:759–772. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Y. Chen, D.H. Kweon, C.S. Kim, and Y.K. Shin. 2002. The four-helix bundle of the neuronal target membrane SNARE complex is neither disordered in the middle nor uncoiled at the C-terminal region. J. Biol. Chem. 277:24294–24298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.