The reliability of gene predictors of cancer outcome has been recently questioned, pointing to deficiencies in experimental design, insufficient statistical power due to small sample size, and flaws in predictor generation and performance assessment, with proposed guidelines to overcome these limitations (Ntzani and Ioannidis, 2003; Michiels et al, 2005, Dupuy and Simon, 2007). Now, in a recent article published in Molecular Systems Biology (Chuang et al, 2007), a complementary strategy has been proposed based on integration of expression profiles with protein interactions, demonstrating that more reproducible and robust predictors can be generated with the additional benefit of including mutated genes which are excluded in the classical analyses, and also providing models for the molecular mechanisms involved in metastasis formation. This is achieved through combination of mRNA expression profiles with curated protein–protein interaction data, which became recently available (Rual et al, 2005), leveraging methods for modular subnetwork identification and biological validation (Segal et al, 2003; Poyatos and Hurst, 2004).
During the past decade, transcriptome analysis has been used increasingly to monitor expression profiles of extensive collections of genes in cancer samples, providing insights into the molecular mechanisms underlying cancer development and outcome. Gene signatures developed from these data sets allowed characterization of cancer types and stages, formation of metastasis or response to therapy with the potential to complement or even outperform traditional cellular, molecular and clinical markers. In early-diagnosed primary breast cancer, gene predictors from several studies appeared capable of informing clinicians on the likelihood of metastasis formation, so that they could eventually restrict exposure of their patients to aggressive chemotherapy after surgical removal of the tumor to those cases with poor prognosis. The expected benefit is to avoid unnecessary secondary effects of the treatment in the large majority of patients for which chemotherapy is not beneficial.
While large-scale clinical trials are underway to establish such predictors as prognosis markers for widespread use in the clinical setting, a number of observations have questioned their validity, stability or robustness. For example, cross-comparison of the predictors from two landmark studies (van't Veer et al, 2002; Wang et al, 2005) revealed a very limited overlap with only 3 genes out of 70 or 76 in common, and much less successful predictions on the other sample collection. Extensive statistical re-analysis of the two data sets suggested that differences in the samples analyzed, the microarray platforms or the data analysis schemes used were not sufficient to account for these discrepancies, arguing for the need of analyzing thousands of samples to obtain largely overlapping gene signatures (Ein-Dor et al, 2006).
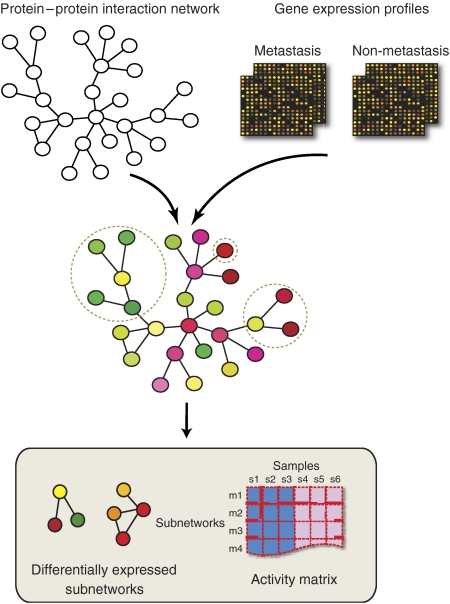
The novel strategy described by Trey Ideker and co-workers (Chuang et al, 2007) is based on the identification of protein interaction subnetworks with coherent expression patterns of their component genes, which can distinguish the samples of patients which developed distant metastasis after surgery from those that did not. This is achieved by overlaying protein–protein interaction data on the gene expression profiles, generating an activity score for protein subnetworks in all patient samples, then computing the mutual information between activity scores and metastasis potential to assess their discriminative potential (Figure 1). The prospect is to use the selected subnetworks as novel markers for prognosis of metastasis formation in newly diagnosed breast cancer patients instead of the predictors based on collections of non-interconnected genes.
Figure 1.
Identification of protein subnetwork markers. Gene expression profiles from metastatic and nonmetastatic tumor sample are overlayed onto a protein–protein interaction network. Iterative exploration of all possible protein subnetworks generates an activity score for protein subnetworks in all patient samples. Computing the mutual information between activity scores and metastasis potential enables the identification of subnetworks with a high discriminative potential. Statistical significance of subnetwork discriminative power is assessed by comparison with three null distributions obtained by randomizing gene labels, network structure and sample labels, respectively. (Figure courtesy of Trey Ideker).
Of course, iterative exploration of the high-dimensional space of all possible protein subnetworks seeded at all nodes of a highly branched and interconnected network requires the use of a greedy algorithm. Moreover, it is essential at all steps of the process to assess statistically the significance and performance of the selected subnetworks for their discriminative power when compared to randomly generated or permutated networks and sample classes on one hand, and to the established predictors on the other hand. This is also computationally demanding, but not a limiting factor of the approach with the ever-increasing high-performance computing power becoming available within laboratory workstations.
The findings that are very encouraging is that subnetwork markers developed by this approach overlap much more extensively and are more accurate in the classification of metastasis than the previous predictors. An added value of the approach is that the selected protein subnetworks are significantly enriched in protein involved in common biological processes, thus providing potential insights into the molecular mechanisms involved in metastasis formation. In addition, the vast majority of the selected subnetworks contain highly interconnected proteins encoded by genes that are not by themselves discriminative, because they are not detected as differentially expressed. These include a significant number of previously identified breast cancer susceptibility genes absent in the established predictors.
The approach proposed by Chuang et al (2007) should now be complemented by integration with other high-precision data sets (Hwang et al, 2005). The reported advances are significant, but they also have limitations: the subnetwork markers selected from the two data sets still overlap only very partially, and contain only a minority of the cancer susceptibility genes. It is well possible that the level and/or modulation of expression of many of the functionally relevant genes are of low magnitude and beyond the detection capabilities of current microarray technologies. This suggests that there is a wealth of unexploited information in these and other similar data sets available in human and other species, and calls for re-analysis of sufficiently powered studies when high-quality protein–protein interaction data is available. Even more profound insights can be expected when novel and more sensitive measurement technologies will become available.
References
- Chuang H-Y, Lee E, Liu Y-T, Lee D, Ideker T (2007) Network-based classification of breast cancer metastasis. Mol Syst Biol 3: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy A, Simon R (2007) Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Nat Cancer Inst 99: 147–157 [DOI] [PubMed] [Google Scholar]
- Ein-Dor L, Zuk O, Domany E (2006) Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA 103: 5923–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Smith JJ, Leslie DM, Weston AD, Rust AG, Ramsey S, de Atauri P, Siegel AF, Bolouri H, AItchison JD, Hood L (2005) A data integration methodology for systems biology. Proc Natl Acad Sci USA 102: 17302–17307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels S, Koscielny S, Hill C (2005) Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet 365: 488–492 [DOI] [PubMed] [Google Scholar]
- Ntzani E, Ioannidis JP (2003) Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet 362: 1439–1444 [DOI] [PubMed] [Google Scholar]
- Poyatos JF, Hurst LD (2004) How biologically relevant are interaction-based modules in protein networks? Genome Biol 5: R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–176 [DOI] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365: 671–679 [DOI] [PubMed] [Google Scholar]