Abstract
The role of estrogen in promoting mammary stem cell proliferation remains controversial. It is unclear if estrogen receptor (ER)–expressing cells have stem/progenitor activity themselves or if they act in a paracrine fashion to stimulate stem cell proliferation. We have used flow cytometry to prospectively isolate mouse mammary ER-expressing epithelial cells and shown, using analysis of gene expression patterns and cell type–specific markers, that they form a distinct luminal epithelial cell subpopulation that expresses not only the ER but also the progesterone and prolactin receptors. Furthermore, we have used an in vivo functional transplantation assay to directly demonstrate that the ER-expressing luminal epithelial subpopulation contains little in vivo stem cell activity. Rather, the mammary stem cell activity is found within the basal mammary epithelial cell population. Therefore, ER-expressing cells of the mammary epithelium are distinct from the mammary stem cell population, and the effects of estrogen on mammary stem cells are likely to be mediated indirectly. These results are important for our understanding of cellular responses to hormonal stimulation in the normal breast and in breast cancer.
Introduction
Estrogen is vital for normal postpubertal mammary development, as well as for growth of the majority of breast cancers (Sternlicht, 2006; Yager and Davidson, 2006). The relationship between mammary stem cells and hormone receptor–expressing cells is therefore a fundamental issue in breast biology. It is known that in both the developing and the adult mammary gland only a subset of cells express the estrogen receptor (ER). However, there are conflicting views as to the role of these cells. It has been proposed that ER-positive cells form a stem cell compartment that is directly stimulated by circulating hormones (Cheng et al., 2004; Clarke et al., 2005). Alternatively, ER-positive cells may stimulate proliferation of a separate stem cell compartment in a paracrine manner (Mallepell et al., 2006).
Evidence that ER-positive cells form a stem cell compartment has come from studies of cell cycling times and proliferation in both human and rodent tissues. ER- and progesterone receptor–positive cells in the mouse mammary epithelium have been identified as cells that retain BrdU (label-retaining cells) in pulse–chase experiments (Zeps et al., 1998, 1999; Welm et al., 2002; Smith, 2005), suggesting that they form a slowly cycling cell compartment. Slow in vivo cycling time is thought to be a property of stem cells (Welm et al., 2002; Clarke et al., 2005). Consistent with the slow cycling times is the observation that, in the normal human adult tissue, ER-positive cells do not express markers of proliferation (Clarke et al., 1997). It has been proposed that ER down-regulation occurs in these cells before the proliferative response, as stimulation with estrogen led to a decrease in ER expression within 4 h in mice (Cheng et al., 2004).
The paracrine stimulation theory is supported by observations that ER-null mammary cells can reconstitute the mammary epithelial network in cleared fat pad transplantation experiments only if cotransplanted with mammary cells from ER wild-type mice (Mallepell et al., 2006). Resolution of this issue requires prospective isolation and functional analysis of the ER-expressing cellular compartment. We have used such an approach to directly demonstrate that the mammary epithelium contains separate hormone-sensing and stem/progenitor compartments.
Results and discussion
Sca-1 and prominin-1 expression define a distinct subpopulation of mouse mammary luminal epithelial cells
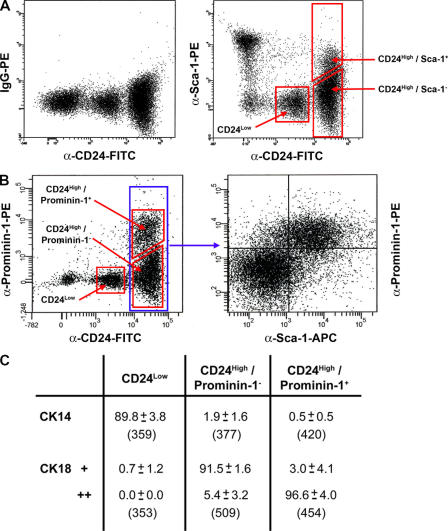
The mouse mammary epithelium consists of two cell compartments, an outer layer of basal epithelial cells, the majority of which are functionally specialized myoepithelial cells, and an inner layer of luminal epithelial cells. These cells can be distinguished by expression of cell type–specific cytoskeletal markers (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200604065/DC1; Smalley et al., 1998). We have previously described CD24 as a marker that allows the separation and isolation of the luminal (CD24High) and basal (CD24Low) compartments of the mouse mammary epithelium (Sleeman et al., 2006). To prospectively isolate further subpopulations of the mammary epithelium, we costained mouse mammary cell preparations with CD24-FITC and two cell-surface markers of mouse hematopoietic stem cells, Sca- and prominin-1 (Fig. 1). Costaining with CD24 and Sca-1 defined a CD24Negative/Sca-1+ nonepithelial population and a CD24High/Sca-1+ luminal epithelial population (Fig. 1 A). Costaining with CD24 and prominin-1 defined only a CD24High/prominin-1+ population. No cells from the nonepithelial compartment stained with this marker. Simultaneous staining with CD24, Sca-1, and prominin-1 revealed an almost complete overlap between CD24High/Sca-1+ and CD24High/prominin-1+ cell populations (Fig. 1 B).
Figure 1.
CD24, Sca-1, and prominin-1 expression define a distinct mammary epithelial cell compartment. (A) Flow cytometric staining patterns of freshly isolated mouse mammary cells stained for CD24 expression (left) and CD24 and Sca-1 expression (right). Only live, single CD45−cells were included in the analysis. (B) Flow cytometric staining patterns of freshly isolated mouse mammary cells triple stained for CD24, prominin-1, and Sca-1 expression. The CD24 and prominin-1 staining pattern is shown on the left, and the Sca-1 and prominin-1 staining pattern of the CD24High fraction is shown only on the right. (C) Analysis of cytoskeletal antigen expression in cells separated by CD24 and prominin-1 expression. Results of three independent cell isolations in which freshly isolated cells were sorted onto slides and single stained for CK14 or CK18. Numbers represent the percentage of positive cells ± the SD for three experiments. The total numbers of cells observed are given below the percentages. Intensity of CK18 staining is indicated by CK18+ (weak) and CK18++ (strong).
To confirm the luminal epithelial nature of the CD24High/prominin-1− and CD24High/prominin-1+ populations, cells from both these compartments and the CD24Low population were sorted onto slides and stained for expression of the basal epithelial cell marker cytokeratin 14 (CK14) and the luminal epithelial cell marker, CK8/18 (CK18). The results (Fig. 1 C) confirmed that the majority of CD24Low cells were CK14+ basal cells and that the CD24High/prominin-1− and CD24High/prominin-1+ populations were CK18+ luminal cells. Unexpectedly, the CD24High/prominin-1+ cells had more intense CK18 expression than the CD24High/prominin-1− cells, suggesting that these were functionally distinct populations. Staining of the epithelial populations defined by CD24 and Sca-1 staining gave essentially identical results (unpublished data).
To confirm that the CD24Low population consisted predominantly of basal myoepithelial cells, CD24Low cells were sorted onto slides and double stained with antibodies against CK14 and α-isoform smooth muscle actin (SMA). As expected, the majority (>90%) were indeed CK14+/SMA+ myoepithelial cells (Fig. S1 B). Keratin-negative cells were also present, and a small number of CK14+/SMA-negative cells were observed.
CD24High/prominin-1+ cells are a specialized hormone-sensing luminal epithelial compartment
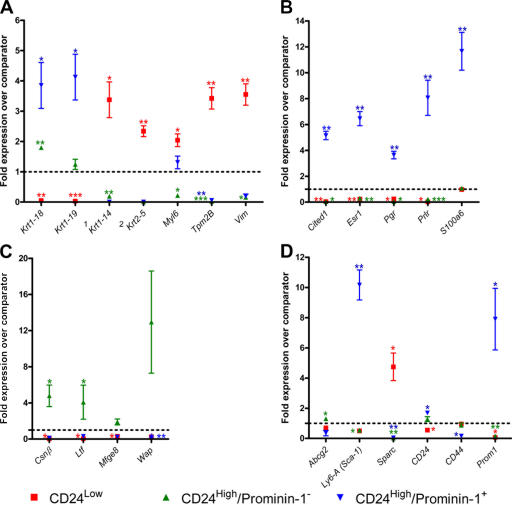
To characterize the potential functional roles of the compartments defined by CD24 and prominin-1 staining, including the expression of hormone receptors, expression of several genes (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200604065/DC1) relative to total mammary epithelial cell expression was analyzed by quantitative real time PCR (qPCR). In the CD24Low population, as expected, there was increased expression of the myoepithelial/basal cytoskeletal markers Krt1-14 (CK14), Krt2-5 (CK5), and Vim. Myl6 and Tpm2B, which are two genes involved in actin–myosin activity, were also up-regulated in this population, reflecting the contractile function of myoepithelial cells. In both CD24High populations, expression of luminal cytoskeletal markers was elevated. CD24High/prominin-1− cells had significantly elevated levels of Krt1-18 (CK18), whereas in CD24High/prominin-1+ cells, expression of both Krt1-18 and Krt1-19 (CK19) was significantly increased (Fig. 2 A).
Figure 2.
qPCR analysis identifies a hormone receptor–expressing cell population. (A) Cytoskeletal genes. 1A P value could not be determined for CD24High/prominin-1+ with CK14, as all three replicates gave a value of 0. 2A P value could not be determined for CD24High/prominin-1− or CD24High/prominin-1+ with CK5, as in both cases two of the three samples gave a value of 0. (B) Hormone response genes. (C) Milk component genes. (D) Miscellaneous genes. Each data point on each graph is a mean ± the SD of fold-relative expression of the gene in three independently isolated samples of the population of interest compared with total mammary epithelial cells. The fold-relative expression for each sample is itself a mean of two independent cDNA syntheses performed on that sample. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Analysis of expression of five genes involved in sensing systemic hormones, Esr (ERα), Pr (progesterone receptor), Prlr (prolactin receptor), Cited1, and S100A6, demonstrated that all of these genes were significantly up-regulated in CD24High/prominin-1+ cells (Fig. 2 B). Four out of the five genes were also significantly down-regulated in the CD24Low and the CD24High/prominin-1− populations. Cited1 is involved in transcriptional coactivation together with ER (Yahata et al., 2001), and S100A6 is a calcium-binding protein that is likely to have several cellular functions, including binding the prolactin receptor (Murphy et al., 1988). The pattern of expression of these five genes strongly suggests that the CD24High/prominin-1+ cell population forms a specific hormone-sensing compartment.
As the ultimate purpose of the mammary epithelium is milk production, the expression of four genes for milk proteins (Csnβ, Ltf, Mfge8, and Wap) was examined (Fig. 2 C). Despite the fact that the cells assayed were harvested from virgin animals, both Csnβ and Ltf were significantly more highly expressed in the CD24High/prominin-1− population, compared with total mammary epithelium. There was also elevated expression of Mfge8 and Wap in these populations, but this did not achieve significance. These analyses therefore confirmed the basal/myoepithelial and luminal identities of the CD24Low and CD24High populations, respectively, and suggested that prominin-1 expression is a marker of two different functional luminal epithelial populations, a hormone-sensing compartment (prominin-1+), and a compartment containing cells involved in milk production (prominin-1−).
As expected, the CD24High/prominin-1+ hormone receptor–expressing population had very strongly up-regulated levels of both Prominin-1 (Prom1) and Sca-1 (Ly6A/Sca-1; Fig. 2 D). CD24 expression was significantly increased in this population, and it significantly decreased in the CD24Low population. CD44, which is a marker of basal cells (Fonseca et al., 2000) and breast cancer stem cells (Al-Hajj et al., 2003), was significantly decreased in CD24High/prominin-1+ luminal cells. Consistent with the pattern of expression of cytoskeletal genes, CD24Low cells were found to have increased expression of Sparc, which is a secreted matrix-binding protein that was previously described as a human breast myoepithelial cell marker and a marker of poor prognosis in breast cancers (Jones et al., 2004). The CD24High/prominin-1− cell population also showed a modest, but significantly higher, level of expression of Abcg2, which is the gene for breast cancer–resistance protein 1. Abcg2 is an ABC transmembrane protein pump that is the main molecular determinant of the side population phenomenon (Smalley and Clarke, 2005), and that is also localized to the terminally differentiated milk-producing cells of the alveolar epithelium during lactation (Jonker et al., 2005). Analysis of gene expression within the epithelial populations defined by CD24 and Sca-1 staining gave essentially identical results (unpublished data).
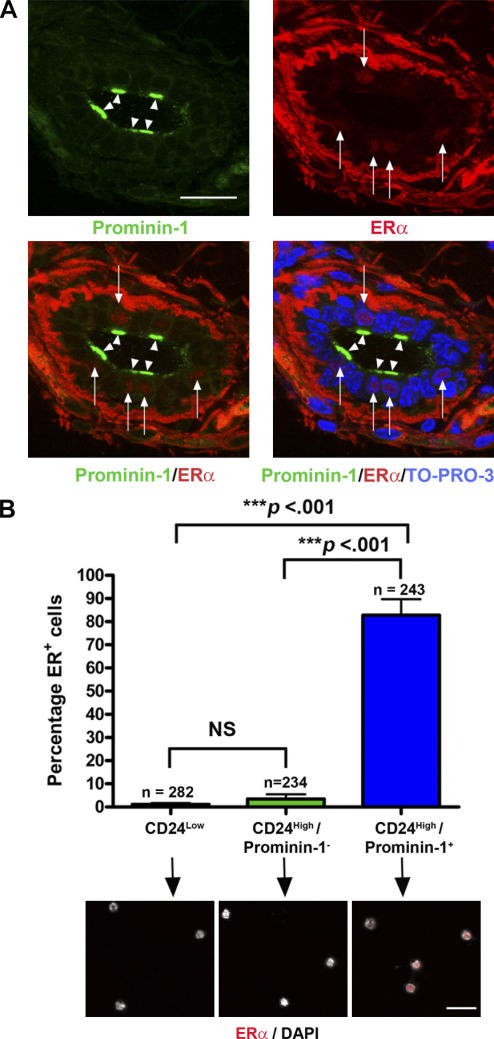
To confirm that prominin-1+ cells in the mouse mammary epithelium were ERα+, frozen sections of adult mouse mammary gland were costained with antibodies against these proteins. The results (Fig. 3 A) showed that prominin-1 is apically localized on a subset of mouse mammary luminal epithelial cells, and also confirmed that the majority of luminal epithelial cells with nuclear ERα staining were prominin-1+. To provide quantification, cells were sorted onto slides, stained for nuclear ERα, and counted. The data (Fig. 3 B) confirmed that the majority (>80%) of CD24High/prominin-1+ cells were ERα+.
Figure 3.
Prominin-1+ cells are ERα+. (A) Frozen section of adult mouse mammary gland stained with anti–prominin-1, anti-ERα, and a nuclear counterstain (TO-PRO-3). The majority of cells with nuclear ERα staining (red; arrows) also show apical prominin-1 staining (green; arrowheads). Heavy background staining in the stroma is caused by the Alexa Fluor 555 antibody used to detect the ERα. (B) To quantify prominin-1/ERα double staining, CD24Low, CD24High/prominin-1−, and CD24High/prominin-1+ cells were sorted onto slides and stained for nuclear ERα expression. The histogram indicates the percentage of ERα+ cells in each population from analysis of three independent sorts and the total numbers of cells counted (n). Examples of cells from the three populations are shown below the corresponding populations. Nuclear ERα staining (red) can only be seen in the CD24High/prominin-1+ population. Significant differences between the populations were determined using a t test of log10-transformed data. NS, not significant. Bars, 30 μm.
CD24High/prominin-1− cells contain the highest proportion of in vitro mammary colony-forming cells
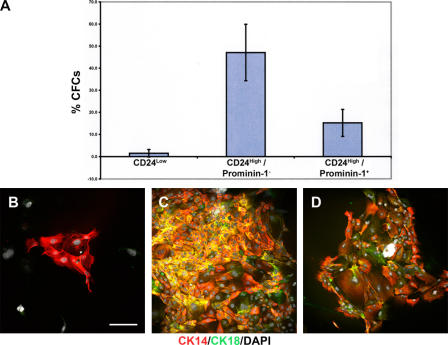
To investigate the in vitro progenitor abilities of the CD24Low, CD24High/prominin-1−, and CD24High/prominin-1+ populations, colony forming assays were performed using freshly isolated single cells sorted into individual wells of 96-well plates to determine the relative proportions of mammary colony-forming cells (CFCs) within the populations. The results (Fig. 4 A) demonstrated that the CD24High/prominin-1− population contained the highest proportion of CFCs, with >40% of cells capable of forming colonies in vitro. The CD24High/prominin-1+ population contained significantly fewer CFCs (15%). The CD24Low population showed very low levels of CFC activity. These data support an in vitro progenitor function for the CD24High/prominin-1− cells.
Figure 4.
CD24High/prominin-1− mammary epithelial cells are enriched for in vitro CFCs. (A) Bar chart indicating colony-forming efficiencies of single cells sorted into individual wells of 96-well plates from CD24Low (fifteen 96-well plates from four sorts), CD24High/prominin-1− (17 96-well plates from 4 sorts), and CD24High/prominin-1+ (17 96-well plates from 4 sorts) populations. Data are the mean percentage cloning efficiencies ± the SD. CFCs, mammary colony forming cells. (B–D) Immunophenotyping of 8–10-d-old colonies cultured on glass coverslips derived from CD24Low (B), CD24High/prominin-1− (C), and CD24High/prominin-1+ (D) cells. Stained for CK14 (red) and CK18 (green) expression and with DAPI (white) to distinguish nuclei. Bar, 150 μm.
To derive qualitative information on the colonies formed in the CFC assays, freshly isolated single cells sorted from the different populations were cultured on coverslips for 10 d, and the colonies generated were stained to assess their CK14 and CK18 expression pattern. Colonies derived from CD24Low CFCs were small and consisted of only CK14+ cells, which is consistent with a basal/myoepithelial origin (Smalley et al., 1998). CFCs from both the CD24High/prominin-1− and CD24High/prominin-1+ populations formed large colonies that contained cells with a heterogeneous staining pattern consisting of both single-stained CK14+ and CK18+ and double-stained CK14+/CK18+ cells, which is consistent with a luminal epithelial cell origin (Fig. 4, B–D; Smalley et al., 1998). These data demonstrate that the mouse mammary epithelium contains at least two different CFCs—basal and luminal.
To determine whether the differences in in vitro colony-forming ability were reflected by the proliferative status of the three populations, the cell cycle profile of freshly isolated CD24Low, CD24High/prominin-1−, and CD24High/prominin-1+ cells was determined. The results (Fig. S1 C) showed that while the majority of cells in all three populations were in G0/G1, the percentage of cells in S phase in both the CD24High compartments was significantly increased compared with the CD24Low cells.
ER-expressing cells of the mouse mammary gland have low in vivo stem/progenitor cell activity
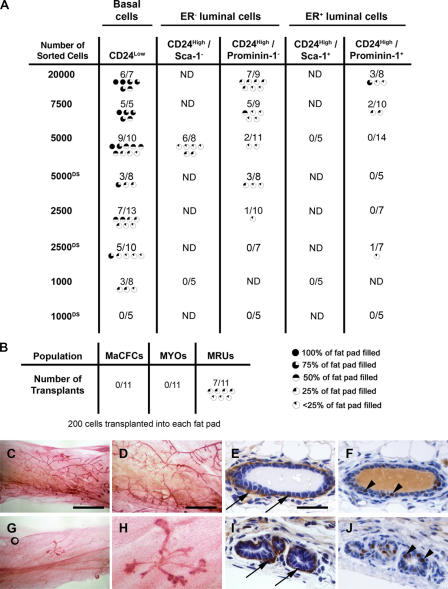
The aforementioned data establish the existence of three distinct cell populations in the mammary epithelium; basal/myoepithelial cells, defined by the CD24Low phenotype, and two distinct CD24High luminal epithelial populations, one of which is specialized for detecting systemic hormonal signals, and the other demonstrating elevated expression of milk protein genes and high in vitro colony-forming activity. The key in vivo functional assay for stem/progenitor cell activity in mammary epithelium is the cleared fat pad transplant, in which cells are transplanted into a mammary fat pad of a 3-wk-old mouse from which the endogenous epithelium has been surgically removed (DeOme et al., 1959; Smith, 1996). Therefore, to determine the in vivo stem cell activity of cells from the different epithelial populations, limiting dilution cleared fat pad transplantation of prospectively isolated basal epithelial, ER− luminal epithelial, and ER+ luminal epithelial cells was performed. Both prominin-1 and Sca-1 staining were used to separate the ER− and ER+ luminal populations in different transplant experiments. In some experiments, cells were sorted and then immediately resorted to achieve very high purity before transplantation (Fig. S1, D–H). Consistent with our previous data (Sleeman et al., 2006), CD24Low basal epithelial cells (Fig. 5) were the most highly enriched for mammary epithelial stem/progenitor activity. The high in vivo growth potential of this population is in contrast to their low in vitro cloning efficiency. Some previous investigators have demonstrated a similar disparity (Stingl et al., 2006), whereas others show good correlation between in vitro and in vivo growth (Shackleton et al., 2006). The reasons for these differences are not clear, but may be caused by differences in sorting strategies, culture conditions, or the ages of mice used to isolate cells.
Figure 5.
Functional assays identify in vivo stem/progenitor activity in CD24Low epithelial cells. (A) Results of cleared fat pad transplantation of CD24Low, CD24High/Sca-1−, CD24High/Sca-1+, CD24High/prominin-1−, and CD24High/prominin-1+ cells. Freshly isolated sorted cells were transplanted at dilutions ranging from 20,000 to 1,000 cells per fat pad. Results from transplants of double-sorted populations are indicated by “DS.” (B) Results of transplantation of 200 mammary colony forming cells (MaCFCs), myoepithelial cells (MYOs), or mammary repopulating cells (MRUs) isolated by CD24 and CD49f staining. For both datasets, the number of successful outgrowths and the number of fat pads transplanted for each population are indicated. Also shown are graphic indications of the extent to which each transplant filled the fat pad. ND, not determined. (C–J) Wholemounts and sections through representative 100% (C–F) and <25% (G–J) transplants. (C, D, G, and H) Carmine-stained wholemounts. (E, F, I, and J) Sections through transplant outgrowths stained for SMA to detect myoepithelial cells (E and I; arrows) or for ERα to detect ER+ cells (F and J; arrowheads). Bars: (C and G) 6 mm; (D and H) 2 mm; (E, F, I, and J) 40 μm.
Both luminal ER− and luminal ER+ populations had rates of in vivo epithelial outgrowth formation much lower than the CD24Low basal cells, with the luminal ER+ populations showing the lowest transplantation activity. Double sorting for very high purity did not prevent these rare outgrowths from occurring. Histological examination showed that outgrowths derived from all populations contained SMA-positive myoepithelial cells, ER+ luminal epithelial cells, and ER− luminal epithelial cells (Fig. 5, C–J). Thus, the in vivo differentiation potential of both the transplantable cells present at high frequency in the CD24Low population and the rare transplantable cells in the luminal populations was similar.
Mouse mammary epithelial cell populations highly enriched for stem cells (mammary-repopulating units [MRUs]), for in vitro colony-forming cells (mammary colony-forming cells [MaCFCs]), and for myoepithelial cells (MYOs) have recently been isolated on the basis of expression of CD24 and CD49f (Stingl et al., 2006). To relate these populations to the cell compartments we have described in this study and in a previous work (Sleeman et al., 2006), we identified cells corresponding to the MRUs, MaCFCs, and MYOs in mammary epithelial cell preparations stained for CD24 and CD49f expression (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200604065/DC1). The identity of the MaCFC and MYO populations was confirmed by in vitro colony-forming assays, qPCR, and CK staining of cells sorted onto slides (unpublished data). Cleared fat pad transplantation activity was used to demonstrate that we had correctly identified the MRUs (Fig. 5 B). Mammary cell preparations were simultaneously stained with multiple antibodies to identify the MaCFCs, MYOs, and MRUs within the CD24Negative, CD24Low, and CD24High populations. The results confirmed that the MRUs fell within the CD24Low population, as did the MYOs. The MaCFCs corresponded to the CD24High luminal population (Fig. S2, A and B).
In conclusion, we have for the first time prospectively isolated mouse mammary luminal ER+ and ER− cells and directly analyzed their cleared fat pad repopulation activity (stem/progenitor cell activity). Our results indicate that the majority of stem/progenitor cell activity in the adult virgin mouse mammary epithelium is located in the basal compartment, confirming and extending previous observations (Shackleton et al., 2006; Sleeman et al., 2006; Stingl et al., 2006). In contrast, the ER+ luminal compartment contains little in vivo stem/progenitor cell activity, indicating that the hormone-sensing and in vivo stem/progenitor activities of the mammary epithelium are properties of distinct, separate cell populations. Gene expression analysis of the ER− and ER+ luminal epithelial compartments revealed evidence of further distinct functional specialization. These results will provide a basis for elucidating the nature of the interaction between ER− and ER+ luminal epithelial cells and basal stem cells in the mammary gland.
Materials and methods
Preparation of single mammary cell suspensions
Mammary epithelial organoids were harvested from fourth mammary fat pads of mature, virgin, female 10–12-wk-old FVB mice and processed to single cells, as previously described (Sleeman, et al., 2006).
Flow cytometry and cell sorting
Cells were stained as previously described (Sleeman et al., 2006) with 0.5 μg/ml anti–CD24-FITC (clone M1/69; BD Biosciences), 0.25 μg/ml anti–CD45-phycoerythrin (PE)-Cy5 (clone 30-F11; BD Biosciences), 0.1 μg/ml anti–prominin-1-PE (clone 13A4; Insight Biotechnology), and 0.1 μg/ml anti–Sca-1-PE and/or anti–Sca-1-allophycocyanin (clone D7; Cambridge Bioscience). 0.01% DAPI or TO-PRO-3 were used to detect dead cells. Nonspecific IgG controls were used for compensation and to set sort gates. Analysis and exclusion of dead cells, CD45+ cells, and nonsingle cells was performed as previously described (Sleeman et al., 2006). For double sorting of populations, cells were isolated after the first round of sorting, pelleted, resuspended in fresh 0.01% TO-PRO-3, and resorted using the same gates.
To investigate whether different fluorochrome conjugates affect the staining profile of CD24, cells at a density of 106 cells/ml were stained with 0.5 μg/ml anti–CD24-FITC (clone M1/69), anti–CD24-PE (clone M1/69; tested at a range of concentrations from 0.2 μg/ml up to 10 μg/ml), 0.25 μg/ml anti–CD24-PE-Cy5 (clone M1/69), or 0.5 μg/ml unconjugated anti-CD24 (clone M1/69), followed by anti–rat–Alexa Fluor 633 (A-21094; 20 μg/ml) in a two-step procedure. To confirm that populations identified with different fluorochrome conjugates were identical, cells at a density of 106 cells/ml were stained simultaneously with 0.5 μg/ml anti–CD24-FITC (clone M1/69) and 0.25 μg/ml anti–CD24-PE-Cy5 (Fig. S2, B and C)
To identify previously described (Stingl et al., 2006) MaCFCs, MYOs, and MRUs (stem cells) and locate them within the three-region CD24 profile described both in this study and in a previous work (Sleeman et al., 2006), cells were stained with anti–CD24-PE (clone M1/69; 1.5 μg/ml; BD Biosciences), anti–CD49f-FITC (clone GoH3; 1:50 vol/vol dilution; BD Biosciences), and 0.25 μg/ml anti–CD45-PE-Cy7 (clone 30-F11; BD Biosciences) together with 0.25 μg/ml anti–CD24-PE-Cy5 (clone M1/69; Insight Biotechnology). 0.01% TO-PRO-3 was used to detect dead cells. Nonspecific IgG controls were used for compensation and to set sort gates. Analysis and exclusion of dead cells, CD45+ cells and nonsingle cells was performed as previously described (Sleeman et al., 2006). MaCFC, MYO, and MRU cells were identified, and their location within the three regions defined by anti–CD24-PE-Cy5 was determined by staining (Fig. S2 A).
Immunofluorescence staining and in vitro analysis
For prominin-1 and ERα staining of frozen sections of mouse mammary gland, small pieces (∼5 mm3) of 10-wk-old mouse mammary fat pads were fixed in 4% paraformaldehyde in PBS for 1 h, infiltrated with 1 M sucrose overnight at 4°C as a cryoprotectant, and snap frozen in isopentane cooled in liquid nitrogen. 10-μm frozen sections were cut and stored at −80°C. Before use, sections were thawed at RT for 15–30 min and stained essentially as previously described (Smalley, et al., 1998) with 5 μg/ml anti–prominin-1 (rat monoclonal clone 13A4; Insight Biotechnology) and 6 μg/ml anti-ERα (mouse monoclonal clone 1D5; Insight Biotechnology). Sections were counterstained with TO-PRO-3 (0.01% in PBS) and mounted in Vectashield H1000 mounting medium (Vector Laboratories). For cytoskeletal marker staining, tissue was snap frozen with no prior fixation. Frozen sections were fixed in 1:1 methanol acetone at −20°C for 5 min and stained with antibodies against 2.1 μg/ml CK14 (mouse IgG3 clone LL002; Lab Vision) and 2 μg/ml CK8/18 (CK18; mouse IgG1 clone 5D3; Vision Biosystems) in addition to DAPI. Secondary antibodies were isotype-specific goat anti–mouse antibodies (A21127 and A21157; Invitrogen) conjugated to Alexa Fluor 488 or 555 fluorochromes. Sections were mounted in Vectashield. Lack of nonspecific staining by secondary antibodies was confirmed using isotype-matched control primary antibodies (Cambridge Bioscience). Lack of cross-reactivity was confirmed with controls incubated with single primary antibodies and both secondary antibodies.
Immunophenotyping of cell populations sorted onto slides was performed as previously described (Sleeman et al., 2006) using the anti-CK14 (LLOO2) and anti-CK18 (5D3) primary antibodies, secondary antibodies, and DAPI, as described in the previous paragraph. Samples were mounted in Vectashield. For immunostaining of clones on coverslips, mouse mammary cells were cultured on coverslips with a feeder layer of irradiated 3T3-L1 preadipocytes in 1:1 DME/F12 (Sigma-Aldrich) with 10% fetal calf serum (Invitrogen), 5 μg/ml insulin (Sigma-Aldrich), 10 ng/ml epidermal growth factor (Sigma-Aldrich), and 10 ng/ml cholera toxin (Sigma-Aldrich). Cultures were maintained at 37°C in a 5% vol/vol CO2/5% vol/vol O2 atmosphere for 8–10 d before fixation in cold (−20°C) 1:1 methanol/acetone and staining (Smalley et al., 1998). Clones were stained with anti-CK14 (LLOO2) and anti-CK18 (5D3) primary antibodies, secondary antibodies, and DAPI, as described in the previous paragraph. Samples were mounted in Vectashield.
All fluorescence samples were examined at room temperature using a microscope (TCS SP2; Leica) with an Acousto-Optical Beam Splitter and lasers exciting at 405, 488, 543, and 633 nm (Leica). PMT levels were set using control samples. Multicolor images were collected sequentially in three or four channels. Images of clones were taken using a 20×/0.70 NA dry HC Plan Apo CS lens (Leica). Images of sections and cells sorted on slides were taken using a 40×/1.25 NA oil HCX Plan Apo lens. Images were captured using the Leica system and Leica TCS image acquisition software. Overlays were generated using TCS software. Photo montages were generated using Photoshop (Adobe), but were not further processed.
Analysis of the in vitro colony-forming potential of mammary cell subpopulations was performed in a 96-well plate format, as previously described (Smalley et al., 2005). The feeder layers, growth media, and conditions were identical to those described in this section for coverslip culture.
Cell cycle analysis
Cells were pelleted and resuspended in ice-cold 70% ethanol and maintained at 4°C for at least 15 min. They were then pelleted and resuspended in PBS containing 40 μg/ml propidium iodide and 100 μg/ml RNase A. Samples were incubated at 37°C for 20 min and analyzed by standard protocols.
qPCR analysis
qPCR reactions were performed as previously described (Sleeman et al., 2006) to determine fold changes in expression of a selection of genes (Table S1) in mammary epithelial cell subpopulations, compared with a leukocyte-depleted, bulk mammary epithelial cell (CD45−/CD24+) comparator sample. Significant deviation of the mean value of the three data points from a fold difference of 1 (no change compared with the comparator sample) was tested using a t test on log10-transformed data.
Cleared mammary fat pad transplantation
Freshly isolated cells were sorted and transplanted (Sleeman et al., 2006). There was no intervening culture period before transplantation. All animal work was approved by the Local Ethics Committee and performed under Home Office approval. 8 wk after transplantation, fat pads were wholemounted and analyzed, as previously described (Sleeman et al., 2006). Failed clears were excluded from the analysis. Wholemounts were examined on a binocular microscope (MZ12.5; Leica) with a Plan 1× lens and a Cold Light Source (Leica). Images were captured with a camera (DFC500; Leica) and IM50 image acquisition software (with auto white balance and auto exposure activated). A proportion of successful transplants had a region of epithelial outgrowth dissected out under the microscope for paraffin embedding by routine methods and routine immunocytochemistry to detect SMA (clone 1A4; Sigma-Aldrich) and ERα (clone 1D5). Images of stained sections were captured on a microscope (DM RA2; Leica) using a 63× oil Plan Apo lens (N/A 1.32), a camera (DFC320; Leica), and IM50 image acquisition software (with auto white balance and auto exposure activated). Photo montages were generated using Photoshop, but were not further processed.
To confirm that differential survival of cells before transplantation did not confound assessment of their relative engraftment potentials, viability assays of CD24Low, CD24High/prominin-1−, and CD24High/prominin-1+ cells were performed by TO-PRO-3 staining and flow cytometric analysis immediately after separation, and again after 3.5 h on ice. The analysis showed that CD24Low cells, which were the most potent at repopulating cleared fat pads (Fig. 5), had a viability immediately after sorting of >70%, dropping to >65% after 3.5 h. CD24High/prominin-1− cells had a viability of >90% immediately after sorting, and this was maintained after 3.5 h on ice. CD24High/prominin-1+ cells had a viability of >75% immediately after sorting, and this was also maintained at >75% after 3.5 h on ice. Therefore, differential sensitivity to the sorting procedure or to being maintained on ice during transplantation could not explain the differences in engraftment potential.
Online supplemental material
Table S1 lists the genes examined by qPCR analysis. Fig. S1 shows the characterization of mouse mammary epithelial cell subpopulations. Fig. S2 shows the identification of MaCFC, Myo, and MRU regions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200604065/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Ian Titley for technical assistance with FACS analysis, Gemma Molyneux for technical assistance with preparation of cell samples, Naheed Kanuga for technical assistance with preparation of frozen sections, and Alan Mackay, Marjan Iravani, Tim Dexter, Anita Grigoriadis, and Haukur Valgeirsson of the Breakthrough Microarray Facility. The authors would also like to thank the Breakthough Histopathology Facility.
This work was funded by Breakthrough Breast Cancer.
Abbreviations used in this paper: CFC, colony-forming cell; CK, cytokeratin; ER, estrogen receptor; MaCFC, mammary colony-forming cell; MRU, mammary-repopulating unit; PE, phycoerythrin; qPCR, quantitative real time PCR; SMA, α-isoform smooth muscle actin.
References
- Al-Hajj, M., M.S. Wicha, A. Benito-Hernandez, S.J. Morrison, and M.F. Clarke. 2003. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 100:3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, G., Z. Weihua, M. Warner, and J.A. Gustafsson. 2004. Estrogen receptors ER alpha and ER beta in proliferation in the rodent mammary gland. Proc. Natl. Acad. Sci. USA. 101:3739–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, R.B., A. Howell, C.S. Potten, and E. Anderson. 1997. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 57:4987–4991. [PubMed] [Google Scholar]
- Clarke, R.B., K. Spence, E. Anderson, A. Howell, H. Okano, and C.S. Potten. 2005. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev. Biol. 277:443–456. [DOI] [PubMed] [Google Scholar]
- DeOme, K.B., L.J. Faulkin Jr., H.A. Bern, and P.B. Blair. 1959. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 19:515–520. [PubMed] [Google Scholar]
- Fonseca, I., J.F. Moura Nunes, and J. Soares. 2000. Expression of CD44 isoforms in normal salivary gland tissue: an immunohistochemical and ultrastructural study. Histochem. Cell Biol. 114:483–488. [DOI] [PubMed] [Google Scholar]
- Jones, C., A. Mackay, A. Grigoriadis, A. Cossu, J.S. Reis-Filho, L. Fulford, T. Dexter, S. Davies, K. Bulmer, E. Ford, et al. 2004. Expression profiling of purified normal human luminal and myoepithelial breast cells: identification of novel prognostic markers for breast cancer. Cancer Res. 64:3037–3045. [DOI] [PubMed] [Google Scholar]
- Jonker, J.W., G. Merino, S. Musters, A.E. van Herwaarden, E. Bolscher, E. Wagenaar, E. Mesman, T.C. Dale, and A.H. Schinkel. 2005. The breast cancer resistance protein BCRP (ABCG2) concentrates drugs and carcinogenic xenotoxins into milk. Nat. Med. 11:127–129. [DOI] [PubMed] [Google Scholar]
- Mallepell, S., A. Krust, P. Chambon, and C. Brisken. 2006. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc. Natl. Acad. Sci. USA. 103:2196–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, L.C., L.J. Murphy, D. Tsuyuki, M.L. Duckworth, and R.P. Shiu. 1988. Cloning and characterization of a cDNA encoding a highly conserved, putative calcium binding protein, identified by an anti-prolactin receptor antiserum. J. Biol. Chem. 263:2397–2401. [PubMed] [Google Scholar]
- Shackleton, M., F. Vaillant, K.J. Simpson, J. Stingl, G.K. Smyth, M.L. Asselin-Labat, L. Wu, G.J. Lindeman, and J.E. Visvader. 2006. Generation of a functional mammary gland from a single stem cell. Nature. 439:84–88. [DOI] [PubMed] [Google Scholar]
- Sleeman, K.E., H. Kendrick, A. Ashworth, C.M. Isacke, and M.J. Smalley. 2006. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 8:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley, M.J., and R.B. Clarke. 2005. The mammary gland “side population”: a putative stem/progenitor cell marker? J. Mammary Gland Biol. Neoplasia. 10:37–47. [DOI] [PubMed] [Google Scholar]
- Smalley, M.J., J. Titley, and M.J. O'Hare. 1998. Clonal characterization of mouse mammary luminal epithelial and myoepithelial cells separated by fluorescence-activated cell sorting. In Vitro Cell. Dev. Biol. Anim. 34:711–721. [DOI] [PubMed] [Google Scholar]
- Smalley, M.J., I. Titley, and A. Ashworth. 2005. An improved definition of mouse mammary epithelial side population cells. Cytotherapy. 7:497–508. [DOI] [PubMed] [Google Scholar]
- Smith, G.H. 1996. Experimental mammary epithelial morphogenesis in an in vivo model: evidence for distinct cellular progenitors of the ductal and lobular phenotype. Breast Cancer Res. Treat. 39:21–31. [DOI] [PubMed] [Google Scholar]
- Smith, G.H. 2005. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 132:681–687. [DOI] [PubMed] [Google Scholar]
- Sternlicht, M.D. 2006. Key stages in mammary gland development: the cues that regulate ductal branching morphogenesis. Breast Cancer Res. 8:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl, J., P. Eirew, I. Ricketson, M. Shackleton, F. Vaillant, D. Choi, H.I. Li, and C.J. Eaves. 2006. Purification and unique properties of mammary epithelial stem cells. Nature. 439:993–997. [DOI] [PubMed] [Google Scholar]
- Welm, B.E., S.B. Tepera, T. Venezia, T.A. Graubert, J.M. Rosen, and M.A. Goodell. 2002. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev. Biol. 245:42–56. [DOI] [PubMed] [Google Scholar]
- Yager, J.D., and N.E. Davidson. 2006. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 354:270–282. [DOI] [PubMed] [Google Scholar]
- Yahata, T., W. Shao, H. Endoh, J. Hur, K.R. Coser, H. Sun, Y. Ueda, S. Kato, K.J. Isselbacher, M. Brown, and T. Shioda. 2001. Selective coactivation of estrogen-dependent transcription by CITED1 CBP/p300-binding protein. Genes Dev. 15:2598–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeps, N., J.M. Bentel, J.M. Papadimitriou, M.F. D'Antuono, and H.J. Dawkins. 1998. Estrogen receptor-negative epithelial cells in mouse mammary gland development and growth. Differentiation. 62:221–226. [DOI] [PubMed] [Google Scholar]
- Zeps, N., J.M. Bentel, J.M. Papadimitriou, and H.J. Dawkins. 1999. Murine progesterone receptor expression in proliferating mammary epithelial cells during normal pubertal development and adult estrous cycle. Association with ERα and ERβ status. J. Histochem. Cytochem. 47:1323–1330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.