Abstract
The tensin family member cten (C-terminal tensin like) is an Src homology 2 (SH2) and phosphotyrosine binding domain–containing focal adhesion molecule that may function as a tumor suppressor. However, the mechanism has not been well established. We report that cten binds to another tumor suppressor, deleted in liver cancer 1 (DLC-1), and the SH2 domain of cten is responsible for the interaction. Unexpectedly, the interaction between DLC-1 and the cten SH2 domain is independent of tyrosine phosphorylation of DLC-1. By site-directed mutagenesis, we have identified several amino acid residues on cten and DLC-1 that are essential for this interaction. Mutations on DLC-1 perturb the interaction with cten and disrupt the focal adhesion localization of DLC-1. Furthermore, these DLC-1 mutants have lost their tumor suppression activities. When these DLC-1 mutants were fused to a focal adhesion targeting sequence, their tumor suppression activities were significantly restored. These results provide a novel mechanism whereby the SH2 domain of cten-mediated focal adhesion localization of DLC-1 plays an essential role in its tumor suppression activity.
Introduction
Focal adhesions are integrin-mediated cell matrix junctions connecting the ECM to the actin cytoskeleton. The ECM proteins bind to the extracellular domains of integrin heterodimers, whereas the actin stress fibers link to integrin cytoplasmic tails via large molecular complexes. These complexes comprise actin-binding/modulating proteins, protein kinases, phosphatases, GTPases, and adaptor proteins (Lo, 2006) and are targets of regulatory signals that control focal adhesions' function, including cell adhesion, migration, proliferation, differentiation, and gene expression (Schwartz et al., 1995; Hynes, 2002). Dysregulation of these components is associated with diseases such as cancer (Lo, 2006).
Tensin is a gene family with four members (tensin1, tensin2, tensin3, and cten), and their encoding proteins are localized to the cytoplasmic side of focal adhesions. Tensin1, the prototype of the family, interacts with actin filaments in multiple ways (Lo et al., 1994) and contains an Src homology 2 (SH2) domain that binds to phosphotyrosine-containing proteins (Davis et al., 1991; Cui et al., 2004), followed by a phosphotyrosine binding (PTB) domain that interacts with the NPXY motif on the β integrin cytoplasmic tails (Calderwood et al., 2003). Tensin2 and -3 have domain structures that are very similar to those of tensin1, although the central regions are diverse (Lo, 2004). On the other hand, cten (C-terminal tensin like) is a distant member of the family with smaller molecular mass, and the only sequence homologous region is the SH2 and PTB domains. The cten gene localizes to chromosome 17q21, a region frequently deleted in prostate cancer (Gao et al., 1995; Hagmann et al., 1996; Williams et al., 1996), and its expression is reduced or absent in prostate cancer (Lo and Lo, 2002), suggesting a role of cten as a tumor suppressor. However, the potential mechanism has not been well understood. In this study, we have identified deleted in liver cancer 1 (DLC-1) as one of the binding partners of cten, mapped the binding sites on cten and DLC-1, and demonstrated the biological relevance of this interaction. Our results provide new insight into how cten may be involved in preventing tumor formation.
Results and discussion
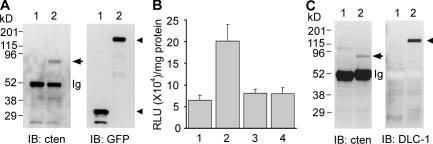
To understand cten's biological function and the potential mechanism involved, we set up experiments to identify cten-associated proteins by yeast two-hybrid assay, mass spectrometry analysis, and candidate screenings. One of the molecules identified is DLC-1, which is a tumor suppressor that regulates actin stress fibers and cell adhesion and inhibits tumor cell growth and migration (Yuan et al., 1998, 2003b; Ng et al., 2000; Goodison et al., 2005; Wong et al., 2005). Its rat homologue, p122RhoGAP (RhoGTPase-activating protein), is isolated as a phospholipase Cδ1–interacting protein (Homma and Emori, 1995) and is localized to caveolae (Yamaga et al., 2004) and focal adhesions (Kawai et al., 2004). To demonstrate the relation between DLC-1 and cten, an expression vector encoding GFP or GFP–DLC-1 was transfected into cten-expressing A549 cells and molecules associated with GFP–DLC-1 or GFP were immunoprecipitated with anti-GFP antibodies. The immunoblot analysis indicated that endogenous cten was present in the GFP–DLC-1–associated complexes, but not in the GFP control (Fig. 1 A). The reciprocal experiment also detected GFP–DLC-1 in the cten immunocomplexes (unpublished data). The interaction was further examined by a luciferase reporter–based mammalian two-hybrid assay. The positive interaction shown by a fourfold enhancement of luciferase activity was detected when DLC-1 and cten were cotransfected into NIH 3T3 cells (Fig. 1 B). Finally, to test the interaction between endogenous DLC-1 and cten, we screened numerous cell lines and found that MLC-SV40 (immortalized normal prostate epithelialcell line) expressed both cten and a low level of DLC-1. This cell line was used for coimmunoprecipitation assay, and the results demonstrated that cten interacted with endogenous DLC-1 (Fig. 1 C).
Figure 1.
Identification of DLC-1 as a cten binding partner. (A) A549 cells were transfected with pEGFP (lane 1) or pEGFP–DLC-1 (lane 2). Cell lysates were coimmunoprecipitated with anti-GFP and analyzed by immunoblotting (IB) with anti-cten (left) or anti-GFP (right). The arrow indicates cten, and arrowheads show GFP and GFP–DLC-1. (B) NIH 3T3 cells in 24-well dishes were cotransfected with pCMV-AD-cten, and pCMV-BD vector with no insert as a negative control (column 1), pCMV-BD-DLC-1 (column 2), pCMV-BD-DLC-1S440A (column 3), or pCMV-BD-DLC-1Y442F (column 4), and together with the reporter plasmid pFR-Luc. The luciferase activities were measured by luminometry and shown as relative light units (RLU) per milligram of cellular proteins. Data are from two independent triplicate experiments. Error bars indicate mean ± SD. (C) MLC SV-40 cell lysates were coimmunoprecipitated with normal rabbit serum (lane 1) or anti–DLC-1 (lane 2) and analyzed by immunoblotting with anti-cten (left) or anti–DLC-1 (right). The arrow indicates cten, and the arrowhead shows DLC-1.
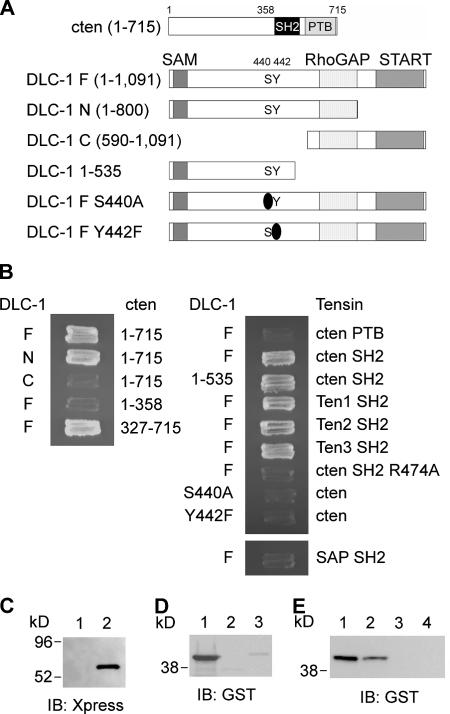
To demonstrate the direct interaction and map the regions responsible for the binding, we have applied yeast two-hybrid assay. As expected, the full-length DLC-1 binds to intact cten (Fig. 2, A and B). With truncated constructs, the interaction regions were initially mapped to the N-terminal half (1–800) of DLC-1 and the C-terminal region (327–715) of cten, which contains the SH2 and PTB domains. We generated and examined constructs containing only the SH2 or PTB domain. Surprisingly, it was the SH2 domain that interacted with DLC-1. Because all tensin members contain the highly conserved SH2 domains (Lo, 2004), we predicted that they were likely to bind to DLC-1 as well. Indeed, DLC-1 interacted with SH2 domains of tensin1, -2, and -3 in the yeast two-hybrid assay (Fig. 2 B). Furthermore, when the arginine residue at the critical position, βB5, in the SH2 domain of cten was mutated into alanine (R474A), it abolished the interaction. Therefore, we have confirmed that the SH2 domain of cten binds to DLC-1. By a similar approach, we have defined the binding region on DLC-1 (1–535; Fig. 2 B).
Figure 2.
Determination of binding regions on cten and DLC-1 and their binding specificities. (A) Schematic diagram of cten and DLC-1 and their segments that were used for mapping the binding sites. (B) AH109 yeast cells transformed with the indicated plasmids and grown on two-dropout plates were restreaked on four-dropout plates. (C) Bacterial lysates containing Xpress–DLC-1 (113–535) were incubated with immobilized GST (lane 1) or GST-cten SH2 (lane 2). After washing, the associated proteins were analyzed by immunoblotting (IB) with anti-Xpress antibody. (D) 0.5 μg of purified GST-cten SH2 (lane 1), GST-Src SH2 (lane 2), and GST-p85 SH2 (lane 3) recombinant proteins were incubated with DLC-1 peptide (CSRLSIY442DNVPG) immobilized on agarose beads. After washing, the associated proteins were analyzed by immunoblotting with anti-GST antibody. (E) 0.5 μg of purified GST-cten SH2 recombinant proteins were incubated with DLC-1 peptide (CSRLSIY442DNVPG; lane 1), tyrosine-phosphorylated DLC-1 peptide (CSRLSIpY442DNVPG; lane 2), EGFR peptide (CSVQNPVY1086HNQP; lane 3), or tyrosine-phosphorylated EGFR peptide (CSVQNPVpY1086HNQP; lane 4) immobilized on agarose beads. After washing, the associated proteins were analyzed by immunoblotting with anti-GST antibody.
The SH2 domain is known as a binding motif for phosphotyrosine-containing peptides. However, yeast cells contain a very low level of, if any, phosphotyrosine. To test that this SH2–DLC-1 interaction is truly independent of tyrosine phosphorylation and to further map the binding region, because shorter fragments of N-terminal DLC-1 displayed self-activation activity in the yeast two-hybrid system, we performed a pull-down assay using recombinant GST-SH2 and Xpress–DLC-1 fragments expressed in bacteria, which contain no tyrosine kinase at all. The result showed that recombinant SH2 remained bound to DLC-1 (113–535; Fig. 2 C). Together with our results using synthetic peptides for binding (see the following paragraph), we have confirmed that the interaction is phosphotyrosine independent.
It has been shown that the interaction of the SH2 domain of SAP (also named SH2D1A), the gene product mutated in X-linked lymphoproliferative syndrome, to lymphocyte coreceptor SLAM is independent of tyrosine phosphorylation (Sayos et al., 1998). In fact, the SH2 domain of SAP interacts with non–tyrosine-phosphorylated peptides containing S/TIYxxI/V (Poy et al., 1999), and we found that there was one such site, 440SIYDNV, in DLC-1. Coincidentally, this site resides in the SH2 binding region (113–535). When either S440 or Y442 was mutated (S440A or Y442F), the interaction was abolished in both mammalian and yeast two-hybrid assays (Fig. 1 B and Fig. 2 B), demonstrating that this is indeed the essential site on DLC-1 for binding to the SH2 domain of cten. Interestingly, although the SAP SH2 domain binds to a similar motif, SAP SH2 domain does not interact with DLC-1 (Fig. 2 B). We further tested whether DLC-1 might be able to interact with other SH2 domain containing proteins, such as Src and p85, by a pull-down assay using the DLC-1 peptide (CSRLSIY442DNVPG)–conjugated beads. As shown in Fig. 2 D, only cten SH2 domain could interact with the DLC-1 peptide. In addition, cten SH2 domain did not bind to an EGFR peptide (CSVQNPVY1086HNQP) regardless of whether Y1086 was phosphorylated (Fig. 2 E). These results demonstrated the binding specificity between the DLC-1 and cten SH2 domain. Furthermore, we tested whether synthetic tyrosine-phosphorylated peptide (CSRLSIpY442DNVPG) interacted with the cten SH2 domain and found that phosphorylation on Y442 slightly reduced the interaction (Fig. 2 E). Because no report had documented the tyrosine phosphorylation of DLC-1 and we did not detect tyrosine phosphorylation of DLC-1 (113–535) when incubated with recombinant Src (unpublished data), the biological relevance of this reduced binding is currently unknown.
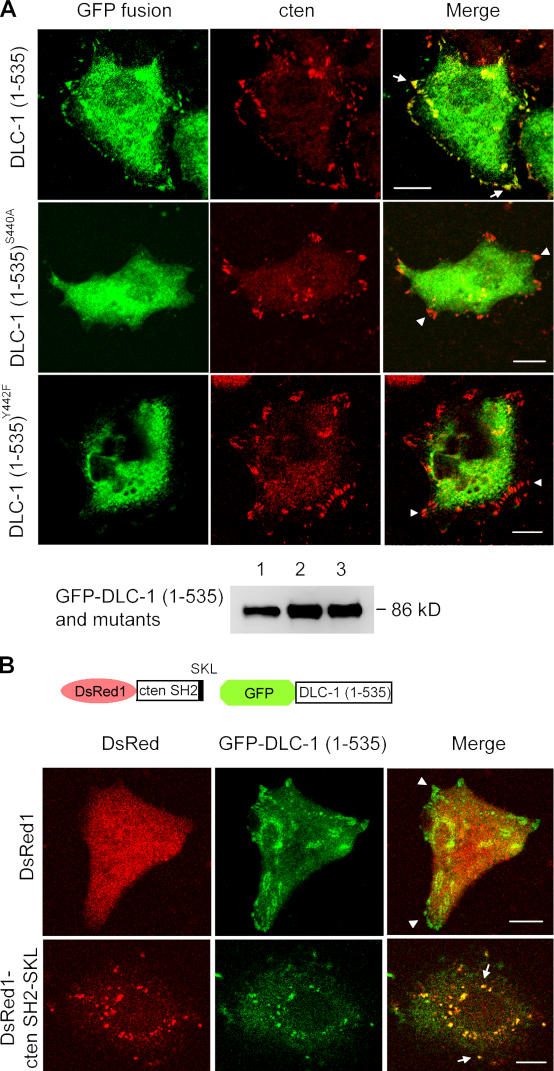
Because both cten and DLC-1 localize to focal adhesions and a previous study found that p122RhoGAP (117–533; Kawai et al., 2004), corresponding to DLC-1 (125–541), contained the focal adhesion targeting site, which overlapped with the SH2 binding site identified in this study, we speculated that the DLC-1 and cten interaction might be responsible for recruiting DLC-1 to focal adhesions. If this is the case, S440A and Y442F DLC-1 mutants would not be able to localize to focal adhesions. In contrast to the colocalization of cten and GFP–DLC-1 (1–535) at focal adhesion sites, the GFP–DLC-1 (1–535)S440A or GFP–DLC-1 (1–535)Y442F was diffusely distributed in the cytoplasm (Fig. 3 A), indicating that the SH2 binding site is essential for DLC-1's focal adhesion localization. The protein expressions of these constructs were confirmed by immunoblotting (Fig. 3 A). To further demonstrate that the cten SH2 domain is crucial for recruiting DLC-1 to a subcellular compartment, we generated a DsRed-cten SH2-SKL construct so that the DsRed-cten SH2 domain would be fused with the peroxisomal targeting peptide, SKL (Gould et al., 1989), at the C terminus when expressed and be targeted to peroxisomes. As shown in Fig. 3 B, although the DsRed was distributed in the cytoplasm, the DsRed-cten SH2-SKL proteins were accumulated at peroxisomes. When these constructs were cotransfected with GFP–DLC-1 (1–535), the DsRed-cten SH2-SKL was able to recruit some GFP–DLC-1 (1–535) to peroxisomes, demonstrating that cten SH2 alone is sufficient for the interaction and recruitment of DLC-1.
Figure 3.
Subcellular localization of DLC-1 and its recruitment by cten SH2. (A) A549 cells grown on coverslips were transfected with pEGFP–DLC-1 (1–535), pEGFP–DLC-1 (1–535)S440A, or pEGFP–DLC-1 (1–535)Y442F. After labeling with anti-cten antibodies followed by Alexa Fluor 594–conjugated secondary antibody, cells were visualized with a confocal microscope. Arrows indicate cten and GFP fusion protein colocalized at focal adhesions. Arrowheads show only cten at focal adhesions. About 100 GFP-positive cells were examined in each transfection. More than 90% of GFP-positive cells showed focal adhesion localization when transfected with GFP–DLC-1 (1–535), and no GFP-positive cells transfected with DLC-1 mutants showed focal adhesion localization. Cell lysates from A549 transiently expressing GFP–DLC-1 (1–535; lane 1), GFP–DLC-1(1–535)S440A (lane 2), or GFP–DLC-1 (1–535)Y442F (lane 3) were immunoprecipitated with anti-GFP and analyzed by immunoblotting with anti-GFP to show similar amounts, and correct sizes of recombinant proteins were expressed. (B) A549 cells grown on coverslips were cotransfected with pDsRed1/pEGFP–DLC-1 (1–535) or pDsRed1-cten SH2-SKL/pEGFP-DLC-1 (1–535). Cells were visualized with a confocal microscope. Note that the DsRed-cten SH2-SKL and GFP–DLC-1 (1–535) were colocalized at peroxisomes (arrows), although some GFP–DLC-1 (1–535) proteins were still detected at focal adhesions (at different focus plane; not depicted), as predicted, because of the presence of endogenous tensins in A549 cells. Arrowheads show only GFP–DLC-1 (1–535) at focal adhesions. Bars, 10 μm.
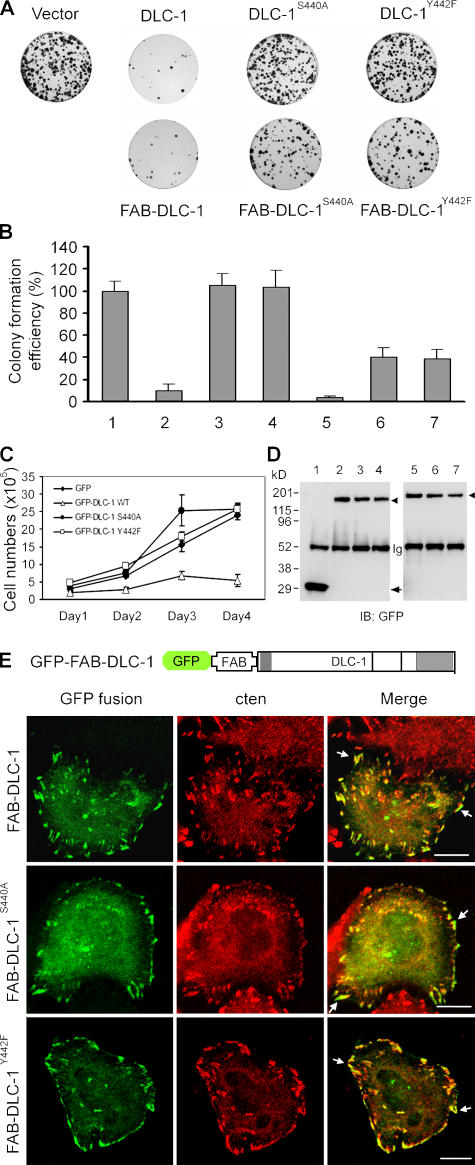
DLC-1 was identified as a candidate tumor suppressor, and its expression was lost or down-regulated in various cancers, including liver, breast, lung, brain, stomach, colon, and prostate, because of either genomic deletion or aberrant DNA methylation (Yuan et al., 1998, 2003a; Ng et al., 2000; Kim et al., 2003; Plaumann et al., 2003; Wong et al., 2003; Song et al., 2006). It has been reported that reexpression of DLC-1 in liver, breast, and lung cancer cell lines inhibits cancer cell growth (Yuan et al., 2003b, 2004; Zhou et al., 2004; Goodison et al., 2005; Wong et al., 2005), supporting its role as a tumor suppressor. DLC-1 contains three conserved domains: the sterile α motif (SAM), RhoGAP, and steroidogenic acute regulatory-related lipid transfer (START) domains (Fig. 2 A). SAM domains have been implicated in protein–protein interactions and are highly versatile in their binding partners. Some SAM domains may bind to each other to form homodimers or polymers, whereas others can interact with other proteins, or even RNA and DNA (Qiao and Bowie, 2005). START domains are predicted to contain a binding pocket for lipids, and modifications in the pocket may determine ligand binding specificity and function (Iyer et al., 2001). RhoGAP domains convert the active GTP-bound Rho proteins to the inactive GDP-bound state and function as negative regulators of RhoGTPases, which are involved in actin cytoskeleton organization, focal adhesion assembly, and cell proliferation (Moon and Zheng, 2003), and dysregulation of Rho activity has been implicated in tumorigenesis (Jaffe and Hall, 2002). A recent study demonstrated that the RhoGAP and START domains of DLC-1 are required for its tumor suppression activity (Wong et al., 2005). However, these two domains are not sufficient because expression of the RhoGAP and START domains alone does not inhibit tumor cell growth (Wong et al., 2005). In fact, the shortest fragment with the suppression activity contains, in addition to the RhoGAP and START domains, a region overlapping with the SH2 binding site, which is critical for focal adhesion localization. We hypothesized that the appropriate focal adhesion localization is essential for DLC-1's functions, including tumor cell suppression activity. To test this, we performed the colony formation assay using MDA-MB-468 breast cancer cell line, in which the growth was suppressed by DLC-1 overexpression (Yuan et al., 2003b). Consistent with a previous report, wild-type DLC-1 was able to suppress MDA-MB-468 cell growth. However, neither GFP–DLC-1S440A nor GFP–DLC-1Y442F could inhibit MDA-MB-468 cell growth (Fig. 4, A and B). In agreement with these results, the growth curve of MDA-MB-468 cells was significantly slower with wild-type DLC-1 (Fig. 4 C). Thus, the SH2 binding site is not only essential for DLC-1's focal adhesion localization but also critical for its tumor suppression activity. To further address the importance of the focal adhesion localization of DLC-1 to its tumor suppression activity, we fused wild-type and mutant DLC-1 with the N-terminal focal adhesion binding (FAB) site (aa 65–360) of chicken tensin1 (Chen and Lo, 2003), which is not conserved in cten. This FAB fusion forced the focal adhesion localization of GFP–FAB–DLC-1, GFP–FAB–DLC-1S440A, and GFP–FAB–DLC-1Y442F (Fig. 4 E). From the colony formation assay, the constitutive focal adhesion localizations of these molecules significantly enhanced the suppression activities of DLC-1S440A and DLC-1Y442F mutants (Fig. 4, A and B). Therefore, the focal adhesion localization of DLC-1 is essential for its tumor suppression activity. Nonetheless, the suppression activities of DLC-1S440A and DLC-1Y442F were not fully restored by linking to FAB. It is possible that the cten–DLC-1 interaction is not just for recruiting DLC-1 to focal adhesions but also for regulating its activity. In addition, because we fused DLC-1 with the N-terminal FAB site of tensin1 and DLC-1 normally binds to the C-terminal SH2 domains of tensins, these FAB fusion mutant proteins were not targeted to the precise position within the focal adhesion complexes. This spatial discrepancy may also contribute to the weaker suppression activities observed in GFP–FAB–DLC-1S440A and GFP–FAB–DLC-1Y442F.
Figure 4.
Colony formation and cell growth assays in MDA-MB-468 cells transfected with wild-type and mutant DLC-1. (A) Cells were transfected with the indicated constructs. After being cultured in media containing 0.8 mg/ml G418 for 2 wk, G418-resistant colonies were stained with crystal violet. (B) The histogram shows the colony formation assay of GFP (column 1), GFP–DLC-1 (column 2), GFP–DLC-1S440A (column 3), GFP–DLC-1Y442F (column 4), GFP–FAB–DLC-1 (column 5), GFP–FAB–DLC-1S440A (column 6), and GFP–FAB–DLC-1Y442F (column 7) from four independent experiments. (C) G418-resistant MDA-MB-468 cells (2 × 105) expressing the indicated proteins were seeded in triplicate in 60-mm dishes. Cells were harvested at 24-h intervals for 4 d, and the numbers of viable cells were counted by trypan blue exclusion assay with a hematocytometer. Error bars indicate mean ± SD. (D) Cell lysates from MDA-MB-468 transfected with GFP vector (lane 1), GFP–DLC-1 (lane 2), GFP–DLC-1S440A (lane 3), GFP–DLC-1Y442F (lane 4), GFP–FAB–DLC-1 (lane 5), GFP–FAB–DLC-1S440A (lane 6), or GFP–FAB–DLC-1Y442F (lane 7) were immunoprecipitated with anti-GFP and analyzed by immunoblotting with anti-GFP to show similar expression levels and correct sizes of recombinant proteins. The arrow indicates GFP, and arrowheads show GFP–DLC-1 and GFP–FAB–DLC-1 fusion proteins. (E) A549 cells grown on coverslips were transfected with pEGFP–FAB–DLC-1, pEGFP–FAB–DLC-1S440A, or pEGFP–FAB–DLC-1Y442F. After labeling with anti-cten antibodies followed by Alexa Fluor 594–conjugated secondary antibody, cells were visualized with a confocal microscope. Arrows indicate cten and GFP fusion protein colocalized at focal adhesions. Bars, 10 μm.
In this study, we have demonstrated that the tumor suppressor DLC-1 interacts with the SH2 domains of cten and other tensins as well. Although the SH2 binding site on DLC-1 also contains a critical tyrosine residue (Y442), the interaction does not rely on the phosphorylation of Y442. However, the phosphorylation of Y442 does reduce the interaction. This is a novel binding feature of tensins' SH2 domains. Furthermore, this interaction is highly specific for the SH2 domain of tensin family, as the SH2 domains of SAP, Src, and p85 all fail to bind to DLC-1. The biological significance of the cten–DLC-1 interaction is illustrated by mislocalization and the loss of tumor suppression activities of DLC-1S440A and DLC-1Y442F mutants. Furthermore, the suppression activities of these mutants could be rescued by tagging with FAB sequence. Therefore, in addition to genomic deletion and promoter hypermethylation, mislocalization of the DLC-1 protein may be another mechanism for acquiring tumorigenicity involving DLC-1 dysregulation. In this regard, further investigations on the DLC-1 protein localization in cancer samples with “normal” DLC-1 expression level are highly warranted. Based on these findings, we propose that DLC-1 is recruited to focal adhesion sites by one or more tensin members, depending on cell types and tissues. At the focal adhesion site, the RhoGAP domain of DLC-1 negatively regulates Rho small GTPase, which organizes actin stress fibers, and focal adhesion turnover, in turn, mediates cell migration and proliferation. When the expression and/or localization of DLC-1 are compromised, the cells are more susceptible for transformation. The fact that DLC-1 is able to bind to all tensins through their SH2 domains may explain why DLC-1 relies on tensin members for its normal localization and function, yet DLC-1–knockout mice (Durkin et al., 2005) displayed a more severe phenotype than tensin1 or -3 single-knockout mice (Lo et al., 1997; Chiang et al., 2005). It may require double or even triple tensin knockout to observe the defect results from mislocalization of DLC-1. On the other hand, recruiting DLC-1 to focal adhesion sites may not be the only function for cten. It is known that activated caspase3 cleaves cten at the DSTD570↓S, site generating two cten fragments: 1–570 and 571–715 (Lo et al., 2005). The later contains the PTB domain alone, which by itself is able to reduce cell growth by inducing apoptosis (Lo et al., 2005). In this case, the loss of cten expression may lead to uncontrolled cell growth and result in cell transformation. Together with our current findings, cten may function as a tumor suppressor in multiple ways.
Materials and methods
Plasmid constructions and mutagenesis
The full-length coding sequence of the DLC-1 gene was amplified from human kidney cDNA. The full-length and truncated fragments of DLC-1 were subcloned in frame into mammalian expression vector pEGFP-C2 (CLONTECH Laboratories, Inc.), yeast expression vector pGBKT7 (CLONTECH Laboratories, Inc.), and mammalian two-hybrid vector pCMV-BD (Stratagene). The full-length and truncated fragments of cten were constructed into pGADT7 (CLONTECH Laboratories, Inc.) and pCMV-AD (Stratagene) for yeast and mammalian two-hybrid analyses, respectively. The cDNA encoding the SH2 domain of SAP was amplified from human thymus cDNA and subcloned into pGADT7. The corresponding coding regions of the SH2 domains of tensin1, tensin2, tensin3, and cten were subcloned into pGADT7 and bacterial expression vector pGEX-5X-1 (GE Healthcare). The region encoding DLC-1 residues 113–535 was inserted into pTrcHis containing His and Xpress tags (Invitrogen). The mutations in DLC-1 (S440A and Y442F) and cten SH2 domain (R474A) were generated by site-directed mutagenesis. The corresponding coding regions of the SH2 domains of Src and p85 were subcloned into pGEX-5X-1. To construct pDsRed1-cten SH2-SKL, SKL residues were introduced into the C terminus of cten SH2 fragment by PCR. The resulting amplified PCR products were then ligated into pDsRed1-C1 (CLONTECH Laboratories, Inc.). The N-terminal FAB site of chicken tensin1 (residues 65–360; Chen and Lo, 2003) was used to construct fusions to the N terminus of DLC-1 in pEGFP–DLC-1, pEGFP–DLC-1S440A, or GFP–DLC-1Y442F plasmids. All constructs were verified by DNA sequencing.
Cell culture and transfection
MLC-SV40 cells, a gift from J. Rhim (Uniform Services University, Bethesda, MD), were cultured in keratinocyte serum-free medium supplemented with antibiotics, 5 ng/ml human recombinant EGF, and 0.05 mg/ml bovine pituitary extract (Invitrogen). A549, NIH3T3, and MDA-MB-468 cells purchased from American Type Culture Collection were cultured in DME supplemented with antibiotics and 10% fetal bovine serum. A549 cells were transfected using Lipofectamine 2000 (Invitrogen), whereas NIH 3T3 and MDA-MB-468 cells were transfected using SuperFect transfection reagent (QIAGEN) according to the manufacturer's instructions.
Immunoprecipitation and immunoblotting
Transiently transfected A549 or MDA-MB-468 cells with GFP fusion constructs were lysed in immunoprecipitation buffer (1% Triton X-100, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatine, and 1 μM PMSF) and cleared by centrifugation at 14,000 g for 15 min at 4°C. The clarified cell lysates were incubated with 1 μg of an anti-GFP goat polyclonal antibody (Rockland) by rotating at 4°C for 1 h, followed by the addition of 30 μl of 50% of protein A–Sepharose slurry (GE Healthcare) for 1 h. The protein A beads were collected by centrifugation and washed with immunoprecipitation buffer. Samples were then boiled in protein loading buffer and subjected to immunoblotting analyses using anti-GFP rabbit polyclonal antibodies (Santa Cruz Biotechnology, Inc.).
For coimmunoprecipitation, 24 h after transfection, A549 cells expressing the GFP or the GFP–DLC-1 constructs were lysed in coimmunoprecipitation buffer (0.1% Triton X-100, 25 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.2 mM EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatine, and 1 μM PMSF). Cell lysates were then sheared by passing through a syringe needle, and the cell debris was removed by centrifugation at 14,000 g for 15 min at 4°C. 1.2 mg of the clarified cell lysates were incubated with 2 μg of an anti-GFP goat polyclonal antibody (Rockland) by rotating at 4°C for 4 h, followed by the addition of protein A–Sepharose. Samples were subjected to immunoblotting analyses using anti-cten (Lo and Lo, 2002) and anti-GFP rabbit polyclonal antibodies (Santa Cruz Biotechnology, Inc.).
A similar approach was performed for coimmunoprecipitation of endogenously expressed DLC-1 in MLC-SV40, except that 3 mg of the clarified MLC-SV40 cell lysates were incubated with 1 μg of an anti–DLC-1 rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc.) by rotating at 4°C overnight. Immunoblotting analyses were performed using anti-cten rabbit polyclonal antibodies and anti–DLC-1 mouse monoclonal antibodies (BD Biosciences).
Mammalian two-hybrid assay
Plasmids (1 μg of each mammalian two-hybrid construct and 0.5 μg of pFR-Luc reporter) were transfected into NIH 3T3 cells using SuperFect. Cells were harvested 24 h after transfection. Firefly luciferase and activities in the cell extracts were determined by the procedure using Luciferase Assay System (Promega) and measured by luminometry.
Yeast two-hybrid analyses
To assay the interaction between DLC-1 and cten, the Saccharomyces cerevisiae strain AH109 was transformed with combinations of cten fragments in the activation domain (AD) plasmid, pGADT7, together with each of the DLC-1 fragments in the DNA-binding domain (DNA-BD) plasmid, pGBKT7. In brief, 5 ml of overnight culture of AH109 in YPD medium was diluted 50-fold and allowed to grow for another 4 h at 30°C. The yeast cells were harvested by centrifugation at 2,000 g for 15 min at room temperature and washed twice with 25 ml of sterile water. The cells were resuspended in 0.5 ml of 0.1 M lithium acetate (LiAc). 100 μl of competent cells were mixed with 600 μl TE-LiAc-PEG (1× TE, 0.1 M LiAc, and 40% polyethylene glycol [mol wt 3,350]), 10 μl of salmon sperm DNA, and 1 μg of each plasmid. After incubation at 30°C for 30 min, 70 μl of DMSO was added to the cells and heat shocked at 42°C for 15 min. The transformation mixture was centrifuged and washed with 1 ml of sterile water. The cell pellets were subsequently resuspended in 1× TE buffer and plated on nutritional selection agar lacking leucine and tryptophan. The resulting colonies were then restreaked on quadruple dropout plates lacking Ade, His, Leu, and Trp.
In vitro pull-down assay
For GST pull-down assay, the cDNAs encoding the SH2 domain of cten were subcloned into pGEX-5X-1 to generate GST fusion proteins (GST-cten SH2). The corresponding coding region of 113–535 amino acids of human DLC-1 was ligated into pTrcHis to express an Xpress-tagged protein, Xpress–DLC-1 (113–535). GST-cten SH2 proteins were expressed in and purified from Escherichia coli using glutathione-agarose (Sigma-Aldrich). 20 μg of GST or GST-cten SH2 on glutathione-agarose beads was mixed with 2 mg of bacterial lysates expressing Xpress–DLC-1 (113–535) in extraction buffer (0.1% Triton X-100, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.2 mM EDTA). After incubation on a rotator for 3 h at 4°C, the slurry was pelleted by centrifugation and washed five times with ice-cold extraction buffer. The pellet was resuspended in protein loading buffer and subjected to immunoblotting analyses using an anti-Xpress mouse monoclonal antibody (Invitrogen).
For peptide pull-down assay, peptides were covalently bound to Sulfolink resin (Pierce Chemical Co.) via terminal cysteine residues following the manufacturer's instructions and used in pull-down assays with eluted GST fusion proteins. GST fusion proteins were purified using glutathione-agarose in PBS containing 0.2% Triton X-100 and eluted by elution buffer (10 mM reduced glutathione in 10 mM Tris-HCl, pH 9.5). Eluted GST fusion proteins were then concentrated in PBS buffer using Centricon (Millipore). 30 μl of 50% slurry of peptide beads (∼10 μg peptides immobilized) were incubated with 0.5 μg GST fusion protein in 1 ml PBS buffer containing 0.1% Triton X-100 by rotating at 4°C for 1 h. After extensive washes, samples were boiled in protein loading buffer and subjected to immunoblotting analyses using anti-GST mouse monoclonal antibodies (Cell Signaling).
Immunofluorescence microscopy
A549 cells grown on glass coverslips were transfected and incubated at 37°C in 5% CO2 for 10–16 h before microscopic imaging. Cells were fixed with methanol at −20°C. After rinsing with PBS, cells were incubated with 1:25 anti-cten rabbit polyclonal antibody for 2 h. Samples were then incubated with 1:800 Alexa Fluor 594–conjugated secondary antibody (Invitrogen) for 1 h and visualized with a confocal microscope (LSM 510; Carl Zeiss MicroImaging, Inc.).
Colony formation assay
MDA-MB-468 cells were seeded in a 6-well plate at 2 × 105 cells per well. 16 h later, 2 μg of pEGFPC2 vector or various DLC-1 constructs (pEGFP–DLC-1, pEGFP–DLC-1S440A, pEGFP–DLC-1Y442F, pEGFP–FAB–DLC-1, pEGFP–FAB–DLC-1S440A, or pEGFP–FAB–DLC-1Y442F) were transfected into the cells. After 48 h, the cells were seeded in a 6-well plate at a density of 104 cells per well and selected by 0.8 mg/ml G418 (Geneticin; Invitrogen) for 2 wk. Colony formation efficiency was determined by counting the G418-resistant colonies stained with crystal violet solution (0.25% crystal violet and 3.7% formaldehyde in 80% methanol).
Cell growth analysis
MDA-MB-468 cells were seeded in a 6-well plate at 2 × 105 cells per well. 16 h later, 2 μg of pEGFPC2 vector or various DLC-1 constructs (pEGFP–DLC-1, pEGFP–DLC-1S440A, or pEGFP–DLC-1Y442F) were transfected into the cells. After 48 h, the cells were seeded in 100-mm dishes and selected by 0.8 mg/ml G418 for 6 d. Cells were then collected and seeded in triplicate in 60-mm dishes at 2 × 105 cell density. Cells were harvested at 24-h intervals for 4 d, and the numbers of viable cells were counted by trypan blue exclusion assay with a hematocytometer.
Acknowledgments
This work is supported by grants from the National Institutes of Health (CA102537) and Shriners Hospital (8580).
Abbreviations used in this paper: cten, C-terminal tensin like; DLC-1, deleted in liver cancer 1; FAB, focal adhesion binding; PTB, phosphotyrosine binding; RhoGAP, RhoGTPase-activating protein; SAM, sterile α motif; SH2, Src homology 2; START, steroidogenic acute regulatory-related lipid transfer.
References
- Calderwood, D.A., Y. Fujioka, J.M. de Pereda, B. Garcia-Alvarez, T. Nakamoto, B. Margolis, C.J. McGlade, R.C. Liddington, and M.H. Ginsberg. 2003. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. USA. 100:2272–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., and S.H. Lo. 2003. Regulation of tensin-promoted cell migration by its focal adhesion-binding and Src homology 2 domains. Biochem. J. 370:1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, M.K., Y.C. Liao, Y. Kuwabara, and S.H. Lo. 2005. Inactivation of tensin3 in mice results in growth retardation and postnatal lethality. Dev. Biol. 279:368–377. [DOI] [PubMed] [Google Scholar]
- Cui, Y., Y.C. Liao, and S.H. Lo. 2004. Epidermal growth factor modulates tyrosine phosphorylation of a novel tensin family member, tensin3. Mol. Cancer Res. 2:225–232. [PubMed] [Google Scholar]
- Davis, S., M.L. Lu, S.H. Lo, S. Lin, J.A. Butler, B.J. Druker, T.M. Roberts, Q. An, and L.B. Chen. 1991. Presence of an SH2 domain in the actin-binding protein tensin. Science. 252:712–715. [DOI] [PubMed] [Google Scholar]
- Durkin, M.E., M.R. Avner, C.G. Huh, B.Z. Yuan, S.S. Thorgeirsson, and N.C. Popescu. 2005. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 579:1191–1196. [DOI] [PubMed] [Google Scholar]
- Gao, X., A. Zacharek, D.J. Grignon, W. Sakr, I.J. Powell, A.T. Porter, and K.V. Honn. 1995. Localization of potential tumor suppressor loci to a < 2 Mb region on chromosome 17q in human prostate cancer. Oncogene. 11:1241–1247. [PubMed] [Google Scholar]
- Goodison, S., J. Yuan, D. Sloan, R. Kim, C. Li, N.C. Popescu, and V. Urquidi. 2005. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 65:6042–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S.J., G.A. Keller, N. Hosken, J. Wilkinson, and S. Subramani. 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108:1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann, W., X. Gao, J. Timar, Y.Q. Chen, A.R. Strohmaier, C. Fahrenkopf, D. Kagawa, M. Lee, A. Zacharek, and K.V. Honn. 1996. 12-Lipoxygenase in A431 cells: genetic identity, modulation of expression, and intracellular localization. Exp. Cell Res. 228:197–205. [DOI] [PubMed] [Google Scholar]
- Homma, Y., and Y. Emori. 1995. A dual functional signal mediator showing RhoGAP and phospholipase C-delta stimulating activities. EMBO J. 14:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687. [DOI] [PubMed] [Google Scholar]
- Iyer, L.M., L. Aravind, and E.V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A.B., and A. Hall. 2002. Rho GTPases in transformation and metastasis. Adv. Cancer Res. 84:57–80. [DOI] [PubMed] [Google Scholar]
- Kawai, K., M. Yamaga, Y. Iwamae, M. Kiyota, H. Kamata, H. Hirata, Y. Homma, and H. Yagisawa. 2004. A PLCdelta1-binding protein, p122RhoGAP, is localized in focal adhesions. Biochem. Soc. Trans. 32:1107–1109. [DOI] [PubMed] [Google Scholar]
- Kim, T.Y., H.S. Jong, S.H. Song, A. Dimtchev, S.J. Jeong, J.W. Lee, N.K. Kim, M. Jung, and Y.J. Bang. 2003. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 22:3943–3951. [DOI] [PubMed] [Google Scholar]
- Lo, S.H. 2004. Tensin. Int. J. Biochem Cell Biol. 36:31–34. [DOI] [PubMed] [Google Scholar]
- Lo, S.H. 2006. Focal adhesions: what's new inside. Dev. Biol. 294:280–291. [DOI] [PubMed] [Google Scholar]
- Lo, S.H., and T.B. Lo. 2002. Cten, a COOH-terminal tensin-like protein with prostate restricted expression, is down-regulated in prostate cancer. Cancer Res. 62:4217–4221. [PubMed] [Google Scholar]
- Lo, S.H., P.A. Janmey, J.H. Hartwig, and L.B. Chen. 1994. Interactions of tensin with actin and identification of its three distinct actin-binding domains. J. Cell Biol. 125:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, S.H., Q.C. Yu, L. Degenstein, L.B. Chen, and E. Fuchs. 1997. Progressive kidney degeneration in mice lacking tensin. J. Cell Biol. 136:1349–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, S.S., S.H. Lo, and S.H. Lo. 2005. Cleavage of cten by caspase-3 during apoptosis. Oncogene. 24:4311–4314. [DOI] [PubMed] [Google Scholar]
- Moon, S.Y., and Y. Zheng. 2003. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13:13–22. [DOI] [PubMed] [Google Scholar]
- Ng, I.O., Z.D. Liang, L. Cao, and T.K. Lee. 2000. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 60:6581–6584. [PubMed] [Google Scholar]
- Plaumann, M., S. Seitz, R. Frege, L. Estevez-Schwarz, and S. Scherneck. 2003. Analysis of DLC-1 expression in human breast cancer. J. Cancer Res. Clin. Oncol. 129:349–354. [DOI] [PubMed] [Google Scholar]
- Poy, F., M.B. Yaffe, J. Sayos, K. Saxena, M. Morra, J. Sumegi, L.C. Cantley, C. Terhorst, and M.J. Eck. 1999. Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol. Cell. 4:555–561. [DOI] [PubMed] [Google Scholar]
- Qiao, F., and J.U. Bowie. 2005. The many faces of SAM. Sci. STKE. 2005:re7. [DOI] [PubMed]
- Sayos, J., C. Wu, M. Morra, N. Wang, X. Zhang, D. Allen, S. van Schaik, L. Notarangelo, R. Geha, M.G. Roncarolo, et al. 1998. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 395:462–469. [DOI] [PubMed] [Google Scholar]
- Schwartz, M.A., M.D. Schaller, and M.H. Ginsberg. 1995. Integrins: emerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 11:549–599. [DOI] [PubMed] [Google Scholar]
- Song, Y.F., R. Xu, X.H. Zhang, B.B. Chen, Q. Chen, Y.M. Chen, and Y. Xie. 2006. High frequent promoter hypermethylation of the deleted in liver cancer-1 gene in multiple myeloma. J. Clin. Pathol. 59:947–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.J., E. Jones, X.L. Zhu, M.R. Steele, R.A. Stephenson, L.R. Rohr, and A.R. Brothman. 1996. Evidence for a tumor suppressor gene distal to BRCA1 in prostate cancer. J. Urol. 155:720–725. [PubMed] [Google Scholar]
- Wong, C.M., J.M. Lee, Y.P. Ching, D.Y. Jin, and I.O. Ng. 2003. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 63:7646–7651. [PubMed] [Google Scholar]
- Wong, C.M., J.W. Yam, Y.P. Ching, T.O. Yau, T.H. Leung, D.Y. Jin, and I.O. Ng. 2005. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 65:8861–8868. [DOI] [PubMed] [Google Scholar]
- Yamaga, M., M. Sekimata, M. Fujii, K. Kawai, H. Kamata, H. Hirata, Y. Homma, and H. Yagisawa. 2004. A PLCdelta1-binding protein, p122/RhoGAP, is localized in caveolin-enriched membrane domains and regulates caveolin internalization. Genes Cells. 9:25–37. [DOI] [PubMed] [Google Scholar]
- Yuan, B.Z., M.J. Miller, C.L. Keck, D.B. Zimonjic, S.S. Thorgeirsson, and N.C. Popescu. 1998. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 58:2196–2199. [PubMed] [Google Scholar]
- Yuan, B.Z., M.E. Durkin, and N.C. Popescu. 2003. a. Promoter hypermethylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet. Cytogenet. 140:113–117. [DOI] [PubMed] [Google Scholar]
- Yuan, B.Z., X. Zhou, M.E. Durkin, D.B. Zimonjic, K. Gumundsdottir, J.E. Eyfjord, S.S. Thorgeirsson, and N.C. Popescu. 2003. b. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 22:445–450. [DOI] [PubMed] [Google Scholar]
- Yuan, B.Z., A.M. Jefferson, K.T. Baldwin, S.S. Thorgeirsson, N.C. Popescu, and S.H. Reynolds. 2004. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 23:1405–1411. [DOI] [PubMed] [Google Scholar]
- Zhou, X., S.S. Thorgeirsson, and N.C. Popescu. 2004. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 23:1308–1313. [DOI] [PubMed] [Google Scholar]