Abstract
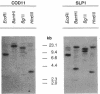
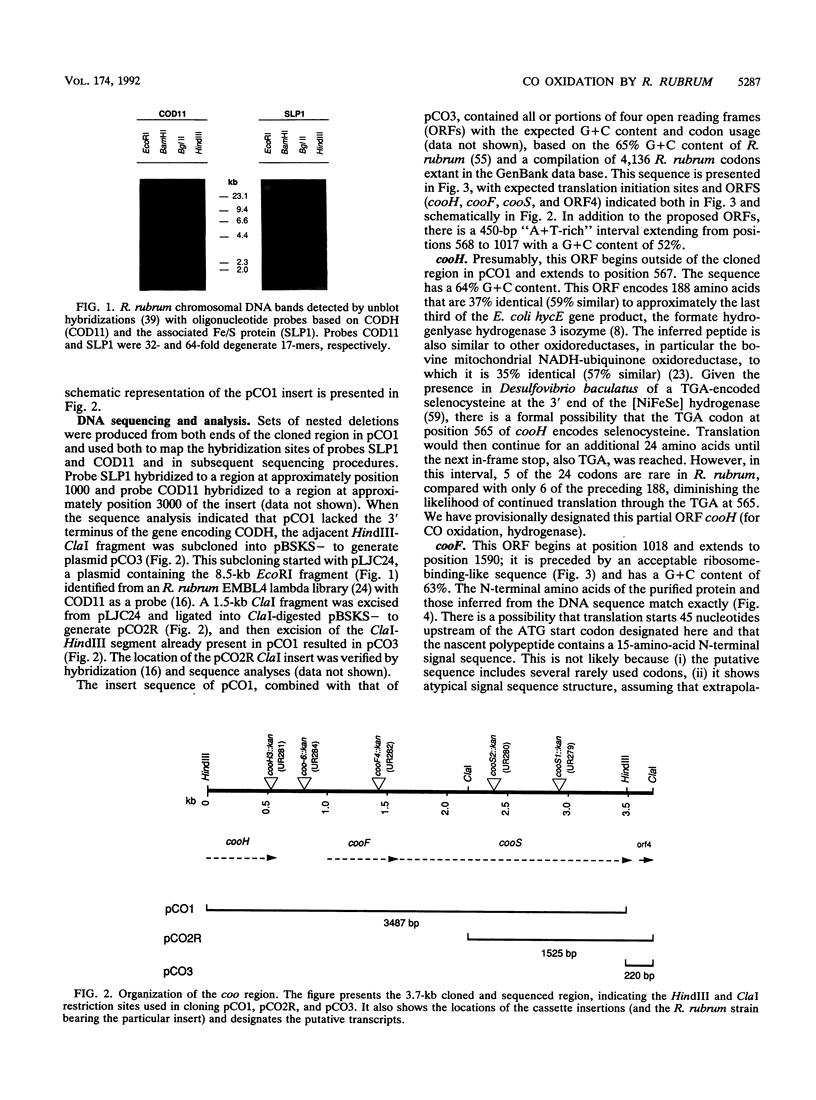
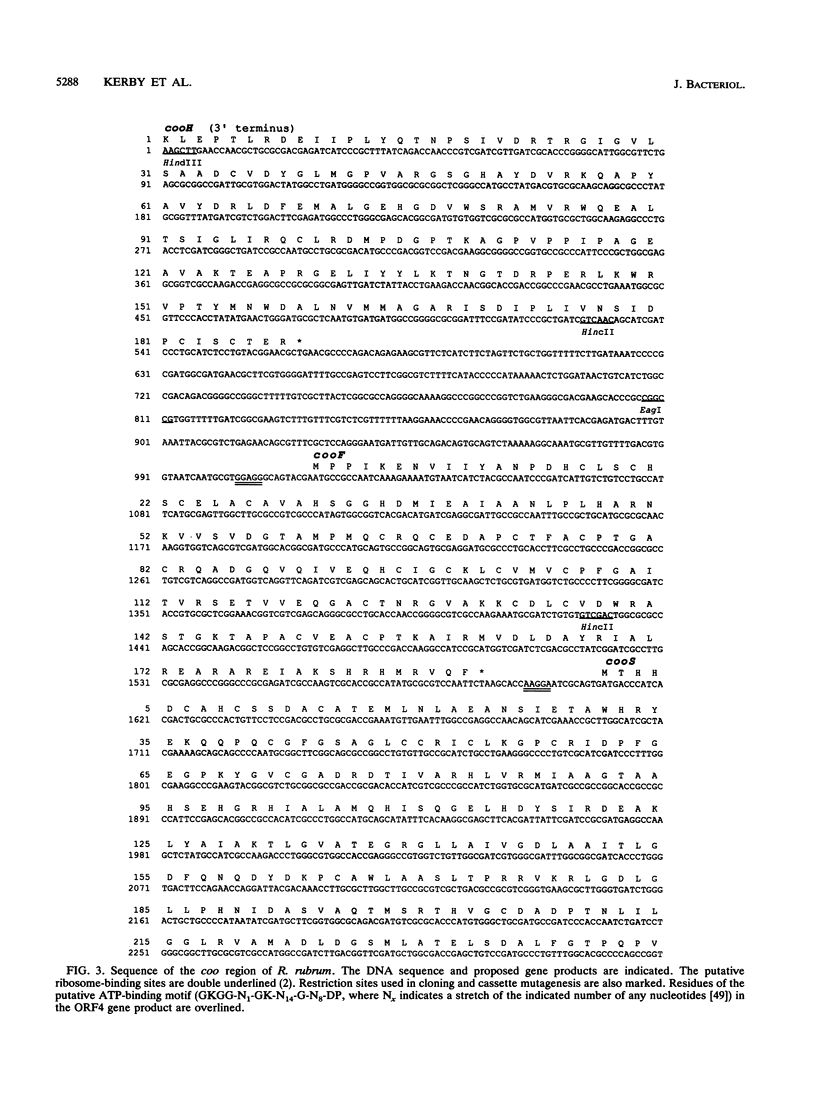
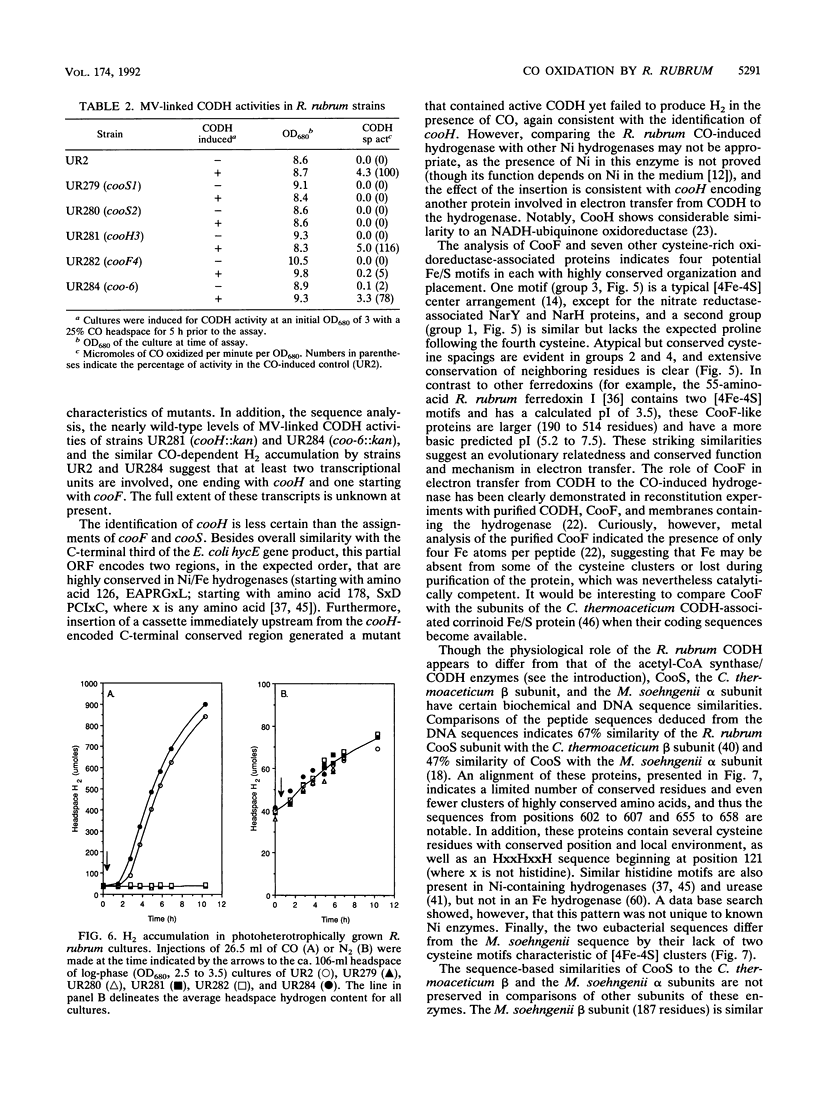
A 3.7-kb DNA region encoding part of the Rhodospirillum rubrum CO oxidation (coo) system was identified by using oligonucleotide probes. Sequence analysis of the cloned region indicated four complete or partial open reading frames (ORFs) with acceptable codon usage. The complete ORFs, the 573-bp cooF and the 1,920-bp cooS, encode an Fe/S protein and the Ni-containing carbon monoxide dehydrogenase (CODH), respectively. The four 4-cysteine motifs encoded by cooF are typical of a class of proteins associated with other oxidoreductases, including formate dehydrogenase, nitrate reductase, dimethyl sulfoxide reductase, and hydrogenase activities. The R. rubrum CODH is 67% similar to the beta subunit of the Clostridium thermoaceticum CODH and 47% similar to the alpha subunit of the Methanothrix soehngenii CODH; an alignment of these three peptides shows relatively limited overall conservation. Kanamycin cassette insertions into cooF and cooS resulted in R. rubrum strains devoid of CO-dependent H2 production with little (cooF::kan) or no (cooS::kan) methyl viologen-linked CODH activity in vitro, but did not dramatically alter their photoheterotrophic growth on malate in the presence of CO. Upstream of cooF is a 567-bp partial ORF, designated cooH, that we ascribe to the CO-induced hydrogenase, based on sequence similarity with other hydrogenases and the elimination of CO-dependent H2 production upon introduction of a cassette into this region. From mutant characterizations, we posit that cooH and cooFS are not cotranscribed. The second partial ORF starts 67 bp downstream of cooS and would be capable of encoding 35 amino acids with an ATP-binding site motif.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews S. C., Harrison P. M., Guest J. R. A molecular analysis of the 53.3 minute region of the Escherichia coli linkage map. J Gen Microbiol. 1991 Feb;137(2):361–367. doi: 10.1099/00221287-137-2-361. [DOI] [PubMed] [Google Scholar]
- Berg B. L., Li J., Heider J., Stewart V. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J Biol Chem. 1991 Nov 25;266(33):22380–22385. [PubMed] [Google Scholar]
- Bilous P. T., Cole S. T., Anderson W. F., Weiner J. H. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol Microbiol. 1988 Nov;2(6):785–795. doi: 10.1111/j.1365-2958.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- Black G. W., Lyons C. M., Williams E., Colby J., Kehoe M., O'Reilly C. Cloning and expression of the carbon monoxide dehydrogenase genes from Pseudomonas thermocarboxydovorans strain C2. FEMS Microbiol Lett. 1990 Aug;58(3):249–254. doi: 10.1016/s0378-1097(05)80003-5. [DOI] [PubMed] [Google Scholar]
- Blasco F., Iobbi C., Giordano G., Chippaux M., Bonnefoy V. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the alpha and beta subunits in iron binding and electron transfer. Mol Gen Genet. 1989 Aug;218(2):249–256. doi: 10.1007/BF00331275. [DOI] [PubMed] [Google Scholar]
- Blasco F., Iobbi C., Ratouchniak J., Bonnefoy V., Chippaux M. Nitrate reductases of Escherichia coli: sequence of the second nitrate reductase and comparison with that encoded by the narGHJI operon. Mol Gen Genet. 1990 Jun;222(1):104–111. doi: 10.1007/BF00283030. [DOI] [PubMed] [Google Scholar]
- Bokranz M., Gutmann M., Körtner C., Kojro E., Fahrenholz F., Lauterbach F., Kröger A. Cloning and nucleotide sequence of the structural genes encoding the formate dehydrogenase of Wolinella succinogenes. Arch Microbiol. 1991;156(2):119–128. doi: 10.1007/BF00290984. [DOI] [PubMed] [Google Scholar]
- Bonam D., Lehman L., Roberts G. P., Ludden P. W. Regulation of carbon monoxide dehydrogenase and hydrogenase in Rhodospirillum rubrum: effects of CO and oxygen on synthesis and activity. J Bacteriol. 1989 Jun;171(6):3102–3107. doi: 10.1128/jb.171.6.3102-3107.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam D., Ludden P. W. Purification and characterization of carbon monoxide dehydrogenase, a nickel, zinc, iron-sulfur protein, from Rhodospirillum rubrum. J Biol Chem. 1987 Mar 5;262(7):2980–2987. [PubMed] [Google Scholar]
- Bonam D., McKenna M. C., Stephens P. J., Ludden P. W. Nickel-deficient carbon monoxide dehydrogenase from Rhodospirillum rubrum: in vivo and in vitro activation by exogenous nickel. Proc Natl Acad Sci U S A. 1988 Jan;85(1):31–35. doi: 10.1073/pnas.85.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam D., Murrell S. A., Ludden P. W. Carbon monoxide dehydrogenase from Rhodospirillum rubrum. J Bacteriol. 1984 Aug;159(2):693–699. doi: 10.1128/jb.159.2.693-699.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschi M., Guerlesquin F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev. 1988 Apr-Jun;4(2):155–175. doi: 10.1111/j.1574-6968.1988.tb02741.x. [DOI] [PubMed] [Google Scholar]
- Bélanger G., Bérard J., Corriveau P., Gingras G. The structural genes coding for the L and M subunits of Rhodospirillum rubrum photoreaction center. J Biol Chem. 1988 Jun 5;263(16):7632–7638. [PubMed] [Google Scholar]
- Böhm R., Sauter M., Böck A. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol. 1990 Feb;4(2):231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggen R. I., Geerling A. C., Jetten M. S., de Vos W. M. Cloning, expression, and sequence analysis of the genes for carbon monoxide dehydrogenase of Methanothrix soehngenii. J Biol Chem. 1991 Apr 15;266(11):6883–6887. [PubMed] [Google Scholar]
- Ensign S. A., Bonam D., Ludden P. W. Nickel is required for the transfer of electrons from carbon monoxide to the iron-sulfur center(s) of carbon monoxide dehydrogenase from Rhodospirillum rubrum. Biochemistry. 1989 Jun 13;28(12):4968–4973. doi: 10.1021/bi00438a010. [DOI] [PubMed] [Google Scholar]
- Ensign S. A., Campbell M. J., Ludden P. W. Activation of the nickel-deficient carbon monoxide dehydrogenase from Rhodospirillum rubrum: kinetic characterization and reductant requirement. Biochemistry. 1990 Feb 27;29(8):2162–2168. doi: 10.1021/bi00460a029. [DOI] [PubMed] [Google Scholar]
- Ensign S. A., Hyman M. R., Ludden P. W. Nickel-specific, slow-binding inhibition of carbon monoxide dehydrogenase from Rhodospirillum rubrum by cyanide. Biochemistry. 1989 Jun 13;28(12):4973–4979. doi: 10.1021/bi00438a011. [DOI] [PubMed] [Google Scholar]
- Ensign S. A., Ludden P. W. Characterization of the CO oxidation/H2 evolution system of Rhodospirillum rubrum. Role of a 22-kDa iron-sulfur protein in mediating electron transfer between carbon monoxide dehydrogenase and hydrogenase. J Biol Chem. 1991 Sep 25;266(27):18395–18403. [PubMed] [Google Scholar]
- Fearnley I. M., Runswick M. J., Walker J. E. A homologue of the nuclear coded 49 kd subunit of bovine mitochondrial NADH-ubiquinone reductase is coded in chloroplast DNA. EMBO J. 1989 Mar;8(3):665–672. doi: 10.1002/j.1460-2075.1989.tb03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Saari L. L., Lowery R. G., Ludden P. W., Roberts G. P. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol Gen Genet. 1989 Aug;218(2):340–347. doi: 10.1007/BF00331287. [DOI] [PubMed] [Google Scholar]
- Gorrell T. E., Uffen R. L. Fermentative metabolism of pyruvate by Rhodospirillum rubrum after anaerobic growth in darkness. J Bacteriol. 1977 Aug;131(2):533–543. doi: 10.1128/jb.131.2.533-543.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Javed A. A., Joshi S. Targeted DNA sequencing: rapid identification of DNA clones by sequencing DNA using mixed oligodeoxynucleotide probes as primers. Biotechniques. 1990 Jul;9(1):28–32. [PubMed] [Google Scholar]
- Jetten M. S., Hagen W. R., Pierik A. J., Stams A. J., Zehnder A. J. Paramagnetic centers and acetyl-coenzyme A/CO exchange activity of carbon monoxide dehydrogenase from Methanothrix soehngenii. Eur J Biochem. 1991 Jan 30;195(2):385–391. doi: 10.1111/j.1432-1033.1991.tb15717.x. [DOI] [PubMed] [Google Scholar]
- Jones W. J., Nagle D. P., Jr, Whitman W. B. Methanogens and the diversity of archaebacteria. Microbiol Rev. 1987 Mar;51(1):135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. Biotechniques. 1990 Dec;9(6):676–679. [PubMed] [Google Scholar]
- Lehman L. J., Roberts G. P. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J Bacteriol. 1991 Sep;173(18):5705–5711. doi: 10.1128/jb.173.18.5705-5711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl L. G. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu Rev Microbiol. 1986;40:415–450. doi: 10.1146/annurev.mi.40.100186.002215. [DOI] [PubMed] [Google Scholar]
- Lu W. P., Ragsdale S. W. Reductive activation of the coenzyme A/acetyl-CoA isotopic exchange reaction catalyzed by carbon monoxide dehydrogenase from Clostridium thermoaceticum and its inhibition by nitrous oxide and carbon monoxide. J Biol Chem. 1991 Feb 25;266(6):3554–3564. [PubMed] [Google Scholar]
- Matsubara H., Inoue K., Hase T., Hiura H., Kakuno T., Yamashita J., Horio T. Structure of the extracellular ferredoxin from Rhodospirillum rubrum: close similarity to clostridial ferredoxins. J Biochem. 1983 May;93(5):1385–1390. doi: 10.1093/oxfordjournals.jbchem.a134273. [DOI] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Peck H. D., Jr, Chatelus C. Y., Choi E. S., Przybyla A. E. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol. 1990 Apr;172(4):1969–1977. doi: 10.1128/jb.172.4.1969-1977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyada C. G., Wallace R. B. Oligonucleotide hybridization techniques. Methods Enzymol. 1987;154:94–107. doi: 10.1016/0076-6879(87)54072-1. [DOI] [PubMed] [Google Scholar]
- Morton T. A., Runquist J. A., Ragsdale S. W., Shanmugasundaram T., Wood H. G., Ljungdahl L. G. The primary structure of the subunits of carbon monoxide dehydrogenase/acetyl-CoA synthase from Clostridium thermoaceticum. J Biol Chem. 1991 Dec 15;266(35):23824–23828. [PubMed] [Google Scholar]
- Mulrooney S. B., Hausinger R. P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990 Oct;172(10):5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale S. W., Clark J. E., Ljungdahl L. G., Lundie L. L., Drake H. L. Properties of purified carbon monoxide dehydrogenase from Clostridium thermoaceticum, a nickel, iron-sulfur protein. J Biol Chem. 1983 Feb 25;258(4):2364–2369. [PubMed] [Google Scholar]
- Reeve J. N., Beckler G. S., Cram D. S., Hamilton P. T., Brown J. W., Krzycki J. A., Kolodziej A. F., Alex L., Orme-Johnson W. H., Walsh C. T. A hydrogenase-linked gene in Methanobacterium thermoautotrophicum strain delta H encodes a polyferredoxin. Proc Natl Acad Sci U S A. 1989 May;86(9):3031–3035. doi: 10.1073/pnas.86.9.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. L., James-Hagstrom J. E., Garvin D. K., Gorst C. M., Runquist J. A., Baur J. R., Haase F. C., Ragsdale S. W. Cloning and expression of the gene cluster encoding key proteins involved in acetyl-CoA synthesis in Clostridium thermoaceticum: CO dehydrogenase, the corrinoid/Fe-S protein, and methyltransferase. Proc Natl Acad Sci U S A. 1989 Jan;86(1):32–36. doi: 10.1073/pnas.86.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Y., Seed B. Electrolyte gradient gels for DNA sequencing. Biotechniques. 1988 Nov-Dec;6(10):942–944. [PubMed] [Google Scholar]
- Silver S., Nucifora G., Chu L., Misra T. K. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989 Feb;14(2):76–80. doi: 10.1016/0968-0004(89)90048-0. [DOI] [PubMed] [Google Scholar]
- Stephens P. J., McKenna M. C., Ensign S. A., Bonam D., Ludden P. W. Identification of a Ni- and Fe-containing cluster in Rhodospirillum rubrum carbon monoxide dehydrogenase. J Biol Chem. 1989 Oct 5;264(28):16347–16350. [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlesky K. C., Ferry J. G. Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J Biol Chem. 1988 Mar 25;263(9):4075–4079. [PubMed] [Google Scholar]
- Thauer R. K., Möller-Zinkhan D., Spormann A. M. Biochemistry of acetate catabolism in anaerobic chemotrophic bacteria. Annu Rev Microbiol. 1989;43:43–67. doi: 10.1146/annurev.mi.43.100189.000355. [DOI] [PubMed] [Google Scholar]
- Uffen R. L. Anaerobic growth of a Rhodopseudomonas species in the dark with carbon monoxide as sole carbon and energy substrate. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3298–3302. doi: 10.1073/pnas.73.9.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uffen R. L. Metabolism of carbon monoxide by Rhodopseudomonas gelatinosa: cell growth and properties of the oxidation system. J Bacteriol. 1983 Sep;155(3):956–965. doi: 10.1128/jb.155.3.956-965.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Menon N. K., LeGall J., Choi E. S., Peck H. D., Jr, Przybyla A. E. Analysis and comparison of nucleotide sequences encoding the genes for [NiFe] and [NiFeSe] hydrogenases from Desulfovibrio gigas and Desulfovibrio baculatus. J Bacteriol. 1989 May;171(5):2894–2899. doi: 10.1128/jb.171.5.2894-2899.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G., Strang J. D., Wilson F. R. Organization of the genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J Bacteriol. 1989 Jul;171(7):3881–3889. doi: 10.1128/jb.171.7.3881-3889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H. G. Life with CO or CO2 and H2 as a source of carbon and energy. FASEB J. 1991 Feb;5(2):156–163. doi: 10.1096/fasebj.5.2.1900793. [DOI] [PubMed] [Google Scholar]