Abstract
Synchronous mitosis is common in multinucleated cells. We analyzed a unique asynchronous nuclear division cycle in a multinucleated filamentous fungus, Ashbya gossypii. Nuclear pedigree analysis and observation of GFP-labeled spindle pole bodies demonstrated that neighboring nuclei in A. gossypii cells are in different cell cycle stages despite close physical proximity. Neighboring nuclei did not differ significantly in their patterns of cyclin protein localization such that both G1 and mitotic cyclins were present regardless of cell cycle stage, suggesting that the complete destruction of cyclins is not occurring in this system. Indeed, the expression of mitotic cyclin lacking NH2-terminal destruction box sequences did not block cell cycle progression. Cells lacking AgSic1p, a predicted cyclin-dependent kinase (CDK) inhibitor, however, showed aberrant multipolar spindles and fragmented nuclei that are indicative of flawed mitoses. We hypothesize that the continuous cytoplasm in these cells promoted the evolution of a nuclear division cycle in which CDK inhibitors primarily control CDK activity rather than oscillating mitotic cyclin proteins.
Introduction
Multinucleated cells are found in a variety of organisms and are integral to processes as diverse as the early development of the fruit fly, bone remodeling, placenta formation, and cancer metastasis. However, information on how multinucleated cells regulate nuclear division is limited to relatively few systems. Fusion experiments with mammalian cells demonstrated that multiple nuclei in a shared cytoplasm synchronize their nuclear division cycles (Rao and Johnson, 1970). These experiments suggested that separate nuclei within a common cytoplasmic environment will alter their normal cell cycle kinetics and align temporally with the other nuclei present. Synchronous mitosis has also been observed in binucleate yeast cells (Hoepfner et al., 2002). Furthermore, in naturally multinucleated cells, mitoses often occur either synchronously, as in the slime mold Physarum polycephalum (Nygaard et al., 1960), or parasynchronously, with a linear wave of nuclear division spreading across a cell, as in the filamentous fungus Aspergillus nidulans (Clutterbuck, 1970).
One hypothesis for the molecular basis for synchronous mitoses in multinucleated cells is that cell cycle regulators shuttle between nuclear and cytoplasmic compartments, which would allow them to diffuse and coordinate multiple nuclei in the same cytoplasm. Thus, the dynamic localization of the cell cycle machinery may promote continual communication between individual nuclei and the cytoplasm and could explain synchronous nuclear division cycles, although in no instance has this hypothesis been tested in multinucleated cells.
In some cases, nuclei appear to behave independently of one another despite sharing a common cytoplasm. In newly fertilized binucleate sea urchin embryos, the two pronuclei have been shown to initiate nuclear envelope breakdown and mitosis at different times if one nucleus is paused as a result of DNA damage (Hinchcliffe et al., 1999). Signaling for cell cycle progression was also nuclear limited in experiments involving binucleate yeast cells in which a single nucleus received DNA damage (Demeter et al., 2000). Similarly, in binucleated cultured marsupial cells, anaphase entry is asynchronous if chromosome attachment is delayed in one spindle (Rieder et al., 1997). Thus, at least for the case of responding to DNA damage or unattached chromosomes, multinucleated cells can undergo asynchronous mitosis. Furthermore, the filamentous fungi Neurospora crassa and Podospora anserina are also thought to have asynchronous mitoses, but in neither case has the phenomenon been studied in depth (Minke et al., 1999; unpublished data). Thus, it seems to be possible for the nucleus itself to control the decision to enter mitosis, although the molecular basis for such nuclear autonomy in any system remains unknown.
In this study, we present evidence for and initial molecular analysis of asynchronous nuclear division cycles in the multinucleated filamentous fungus Ashbya gossypii. This fungus shares a common ancestor with Saccharomyces cerevisiae before the duplication of the budding yeast genome (Dietrich et al., 2004; Kellis et al., 2004). Over 90% of the A. gossypii genome contains syntenic homologues in budding yeast, and, in particular, all cell cycle control proteins described for budding yeast are present with homology ranging from 20 to 95% identity (Dietrich et al., 2004). Despite this conservation of cell cycle gene sets, unlike budding yeast, A. gossypii cells grow exclusively as hyphae containing many nuclei in linear arrays within the same cytoplasm. We predict that these hyphal cells undergo a closed mitosis like many fungi, including S. cerevisiae, and there is evidence that the hyphae are compartmentalized with partial septa that contain pores large enough for nuclei to pass between compartments (Alberti-Segui et al., 2001).
We have generated nuclear pedigrees with time-lapse video microscopy showing that nuclei in A. gossypii cells divide asynchronously in the same cytoplasm. We analyzed this apparent nuclear autonomous behavior and evaluated how periodic cell cycle proteins act in such a system in which nuclei are not in sync with each other. Our experiments showed that both G1 and mitotic cyclins are present across all cell cycle stages. We present evidence that the continuous cytoplasm in these cells permits proteins to diffuse so that nuclei continually receive proteins from the cytosol, which are expressed from neighboring nuclei. These data have led us to hypothesize that differences in cell architecture and spatial organization may have selected for alternative modes of cell cycle control during the evolution of S. cerevisiae and A. gossypii. When combined, our experiments suggest that cyclin-dependent kinase (CDK) inhibitors rather than cycles of accumulation and complete destruction of the cyclins provide the primary source of oscillation in the cell cycle machinery of A. gossypii.
Results
Mitosis is asynchronous in A. gossypii cells
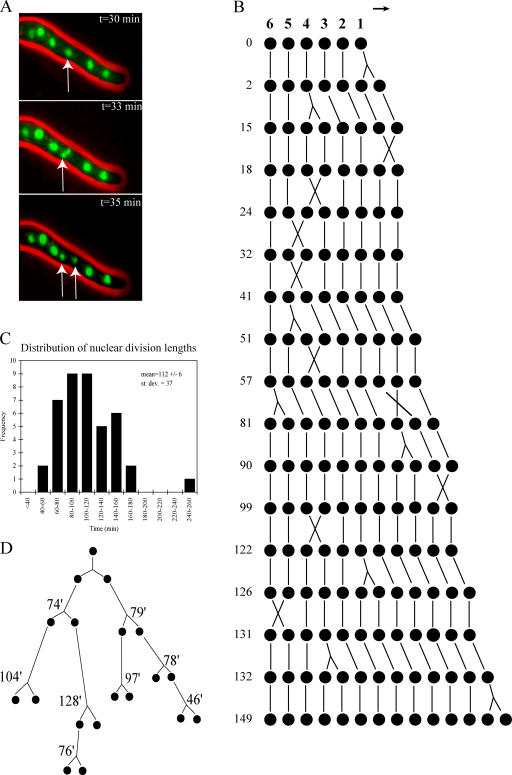
Growth and nuclear dynamics were followed in the growing tip region of A. gossypii cells using in vivo fluorescence and time-lapse video microscopy. Nuclei were readily visualized using a GFP-tagged histone (H4) protein, AgHHF1-GFP. As can be observed in the accompanying video and has been reported previously, these nuclei oscillated rapidly and tumble through the cytoplasm in three dimensions, frequently exchanging places through by-passing one another (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200507003/DC1; Alberti-Segui et al., 2001). Multiple mitoses were easily observed in the growing tip compartment, and, remarkably, in most cases, nuclei divided asynchronously, independently of neighboring nuclei in close physical proximity (Fig. 1, A and B; and Video 1). The distance between nuclei varied between <1.0 μm, when nuclei pass each other, and 7 μm, with a mean of 3.6 ± 0.1 μm. Nuclear division occurred irrespective of the distance from the hyphal tip, and no spatial patterns of mitosis could be discerned from the time-lapse data.
Figure 1.
Mitoses are asynchronous in multinucleated A. gossypii cells. (A) Individual frames from a time-lapse video recording the growth of a strain (HHF1-GFP) with histone-GFP–labeled nuclei. The nuclei indicated by the arrows divide independently from their neighbors. Pictures were acquired every 30 s. (B) Example of a nuclear pedigree demonstrating the capacity for all nuclei to divide. Six starting parent nuclei were followed through subsequent nuclear divisions. Numbers are time in minutes from start of time-lapse acquisition. A total of 11 such pedigrees were performed on independent hyphae. In the pedigree diagram, X is a by-passing event, an upside-down Y is a mitosis, and the arrow denotes the direction of growth. (C) Histogram summarizing variability in nuclear division cycle length from 41 mitoses observed in time-lapse videos of HHF1-GFP strains. (D) Nuclear lineage demonstrating variability in nuclear division cycle length within related nuclei, where the time in minutes represents the time between nuclear divisions.
One way for cells to establish asynchrony would be for some nuclei to be arrested in G0 and act as “insulators” or barriers between actively dividing nuclei. To investigate this possibility, we generated a series of nuclear pedigrees in which nuclei in living cells were followed through several generations of division and were also charted as to their relative positions in the cell (Fig. 1 B). In all pedigrees examined, most nuclei divided, yielding daughter nuclei that also divided (n = 11 independent pedigrees). The mean time between divisions of a nucleus, which represents the total nuclear division cycle length, was 112 min but varied greatly from 46 to 250 min (n = 41 mitoses; Fig. 1 C). Interestingly, variability in cell cycle length was observed within a single lineage starting from one nucleus. Daughter nuclei produced from the same mitosis do not necessarily divide again in the same time interval (Fig. 1 D). Frequently, daughter nuclei by-passed neighboring nuclei soon after mitosis was finished (X on pedigree in Fig. 1 B); however, asynchrony was still observed in the absence of such by-passing events. Furthermore, sudden increases in Hhf1-GFP intensity, which may correlate with S phase, generally occurred at different times in sister nuclei produced from the same mitosis. Thus, most nuclei appear to be capable of division but with variable timing, and, surprisingly, they act independently of neighboring nuclei.
Neighboring nuclei are in different cell cycle stages
The time-lapse data lead to the prediction that adjacent nuclei are in different phases of their division cycle at the same time. To determine the cell cycle stages of neighboring nuclei, we visualized tubulin (Fig. 2 A) in fixed cells and spindle pole bodies (SPBs; fungal equivalent of the centrosome; Fig. 2 B) in living cells. Overall, 62 ± 4.8% of nuclei had a single SPB (noted as 1 in figures), 18 ± 2.6% had duplicated, adjacent SPBs (noted as 2 in figures; scored based on either two distinct dots or a brighter and expanded area of fluorescent signal compared with 1), and 20 ± 2.7% showed separated SPBs and a spindle (noted as Meta for metaphase and Ana for anaphase in figures; Fig. 2, A and B; n > 1,000). Similar proportions were observed by evaluating microtubules in fixed cells with a tubulin antibody (Fig. 2 A) and observing SPBs with SPC42-GFP (Fig. 2 B). We found that only 35% of nuclei (n > 1,000) were in the same spindle stage as their neighbors as judged by SPB morphology, and these were nearly all nuclei with only a single SPB. Focusing specifically on nuclei with mitotic spindles and separated SPBs, >80% were beside a neighbor with only a single or just duplicated SPB, showing that even actively dividing nuclei do not seem to influence neighbors in different cell cycle stages.
Figure 2.
Neighboring nuclei are in different cell cycle stages. Spores were grown overnight at 30°C until young mycelium were formed that contained ∼75–100 nuclei. In all panels, arrows highlight neighboring nuclei in different spindle stages. 1 indicates nuclei with a single SPB; 2 indicates nuclei with adjacent, duplicated SPBs or SPBs that are larger in diameter and ≥2× brighter than single SPBs; Meta indicates likely metaphase nuclei with a spindle aligned across the middle of the DNA; Ana indicates anaphase nuclei in which the spindle connects two separated DNA signals. (A) Tubulin visualization in wild-type cells by immunofluorescence using antitubulin antibody for spindle observation. (B) SPC42-GFP (ASG38) visualization for SPB observation. Proportions are the percentage of nuclei with one, two adjacent, or two separated metaphase/anaphase SPBs (n > 1,000). (C) Wild-type spores were grown for 12 h until a bipolar germling stage and incubated with nocodazole for 4 h. Cells were then washed before release into fresh media and processed at the indicated time points for tubulin immunofluorescence. Nuclei are in blue, and microtubules or SPBs are in green. Bars (A and C), 10 μm; (B) 5 μm.
Four-dimensional time-lapse microscopy of hyphae expressing SPC42-RFP and HHF4-GFP revealed that one nucleus frequently underwent SPB duplication, whereas neighboring nuclei remained with a single SPB (Fig. S1 A, available at http:// www.jcb.org/cgi/content/full/jcb.200507003/DC1). Nuclei spent an extended period with duplicated but not separated SPBs (from 20 min to >1 h, with a mean time of 47 ± 6 min; n = 11 nuclei). Thus, nuclei in A. gossypii appear to transit the different stages of the cell cycle regardless of the state of their neighbors, and many nuclei pause for extended time periods before or during bipolar spindle assembly.
Asynchrony returns rapidly after artificial synchronization
The asynchrony described in Figs. 1 and 2 could come about in either of two ways: each nucleus may independently modulate the lengths of the division cycle phases as suggested by the pedigrees, or all nuclei could undergo a standard cell cycle, but because the nuclei move around, each nucleus is out of phase with respect to its neighbors. To address these possibilities, we used the microtubule poison nocodazole to arrest nuclei in mitosis and synchronize this normally asynchronous system. If nuclei have a standard cycle, synchrony should persist after release from the arrest. All cytoplasmic microtubules were depolymerized by nocodazole, producing tubulin staining limited to SPBs that were used to determine cell cycle stage. After a 4-h incubation in the drug, >70% of the nuclei appeared in synchronous arrest with adjacent, bright SPBs, whereas control cells treated with DMSO still displayed asynchrony similar to that of wild-type cells (n = 200 nuclei for each condition; Fig. 2 C, Fig. S1 B, and not depicted). Cells were released from arrest into fresh media, and nuclear division cycle progression was followed. Initially after release (15-, 30-, and 45-min time points), the extensive network of cytoplasmic microtubules was repolymerized before recovery of nuclear microtubules. By 1 h after release, the cytoplasmic networks of microtubules were largely rebuilt, and the majority of nuclei was in synchrony and had extended spindles indicative of anaphase/telophase. However, by 2 h after the release from nocodazole, only 30% of the nuclei were in the same spindle stages as their immediate neighbors, much like the starting cell population, indicating that asynchrony was quickly restored (Fig. 2 C). These data suggest that each nucleus seems to have an autonomous clock that functions independently of nuclei in their vicinity.
In the process of analyzing these arrest–release experiments, it became clear that hyphal morphogenesis proceeded normally even when the nuclear cycle was blocked (Fig. S1 B). This led to a decrease in the nucleocytoplasmic ratio, and the distance between nuclei ranged from 4 to >30 μm, with a mean of 22 ± 2 μm. Within ∼4 h after recovery (roughly two cell cycle lengths), a nearly normal density of nuclei was restored (5.0 μm between nuclei), suggesting that the nucleocytoplasmic ratio is monitored and that the cell cycle length and/or growth rate can respond to alterations in this ratio.
G1 and mitotic B-type cyclins are nuclear and present across all spindle stages
Are there clues in the A. gossypii genome to explain the molecular basis for nuclear asynchrony? The A. gossypii genome shows striking synteny patterns and similarity in gene set to S. cerevisiae despite their different lifestyles and growth form. Whole genome comparisons between the two organisms strongly suggest that they diverged before the duplication of the budding yeast genome, and, thus, the A. gossypii genome lacks many gene pairs that are retained in budding yeast after this duplication (Dietrich et al., 2004). A. gossypii genes are named based on their syntenic homologue in S. cerevisiae, and in the cases where a syntenic gene pair exists in yeast, the single homologue in A. gossypii is called by both names separated by a slash. We examined homologues of the core cell cycle machinery in A. gossypii and found that the entire gene set is present at syntenic positions, with amino acid identity ranging from 31 to 86% (Table I). Thus, A. gossypii and S. cerevisiae share similar transcription factors, cyclins, the CDK, inhibitors, activators, and degradation machinery involved in cell cycle control.
Table I.
Amino acid sequence comparison of a subset of cell cycle control proteins
| S. cerevisiae cell cycle protein | A. gossypii homologous protein | Percent identity |
|---|---|---|
| Cyclins and CDK | ||
| Cdc28 | Cdc28 | 86 |
| Cln3 | Cln3 | 34 |
| Cln1 | Cln1/2 | 63 |
| Cln2 | Cln1/2 | 60 |
| Clb5 | Clb5/6 | 44 |
| Clb6 | Clb5/6 | 39 |
| Clb3 | Clb3/4 | 51 |
| Clb4 | Clb3/4 | 52 |
| Clb1 | Clb1/2 | 56 |
| Clb2 | Clb1/2 | 65 |
| Degradation machinery | ||
| Cdc4 | Cdc4 | 52 |
| Cdc34 | Cdc34 | 82 |
| Skp1 | Skp1 | 87 |
| Cdc53 | Cdc53 | 64 |
| Grr1 | Grr1 | 50 |
| Cdh1 | Cdh1 | 66 |
| Cdc20 | Cdc20 | 59 |
| Cdc16 | Cdc16 | 53 |
| Apc1 | Apc1 | 35 |
| CDK inhibitors and regulators | ||
| Sic1 | Sic1 | 37 |
| Far1 | Far1 | 31 |
| Hsl7 | Hsl7 | 45 |
| Hsl1 | Hsl1 | 43 |
| Mih1 | Mih1 | 34 |
| Swe1 | Swe1 | 52 |
| Cdc5 | Cdc5 | 73 |
| Transcription factors | ||
| Swi6 | Swi6 | 49 |
| Swi4 | Swi4 | 43 |
| Mbp1 | Mbp1 | 53 |
| Mcm1 | Mcm1 | 54 |
| Swi5 | Swi5/Ace2 | 35 |
| Ace2 | Swi5/Ace2 | 35 |
| Fkh1 | Fkh1/2 | 46 |
| Fkh2 | Fkh1/2 | 50 |
Percentage of amino acid sequence identity between selected S. cerevisiae cell cycle genes and the syntenic A. gossypii homologues. Genes that are duplicated in S. cerevisiae but present in a single copy in A. gossypii are named using both yeast homologues (e.g., AgClb1/2p).
How might this conserved network of cell cycle regulators be constructed to direct asynchronous mitosis in a multinucleated cell? Cyclin proteins are integral to the cell cycle clock in eukaryotic cells and, when complexed to a CDK, drive cell cycle progression (Murray, 2004). Periodic transcription and degradation of cyclins is believed to be a central mode of oscillation that is critical for orderly cell cycle progression. Upstream sequences of AgCLN1/2 and AgCLB1/2 showed the conservation of transcriptional regulatory elements, including some Swi4/6 cell cycle box-binding factor and Mbp1–Swi6 cell cycle box-binding factor transcription factor–binding sites in the AgCLN1/2 promoter and Fkh1p, Fkh2p, and Mcm1p sites in the AgCLB1/2 promoter (consensus sequences summarized in Kellis et al., 2003). Furthermore, protein sequences have degradation motifs, including possible PEST sequences in AgCln1/2p and the destruction (D)-box and KEN-box motifs in AgClb1/2p (Fig. 3; Glotzer et al., 1991; Holloway et al., 1993; Yamano et al., 1996; Pfleger and Kirschner, 2000). The AgClb1/2p sequence has one additional NH2-terminal D box, one of two nuclear export sequences (NESs) that is compared with ScClb2p, and one NLS (Hood et al., 2001).
Figure 3.
Comparison of A. gossypii and S. cerevisiae cyclins. Domain comparison between AgCln1/2p, ScCln1p, and ScCln2p and, below, between AgClb1/2p, ScClb1p, and ScClb2p. The amino acid positions noted for motifs are for the A. gossypii homologue. The cyclin box was identified by a ProSite scan, and the PEST motif is based on Salama et al. (1994). The identity within the PEST domain is 30%, although all key Cdc28p phosphorylation sites are conserved. The D box (db1 [indicated by 1]; Ag aa 28–39; Glotzer et al., 1991), the COOH-terminal cyclin box (ProSite scan), one of two KEN boxes (Pfleger and Kirschner, 2000; Hendrickson et al., 2001), the NLS, and one of two NESs (Hood et al., 2001) are conserved in the A. gossypii CLB1/2 sequence. AgCLB1/2 appears to have an additional putative D-box sequence at aa 84–96 (db2; indicated by 2) and a more NH2-terminal KEN box at aa 6–8.
Based on the homology and similar domain composition of the A. gossypii cyclins compared with those of budding yeast, we would predict that individual nuclei are expressing and degrading distinct cyclins depending on their cell cycle stage. Given that protein translation occurs in a common cytoplasm, however, it is unclear how independent transcriptional programs and oscillating protein levels are achieved by neighboring nuclei that are out of phase with each other. In an effort to determine whether such periodic expression patterns could be locally established in a syncytial cell, the A. gossypii cyclins were localized.
The predicted G1 cyclin AgCLN1/2 and the B-type cyclin AgCLB1/2 were epitope tagged at their endogenous loci with 13 copies of the c-myc epitope to evaluate cyclin protein distribution in the cell. These epitope-tagged strains displayed normal growth and wild-type levels of nuclear asynchrony, and proteins of the proper size were recognized on an anti-myc Western blot (Fig. 4 A and not depicted).
Figure 4.
Cyclins are nuclear and present throughout all nuclear division stages. (A) Western blot on whole cell extracts showing recognition of epitope-tagged cyclins at 80 kD (arrow) with anti-myc antibody (ASG43 and ASG60) and the absence of a band in an untagged wild-type control strain. The load control is a nonspecific cross-reacting band marked by an asterisk, and the molecular mass marker is given in kilodaltons. All images in B–D are derived from stacks of images acquired in the Z axis and compressed to capture all possible signals in the hyphae. (B) Wild-type, untagged A. gossypii cells were processed for immunofluorescence and stained with the myc antibody to show the level of background fluorescence with this antibody. (C) Cyclins AgCln1/2-13mycp (ASG60) and AgClb1/2-13mycp (ASG43) were visualized along with tubulin by immunofluorescence. Individual nuclei are shown representing different spindle stages. All images were acquired and processed under identical conditions. (D) Images showing intact hyphae of AgCln1/2-13mycp and AgClb1/2-13mycp strains prepared as in C. Arrows highlight nuclei of different cell cycle stages as described for Fig. 2. In overlays, DNA is blue, tubulin is green, and cyclins are red, leading to purple nuclei containing cyclins. Bars (B and D), 10 μm; (C) 5 μm.
Cyclin protein localization in the cell was determined using indirect immunofluorescence. The myc antibody did not show any signal in untagged wild-type strains (Fig. 4 B). As would be predicted, AgCln1/2p was concentrated in nuclei with a single SPB and was diffusely present in the cytoplasm (Fig. 4, C and D; left). Surprisingly, however, this G1 cyclin homologue is also readily visible in nuclei with mitotic spindles. Additionally, AgCln1/2p was observed at many hyphal tips, which are sites of polarized growth (Fig. 4 D, left). Similarly, the B-type cyclin homologue AgClb1/2p was concentrated in nuclei with two SPBs and metaphase spindles as expected but, remarkably, was also clearly present in nuclei with a single SPB and in anaphase nuclei (Fig. 4, C and D; right).
Both types of cyclins seem to be present throughout all cell cycle stages (Fig. 4, C and D). AgClb1/2p was found in 91% of nuclei in a mycelium, and AgCln1/2p was observed in 74% of nuclei (n > 200 nuclei). Thus, the proportion of nuclei with a given cyclin is greater than the proportion of nuclei in the corresponding spindle stage (on average only 20% nuclei are in mitosis, yet >90% have mitotic cyclin). Furthermore, nuclei lacking either type of cyclin signal were not in any specific cell cycle stage as determined by spindle appearance, suggesting that these unstained nuclei are results of technical limitations in resolving all proteins by immunofluorescence rather than a large oscillation in cyclin levels in specific stages of the cell cycle (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200507003/DC1). There was some variability in the intensity of signals between nuclei, but this also did not correlate with cell cycle stage, and we predict this is caused by variability inherent in immunofluorescence in fungal cells where there is irregularity in digestion of the cell wall. Thus, the cyclins in A. gossypii do not display complete degradation in sync with cell cycle progression but, instead, are found in most nuclei regardless of their cell cycle stage, suggesting that many nuclei have different types of cyclins present at the same time.
Mitotic cyclins are present in nuclei where they were not transcribed
Synchrony in multinucleated systems is hypothesized to depend upon the free diffusion of mitotic regulators in the cytoplasm. Could asynchrony be established simply by limiting diffusion and concentrating cell cycle factors in nuclei? If so, the nuclear limited localization of mitotic cyclins in A. gossypii cells could compartmentalize the cell and prevent communication between individual nuclei leading to asynchrony. It is unclear, however, if nascent cyclin proteins expressed from a single nucleus and translated in the common cytoplasm are free to diffuse and enter neighboring nuclei of different cell cycle stages. To evaluate the basis of the ubiquitous nuclear cyclin localization, we generated heterokaryon cells in which only a subset of nuclei express a tagged version of the cyclin. We then asked whether the nuclei that do not themselves contain an epitope-tagged cyclin gene have detectable levels of tagged cyclin proteins expressed from neighboring nuclei.
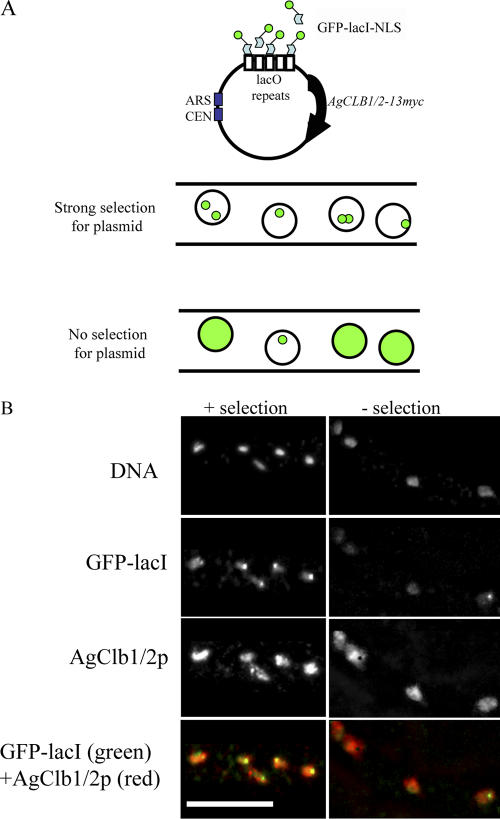
AgCLB1/2-13myc was expressed from an autonomous replicating sequence plasmid that also contained lac operator repeats (lacO) and was put into an A. gossypii strain that had GFP-lacI-NLS integrated in the genome (Fig. 5 A; Belmont and Straight, 1998). Autonomous replicating sequence–containing plasmids are maintained by A. gossypii in a heterokaryon state such that only a subset of nuclei ever contain a plasmid under limited selection pressure, and plasmids are readily lost from nuclei after the removal of all selection (Fig. 5 A). Nuclei containing the plasmid and, thus, also the tagged version of AgCLB1/2 were visible by a bright “dot” of GFP-lacI-NLS where it concentrates as a result of the lacO repeats present on the plasmid (Fig. 5 B, left). When all selection for the plasmid was removed, the bright dots of GFP signal were lost from many nuclei so that only a fraction (28%; n = 300 nuclei) of nuclei retained a plasmid (5 h after release from selection; Fig. 5 B, right). We then asked whether the AgClb1/2-13myc protein was visible in nuclei that did not appear to contain a plasmid. Tagged cyclin proteins were detected in nearly all nuclei regardless of whether they contained the plasmid expressing the epitope-tagged gene. In hyphae in which very few GFP dots were visible, AgClb1/2-13myc protein was still apparent in many nuclei. Thus, nuclei that clearly lacked the plasmid containing the tagged gene had tagged protein product. This product most likely derived from neighboring nuclei that still possessed a plasmid or, in some cases, may still contain a reservoir of stable protein. This experiment suggests that mitotic cyclins are freely diffusing, taken up by, and maintained in neighboring nuclei.
Figure 5.
Cyclins are present in nuclei that are distinct from the site of expression. (A, top) Centromere-based plasmids containing an epitope-tagged AgCLB1/2 gene, lacO repeats, and a dominant drug resistance marker (GEN3) were constructed (pAKH38) that can replicate and segregate in A. gossypii nuclei. (bottom) A. gossypii GFP-lacI-NLS cell strains were transformed with the plasmid containing the AgCLB1/2-13myc and the lacO repeats. In the presence of the G418 drug, many nuclei show one or two tight dots of GFP signal that are indicative of plasmid. When the selective drug pressure is removed, within 5 h, the GFP signal is lost in the majority of nuclei, and GFP-lacI appears as a diffuse signal in the nucleoplasm, which is indicative of no plasmid. CEN, centromeric. (B) GFP-lacI-NLS + placO-CLB1/2-13myc (AKHAg28) were grown overnight for 16 h in 200 μg/ml G418 drug. Half of the cells were fixed before washing out the drug (left), and the remaining half of the cells were resuspended in fresh media lacking the selective drug and allowed to grow for 5 h before fixation (right). Cells were processed for immunofluorescence to visualize GFP-lacI-NLS and AgClb1/2-13mycp. Bar, 10 μm.
Asynchrony persists when cyclins are displaced from nuclei with an exogenous NES
To further address how mitotic cyclin nuclear localization contributes to asynchrony, we shifted a proportion of the cyclin protein out of the nucleus with the addition of a NES to the AgClb1/2-13myc protein (Edgington and Futcher, 2001). If nuclear sequestration of newly made cyclin protein is important for nuclear autonomy, we would predict that the displacement of more mitotic cyclin into the cytoplasm, where it could potentially more freely diffuse, would lead to an increase in synchrony of nuclear division. The additional NES led to an increase in the cytoplasmic levels of AgClb1/2-13myc protein as visible by immunofluorescence but still left sufficient protein in the nucleus to promote normal growth and nuclear division. There was no increase in cell cycle synchrony observed despite the increase in cytoplasmic levels of cyclin proteins between cells with an inactive NES or active NES fusion protein (Fig. 6 A). Interestingly, however, these cells did show a higher proportion of nuclei with duplicated SPBs or mitotic spindles, suggesting that cyclin protein concentration gradients were altered by this displacement, leading to an increase in the frequency of mitosis (Fig. 6 B). Thus, nuclear asynchrony or autonomy seems to be established independently of the origins, levels, and localization of mitotic cyclins.
Figure 6.
Displacement of AgClb1/2p to the cytoplasm with an exogenous NES. (A) AgCLB1/2-NESa-Myc (AKHAg19, active NES) or AgCLB1/2-NESi-Myc (AKHAg22, inactive NES control) were grown for 16 h and processed for antitubulin and anti-myc immunofluorescence. Images were captured and processed identically to compare protein intensities within the figure. Bar, 10 μm. (B) Percentage of nuclei in each spindle stage based on tubulin staining. n > 200 nuclei scored for each strain. M, mitotic.
Mitotic cyclin degradation does not correlate with mitotic exit
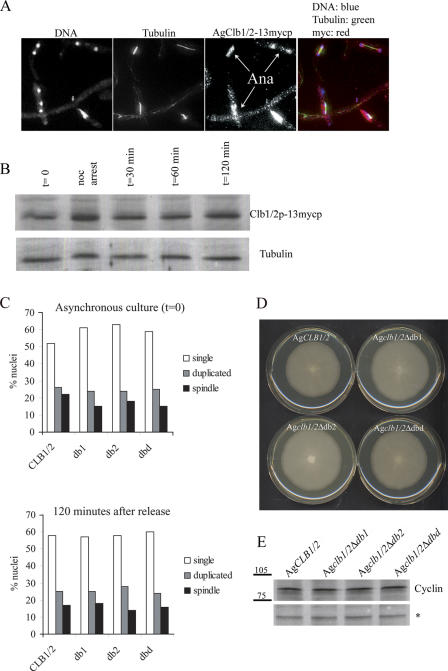
Cyclin proteins appear to be present in all cell cycle stages, which raises the question of how nuclei cycle with accuracy. To further examine the possible persistence of a pool of mitotic cyclin protein across all cell cycle stages, B-type cyclins were evaluated during a nocodazole arrest and release experiment. This allowed AgClb1/2p localization and protein levels to be followed through a synchronous mitotic exit, the time period when mitotic cyclins are predicted to be degraded. Mitotic cyclins were readily observed both by immunofluorescence and by Western blots on whole cell extracts even in cells where the majority (73%) of nuclei had extended spindles and separated DNA indicative of late anaphase/telophase (Fig. 7, A and B). This suggests that B-type cyclin degradation does not correlate with mitotic exit and that any regulative degradation, if present, must occur on only a small fraction of the total protein. Thus, mitotic cyclins seem to persist across the mitosis–G1 transition and potentially may be inactivated for G1 by direct inhibition rather than degradation. When AgCLB1/2 was expressed in yeast cells either from its own promoter or from a GAL promoter, yeast cells showed a delay in telophase, further suggesting that the A. gossypii protein may be somewhat intrinsically stable compared with the yeast cyclins (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200507003/DC1).
Figure 7.
AgClb1/2p levels do not diminish at mitotic exit, and cyclin mutants lacking D-box sequences have normal nuclear division. (A) Mitotic AgClb1/2-13mycp was visualized in young germling cells (ASG43) that were arrested and released from nocodazole. Clb1/2-13myc protein was evaluated by immunofluorescence and (B) on a Western blot, where tubulin was used as a loading control. Arrows highlight anaphase/telophase nuclei with mitotic cyclin. In overlays, DNA is blue, tubulin is green, and cyclins are red, leading to purple nuclei containing cyclins. (C) Wild-type strains were transformed with plasmids containing either AgCLB1/2, Agclb1/2-db1 (pAKH47), Agclb1/2-db2 (pAKH45), or Agclb1/2-dbd (pAKH46) and grown in liquid media containing G418 to maintain the plasmid in most nuclei. A subset of asynchronous cultures grown for 16 h were fixed and processed for antitubulin immunofluorescence, and nuclei were evaluated according to spindle stage (t = 0). The remaining cultures were then arrested and released from nocodazole, and percent nuclei in mitosis were evaluated at 120 min, the point at which wild-type cultures have just returned to normal asynchronous levels of mitosis. (D) Spores from cells containing AgCLB1/2, Agclb1/2-db1, Agclb1/2-db2, or Agclb1/2-dbd were inoculated on plates, and radial growth was evaluated after 5 d at 30°C. Similar growth rates were also observed when alleles are integrated in the genome at the ADE2 locus (not depicted). (E) Spores from cells containing AgCLB1/2-13myc, Agclb1/2-db1-13myc, Agclb1/2-db2-13myc, or Agclb1/2-dbd-13myc were inoculated into liquid media containing 150 μg/ml G418 for selection, grown for 16 h at 30°C, and lysed and processed for an anti-myc Western blot. The asterisk is a nonspecific cross-reacting band for a loading control. Molecular mass is shown in E in kilodaltons.
Mitotic cyclin D-box mutants do not show altered cell cycle progression
These localization and protein level data suggest that cyclin levels do not dramatically fluctuate in a cell cycle–dependent manner in these syncytial cells. The AgClb1/2p homologue, which is an essential gene in A. gossypii, does contain two NH2-terminal D-box sequences that, in other systems, have been shown to lead to anaphase-promoting complex-mediated degradation by the proteosome (Glotzer et al., 1991). In Xenopus laevis, yeast, and Drosophila melanogaster cells, the expression of cyclins lacking the D boxes leads to cell cycle arrest as a result of an inability to exit mitosis (Vodermaier, 2001). If some amount of cyclin destruction is required in vegetatively growing A. gossypii cells, Agclb1/2 D-box mutants should display a clear cell cycle block or delay. The expression of mutants in which either D-box was deleted individually (Agclb1/2-db1 and Agclb1/2-db2) or both were deleted (Agclb1/2-dbd) led to no growth defect or alterations in nuclear division based on the proportion of nuclei in different cell cycle stages and levels of asynchrony (Fig. 7, C [top] and D; n > 200 cells scored for each strain). To determine whether these mutant strains were delayed in exiting mitosis, cells were synchronized with nocodazole and released to observe a synchronous mitosis in the population. (As with Fig. 2, in the first hour after release from nocodazole, cells rebuild cytoplasmic microtubules before restoring nuclear microtubules). When compared with cells with only wild-type AgCLB1/2, both the single and double D-box mutants showed similar rates of return to asynchrony and similar proportions of nuclei in mitosis after 120 min, suggesting no significant delay in mitotic exit (Fig. 7 C, t = 120 min). Surprisingly, protein levels of AgClb1/2-db1p, -db2p, and -dbdp were comparable with wild-type AgClb1/2p (Fig. 7 E). We predict that the D-box sequences may be required in a different growth stage but are dispensable for normal nuclear division during vegetative growth.
AgSic1p contributes to accurate nuclear division
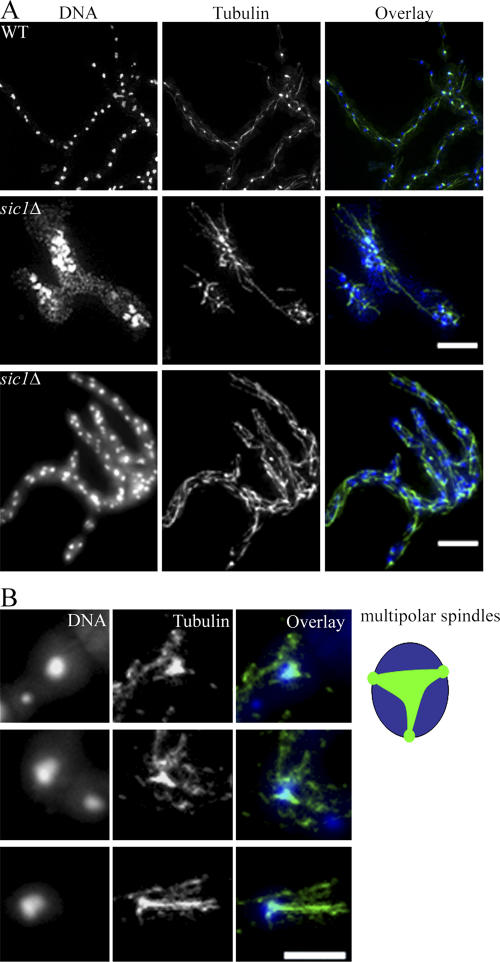
Normal mitotic progression does not appear to require sharp degradation of the mitotic cyclin pools, yet nuclear division is apparently accurate and faithful in A. gossypii. What prevents premature entry into mitosis given the presence of mitotic phase–promoting cyclin early in the cell cycle? One possibility is that mitotic CDK inhibitors such as AgSwe1p and AgSic1p are acting in a nuclear-autonomous fashion and repress any CDK–Clb1/2p complexes present in G1 nuclei. Although mutants lacking AgSWE1 showed minimal mitotic defects (unpublished data), 35% of Agsic1Δ cells stopped growth with 10–20 nuclei, aberrant multipolar spindles, and irregular nuclear distribution and density (Fig. 8, A [middle] and B). A proportion of Agsic1Δ cells (55%) formed a developed mycelium, but, out of these, the majority ultimately arrested growth with up to 100 densely packed nuclei (Fig. 8 A, bottom). About 10% of mutant spores produced viable and sporulating mycelia, potentially as a result of the acquisition of a suppressing mutation or the production of multinucleated spores during sporulation that contain a wild-type nucleus. Many cells in an Agsic1Δ mutant population show defects in nuclear division, suggesting that AgSic1p may help limit the premature activity of mitotic CDK complexes.
Figure 8.
Agsic1Δ mutants arrest growth with abnormal nuclear distribution and spindle defects. (A) Agsic1Δ mutants (ASG65) were germinated in the presence of 50 μg/ml CloNAT to select for cells with the gene disrupted, were fixed and processed for antitubulin immunofluorescence, and compared with wild-type cells. The middle panels show the typical arrest phenotype of bipolar germlings, and the bottom panels show cells that arrest as a small mycelia and high nuclear density. Bars, 10 μm. (B) Higher magnification of nuclei showing multipolar spindle defects observed in Agsic1Δ cells. Nuclei are in blue, and microtubules are in green. Bar, 5 μm.
AgSic1p localization changes across the nuclear division cycle
If AgSic1p is a source of AgCDK/Clb1/2p oscillation, the protein may vary in localization and/or abundance across different stages of the cell cycle. We localized AgSic1p-13myc protein by immunofluorescence and evaluated its behavior during different nuclear division cycle stages to examine whether this predicted CDK inhibitor may oscillate in any way. We found that AgSic1p undergoes notable changes to its sub-nuclear localization, correlating with different stages of nuclear division (Fig. 9). In nuclei with a single SPB, AgSic1p was present throughout the nucleoplasm. Interestingly, the AgSic1p was not uniformly distributed; rather, there were foci of intense signal in addition to a more diffuse signal across the nucleus. In nuclei with duplicated SPBs, the nucleoplasm signal weakened, and some protein was also observed on short spindles. When metaphase spindles were present, AgSic1p was barely detectable in the nucleoplasm, and, occasionally, a low level of AgSic1p was apparent on spindles. In anaphase, as soon as any separation was apparent between the stained DNA, AgSic1p reappeared on elongating spindles as an intense signal. AgSic1p was observed on spindles throughout late anaphase/telophase as well as in the nucleoplasm of the daughter nuclei. These dynamics would suggest that AgSic1p's subnuclear localization is regulated to alter either its own activity or its contact with interacting proteins through different stages of the nuclear cycle.
Figure 9.
AgSic1p localization varies according to nuclear division cycle stage, and summary of A. gossypii nuclear division cycle. (A) AgSic1-13mycp (ASG61) cells were grown for 18 h and processed for antitubulin and anti-myc immunofluorescence. All images in the AgSic1p panel were taken with identical exposure times and processed in parallel with identical processing conditions. 82% of one-SPB nuclei, 76% of two-SPB nuclei, 75% of metaphase nuclei, 80% of early anaphase nuclei, and 79% of late anaphase/telophase nuclei showed signal as depicted by these representative nuclei (n = 400 total nuclei). (B) Summary of asynchronous nuclear division and localization of cell cycle regulators.
Discussion
In this study, we provide evidence for nuclear-autonomous division in multinucleated fungal cells. Nuclear pedigrees and observations of spindle morphology in neighboring nuclei demonstrate that nuclei divide asynchronously in these cells and display independence in a common cytoplasm. Cyclin proteins appear to be highly enriched in nuclei but able to diffuse in the cytoplasm without disturbing asynchrony. Remarkably, the mitotic cyclin protein is present in all stages of the nuclear division cycle, suggesting that oscillation of CDK activity is not primarily generated by periodic expression and complete degradation of cyclins (Fig. 9 B). From our experiments, we propose that the syncytial nature of these cells has favored the evolution of a cell cycle oscillator built primarily upon the action of CDK inhibitors rather than control of cyclin protein abundance. These data raise two unique problems for these cells: how does cell cycle progression occur accurately given the apparent lack of coordinated oscillation in cyclin proteins, and how do nuclei behave independently in the same cytoplasm?
Current models of cell cycle control are rooted in the principle of a biochemical oscillator built through the intertwined regulation of synthesis and destruction of many proteins (Murray, 2004). In yeast, ∼800 genes display periodic expression over the cell cycle, including cyclins, CDK inhibitors, degradation machinery, and transcription factors, and many of these proteins similarly have periodic destruction patterns (Spellman et al., 1998). Extensive experimental data combined with recent mathematical models have shown that this choreographed synthesis and destruction program enables the cell cycle to function as a bistable system that alternates between two stable but mutually exclusive and irreversible states, G1 and S/G2/mitosis phases (Chen et al., 2000; Cross et al., 2002). In yeast, the basis of this bistability is thought to originate from essentially two redundant, although distinct, biochemical oscillators: one rooted in a negative feedback loop that triggers mitotic cyclin (ScClb2p) degradation and the other, which is a “relaxation oscillator” that ensures alternating states of high Clb2p/low Sic1p and low Clb2p/high Sic1 activity (Cross, 2003). Only one of the two oscillators needs to be present for yeast to divide. Importantly, however, both oscillators require fluctuating protein levels of either Clb2p or Sic1p to properly function and maintain the bistable nature of the system (Cross, 2003).
It appears as though the cell cycle oscillators in A. gossypii may not require temporally regulated synthesis and complete degradation of at least some key cell cycle control proteins. Conceivably, there is highly localized degradation of a small but functionally significant fraction of cyclin proteins that has escaped our detection in these studies. However, lack of a dominant phenotype in cells expressing cyclin D-box mutants suggests that regulated proteolysis of cyclins is not critical for regulating mitosis during normal vegetative growth. The continuous cytoplasmic streaming of a syncytial cell complicates the establishment of an oscillating protein gradient through degradation alone because new protein is always supplied by the cytoplasm. Thus, multinucleated cells like A. gossypii may have evolved to favor the modulation of CDK activity by inhibitors to generate bistability in the cell cycle circuit. Although at least the AgCln1/2p and AgClb1/2p cyclins are consistently present in nuclei throughout the cell cycle, we predict that they cannot be simultaneously active. In these cells, the feedback loops that run the cell cycle oscillator would lead to changes in activity instead of changes in protein synthesis and degradation. In fact, a bistable cell cycle oscillator has been shown to function with constant levels of mitotic cyclins in Xenopus egg extracts simply through modulating the activity of Cdc2 (CDK; Pomerening et al., 2003). Our data suggest that Sic1p acts in A. gossypii cells to ensure alternating states of CDK activity even in the presence of relatively constant AgClb1/2p levels.
Nuclear-autonomous regulation of a single factor such as the inhibitor AgSic1p could be sufficient to provide the necessary oscillation in CDK activity for DNA synthesis, spindle construction, and chromosome segregation without great changes in cyclin levels. Thornton and Toczyski (2003) recently presented evidence in S. cerevisiae that oscillation in CDK activity could be generated without the degradation of mitotic cyclins simply by the overexpression of ScSic1p (Thornton et al., 2004). In these cells, ScClb2p was modified to be present in G1 phase, as is observed in the normal A. gossypii cycle, but was held inactive by an overabundance of Sic1p. Furthermore, ScSic1p becomes essential in yeast cells overexpressing ScClb2p (Cross et al., 2005). Clearly, AgSic1p makes a vital contribution to accurate division in these multinucleated cells and may prevent G1 nuclei with AgClb1/2p from entering mitosis before the completion of S phase. The AgSic1p homologue shows relatively low identity to the yeast homologue, suggesting it could have diverged to be more stable and active and may act as a more potent CDK inhibitor. Additionally, AgSic1p subnuclear localization dynamics present the possibility that it is tightly regulated in the nuclear cycle. In the middle of mitosis, when CDK/Clb activity is at a peak, AgSic1p levels diminish in the nucleus potentially as a result of either nuclear autonomous degradation or transient nuclear export. The rapid accumulation of AgSic1 shortly thereafter in early anaphase, however, argues that nuclear import rather than new synthesis would replenish the nuclear protein pool. Thus, we hypothesize that multinucleated cells such as A. gossypii may generate oscillation in CDK activity through posttranslational regulation of inhibitor and cyclin protein activity rather than through tightly controlled abundance. It is conceivable that other cyclins in the system, such as AgClb3/4p or AgClb5/6p, may fluctuate, but this will be tested in future experiments.
How does each nucleus cycle independently? One simple possibility is that asynchrony or nuclear autonomy could be generated as a result of the nature of the cell cycle circuits in A. gossypii, which, as discussed above, may be subtly different than those used in uninucleated cells. Interestingly, recent modeling and experimental work by Pomerening et al. (2005) in Xenopus egg extracts in which a positive feedback loop in the Cdc2/anaphase-promoting complex circuit is blocked (producing damped oscillations in CDK activity) led to asynchrony among nuclei in the extracts. In A. gossypii, the core circuit regulating CDK oscillations may be wired in such a way that transitions between division stages are gradual (which happens when positive feedback loops are repressed in the Xenopus model), making nuclei divide out of sync. Alternatively, cell cycle stage identity is given by periodically transcribed genes that may produce proteins that are translated in a common cytoplasm but, unlike the cyclins, could be directed back to their transcriptional “mother” nucleus. This factor could be a direct regulator of Sic1p that ensures temporal regulation of the localization and, potentially, activity of the CDK inhibitor.
What are possible mechanisms to limit the presence of such an “identity” factor to a specific stage nucleus? Potentially, mRNAs could be targeted to a domain of the ER adjacent to nuclear pores to facilitate rapid uptake into nuclei after translation, or the newly translated proteins could be immediately complexed with nuclear import factors. Conceivably, such a protein could actually be translated within the nuclei, thereby avoiding posttranslational diffusion within the cytoplasm (Iborra et al., 2001). Alternatively, perhaps such a factor is constitutively transcribed but would be selectively blocked in certain cell cycle stages by small regulatory RNA molecules that exist only within the nucleus. The nuclear pore complex itself could be remodeled in a cell cycle–dependent manner such that certain proteins are only allowed to enter nuclei of a particular cell cycle phase (Makhnevych et al., 2003; De Souza et al., 2004). Finally, this could be a factor that is associated with some cell cycle event, such as DNA replication, and could be activated in a nuclear-autonomous manner only upon completion of the event in an individual nucleus. Thus, there are many ways to conceive of how the nuclei could maintain independent cell cycle stage identity within the same space.
Why would a multinucleated cell evolve mechanisms to ensure asynchronous nuclear division? What advantage does a nuclear-autonomous cell cycle bring to a multinucleated cell? Such cells may limit mitosis so that it does not occur synchronously to prevent a sudden doubling of the nucleocytoplasmic ratio. A nuclear-autonomous cell cycle is a possible way to maintain the balance of nuclei in the cell, ensuring that large fluctuations in the nuclear number do not occur without concurrent growth. A nuclear-autonomous cycle also enables nuclei in a specific subcellular location to divide, such as near a branch point or at a site rich in nutrients, without having to duplicate or transport more distant nuclei. Thus, asynchronous mitosis enables conservative and more spatially directed control of mitosis. When combined, these experiments demonstrate that the basic eukaryotic cell cycle control network may have evolved in diverse ways to accommodate the unique geometry of a multinucleated cell. Given the evolutionary relationship between budding yeast and A. gossypii, it is clear that dramatic differences in cell behavior can be achieved through modulating a very similar set of proteins.
Materials and methods
Media, strains, and plasmid construction
A. gossypii media, culturing, and transformation protocols are described in Wendland et al. (2000) and Ayad-Durieux et al. (2000), and strains are listed in Table II. Nocodazole stocks were 3 mg/ml in DMSO and used at a final concentration of 15 μg/μl (Sigma-Aldrich). Standard methods were used for culturing S. cerevisiae and plasmid manipulations (Sambrook, 2001). Plasmids generated and used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200507003/DC1). PCR was performed using standard methods with Taq polymerase from Roche, oligonucleotides were synthesized at MWG, and all restriction enzymes came from New England Biolabs, Inc. or Roche. Oligonucleotide primers are listed in Table S2.
Table II.
A. gossypii strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| WT | leu2Δ thr4Δ | (Altmann-Johl and Philippsen,1996) |
| HHF1-GFP | pHHF1-GFP::GEN3 leu2Δ thr4Δ | (Alberti-Segui et al., 2001) |
| ASG38 |
SPC42-GFP::GEN3/SPC42 leu2Δ
thr4Δ |
This study |
| ASG43 | CLB1/2-13myc::GEN3 leu2Δ thr4Δ | This study |
| ASG46 | ade2::HHF1-GFP::ADE2 leu2Δ thr4Δ | This study |
| ASG50 |
ade2::HHF1-GFP::ADE2 leu2Δ
thr4Δ
SPC42-RFP::GEN3/SPC42 |
This study |
| ASG60 | CLN1/2-13myc::GEN3 leu2Δ thr4Δ | This study |
| ASG61 | SIC1-13myc::GEN3 leu2Δ thr4Δ | This study |
| ASG65 | sic1ΔNAT1 leu2Δ thr4Δ | This study |
| AKHAg19 | CLB1/2-NESa-13myc-GEN3 | This study |
| AKHAg22 | CLB1/2-NESi-13myc-GEN3 | This study |
| AKHAg26 | ade2::GFP-lacI-NLS::ADE2 | This study |
| AKHAg28 |
ade2::GFP-lacI-NLS::ADE2+placO-
CLB1/2-13myc |
This study |
| AKHAg29 | ade2::GFP-lacI-NLS::ADE2+placO-CLB1/2-13myc | This study |
All ASG and AKHAg strains are A. gossypii derived from the wild-type (ΔlΔt) strain. Individual nuclei are haploid, and most strains are homokaryotic (all nuclei have same genotype). In cases of heterokaryons (strains with mixed genotypes for a given locus), genotypes of the two different types of nuclei are written, separated by a plus sign.
The wild-type A. gossypii strain was transformed with the pHHF4-GFP to make the AgHHF1 strain (Alberti-Segui et al., 2001). This strain was always grown under selective conditions with 200 μg/ml G418 (Calbiochem/Merck) to maintain the plasmid during time-lapse video microscopy for pedigree analyses. SPC42 was tagged at the COOH terminus with GFP at its genomic locus using PCR-mediated gene targeting with oligonucleotides SPC42-GFPS1 and SPC42-GFPS2 and the pGUG template to generate ASG38, which was verified using oligonucleotides SPC42 I and GG2.
The 13-myc-GEN cassette for A. gossypii was generated by digesting pFA6-13myc with BglII and ligating the gel-purified 3.2-kb fragment to the 1.7-kb fragment generated from digesting pGEN3 with EcoRV–BamHI. To COOH-terminally tag the cyclin genes and AgSic1p, this 13-myc cassette was amplified with oligonucleotides Clb2-MycF and Clb2-MycR for AgCLB1/2, Cln2-MycF and Cln2-MycR for AgCLN1/2, and Sic1-MycF and Sic1-MycR for AgSIC1. The resulting PCR products were cotransformed in yeast with the plasmid pAG7578 (for CLB1/2) or pAG5016 (for CLN1/2) to produce pCLB2-13myc and pCLN2-13myc. AgSIC1 was tagged at its endogenous locus by direct transformation with the PCR fragment. pCLB2-13myc was digested with BglI and NcoI, and pCLN2-13myc was digested with ApaI before the transformation of A. gossypii cells to tag the endogenous cyclin genes, creating ASG43 and ASG60, which were verified with oligonucleotides Clb1-Myc-I or Cln2-Myc-I and MycI.
To generate the ASG46 strain, the plasmid pHHF1 [29] was digested with BglII and SacI. Blunt ends were generated using the 5′–3′ exonuclease activity of Vent polymerase. The 1,747-bp fragment was subcloned into pAIC opened at the ScaI site (Knechtle, 2002). The new plasmid pHPH001 carried a COOH-terminal fusion of GFP (S65T) to the ORF of histone H4 (AgHHF1) with flanking homologies to the AgADE2 locus. HHF1-GFP was under the control of the AgHHF1 promoter and was terminated by ScADH1-T. 5 μg pHPH001 was isolated from Escherichia coli, digested with EcoRI–HindIII, and transformed into the partially deleted AgADE2 locus of the Agade2Δ1 strain (Knechtle, 2002). Transformants were obtained on minimal medium lacking adenine (ASG46). This strain was then used as the basis for constructing ASG50, which contains HHF4-GFP and SPC42-RFP.
The RFP-tagging cassette for A. gossypii was generated by digesting pRedStar2 (provided by M. Knop, European Molecular Biology Laboratory, Heidelberg, Germany; Maekawa et al., 2003) with BamHI and SphI and ligating the purified 3-kb fragment to the 1.5-kb fragment generated by digesting pGEN3 with BamHI and SphI. This was used as a template to amplify RFP-GEN3 using oligonucleotides with homology to the COOH terminus of AgSPC42, SPC42RedS1, and SPC42RedGS2. The resulting PCR product was cotransformed into yeast cells together with pAG19228 to generate pAG-SPC42RFP, which was then digested with XbaI and MscI to transform strain ASG46 and tag the endogenous SPC42 gene with RFP to produce ASG50, which was verified with oligonucleotides Red I and SPC42 I.
A. gossypii deletion mutants were made using pairs of gene name S1/S2 oligonucleotides for each locus, which contained 45-bp homology upstream and downstream of the ORFs. Wild-type A. gossypii cells were transformed with PCR products amplified off the pGEN3 template with the S1/S2 oligonucleotide pairs. The primary transformation produces heterokaryon cells, which have a mixture of wild-type and transformed nuclei; thus, even mutations in essential genes produce viable transformants. Transformants were verified with oligonucleotide gene name G1/G4/I, and three independent transformants were characterized for each mutant. To evaluate phenotypes of lethal mutants, the heterokaryon was sporulated to produce uninucleated spores, which were then germinated under selective conditions.
The clb1/2Δdb mutants were made using an overlap PCR approach, which deleted amino acids 27–36 (db1) and/or amino acids 84–96 (db2) in AgClb1/2. To generate pAKH47 (db1), pCLB2-13myc was used as a template for a PCR reaction with clb2dbA and clb2dbB to produce a 643-bp product and a second reaction with clb2dbC and clb2dbD to create a 537-bp product. Oligonucleotide clb2dbA was homologous to a region 500 bp upstream of the D box, and clb2dbD was homologous to a region 600 bp downstream of the D box. Oligonucleotide clb2dbB contained homology to CLB2 immediately upstream of the D box but lacked the D-box sequences between nt 81 and 108. Additionally, the 3′ end of clb2dbB has homology to the region immediately downstream of the D box, which is also present in the 5′ end of oligonucleotide clb2dbC. This overlapping homology was then used in a second PCR reaction involving the two products of A + B and C + D and additional clb2dbA and clb2dbD primers to create a 1.1-kb fragment containing an in-frame deletion of the D box (db1) and part of the AgCLB1/2 ORF. To generate a full-length db mutant, this 1.1-kb fragment was cotransformed into yeast strains with pCLB2-13myc, which had been digested with NheI located just upstream of the D box. The gap-repaired plasmid containing the deleted D box was confirmed by sequencing and was called pclb2Δdb1-13myc.
Similarly, to generate pAKH45 (db2), oligonucleotides clb2dbA and clb2db2B were used with pCLB2-13myc as a template in the first reaction, and clb2db2C and clb2dbD were used in the second reaction. Clb2db2B contains sequence just upstream of D box 2 and lacks the sequence from nt 248–287 corresponding to db2. Additionally, the 3′ end of clb2db2B has homology to the region immediately downstream of the D box 2, which is also present in the 5′ end of oligonucleotide clb2db2C. The full-length mutant lacking db2 was generated as for db1, but the pCLB2-13myc plasmid was digested with Aar1 for cotransformation in yeast. To delete both D boxes (pAKH46), a similar attempt was chosen, but pAKH45 was used as template for both PCR reactions, using clb2dbA and clb2db1B for the first and clb2db1C and clb2dbD as primers for the second PCR reaction, as described for constructing the db1 mutant.
The GFP-lacI-NLS integration cassette for A. gossypii was generated with pLKL55Y, which was provided by C. Pearson and K. Bloom (University of North Carolina, Chapel Hill, NC). pLKL55Y was digested with Sap1 and HindIII to generate a 2-kb fragment, and blunt ends were generated with Vent polymerase and ligated into the Sca1 site of the pAIC plasmid. This produced two plasmids, pAKH35 and pAKH36, which were verified by sequencing and differ only in the orientation of lacI-NLS-GFP. pAKH35 and pAKH36 were digested with HindIII–Not1 and transformed into the partially deleted AgADE2 locus of the Agade2Δ1 A. gossypii strain (Knechtle, 2002) to generate AKHAg26 and AKHAg27, which contain the lacI-NLS-GFP integrated at the ADE2 locus. These strains were verified by PCR and showed diffuse, nuclear GFP signal.
To generate the plasmid containing the lacO repeats and AgCLB1/ 2-13myc (pAKH38), pLKL60Y (provided by C. Pearson and K. Bloom) was cut with Pac1 and BamHI to generate a 1.5-kb fragment containing 32 lacO repeats, and blunt ends were generated using Vent polymerase. pUC21 was linearized with the blunt end cutter HincII and ligated with the lacO fragment, resulting in pAKH37. pAKH38 was generated by digesting pAKH37 with KpnI and Nde1 and ligating this 1.6-kb fragment to the 9,535-bp fragment generated from digesting pCLB2-13myc with KpnI–Nde1. pAKH38 was sequenced and transformed as plasmid into AKHAg26 and AKHAg27, resulting in AKHAg28 and AKHAg29, respectively.
The forced localization cassettes were built using p306-NESa and p306-NESi, which were provided by N.P. Edgington and B. Futcher (SUNY Stony Brook, Stony Brook, NY). The NESa and NESi of the plasmids p306-NESa and p306-NESi were PCR amplified with primers NES-P1-Nde1 and NES-a-P2-SmaI or NES-i-P2-SmaI containing the restriction sites Nde1 and Sma1. The 156-bp PCR product was cut with Nde1 and Sma1, resulting in a 130-bp fragment, and was ligated into the Nde1- and Sma1-cut pAG13-myc. This gave rise to pAKH20 (containing NESa) and pAKH21 (containing NESi), which were both verified by sequencing.
For construction of the forced localization cassettes of the mitotic cyclin AgCLB1/2, the pAKH20 and pAKH21 cassettes were amplified with the oligonucleotides Clb-NES-P1 and Clb-Myc-P2, each giving rise to a 2,800-bp fragment. These PCR fragments were cotransformed into yeast cells together with pAG7578 to generate pAKH25 and pAKH26. pAKH25 and pAKH26 were digested with Stu1, NcoI, and BsrG1, and the resulting DNA was transformed into A. gossypii cells, resulting in the strains AKHAg19 and AKHAg22. These new strains were verified using oligonucleotide CLB1/2-I1 and G3.
Immunofluorescence, Hoechst staining, and Western blotting
A. gossypii cells were processed for immunofluorescence as described for yeast cells (Pringle et al., 1991) with slight modifications. Young mycelium containing ∼75–100 nuclei were fixed for 1.5 h in 3.7% formaldehyde (Fluka) and digested in 1.0 mg/ml zymolyase + 1% β-meracptoethanol for 30–45 min before antibody incubation. Anti-myc and tubulin stainings were performed sequentially so as to limit cross reactivity, beginning with mouse anti-myc (Santa Cruz Biotechnology, Inc.) at 1:100, AlexaFluor488 anti–mouse (Invitrogen) at 1:200, rat antitubulin (YOL34; Serotec) at 1:50, and AlexaFluor568 anti–rat (Invitrogen) at 1:200 with Hoechst (Invitrogen) dye to visualize nuclei at 5 μg/ml. Rabbit anti-GFP (Invitrogen) was used to detect GFP-lacI-NLS at a 1:100 dilution, and AlexaFluor488 anti–rabbit was used at a 1:200 dilution (Invitrogen). Antibody dilutions and washes were made with PBS + 1.0 mg/ml BSA.
When only nuclei were visualized, cells were fixed for 1 h in 3.7% formaldehyde in growth media at growth temperature, washed once in 1× PBS, and resuspended in a 5.0-μg/ml solution of Hoechst dye. Cells were incubated with dye for 30 min at room temperature and washed three times in 1× PBS.
Western blotting was performed using standard conditions and nitrocellulose membranes, blocking with 3% milk in 0.1% PBS-Tween (Sambrook, 2001). Mouse anti-myc was used at 1:2,000, and HRP anti–mouse (Jackson ImmunoResearch Laboratories) was used at 1:10,000. ECL chemiluminescence solution (GE Healthcare) was used to develop the Western blots.
Microscopy
The microscope used for all fixed cell images (immunofluorescence and Hoechst stainings with cells mounted in standard fluorescent mounting medium containing 1 mg/ml p-phenylenediamine in 90% glycerol) was essentially as described in Hoepfner et al. (2000) and consisted of an imaging microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) with plan Neofluar 100× Ph3 NA 1.3 objectives. It was equipped with a 75 WXBO and a 100 WHBO illumination source controlled by a shutter and filter wheel system (MAC2000; Ludl Electronics). The camera was a back-illuminated cooled CCD camera (TE/CCD-1000PB; Princeton Instruments). The following filter sets for different fluorophores were used: No. 10 for AlexaFluor488 and No. 20 for rhodamine/AlexaFluor568 (Carl Zeiss MicroImaging, Inc.); and No. 41018 for GFP (Chroma Technology Corp.). The excitation intensity was controlled with different neutral density filters (Chroma Technology Corp.). MetaMorph 4.6r9 software (Universal Imaging Corp.) controlled the microscope, camera, and Ludl controller and was used for processing images. The microscope used for time-lapse images was a similarly equipped microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) with a plan Neofluar 63× NA 1.25 or 100× (same as above) objective. The 100× objective was driven by a PIFOC P721.10 objective nanopositioner (Piezo device) to move quickly through multiple planes in the Z axis (Physik Instrumente). A Micromax back-illuminated CCD camera (512BFT; Princeton Instruments) was used for rapid acquisition and transfer of images. Stacks of tubulin images were processed using the “no-neighbors” deconvolution within MetaMorph, and, like all other fixed specimens, the Z-stacks were compressed using stack arithmetic, brightness and contrast were adjusted, and images were overlayed using color align.
For time-lapse videos, spores were grown 12–16 h in liquid media at 30° under selective conditions to maintain the HHF1-GFP plasmid. Cells were washed and resuspended in 1/4× AFM + G418 + 1× glucose to limit fluorescent background from the media during time-lapse acquisition. Glass slides containing a concave well in the middle were filled with hot 1/4× AFM + 1× glucose + 1% agarose, which solidified and provided a growth surface for the prepared cells. 8 μl of washed cells were placed on the agarose pad, and a coverslip was gently placed on top of the cell suspension. Slides were then placed in a moist chamber and incubated at 30°C for 30 min to allow cells to adapt to the solid substrate.
Time-lapse conditions used XBO fluorescence at 5–10% intensity and exposures of 100 ms that were acquired every 30 s. For HHF1-GFP videos, a 63× objective was used, and a single-phase image with transmitted light was taken followed by 5 × 0.5–μm images in the Z axis with the EGFP filter and fluorescent light. For HHF1-GFP SPC42-RFP videos, a single Hhf1-GFP image was acquired followed by 16 × 0.5–μm images in the Z axis with the rhodamine filter to capture the dynamics of Spc42-RFP using the 100× objective. Focusing was automatically performed using the find focus function in MetaMorph software, and for HHF1-GFP SPC42-RFP videos, a Piezo motor directed the objective to allow more rapid movement through the Z axis than with the motorized stage. Images were acquired at a constant ambient temperature of 25–27°C.
Time-lapse images were processed in MetaMorph to construct videos. Phase images were false colored red to provide an outline of the cell, and the GFP signal was colored green. Fluorescent images were flattened into a single plane, and phase and fluorescent images were assembled into separate stacks, which were then aligned using the color align function into a single stack. This then compressed the phase and fluorescent images into a single plane for each time point. This stack was then made into a video in which each image plays for 1/6 s.
Online supplemental material
Figs. S1 shows still images from a time-lapse video observing SPBs and nuclei. Fig. S2 shows the categorization of nuclei that lack detectable cyclin signal according to nuclear cycle stage. Fig. S3 shows that AgCLB1/2 causes a transient delay in the budding yeast cell cycle. Table S1 presents the plasmids that were used in this study. Table S2 presents the oligonucleotide primers used in this study. Video 1 shows the dynamics and divisions of nuclei in a growing A. gossypii hyphal tip. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200507003/DC1 .
Supplementary Material
Acknowledgments
We would like to thank Hanspeter Helfer for constructing the integrated HHF1-GFP strain and for his image processing expertise; Hans-Peter Schmitz, Sophie Lemire-Brachat, and Zaki Sellam for assistance with bioinformatics; and Pierre Philippe Laissue for providing microscopy suggestions. We appreciated critical comments on this manuscript from Mike Hall, Jacob Harrrison, Hanspeter Helfer, Stephen Helliwell, Javier Irazoqui, Philipp Knechtle, Lukasz Kozubowski, Daniel Lew, Hans-Peter Schmitz, Chandra Theesfeld, and Kelly Tatchell. We also thank Cristine Alberti-Segui for preliminary observations of nuclear dynamics and useful discussions. Plasmids and strains were provided by Michael Knop, Kerry Bloom, and Bruce Futcher.
This work was supported by a National Science Foundation postdoctoral fellowship and Roche Research Foundation grant to A.S. Gladfelter and a Swiss National Fund grant to A.S. Gladfelter and P. Philippsen.
A.S. Gladfelter's present address is Department of Biology, Dartmouth College, Hanover, NH 03755.
Abbreviations used in this paper: CDK, cyclin-dependent kinase; NES, nuclear export sequence; SPB, spindle pole body.
References
- Alberti-Segui, C., F. Dietrich, R. Altmann-Johl, D. Hoepfner, and P. Philippsen. 2001. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 114:975–986. [DOI] [PubMed] [Google Scholar]
- Altmann-Johl, R., and P. Philippsen. 1996. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250:69–80. [DOI] [PubMed] [Google Scholar]
- Ayad-Durieux, Y., P. Knechtle, S. Goff, F. Dietrich, and P. Philippsen. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113:4563–4575. [DOI] [PubMed] [Google Scholar]
- Belmont, A.S., and A.F. Straight. 1998. In vivo visualization of chromosomes using lac operator-repressor binding. Trends Cell Biol. 8:121–124. [DOI] [PubMed] [Google Scholar]
- Chen, K.C., A. Csikasz-Nagy, B. Gyorffy, J. Val, B. Novak, and J.J. Tyson. 2000. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol. Biol. Cell. 11:369–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck, A.J. 1970. Synchronous nuclear division and septation in Aspergillus nidulans. J. Gen. Microbiol. 60:133–135. [DOI] [PubMed] [Google Scholar]
- Cross, F.R. 2003. Two redundant oscillatory mechanisms in the yeast cell cycle. Dev. Cell. 4:741–752. [DOI] [PubMed] [Google Scholar]
- Cross, F.R., V. Archambault, M. Miller, and M. Klovstad. 2002. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell. 13:52–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F.R., L. Schroeder, M. Kruse, and K.C. Chen. 2005. Quantitative characterization of a mitotic cyclin threshold regulating exit from mitosis. Mol. Biol. Cell. 16:2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, C.P., A.H. Osmani, S.B. Hashmi, and S.A. Osmani. 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14:1973–1984. [DOI] [PubMed] [Google Scholar]
- Demeter, J., S.E. Lee, J.E. Haber, and T. Stearns. 2000. The DNA damage checkpoint signal in budding yeast is nuclear limited. Mol. Cell. 6:487–492. [DOI] [PubMed] [Google Scholar]
- Dietrich, F.S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 304:304–307. [DOI] [PubMed] [Google Scholar]
- Edgington, N.P., and B. Futcher. 2001. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 114:4599–4611. [DOI] [PubMed] [Google Scholar]
- Glotzer, M., A. Murray, and M. Kirschner. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138. [DOI] [PubMed] [Google Scholar]
- Hendrickson, C., M.A. Meyn III, L. Morabito, and S.L. Holloway. 2001. The KEN box regulates Clb2 proteolysis in G1 and at the metaphase- to-anaphase transition. Curr. Biol. 11:1781–1787. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., E.A. Thompson, F.J. Miller, J. Yang, and G. Sluder. 1999. Nucleo-cytoplasmic interactions that control nuclear envelope breakdown and entry into mitosis in the sea urchin zygote. J. Cell Sci. 112:1139–1148. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., A. Brachat, and P. Philippsen. 2000. Time-lapse video microscopy analysis reveals astral microtubule detachment in the yeast spindle pole mutant cnm67. Mol. Biol. Cell. 11:1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner, D., F. Schaerer, A. Brachat, A. Wach, and P. Philippsen. 2002. Reorientation of mispositioned spindles in short astral microtubule mutant spc72Delta is dependent on spindle pole body outer plaque and Kar3 motor protein. Mol. Biol. Cell. 13:1366–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway, S.L., M. Glotzer, R.W. King, and A.W. Murray. 1993. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 73:1393–1402. [DOI] [PubMed] [Google Scholar]
- Hood, J.K., W.W. Hwang, and P.A. Silver. 2001. The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J. Cell Sci. 114:589–597. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., D.A. Jackson, and P.R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science. 293:1139–1142. [DOI] [PubMed] [Google Scholar]
- Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E.S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 423:241–254. [DOI] [PubMed] [Google Scholar]
- Kellis, M., B.W. Birren, and E.S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 428:617–624. [DOI] [PubMed] [Google Scholar]
- Knechtle, P. 2002. AgSPA2 and AgBOI control landmarks of filamentous growth in the filamentous ascomycete Ashbya gossypii. Ph.D. thesis. Biozentrum Universität Basel, Basel, Switzerland. 69 pp.
- Maekawa, H., T. Usui, M. Knop, and E. Schiebel. 2003. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22:438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych, T., C.P. Lusk, A.M. Anderson, J.D. Aitchison, and R.W. Wozniak. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 115:813–823. [DOI] [PubMed] [Google Scholar]
- Minke, P.F., I.H. Lee, and M. Plamann. 1999. Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet. Biol. 28:55–67. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. 2004. Recycling the cell cycle: cyclins revisited. Cell. 116:221–234. [DOI] [PubMed] [Google Scholar]
- Nygaard, O.F., S. Guttes, and H.P. Rusch. 1960. Nucleic acid metabolism in a slime mold with synchronous mitosis. Biochim. Biophys. Acta. 38:298–306. [DOI] [PubMed] [Google Scholar]
- Pfleger, C.M., and M.W. Kirschner. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pomerening, J.R., E.D. Sontag, and J.E. Ferrell Jr. 2003. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat. Cell Biol. 5:346–351. [DOI] [PubMed] [Google Scholar]
- Pomerening, J.R., S.Y. Kim, and J.E. Ferrell Jr. 2005. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 122:565–578. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., A.E.M. Adams, D.G. Drubin, and B.K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565–602. [DOI] [PubMed] [Google Scholar]
- Rao, P.N., and R.T. Johnson. 1970. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 225:159–162. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., A. Khodjakov, L.V. Paliulis, T.M. Fortier, R.W. Cole, and G. Sluder. 1997. Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc. Natl. Acad. Sci. USA. 94:5107–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama, S.R., K.B. Hendricks, and J. Thorner. 1994. G1 cyclin degradation: the PEST motif of yeast CLN2 is necessary, but not sufficient, for rapid protein turnover. Mol. Cell. Biol. 14:7953–7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D.W. Russell. 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Spellman, P.T., G. Sherlock, M.Q. Zhang, V.R. Iyer, K. Anders, M.B. Eisen, P.O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 9:3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, B.R., and D.P. Toczyski. 2003. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 5:1090–1094. [DOI] [PubMed] [Google Scholar]
- Thornton, B.R., K.C. Chen, F.R. Cross, J.J. Tyson, and D.P. Toczyski. 2004. Cycling without the cyclosome: modeling a yeast strain lacking the APC. Cell Cycle. 3:629–633. [PubMed] [Google Scholar]
- Vodermaier, H.C. 2001. Cell cycle: waiters serving the destruction machinery. Curr. Biol. 11:R834–R837. [DOI] [PubMed] [Google Scholar]
- Wendland, J., Y. Ayad-Durieux, P. Knechtle, C. Rebischung, and P. Philippsen. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 242:381–391. [DOI] [PubMed] [Google Scholar]
- Yamano, H., J. Gannon, and T. Hunt. 1996. The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J. 15:5268–5279. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.