Abstract
Proteins that are unfolded or misfolded in the endoplasmic reticulum (ER) must be refolded or degraded to maintain the homeostasis of the ER. Components of both productive folding and ER-associated degradation (ERAD) mechanisms are known to be up-regulated by the unfolded protein response (UPR). We describe two novel components of mammalian ERAD, Derlin-2 and -3, which show weak homology to Der1p, a transmembrane protein involved in yeast ERAD. Both Derlin-2 and -3 are up-regulated by the UPR, and at least Derlin-2 is a target of the IRE1 branch of the response, which is known to up-regulate ER degradation enhancing α-mannosidase–like protein (EDEM) and EDEM2, receptor-like molecules for misfolded glycoprotein. Overexpression of Derlin-2 or -3 accelerated degradation of misfolded glycoprotein, whereas their knockdown blocked degradation. Derlin-2 and -3 are associated with EDEM and p97, a cytosolic ATPase responsible for extraction of ERAD substrates. These findings indicate that Derlin-2 and -3 provide the missing link between EDEM and p97 in the process of degrading misfolded glycoproteins.
Introduction
The ER provides an environment that facilitates the folding and assembly of newly synthesized secretory and transmembrane proteins and actively participates in the quality control of these proteins. By these means, correctly folded proteins are allowed to exit the ER to reach their final destination, whereas incompletely folded or misfolded molecules are retained in the ER. Quality control in the ER is achieved by two independent mechanisms, the productive folding mechanism, which uses various molecular chaperones and folding enzymes localized in the ER (ER chaperones), and the ER-associated degradation (ERAD) mechanism, by which unfolded or misfolded proteins are retrotranslocated from the ER to the cytoplasm to be degraded by the ubiquitin-dependent proteasome system (Gething and Sambrook, 1992; Helenius et al., 1992; Kopito, 1997; Kaufman, 1999).
Under a variety of conditions collectively termed ER stress, however, quality control becomes inefficient, resulting in the accumulation of unfolded proteins in the ER. Essentially, all eukaryotic cells are equipped with a system to cope with protein unfolding or misfolding in the ER. Interestingly, the strategy taken is well conserved from yeast to humans: components of both the productive folding and ERAD mechanisms are induced in response to ER stress by a transcriptional program termed the unfolded protein response (UPR), leading to accelerated refolding and degradation of unfolded proteins, respectively (Mori, 2000; Patil and Walter, 2001; Harding et al., 2002; Schroder and Kaufman, 2005). Nonetheless, an as-yet-unanswered critical question is how eukaryotic cells maintain the homeostasis of the ER by balancing the refolding and degradation mechanisms to counteract ER stress or, in other words, how proteins to be refolded are discriminated from those to be degraded.
Transmembrane proteins are capable of transmitting signals across the membrane, and eukaryotic cells indeed use some of these molecules as sensors and transducers of ER stress for the UPR. Among these, Ire1p/Ire-1/IRE1 is an ER membrane–bound endoribonuclease conserved from yeast to humans. IRE1 initiates unconventional (frame switch) splicing in response to ER stress and has the substrates yeast HAC1 mRNA and metazoan XBP1 mRNA encoding the UPR-specific transcription factors Hac1p and XBP1, respectively. As a result of frame-switch splicing, the DNA binding domain and activation domain of these substrates are joined to produce a transcription factor capable of activating transcription efficiently (Kaufman et al., 2002; Mori, 2003). A second transmembrane UPR signal transducer is pek-1/PERK, an ER membrane–bound protein kinase conserved in metazoan cells. PERK activated in response to ER stress phosphorylates the α subunit of eukaryotic initiation factor 2, resulting in a general attenuation of translation (Ron, 2002). The third UPR transducer is ATF6, an ER membrane–bound transcription factor whose involvement in the UPR has to date been clarified for mammalian cells only. Upon ER stress, ATF6 is subjected to regulated intramembrane proteolysis, leading to liberation of the cytosolic transcription factor domain, which translocates into the nucleus to activate transcription (Mori, 2003). We are currently investigating the roles of the IRE1–XBP1 and ATF6 pathways in the homeostasis of the ER and have proposed a time-dependent phase-transition model based on differential properties of the two signaling pathways (Yoshida et al., 2003; see Discussion).
Various components of ERAD have also been identified (Wilhovsky et al., 2000; Tsai et al., 2002). Among these, ER degradation enhancing α-mannosidase–like protein (EDEM) is a key component, as it targets misfolded glycoproteins to the proteasome, presumably by directly recognizing a signal that is present in misfolded glycoproteins, such as Man8GlcNAc2 structure (Hosokawa et al., 2001). Transcription of EDEM is ER stress inducible, and its induction depends on the IRE1–XBP1 pathway (Yoshida et al., 2003). The mammalian genome contains two further EDEM-like molecules, designated EDEM2 and -3, which are also involved in the ERAD of misfolded glycoproteins and are ER stress inducible in a manner dependent on the IRE1–XBP1 pathway (Mast et al., 2004; Olivari et al., 2005; Molinari, M., personal communication).
Recently, two groups independently identified an interesting human protein that shows weak homology to yeast Der1p, a protein involved in yeast ERAD (Hiller et al., 1996; Knop et al., 1996), and designated it Derlin-1 (Der1p-like protein; Fig. 1 C; Lilley and Ploegh, 2004; Ye et al., 2004). This protein is involved in a process similar to the ERAD used by human cytomegalovirus to escape from the immune system (Wiertz et al., 1996a). In this process, the US11 protein of human cytomegalovirus causes retrotranslocation of major histocompatibility complex class I heavy chain (HC) from the ER to the cytosol for degradation by the proteasome, in which the cytosolic ATPase p97 plays an essential role in extracting class I HC through the ER membrane (Ye et al., 2001). Given that Derlin-1 interacts with functional US11 but not with nonfunctional US11 and that it recruits p97 to the ER membrane, Derlin-1 is considered to provide the missing link between events on the lumenal side of the ER (recognition of class I HC by US11) and those on the cytosolic side (extraction catalyzed by p97; Lilley and Ploegh, 2004; Ye et al., 2004). The mammalian genome contains two additional Der1p-homologous proteins, designated Derlin-2 and -3. It was recently shown that Derlin-2 forms a complex not only with p97 but also with mammalian homologues of yeast Hrd1p and -3p, proteins involved in yeast ERAD (Wilhovsky et al., 2000), similar to Derlin-1 (Lilley and Ploegh, 2005; Ye et al., 2005). However, it is not known whether Derlin-2 or -3 is required for ERAD.
Figure 1.
Identification of Derlin-2 and -3. (A) The levels of CGI-101, BiP, and β-actin mRNA are determined by microarray analysis in HeLa cells treated with or without 2 μg/ml tunicamycin for 8 h. Fold induction was determined, with the means from six independent experiments presented with SDs (error bars). CGI-101 is identical to Derlin-2. (B) Hydropathy plots of human Derlin-2 (CGI-101), -3 (FLJ43842), and -1 are shown. Hydrophobicity and hydrophilicity (expressed by positive and negative numbers, respectively) of the amino acid sequences of three human Derlins were obtained according to the method of Kyte and Doolittle (1982). Black bars mark hydrophobic regions that span the membrane. (C) Amino acid sequence alignment of human Derlin-2, Derlin-3 tv1, Derlin-3 tv2, Derlin-1, and yeast Der1p is shown. Identical amino acids are indicated by white letters in black boxes. Two transcriptional variants for Derlin-3 are deposited in the data bank. Derlin-3 tv1 lacks the 30 COOH-terminal amino acids present in Derlin-3 tv2.
We demonstrate that Derlin-2 and -3, proteins regulated by the UPR, are functional components of ERAD for misfolded glycoproteins and that Derlin-2 at least is a target of the IRE1–XBP1 pathway, similar to EDEM. We also discuss the role of the mammalian UPR in distinguishing proteins to be refolded from those to be degraded.
Results
Derlin-2 and -3 are novel genes regulated by the UPR
We conducted a microarray analysis to identify novel ER stress–inducible genes that might be involved in determining the fate of proteins that are unfolded or misfolded in the ER. Total RNA was isolated from HeLa cells that had been treated for 8 h with or without tunicamycin, an inhibitor of protein N-glycosylation known to cause ER stress (Kaufman, 1999), and the difference in expression levels was determined using the DNA chip cDNA Microarray kit human I carrying 12,814 genes. Results showed ∼300 genes that were induced more than twofold, among which we focused on one, CGI-101, because of its 2.9-fold induction by tunicamycin treatment (Fig. 1 A),the ability of hydrophobic stretches in its translational product to span the ER membrane several times (Fig. 1 B), and, most important, the weak homology it exhibited to yeast Der1p (Fig. 1 C). Although the role of Der1p in ERAD remains poorly understood, these results implied that CGI-101 may be a gene that connects the UPR with ERAD. Our database search also indicated that the human genome contains a gene somewhat similar to CGI-101 (designated FLJ43842; available from GenBank/EMBL/DDBJ under accession no. FLJ43842), the transcription of which we subsequently found to be induced in response to ER stress (see Fig. 3 B).
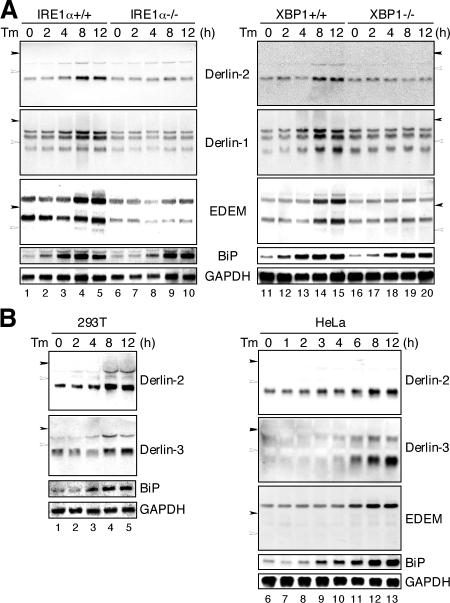
Figure 3.
Involvement of the IRE1–XBP1 pathway in the induction of Derlin-1 and -2 in response to ER stress. (A) IRE1α+/+, IRE1α−/−, XBP1+/+, and XBP1−/− MEFs were treated with 10 μg/ml tunicamycin (Tm) for the indicated periods. Total RNAs were isolated and analyzed by Northern blot hybridization using a DIG-labeled cDNA probe specific to mouse Derlin-2, Derlin-1, EDEM, BiP, or GAPDH. Closed and open arrowheads indicate the migration positions of 28S ribosomal RNA (4.7 kb) and 18S ribosomal RNA (1.9 kb), respectively. (B) 293T or HeLa cells were treated with 2 μg/ml tunicamycin (Tm) for the indicated periods. Total RNAs were analyzed as in A using a DIG-labeled cDNA probe specific to human Derlin-2, Derlin-3, EDEM, BiP, or GAPDH.
During the course of our subsequent analyses, proteins encoded by CGI-101 and FLJ43842 turned out to be identical to Derlin-2 and -3, respectively (see Introduction). We decided to follow this nomenclature hereafter and continued to investigate their functions, which remain largely uncharacterized. A database search indicated the presence of two variants of Derlin-3, probably resulting from alternative splicing. We refer to these short and long forms as Derlin-3 transcriptional variant (tv) 1 and 2, respectively; tv2 has 30 more amino acids at the COOH terminus than does tv1 (Fig. 1 C). Derlin-2 and -3 show ∼75% identity, whereas that between Derlin-1 and -2 or Derlin-1 and -3 is ∼30%.
Transcript structures for Derlin-1, -2, and -3 deposited in the data bank are schematically shown in Fig. 2 A. Northern blot hybridization revealed that both a short form and a long form of Derlin-2 mRNA are expressed in both human and mouse and that in both species the short form is more abundant than the long form (see Figs. 2 and 3). Similarly, short and long Derlin-3 mRNAs also exist, with the short form being more abundant in most human tissues; however, neither short nor long Derlin-3 mRNA was detected in mouse embryonic fibroblasts (MEFs; unpublished data). As shown in Fig. 2 (B and C), Derlin-1 mRNA was expressed ubiquitously. Expression of Derlin-2 mRNA also appeared ubiquitous; the apparent discrepancy in the level of Derlin-2 mRNA in skeletal muscle between Fig. 2 B (lane 6) and Fig. 2 C (lane 3) is likely attributable to a difference in the amount of mRNA loaded (compare the levels of β-actin mRNA). In contrast, expression of Derlin-3 mRNA was restricted to several tissues. Both Derlin-2 and -3 mRNAs were detected abundantly in placenta and pancreas (Fig. 2 B), whereas Derlin-3 mRNA was much more abundant than Derlin-2 mRNA in spleen and small intestine (Fig. 2 C). The biological significance of these findings remains a subject for future investigation. We conclude that the summed expression of Derlin-2 and -3 mRNAs is comparable with that of Derlin-1 mRNA.
Figure 2.
Structure and tissue distribution of Derlin-2 and -3 mRNA. (A) Schematic structures of transcripts deposited in the data bank for human (available from GenBank/EMBL/DDBJ under accession no. NM_024295) and mouse (NM_024207) Derlin-1, human (NM_016041) and mouse (NM_033562) Derlin-2, and human (NM_001002862) and mouse (NM_024440) Derlin-3 are shown. Black underlines indicate the respective region of cDNA probe used for Northern blot analysis. (B) A nylon membrane onto which ∼2 μg each of poly A+ RNA prepared from eight human tissues was blotted after separation through gel electrophoresis (Human MTN blot) was hybridized with a DIG-labeled cDNA probe specific to human Derlin-1, Derlin-2, Derlin-3, or β-actin. Migration positions of molecular weight markers are indicated on the left of each panel. (C) A second nylon membrane onto which ∼1 μg each of poly A+ RNA prepared from 12 human tissues was blotted after separation through gel electrophoresis (Human 12-lane MTN blot) was hybridized as in B.
Derlin-1, -2, and probably -3 are targets of the IRE1–XBP1 pathway
We asked whether the transcriptional induction of Derlin-2 in response to ER stress depends on the IRE1–XBP1 pathway, similar to that observed for EDEM. Northern blot hybridiza- tion was performed for total RNA isolated from IRE1α+/+, IRE1α−/−, XBP1+/+, and XBP1−/− MEFs that had been treated with 10 μg/ml tunicamycin. Consistent with our previous results (Yoshida et al., 2003), mRNA encoding binding protein (BiP), a major ER chaperone, was similarly induced in response to tunicamycin treatment in both wild-type cells and cells lacking IRE1α or XBP1, whereas induction of EDEM mRNA observed in wild-type cells was abolished in cells lacking IRE1α or XBP1 (Fig. 3 A). We found that Derlin-2 mRNA was induced in response to tunicamycin treatment, albeit less efficiently than BiP mRNA (Fig. 3 A, lanes 1–5 and 11–15), consistent with the results of microarray analysis shown in Fig. 1 A. We also found that Derlin-1 mRNA was ER stress inducible (Fig. 3 A, lanes 1–5 and 11–15). Importantly, induction of Derlin-1 or -2 observed in wild-type cells was greatly attenuated in cells lacking IRE1α or XBP1 (Fig. 3 A, lanes 6–10 and 16–20). These results indicated that Derlin-1 and -2 are targets of the IRE1–XBP1 pathway of the UPR.
Derlin-3 mRNA was induced in tunicamycin-treated human cells, such as 293T, and HeLa cells, and the extent of induction was greater than that of Derlin-2 mRNA (Fig. 3 B). However, the dependence of Derlin-3 mRNA induction on the IRE1–XBP1 pathway could not be examined because Derlin-3 mRNA was not detected in MEFs (unpublished data). Nonetheless, Derlin-3 is considered to be a target of the IRE1–XBP1 pathway because the induction time course of Derlin-3 mRNA was similar to that of Derlin-2 and EDEM mRNA but clearly slower than that of BiP mRNA (Fig. 3 B), reflecting the differences in DNA binding specificity and activation mechanisms between ATF6 and XBP1 (see Discussion). These results suggested that, similar to Derlin-1 and EDEM, Derlin-2 and -3 might be components of ERAD.
Derlin-2 and -3 are ER proteins
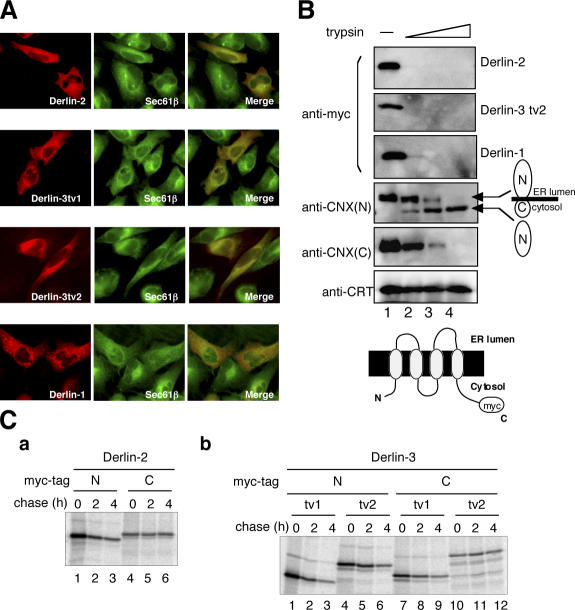
To determine the localization of Derlin-2 and -3, their NH2-terminal c-myc epitope–tagged versions were transfected into HeLa cells. An indirect immunofluorescence analysis of transfected cells revealed that Derlin-2 or -3 was colocalized with Sec61β (Fig. 4 A), a component of the translocon in the ER (Hartmann et al., 1994). It should be noted that overexpression of Derlin-1 created dot-like structures around the ER as reported previously (Lilley and Ploegh, 2004; Ye et al., 2004), whereas that of Derlin-2 or -3 did not do so, although their expression levels appeared comparable when detected with anti-myc antibody (Fig. 4 B and see Fig. 7A).
Figure 4.
Characterization of Derlin-2 and -3. (A) HeLa cells were trans-fected with plasmid to express each Derlin tagged with the c-myc epitope at the respective NH2 terminus. Transfected cells were fixed and stained with anti-myc and anti-Sec61β antibodies. (B) HEK293 cells were trans-fected with plasmid to express each Derlin tagged with the c-myc epitope at the respective COOH terminus. Postnuclear supernatant of transfected cells was incubated with increasing amounts of trypsin (0, 4, 8, and 16 μg for lane 1, 2, 3, and 4, respectively) for 15 min at 4°C. Immunoblotting analysis of the samples was performed using anti-myc, anti–NH2 terminus of calnexin (CNX[N]), anti–COOH terminus of calnexin (CNX[C]), and anti-calreticulin (CRT) antibodies. (C) HEK293 cells were transfected with plasmid to express Derlin-2 (a) or Derlin-3 tv1 or tv2 (b) tagged with the c-myc epitope at either the NH2 (N) or COOH (C) terminus. 36 h later, transfected cells were pulse labeled with 35S-methionine and cysteine for 15 min and then chased for the indicated periods. Cells were lysed with buffer containing 1% NP-40 and subjected to immunoprecipitation analysis using anti-myc antibody.
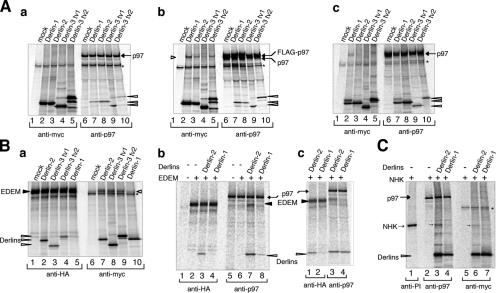
Figure 7.
Association of Derlin-2 and -3 with p97, EDEM, and NHK. (A) HEK293 cells were mock-transfected or transfected with plasmid to express each Derlin tagged with the c-myc epitope at the respective COOH terminus alone (a) or with plasmid to express FLAG-tagged p97 (b). HEK293 cells were also transfected with plasmid to express each Derlin tagged with the c-myc epitope at the respective NH2 terminus (c). 24 h later, transfected cells were labeled with 35S-methionine and cysteine for 1 h, lysed with buffer containing 1% NP-40, and subjected to immunoprecipitation analysis using anti-myc or anti-p97 antibody as indicated. Migration positions of endogenous and FLAG-tagged p97 are marked. The asterisk shows a nonspecific band. The short open arrowhead indicates p97 coimmunoprecipitated with Derlins, whereas long open arrowheads indicate Derlins coimmunoprecipitated with p97. (B, a) HEK293 cells were mock-transfected or transfected with plasmid to express each Derlin tagged with the c-myc epitope at the respective NH2 terminus together with plasmid to express HA-tagged EDEM. 24 h later, transfected cells were labeled with 35S-methionine and cysteine for 2 h, lysed, and subjected to immunoprecipitation analysis using anti-myc or anti-HA antibody as indicated. The migration position of EDEM is marked. The asterisk shows a nonspecific band. The short open arrowhead indicates EDEM coimmunoprecipitated with Derlins, whereas long open arrowheads indicate Derlins coimmunoprecipitated with EDEM. (b and c) HEK293 cells were transfected with (+) or without (−) plasmid to express HA-tagged EDEM with or without plasmid to express Derlin-1 or -2 tagged with the c-myc epitope at the respective NH2 terminus. Transfected cells pulse labeled and then lysed as in a were subjected to immunoprecipitation analysis using anti-HA or anti-p97 antibody as indicated. (c) The amount of Derlin-1 expression plasmid to transfect HEK293 cells was twice that of Derlin-2 expression plasmid. Migration positions of p97, EDEM, and Derlins coimmunoprecipitated with EDEM or p97 are indicated. (C) HEK293 cells were transfected with (+) plasmid to express NHK and with or without plasmid to express Derlin-1 or -2 tagged with the c-myc epitope at the respective NH2 terminus. Transfected cells were pulse labeled and then lysed as in panel a. Equal amounts of cell lysate were subjected to immunoprecipitation analysis using anti–α1-PI, anti-p97, or anti-myc antibody as indicated. Migration positions of p97, NHK, and Derlins are indicated.
Derlin-1 is considered to be a protein that spans the ER membrane four times, with both its NH2 and COOH termini facing the cytosol (Lilley and Ploegh, 2004; Ye et al., 2004), and Derlin-2 and -3 contain four hydrophobic stretches at positions similar to that of Derlin-1 (Fig. 1 B). To determine actual topology, Derlin-2 or -3 tagged with the c-myc epitope at the respective COOH terminus was transfected into human embryonic kidney (HEK) 293 cells and microsomes isolated from transfected cells were digested with trypsin. Calnexin, which is a type I transmembrane protein in the ER, and calreticulin, which is a soluble lumenal protein, were used as controls. As shown in Fig. 4 B, the COOH termini of Derlin-2 and -3 were not protected against trypsin digestion, as was also the case for Derlin-1 and calnexin but not for calreticulin and the NH2 terminus of calnexin, which remained intact.
Similar protease-sensitivity experiments conducted on Derlin-2 and -3 tagged with the c-myc epitope at the respective NH2 terminus gave rise to complicated results (unpublished data). The mode of insertion into the ER membrane may not have been uniform when NH2-terminally tagged Derlin-2 or -3 was overexpressed, even though they were functional (see the following section). This notion was supported by the finding from the pulse-chase analysis that NH2-terminally tagged proteins were much more unstable than COOH-terminally tagged proteins, albeit with more abundant expression (Fig. 4 C). Formal conclusion that endogenous Derlin-2 and -3 are four-transmembrane proteins in the ER with both their NH2 and COOH termini facing the cytosol thus requires analysis of their topology by raising antibodies against their NH2 and COOH termini.
Derlin-2 and -3 are required for ERAD
We next examined whether Derlin-2 and -3 are indeed involved in ERAD using two approaches, overexpression and knockdown experiments. Because a previous study indicated that Derlin-2 does not participate in US11-mediated degradation of class I HC, a transmembrane protein in the ER (Lilley and Ploegh, 2004), we chose as substrate a soluble lumenal protein misfolded in the ER, the null Hong Kong (NHK) mutant of α1-proteinase inhibitor (α1-PI, also called α1-antitrypsin; Sifers et al., 1988). NHK glycosylated and misfolded in the lumen of the ER is recognized by EDEM for destruction by the proteasome in the cytosol (Hosokawa et al., 2001). Pulse-chase experiments followed by immunoprecipitation analysis with antibody against α1-PI showed that NHK was degraded with a half-life of <3 h (Fig. 5 A, lanes 1–3). Importantly, overexpression of Derlin-2 or -3 accelerated the degradation of NHK to a half-life of ∼1.5 h (Fig. 5 A, lanes 4–9). This acceleration was comparable with that observed with overexpression of EDEM (Hosokawa et al., 2001). Interestingly, both Derlin-2 and -3 were coimmunoprecipitated with NHK when cells were lysed with buffer containing 1% NP-40 (Fig. 5 A), and this association was lost when they were lysed with buffer containing 1% SDS (not depicted). Thus, as in the case of Derlin-1, Derlin-2 and -3 interact with the substrate to be degraded.
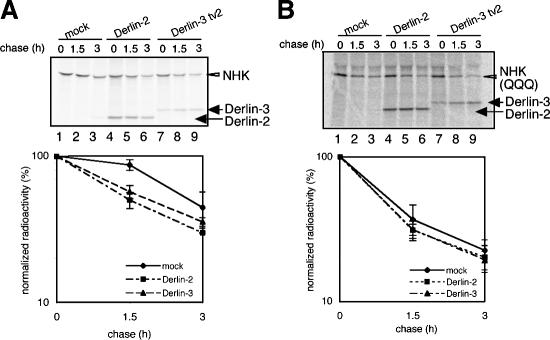
Figure 5.
Effects of overexpression of Derlin-2 and -3 on degradation of NHK and NHK(QQQ). (A and B) HEK293 cells were mock-transfected or transfected with plasmid to express Derlin-2 or Derlin-3 tv2 tagged with the c-myc epitope at the respective NH2 terminus together with plasmid to express NHK (A) or NHK(QQQ) (B). Transfected cells were pulse-chased and subjected to immunoprecipitation analysis using anti–α1-PI antibody as in Fig. 4 C. Migration positions of NHK, NHK(QQQ), Derlin-2, and Derlin-3 are indicated. The radioactivity of each NHK band was determined and expressed as relative to the summation of radioactivity of nine NHK bands obtained in each experiment. The relative radioactivity of each band was normalized with the value at chase period 0 h. The means from three independent experiments with SDs (error bars) are plotted against the chase period (bottom).
We then asked whether overexpression of Derlin-2 and -3 also affects degradation of nonglycoproteins misfolded in the ER. A previously modified NHK designated NHK(QQQ), in which all asparagines of three potential N-glycosylation sites are mutated to glutamine, was shown to degrade quite rapidly (Hosokawa, N., personal communication) with a half-life of ∼1 h, which is even shorter than that of NHK (Fig. 5 B, lanes 1–3). Results showed that overexpression of Derlin-2 or -3 did not significantly accelerate the degradation of NHK(QQQ) (Fig. 5 B, lanes 4–9), indicating that Derlin-2 and -3 induced by the UPR assist in the degradation of misfolded glycoproteins but not in that of misfolded nonglycoproteins. Unexpectedly, both Derlin-2 and -3 coimmunoprecipitated with NHK(QQQ) (Fig. 5 B), indicating that the binding activity of Derlin-2 and -3 is not simply correlated with their degradation activity. In this connection, we also found that EDEM located upstream of Derlin-2 and -3 in the degradation process (see the following section) can bind to NHK and NHK(QQQ) with similar efficiency (unpublished data).
We then examined the effect of reducing Derlin-2 and -3 levels on the degradation of NHK. Double-stranded oligonucleotides corresponding to two regions each of Derlin-2 or -3 were inserted into the mammalian expression vector pSUPER to produce short hairpin RNAs (shRNAs) that act as short interfering RNA–like molecules in transfected cells (Brummelkamp et al., 2002). The knockdown efficiencies of these constructs were determined by Northern blot hybridization. As shown in Fig. 6 A, they effectively suppressed expression of Derlin-2 or -3 without affecting that of other Derlins significantly. We therefore transfected HEK293 cells with these pSUPER derivatives together with the plasmid for expression of NHK. Pulse-chase experiments revealed that degradation of NHK became inefficient in cells with reduced levels of Derlin-2 or -3 (Fig. 6 B), indicating that they are functional components of ERAD for misfolded glycoprotein. We obtained no indication from reporter assays that the UPR was activated in these knockdown cells (unpublished data).
Figure 6.
Effects of knockdown of Derlin-2 and -3 on degradation of NHK. (A) HEK293 cells were untransfected or transfected with the shRNA vector pSUPER alone or pSUPER carrying a sequence corresponding to a part of Derlin-2 or -3. Total RNAs were isolated 64 h after transfection and analyzed by Northern blot hybridization using a DIG-labeled cDNA probe specific to human Derlin-1, Derlin-2, Derlin-3, or GAPDH. (B) HEK293 cells were transfected with pSUPER alone or pSUPER carrying a sequence corresponding to a part of Derlin-2, Derlin-3, or both together with plasmid to express NHK. Pulse-chase and subsequent immunoprecipitation were performed as in Fig. 5 A 64 h after transfection. The results of three independent experiments are shown (left). Normalized radioactivity of each NHK band was determined and is presented as in Fig. 5 A (right). Error bars depict means ± SD. (C) HEK293 cells were transfected with (+) or without (−) plasmid to express Derlin-1 or -2 tagged with the c-myc epitope at the respective NH2 terminus with (+) or without (−) plasmid to express Derlin-3 tv2 tagged with the HA epitope at the NH2 terminus. 36 h later, transfected cells were labeled with 35S-methionine and cysteine for 2 h, lysed with buffer containing 1% NP-40, and subjected to immunoprecipitation analysis using anti-myc or anti-HA antibody as indicated. Migration positions of Derlin-1, Derlin-2, and Derlin-3 tv2 are marked. (D) HEK293 cells were transfected with pSUPER alone or pSUPER derivatives as in B together with plasmid to express the wild-type (wt) α1-PI. Pulse-chase and subsequent immunoprecipitation from cell lysate as well as from media were performed as in B. Migration positions of high-mannose and complex wild-type α1-PI are indicated.
Because a reduction in the expression of Derlin-2 or -3 alone effectively blocked the degradation of NHK, we explored the possibility that Derlin-2 and -3 are heterooligomerized. Results of cotransfection experiments clearly showed coimmunoprecipitatation of HA-tagged Derlin-3 with myc-tagged Derlin-2 and that of myc-tagged Derlin-2 with HA-tagged Derlin-3 in a fairly stoichiometric manner (Fig. 6 C, lanes 6 and 8, respectively). On the other hand, Derlin-1 was found to be poorly associated with Derlin-3 (Fig. 6 C, lanes 2 and 4), suggesting that Derlin-2 and -3 may constitute a group distinct from Derlin-1, as suggested from the distances in the phylogenic tree (Lilley and Ploegh, 2004). As a reduction in the expression of Derlin-2, Derlin-3, or both had little effect on the synthesis and secretion of the wild-type α1-PI (Fig. 6 D), we concluded that they are required for the ERAD of a soluble misfolded glycoprotein but not for the maturation of a correctly foldable protein.
Derlin-2 and -3 are associated with p97 and EDEM
We next determined whether Derlin-2 and -3 are able to associate with known components of ERAD, such as p97, by pulse labeling HEK293 cells expressing COOH-terminal c-myc–tagged versions of Derlin-2 or -3 by transfection, followed by immunoprecipitation. Analysis with anti-p97 antibody revealed that every Derlin coimmunoprecipitated with p97 (Fig. 7 A, a, lanes 7–10). In contrast, analysis with anti-myc antibody did not detect significant p97 coimmunoprecipitation with any of the Derlins (Fig. 7 A, a, lanes 2–5). However, when FLAG-tagged p97 was overexpressed by cotransfection, each Derlin coimmunoprecipitated with p97 (Fig. 7 A, b, lanes 7–10) and FLAG-p97 coimmunoprecipitated with each Derlin (Fig. 7 A, b, lanes 2–5). Further, coimmunoprecipitation of each Derlin with p97 was observed when each Derlin was tagged with the c-myc epitope at the respective NH2 terminus (Fig. 7 A, c, lanes 7–10). These results are consistent with those of previous studies demonstrating the association of Derlin-1 and -2 with p97 (Lilley and Ploegh, 2004, 2005; Ye et al., 2004, 2005).
We then examined whether Derlins are also able to associate with EDEM. A similar immunoprecipitation analysis was performed in HEK293 cells expressing HA-tagged EDEM and one of the three Derlins tagged with the c-myc epitope at the respective NH2 terminus by transfection. Immunoprecipitation with both anti-myc and anti-HA antibodies revealed an association between EDEM and Derlin-2 and -3 (Fig. 7 B, a, lanes 2–4 and 7–9) but not Derlin-1 (Fig. 7 B, a, compare lanes 5 and 10 with lanes 1 and 6, respectively), indicating a functional difference between Derlin-1 and a group consisting of Derlin-2 and -3. This notion was further confirmed by the next series of immunoprecipitation analyses. Immunoprecipitation with anti-p97 antibody revealed no association between EDEM and p97, even though HA-tagged EDEM was overexpressed in HEK293 cells by transfection (Fig. 7 B, b, compare lane 6 with 5). Importantly, however, immunoprecipitation with anti-p97 antibody did reveal that there was an association between EDEM and p97 when Derlin-2 was overexpressed simultaneously with EDEM (Fig. 7 B, b, compare lane 7 with 6). In contrast, cooverexpression of Derlin-1 did not mediate the association of p97 with EDEM (Fig. 7 B, b, compare lane 8 with 6). As the level of Derlin-1 coimmunoprecipitated with p97 was lower than that of Derlin-2 in this experiment (Fig. 7 B, b, compare lane 8 with 7), we increased the amount of Derlin-1–expressing plasmid DNA in the next experiment. As a result, the level of Derlin-1 coimmunoprecipitated with p97 became comparable with that of Derlin-2 (Fig. 7 B, c, compare lane 4 with 3). Nevertheless, only Derlin-2 mediated the association of EDEM with p97 (Fig. 7 B, c, compare lane 3 with 4).
We further examined whether Derlin-2 mediates association of NHK with p97. Immunoprecipitation analysis was performed in HEK293 cells expressing NHK and Derlin-2 or -1 tagged with the c-myc epitope at the respective NH2 terminus by transfection. Immunoprecipitation with anti-myc antibody revealed coimmunoprecipitation of NHK with Derlin-2 but not with Derlin-1 (Fig. 7 C, compare lanes 6 and 7 with 5). Immunoprecipitation with anti-p97 antibody showed no direct interaction between p97 and NHK (Fig. 7 C, lane 2). Importantly, however, p97 became associated with NHK when Derlin-2 was overexpressed simultaneously (Fig. 7 C, lane 3) and the NHK–Derlin-2 complex recovered with anti-p97 antibody was quantitatively similar to that recovered with anti-myc antibody (Fig. 7 C, lanes 3 and 6, respectively), strongly indicating that Derlin-2 associated with p97 was the same molecule as that associated with NHK. In contrast, Derlin-1 failed to mediate the association of NHK with p97 (Fig. 7 C, lane 4).
These results prompted us to examine the effect of a reduction in the level of Derlin-1 on the degradation of NHK. It was recently reported that a sequence corresponding to a part of Derlin-1 successfully decreased the level of Derlin-1 without affecting that of Derlin-2 when incorporated into the shRNA vector (Lilley and Ploegh, 2005). We confirmed this observation by immunoblotting in our system (Fig. 8 A, lane 3). Similarly, our shRNA vector for Derlin-2 succeeded in decreasing the level of Derlin-2 without affecting that of Derlin-1 (Fig. 8 A, lane 4). Interestingly, knockdown of Derlin-1 had little effect on the degradation of NHK, whereas that of Derlin-3 reproducibly blocked it (Fig. 8 B). On the basis of these results, we concluded that Derlin-2 and -3, but not Derlin-1, function downstream of EDEM and are required for the degradation of NHK, a misfolded glycoprotein substrate of ERAD, although we cannot formally rule out the possibility that the residual amount of Derlin-1 in Derlin-1 knocked down cells (Fig. 8 A, lane 3) is sufficient for the degradation of NHK.
Figure 8.
Effects of knockdown of Derlin-1 on degradation of NHK. (A) HEK293 cells were untransfected or transfected with pSUPER alone or pSUPER carrying a sequence corresponding to a part of Derlin-1, -2, or -3. 64 h later, transfected cells were subjected to immunoblotting analysis using antibody against Derlin-1, Derlin-2, or GAPDH. (B) HEK293 cells were transfected with pSUPER alone or pSUPER carrying a sequence corresponding to a part of Derlin-1 or -3 together with plasmid to express NHK. 64 h later, transfected cells were pulse-chased, lysed, and subjected to immunoprecipitation as in Fig. 6 B (left). The results of three independent experiments are shown. Normalized radioactivity of each NHK band was also determined and is presented as in Fig. 5 A (right). Error bars depict mean ± SD.
Discussion
Proteins misfolded in the ER are degraded by the proteasome in the cytosol via a process called ERAD, in which misfolded proteins are recognized inside the ER lumen, targeted to a channel, extracted from the ER membrane, and delivered to the proteasome (Wilhovsky et al., 2000; Tsai et al., 2002). Misfolded glycoproteins are recognized by presentation of an N-linked oligosaccharide processing intermediates, such as Man8GlcNAc2 (Jakob et al., 1998), which acts as a motif that triggers binding to EDEM (Hosokawa et al., 2001). In this way, glycoproteins that are unable to fold properly are transferred from calnexin, a glucoprotein-specific molecular chaperone, to EDEM (Molinari et al., 2003; Oda et al., 2003). Subsequently, the cytosolic multifunctional protein p97 acts to extract the glycoprotein from the ER membrane (Dai and Li, 2001; Ye et al., 2001; Braun et al., 2002; Kobayashi et al., 2002; Meyer et al., 2002). However, the events that link these two steps have remained undefined.
The recent identification of Derlin-1 has brought new insights into the molecular mechanism of the US11-mediated retrotranslocation of class I HC, providing a missing link between events in the ER and those in the cytosol (Lilley and Ploegh, 2004; Ye et al., 2004). Further, the findings that Derlin-1 binds to misfolded and ubiquitinated proteins and that RNA interference of Derlin-1 in Caenorhabditis elegans evokes ER stress suggest a more generalized role of Derlin-1 in ERAD (Ye et al., 2004). Recent studies demonstrated that Derlin-1 is associated with ubiquitin ligases such as HRD1 and gp78 via binding to p97 and vasolin-containing protein (VCP)–interacting membrane protein and that Derlin-2 also forms such a multiprotein complex, consisting of p97, VCP-interacting membrane protein, HRD1, and HRD3/SEL1L (Lilley and Ploegh, 2005; Ye et al., 2005). However, it remains to be determined whether Derlin-2 is required for ERAD.
We show that the properties of Derlin-2 and -3 are exactly those expected for proteins able to provide the missing link between EDEM and p97 in the process of degrading glycoproteins misfolded in the ER. Derlin-2 and -3 are transmembrane proteins that span the ER membrane multiple times (Figs. 1 and 4) and are required for the ERAD of misfolded glycoprotein (Figs. 5 and 6). They are associated with the degradation substrate, p97, and EDEM (Figs. 5 and 7). p97 and EDEM form a complex only in the presence of Derlin-2 (Fig. 7 B), and Derlin-2 mediates the association of p97 with the degradation substrate (Fig. 7 C). In contrast, Derlin-1 does not mediate the association of p97 with either EDEM or the degradation substrate (Fig. 7, B and C). This specificity might be attributable to differences in amino acid sequences between Derlin-1 and -2 (Fig. 1 C) or, as Derlin-1 and -2 or -3 appear to be associated with different cellular proteins (Fig. 7 A), to differences in accessory proteins.
Interestingly, MEFs express only Derlin-2 mRNA, whereas human cells express both Derlin-2 and -3 mRNAs (Figs. 2 and 3 and not depicted). In addition, Derlin-2 and -3 mRNAs are distributed differently in various human tissues (Fig. 2). The finding that the knockdown of Derlin-2 or -3 alone effectively blocked the degradation of a misfolded glycoprotein (Fig. 6 B) suggested that they may form heterooligomers to function when expressed simultaneously, and this was indeed shown to be the case (Fig. 6 C). Similarly, they may form homooligomers when expressed singularly. It is of great interest whether Derlin-2 and -3 themselves can form a channel through which misfolded glycoproteins move from the ER lumen to the cytosol, as has been proposed for Derlin-1 (Lilley and Ploegh, 2004; Ye et al., 2004). We are also aware of a recent study showing that the proteasome binds directly to Sec61 (Kalies et al., 2005), supporting the idea that misfolded proteins are retrotranslocated from the ER to the cytosol through the translocon as originally proposed (Wiertz et al., 1996b). Thus, it appears that we are a far way from any conclusive determination on this issue.
The UPR consists of transcriptional control only in yeast, and the Ire1p-mediated program covers transcriptional induction of not only ER chaperones but also components of ERAD in response to ER stress (Kaufman, 1999; Mori, 2000; Patil and Walter, 2001). On this basis, the UPR plays little role in determining the fate of unfolded or misfolded proteins in yeast ER. The activated refolding system and degradation system may deal with unfolded proteins in a competitive manner. In contrast, metazoan cells have developed the ability to attenuate translation via activation by PERK in response to ER stress, thereby decreasing the burden on the ER (Ron, 2002). Mammals have further evolved two signaling pathways, namely the ATF6 and IRE1–XBP1 pathways, for transcriptional induction of ER chaperones and components of ERAD in response to ER stress (Mori, 2003). The transcription factor ATF6 is activated by a posttranslational mechanism, whereas the transcription factor XBP1 is activated by a posttranscriptional mechanism, causing the active form of ATF6 to be detected in cell extracts earlier than the active form of XBP1. In addition, XBP1 has broader target specificity than ATF6. Thus, the transcription of genes unaffected by the active form of ATF6 can be later induced by the active form of XBP1, allowing a time-dependent decision (Yoshida et al., 2001).
Accumulating evidence indicates that the ATF6 pathway mainly controls the expression of ER chaperones, whereas the IRE1–XBP1 pathway regulates the expression of not only ER chaperones but also components of ERAD such as EDEM, EDEM2, and HRD1 (Kaneko et al., 2002; Okada et al., 2002; Lee et al., 2003; Yoshida et al., 2003; Olivari et al., 2005). Our present analysis adds Derlin-1, -2, and probably -3 to the list of targets of the IRE1–XBP1 pathway (Fig. 3 and Table I). We have also found that the misfolded glycoprotein NHK was degraded more slowly in IRE1α−/− cells than in IRE1α+/+ cells (Yoshida et al., 2003), whereas the misfolded nonglycoprotein NHK(QQQ) was degraded at similar rates in both cell types (unpublished data). These results strongly support our time-dependent phase-transition model for determining the fate of proteins that are unfolded or misfolded in the mammalian ER, at least as far as glycoproteins are concerned, in which the ATF6-mediated unidirectional phase (refolding only) is shifted to the XBP1-mediated bidirectional phase (refolding plus degradation) depending on the quality, quantity, or both of unfolded or misfolded proteins in the ER (Yoshida et al., 2003). The availability of multiple pathways for transcriptional control confers diversity for mammalian cells to adjust to the accumulation of unfolded proteins in the ER.
Table I.
List of mammalian ERAD components regulated by the IRE1–XBP1 pathway
| Component | Induction upon ER stress |
Dependence on the IRE1–XBP1 pathway |
Reference |
|---|---|---|---|
| ER α1, 2-mannosidase I | no | — | Hosokawa et al., 2001 |
| EDEM | yes | yes | Hosokawa et al., 2001; Yoshida et al., 2003 |
| EDEM2 | yes | yes | Olivari et al., 2005 |
| EDEM3 | yes | yes | Molinari, M., personal communication |
| HRD1 | yes | yes | Kaneko et al., 2002 |
| HRD3/SEL1 | yes | no | Kaneko and Nomura, 2003 |
| Derlin-1 | yes | yes | Lilley and Ploegh, 2005; this study |
| Derlin-2 | yes | yes | Lilley and Ploegh, 2005; this study |
| Derlin-3 | yes | yesa | this study |
Presumed but not directly demonstrated (Fig. 3).
Materials and methods
Microarray analysis
Total RNA was isolated from HeLa cells that had been untreated or treated with 2 μg/ml tunicamycin for 8 h by the acid guanidium/phenol/chloroform method using ISOGEN (Nippon Gene) and was further purified using RNeasy Midi (QIAGEN). 20-μg aliquots of total RNA from tunicamycin-treated and untreated cells were labeled with cyanine 3– and cyanine 5–2′-deoxycytidine 5′-triphosphate, respectively, using a Direct-Label cDNA synthesis kit (Agilent Technologies) in three of six independent analyses. In the other three analyses, the labeling dyes were swapped to avoid false induction resulting from biased labeling. Labeled cDNAs were mixed and hybridized with a human 1 cDNA microarray kit (Agilent Technologies), on which 12,814 human cDNA clones were spotted. Fluorescence intensities of cyanine 3 and 5 were determined for each spot using a GenePix 4000B and GenePix Pro 4.0 software (Axon Instruments, Inc.) and normalized so that the sum of cyanine 3 intensities of all spots was equal to that of cyanine 5 intensities of all spots. Fold induction caused by tunicamycin treatment was defined as the ratio of normalized intensity for treated cells to that for untreated cells.
cDNA cloning and plasmid construction
Recombinant DNA techniques were performed according to standard procedures (Sambrook et al., 1989). Human Derlin-1 and -2 partial cDNAs containing the respective complete open reading frame were obtained by reverse transcription–coupled polymerase chain reaction using total RNA isolated from HEK293T cells and the following primers: 5′-ATCTTGGCTACCTGTGGGTCGAAGATGTCG-3′ and 5′-AACGCAGTTGTTAAGTGCACCCAGCACTGG-3′ for Derlin-1 and 5′-GGAAGATGGCGTACCAGAGC-3′ and 5′-GCTGCTTTAACCTCCAAGCC-3′ for Derlin-2. Mouse Derlin-2 partial cDNA containing the complete open reading frame was obtained similarly using total RNA isolated from MEF and the primers 5′-GGAAAGATGGCGTACCAGAGC-3′ and 5′-TCTCGTAATTGGCACCGCTG-3′. cDNAs for human Derlin-3, mouse Derlin-1, and mouse Derlin-3 were purchased as IMAGE cDNA clones (IMAGE ID 30338943, 2655640, and 3582647, respectively) from Open Biosystems. pCMV-Myc (CLONTECH Laboratories, Inc.) and pcDNA3.1/Myc-His(+)A (Invitrogen) were used to express each Derlin tagged with the c-myc epitope at the NH2 and COOH terminus, respectively. pCMV-SPORT2-EDEM-HA to express HA-tagged EDEM as well as pREP9–α1-PI and pREP9-NHK to express the wild-type α1-PI and its NHK mutant were described previously (Hosokawa et al., 2001; Yoshida et al., 2003). Plasmids to express NHK(QQQ) and FLAG-tagged p97 were gifts of N. Hosokawa (Kyoto University, Kyoto, Japan) and M. Tagaya (Tokyo University of Pharmacy and Life Science, Tokyo, Japan) respectively.
Cell culture and transfection
HEK293, HEK293T, and HeLa cells, as well as IRE1α+/+, IRE1α−/− (Lee et al., 2002), XBP1+/+, and XBP1−/− (gifts of L. Glimcher, Harvard University, Cambridge, MA) MEFs, were cultured at 37°C in a humidified 5% CO2/95% air atmosphere in Dulbecco's modified Eagle's medium (glucose at 4.5 g/liter) supplemented with 10% fetal bovine serum, 2 mM glutamine, and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). Transfection was performed using Fugene6 (Roche) and Lipofectamine 2000 (Invitrogen) transfection reagents according to the manufacturers' instructions.
Northern blot hybridization
Total RNA was isolated from cultured cells using ISOGEN. 5 μg each of total RNA were subjected to 1.2% agarose gel electrophoresis containing 2.2 M formaldehyde and transferred to a nylon membrane. RNA isolated from human tissues was separated by electrophoresis and blotted on a membrane (Human MTN blot and Human 12-lane MTN blot; CLONTECH Laboratories, Inc.). Digoxigenin (DIG)-labeled cDNA probes were prepared using polymerase chain reaction according to the manufacturer's instruction (Roche) and hybridized with RNA blotted on a membrane in an ultrasensitive hybridization buffer (ULTRAhyb; Ambion). Subsequent reaction with anti-DIG antibody (Roche) and treatment with chemiluminescent detection reagent CDP-star (GE Healthcare) were performed according to the manufacturers' specification. Chemiluminescence was visualized using an LAS-1000plus LuminoImage analyzer (FujiFilm).
Indirect immunofluorescence
HeLa cells grown directly on slide glass in a 24-well plate were transfected with plasmid to express each Derlin tagged with the c-myc epitope at the respective NH2 terminus. 36 h later, cells were fixed with 4% formaldehyde at room temperature for 10 min, permeabilized with 0.2% Triton-X 100 in PBS, reacted with mouse anti-myc antibody (Santa Cruz Biotechnology, Inc.) and rabbit anti-Sec61β antibody (Upstate Biotechnology) for 1 h, and incubated with FITC-conjugated anti–rabbit IgG antibody (MP Biomedicals) or rhodamine-conjugated anti–mouse IgG antibody (Cappel). Fluorescence was visualized using a laser-scanning microscope (Eclipse E800; Nikon).
Protection assay and immunoblotting
HEK293 cells were transfected with plasmid to express each Derlin tagged with the c-myc epitope at the respective COOH terminus. Transfected cells were homogenized in sucrose buffer (50 mM Tris-Cl, pH 8.0, containing 0.32 M sucrose, 1 mM dithiothreitol, and 1 mM EDTA). Postnuclear supernatant obtained after 1,000 g centrifugation was treated with various concentrations of trypsin at 4°C for 15 min. Digestion was terminated by adding SDS sample buffer and boiling for 15 min. Immunoblotting was performed using antibodies against myc (Santa Cruz Biotechnology, Inc.), calnexin (StressGen Biotechnologies), and calreticulin (Affinity BioReagents, Inc.) and by reaction with Western blotting luminol reagent (Santa Cruz Biotechnology, Inc.). Chemiluminescence was visualized using a LAS-1000plus LuminoImage analyzer. Antibodies against Derlin-1 and -2 were purchased from MBL International Corporation, and anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was obtained from Trevigen.
Metabolic labeling and immunoprecipitation
Cells were incubated for 30 min in methionine- and cysteine-free Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 2 mM glutamine and 10% dialyzed fetal bovine serum. Cells were then pulse labeled with 4.1 Mbq/dish EXPRE35S35S protein labeling mixture (NEN Life Science Products) and chased in fresh complete medium. Cells were lysed in buffer A (50 mM Tris-Cl, pH 8.0, containing 1% NP-40, 150 mM NaCl, and protease inhibitors). Immunoprecipitation was performed with anti–α1-PI antibody (DakoCytomation), anti-myc antibody (Santa Cruz Biotechnology, Inc.), anti-HA antibody (Santa Cruz Biotechnology, Inc.), or anti-VCP antibody (Affinity BioReagents, Inc.) and protein A– or G–coupled Sepharose beads (GE Healthcare). Beads were washed in high ionic buffer (50 mM Tris-Cl, pH 8.0, containing 1% NP-40 and 150 mM NaCl) twice, washed in low ionic buffer (10 mM Tris-Cl buffer, pH 7.4), and boiled in Laemmli's sample buffer. Immunoprecipitates were subjected to SDS-PAGE, and radioactive bands were analyzed using a FLA-3000G FluoroImage analyzer (FujiFilm). Cells were also lysed in buffer A plus 1% SDS, boiled for 5 min, diluted with four volumes of buffer A, and centrifuged at 13,000 g. The resulting supernatant was used for subsequent immunoprecipitation.
Knockdown of Derlin-2 and -3
Double-stranded oligonucleotides corresponding to five regions each of human Derlin-2 and -3 were inserted into the shRNA expression vector pSUPER (OligoEngine) between the BglII and HindIII sites. HEK293 cells were transfected with 4 μg of pSUPER or pSUPER derivatives for Northern blot hybridization. Two regions each were found to decrease Derlin-2 and -3 mRNA levels effectively, i.e., sh-Derlin-2 (1), 441–459; sh-Derlin-2 (2), 737–755; sh-Derlin-3 (1), 2160–2178; and sh-Derlin-3 (2), 3088–3106, where A of the initiation codon ATG is set as 1. The sequence to knockdown Derlin-1 was obtained from a recent study (Lilley and Ploegh, 2005).
Acknowledgments
We thank Ms. Kaoru Miyagawa for technical and secretarial assistance. We are grateful to people in Radioisotope Research Center of Kyoto University for their help in our analyses using radioisotopes.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (14037233 and 15GS0310 to K. Mori and 17770158 to T. Okada). Y. Oda is the recipient of a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists (00894).
Y. Oda and T. Okada contributed equally to this paper.
Abbreviations used in this paper: α1-PI, α1-proteinase inhibitor; BiP, binding protein; DIG, digoxigenin; EDEM, ER degradation enhancing α-mannosidase–like protein; ERAD, ER-associated degradation; GAPDH, glyceraldehyde- 3-phosphate dehydrogenase; HC, heavy chain; HEK, human embryonic kidney; MEF, mouse embryonic fibroblast; NHK, null Hong Kong; shRNA, short hairpin RNA; tv, transcriptional variant; UPR, unfolded protein response; VCP, vasolin-containing protein.
References
- Braun, S., K. Matuschewski, M. Rape, S. Thoms, and S. Jentsch. 2002. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. [DOI] [PubMed] [Google Scholar]
- Dai, R.M., and C.C. Li. 2001. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3:740–744. [DOI] [PubMed] [Google Scholar]
- Gething, M.J., and J. Sambrook. 1992. Protein folding in the cell. Nature. 355:33–45. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575–599. [DOI] [PubMed] [Google Scholar]
- Hartmann, E., T. Sommer, S. Prehn, D. Gorlich, S. Jentsch, and T.A. Rapoport. 1994. Evolutionary conservation of components of the protein translocation complex. Nature. 367:654–657. [DOI] [PubMed] [Google Scholar]
- Helenius, A., T. Marquardt, and I. Braakman. 1992. The endoplasmic reticulum as a protein folding compartment. Trends Cell Biol. 2:227–231. [DOI] [PubMed] [Google Scholar]
- Hiller, M.M., A. Finger, M. Schweiger, and D.H. Wolf. 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 273:1725–1728. [DOI] [PubMed] [Google Scholar]
- Hosokawa, N., I. Wada, K. Hasegawa, T. Yorihuzi, L.O. Tremblay, A. Herscovics, and K. Nagata. 2001. A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob, C.A., P. Burda, J. Roth, and M. Aebi. 1998. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 142:1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies, K.U., S. Allan, T. Sergeyenko, H. Kroger, and K. Romisch. 2005. The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane. EMBO J. 24:2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, M., and Y. Nomura. 2003. ER signaling in unfolded protein response. Life Sci. 74:199–205. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., M. Ishiguro, Y. Niinuma, M. Uesugi, and Y. Nomura. 2002. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532:147–152. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211–1233. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J., D. Scheuner, M. Schroder, X. Shen, K. Lee, C.Y. Liu, and S.M. Arnold. 2002. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 3:411–421. [DOI] [PubMed] [Google Scholar]
- Knop, M., A. Finger, T. Braun, K. Hellmuth, and D.H. Wolf. 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., K. Tanaka, K. Inoue, and A. Kakizuka. 2002. Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 277:47358–47365. [DOI] [PubMed] [Google Scholar]
- Kopito, R.R. 1997. ER quality control: the cytoplasmic connection. Cell. 88:427–430. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and R.F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132. [DOI] [PubMed] [Google Scholar]
- Lee, A.H., N.N. Iwakoshi, and L.H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R.J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, B.N., and H.L. Ploegh. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 429:834–840. [DOI] [PubMed] [Google Scholar]
- Lilley, B.N., and H.L. Ploegh. 2005. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 102:14296–14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast, S.W., K. Diekman, K. Karaveg, A. Davis, R.N. Sifers, and K.W. Moremen. 2004. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 15:421–436. [DOI] [PubMed] [Google Scholar]
- Meyer, H.H., Y. Wang, and G. Warren. 2002. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21:5645–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari, M., V. Calanca, C. Galli, P. Lucca, and P. Paganetti. 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 299:1397–1400. [DOI] [PubMed] [Google Scholar]
- Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 101:451–454. [DOI] [PubMed] [Google Scholar]
- Mori, K. 2003. Frame switch splicing and regulated intramembrane proteolysis: key words to understand the unfolded protein response. Traffic. 4:519–528. [DOI] [PubMed] [Google Scholar]
- Oda, Y., N. Hosokawa, I. Wada, and K. Nagata. 2003. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 299:1394–1397. [DOI] [PubMed] [Google Scholar]
- Okada, T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366:585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari, S., C. Galli, H. Alanen, L. Ruddock, and M. Molinari. 2005. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 280:2424–2428. [DOI] [PubMed] [Google Scholar]
- Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349–356. [DOI] [PubMed] [Google Scholar]
- Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Invest. 110:1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Schroder, M., and R.J. Kaufman. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74:739–789. [DOI] [PubMed] [Google Scholar]
- Sifers, R.N., S. Brashears-Macatee, V.J. Kidd, H. Muensch, and S.L. Woo. 1988. A frameshift mutation results in a truncated α1-antitrypsin that is retained within the rough endoplasmic reticulum. J. Biol. Chem. 263:7330–7335. [PubMed] [Google Scholar]
- Tsai, B., Y. Ye, and T.A. Rapoport. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 3:246–255. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., T.R. Jones, L. Sun, M. Bogyo, H.J. Geuze, and H.L. Ploegh. 1996. a. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 84:769–779. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T.R. Jones, T.A. Rapoport, and H.L. Ploegh. 1996. b. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 384:432–438. [DOI] [PubMed] [Google Scholar]
- Wilhovsky, S., R. Gardner, and R. Hampton. 2000. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol. Biol. Cell. 11:1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y., H.H. Meyer, and T.A. Rapoport. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Y. Shibata, C. Yun, D. Ron, and T.A. Rapoport. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 429:841–847. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Y. Shibata, M. Kikkert, S. van Voorden, E. Wiertz, and T.A. Rapoport. 2005. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 102:14132–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., T. Matsui, N. Hosokawa, R.J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 4:265–271. [DOI] [PubMed] [Google Scholar]