Abstract
Polyglutamine diseases are inherited neurodegenerative diseases caused by the expanded polyglutamine proteins (polyQs). We have identified a novel guanosine triphosphatase (GTPase) named CRAG that contains a nuclear localization signal (NLS) sequence and forms nuclear inclusions in response to stress. After ultraviolet irradiation, CRAG interacted with and induced an enlarged ring-like structure of promyelocytic leukemia protein (PML) body in a GTPase-dependent manner. Reactive oxygen species (ROS) generated by polyQ accumulation triggered the association of CRAG with polyQ and the nuclear translocation of the CRAG–polyQ complex. Furthermore, CRAG promoted the degradation of polyQ at PML/CRAG bodies through the ubiquitin–proteasome pathway. CRAG knockdown by small interfering RNA in neuronal cells consistently blocked the nuclear translocation of polyQ and enhanced polyQ-mediated cell death. We propose that CRAG is a modulator of PML function and dynamics in ROS signaling and is protectively involved in the pathogenesis of polyglutamine diseases.
Introduction
Polyglutamine diseases are inherited neurodegenerative diseases caused by the expansion of the polyglutamine tract (Ross, 1997; Orr, 2001). Expansion of polyglutamine repeats alters the conformation or results in the misfolding of the disease-associated protein, thereby conferring a toxic gain of function that is selectively deleterious to neurons (Sato et al., 1999; Yoshizawa et al., 2000). Nuclear inclusions (NIs) with ubiquitination formed by the disease protein are a common pathological feature of polyglutamine diseases. Indeed, NIs have been observed in at least six polyglutamine diseases and many transgenic animal models and thus represent a common hallmark of polyglutamine diseases. Nuclear translocation of expanded polyglutamine protein (polyQ) was promoted efficiently in neuronal cells, suggesting that some neuronal factors containing a nuclear localization signal (NLS) may regulate the nuclear translocation of polyQ. Furthermore, polyQ nuclear aggregates have recently been demonstrated to interact with and alter the nuclear structures of the promyelocytic leukemia protein (PML), a major component of nuclear bodies (Takahashi et al., 2003). We identified a novel guanosine triphosphatase (GTPase), CRAG, which associated with PML and formed NIs in response to various stresses. We report that CRAG is involved in the mechanisms underlying nuclear translocation, ubiquitination, and inclusion body formation of polyQ.
Results and discussion
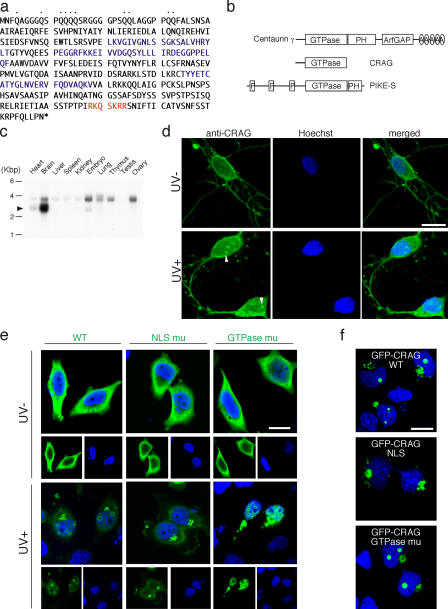
By screening for signaling targets of repulsive axon guidance factors, semaphorins (described in Materials and methods), we identified a novel GTPase and named it CRAG, after collapsin response mediator protein (CRMP)–associated molecule (CRAM [CRMP-5])–associated GTPase. The full-length CRAG cDNA presents an open reading frame of 369 amino acid residues containing a glutamine-rich domain at the NH2 terminus, a Ras homology domain in the middle, and an NLS at the COOH terminus (Fig. 1 a). Fig. 1 b indicates a comparison of structure between CRAG and other related GTPase proteins. The amino acid sequence of CRAG shows 95% identity with centaurin-γ3 and 43% identity with the nuclear GTPase phosphatidylinositol 3-kinase enhancer, short isoform (alignment is not depicted; Ye et al., 2000). Analysis of human genomic databases suggests that CRAG may be an alternative splicing variant of centaurin-γ3. Northern blot analysis indicated that the CRAG gene was dominantly expressed in brain and slightly in heart (Fig. 1 c). A band higher than CRAG that is present in various tissues may be centaurin-γ3. Immunohistochemical analysis revealed a diffuse cytoplasmic distribution of CRAG in rat hippocampal neurons at rest (Fig. 1 d, top). Upon UV irradiation, an NI formed of CRAG was detected at 10 min (Fig. 1 d). These NIs exhibited a doughnut shape under the large-scale microscopic analysis (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200505079/DC1). This phenomenon was reproduced in UV-stimulated HeLa cells expressing HA-tagged CRAG (HA-CRAG) wild type (WT) or GTPase-deficient mutants (S114N; Fig. 1 e). In contrast, NLS-disrupted mutants of CRAG (KR342-343EE within the NLS motif) formed inclusions but were cytosolic even after UV stimulation. These results demonstrated that NLS, but not GTPase activity, was required for nuclear translocation and NI formation by CRAG.
Figure 1.
NI body formation of CRAG by response to UV irradiation. (a) Amino acid sequence of CRAG. CRAG contains a Q-rich domain (dots), a Ras homology domain (blue), and an NLS sequence (red). (b) Comparison of CRAG with the related GTPase proteins centaurin-γ3 and phosphatidylinositol 3-kinase enhancer, short isoform. A, ankyrin repeat; P, proline-rich domain; PH, pleckstrin homology domain. (c) Northern blot analysis of CRAG in mouse tissues. The arrowhead indicates the position of CRAG mRNA. (d) NIs of CRAG in UV-irradiated hippocampal neurons. Cells were stimulated with or without a pulse of UV irradiation at 100 J/m2 and after 10 min fixed and immunostained with anti-CRAG antibody (green) and Hoechst 33258 (blue). The arrowheads indicate CRAG inclusions in the nucleus. (e) NLS-dependent and GTPase-independent NIs of CRAG. HeLa cells expressing HA-CRAG of WT, NLS, or GTPase mutants were treated with or without a pulse of UV irradiation at 200 J/m2. After 2 h, cells were fixed and immunostained with anti-HA antibody (green) and Hoechst 33258 (blue). (f) Spontaneous NIs of GFP-CRAG. HeLa cells expressing GFP-CRAG WT and NLS- and GTPase-deficient mutants were stained with Hoechst 33258 (blue). Bars, 20 μm.
We found that GFP fused to the NH2 terminus of CRAG WT or GTPase mutants spontaneously translocated to the nucleus and formed NI without any stimulation. The absence of nuclear localization of NLS-disrupted mutants of GFP-CRAG also supported an NLS-dependent NI formation by CRAG (Fig. 1 f). This suggested that GFP fusion at the NH2 terminus of CRAG caused a drastic conformational change in CRAG and converted it from an inactive to an active form. An in vitro GTPase assay revealed CRAG activation by UV irradiation and spontaneous activation of GFP-CRAG (unpublished data). Collectively, these data demonstrate that CRAG is a unique GTPase that forms NI under UV stress in an NLS-dependent and GTPase-independent manner, and a conformational change in CRAG may induce its intrinsic activity to form NI in an NLS-dependent manner.
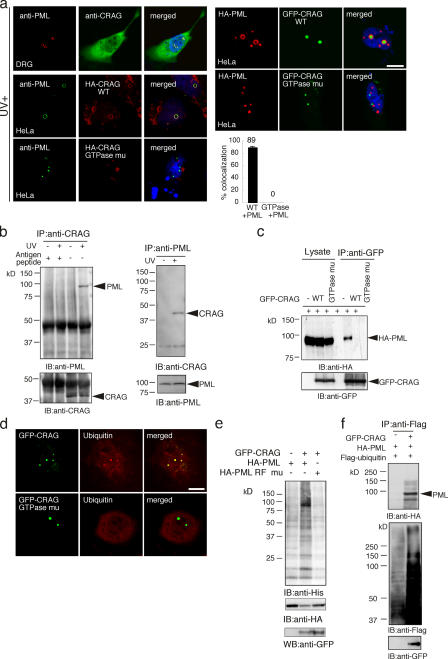
Several nuclear domains have been reported, including PML body, a major component of nuclear bodies (Hodges et al., 1998; Lamond and Earnshaw, 1998). Merged images demonstrated the colocalization of endogenous CRAG and PML bodies with an enlarged ring-like structure in a UV- stimulated dorsal root ganglion (DRG) neuron (Fig. 2 a and Fig. S1 B). Similar results were obtained in UV-stimulated HeLa cells expressing HA-CRAG or unstimulated cells expressing GFP-CRAG. In contrast, HA-CRAG GTPase mutants or GFP-CRAG GTPase mutants did not colocalize with PML. Statistical analysis indicated that GTPase activity was essential for colocalization of GFP-CRAG with PML. A coimmunoprecipitation assay demonstrated that endogenous CRAG associated with PML in UV-stimulated neurons (Fig. 2 b). No association of CRAG with PML by antigen peptide against anti-CRAG antibody demonstrated the specificity of anti-CRAG antibody. Furthermore, in HeLa cell expression system, GFP-CRAG associated with HA-PML in a GTPase-dependent manner (Fig. 2 c).
Figure 2.
CRAG modulates PML function and dynamics. (a) Active CRAG colocalized with and induced a large ring-like structure of PML body. UV-stimulated DRG neurons were immunostained with anti-PML (red), anti-CRAG (green), and Hoechst 33258 (blue). HeLa cells expressing HA-CRAG WT or GTPase mutants were immunostained with anti-PML (green), anti-HA (red), and Hoechst 33258 (blue). Likewise, unstimulated HeLa cells coexpressing HA-PML with GFP-CRAG WT or GTPase mutants were immunostained with anti-HA (red) and Hoechst 33258 (blue). Measurement of colocalization was performed by counting the cells showing colocalization of GFP-CRAG to PML on 100 cells. The percentages are calculated from three independent experiments. Error bars represent SD. n = 3.(b) Association of endogenous CRAG with PML in response to UV stimulation. Lysates of control or UV-irradiated hippocampal neurons (100 J/m2 for 10 min) were immunoprecipitated with anti-CRAG in the presence or absence of 10 μg/ml antigen peptide against anti-CRAG or with anti-PML antibody. Immunoprecipitates (IP) were immunoblotted with the indicated antibodies. (c) GFP-CRAG associates with PML in a GTPase-dependent manner. Lysates or immunoprecipitates with anti-GFP antibody from HeLa cells transfected with indicated vectors were immunoblotted with anti-HA (top) or anti-GFP antibody (bottom). (d) Colocalization of GFP-CRAG WT, but not GTPase mutants, with ubiquitin signals. HeLa cells expressing GFP-CRAG WT or GTPase mutants were immunostained with anti-ubiquitin antibody (red). (e) GFP-CRAG induced ubiquitin ligase activity in PML immunoprecipitates. Immunoprecipitates of HA-PML WT or HA-PML RING-finger (RF) mutants from HeLa cells cotransfected with or without GFP-CRAG were used for an in vitro ubiquitin ligase assay containing His-tagged ubiquitin (described in Materials and methods) and immunoblotted with the indicated antibodies. (f) GFP-CRAG enhanced in vivo ubiquitination. Ubiquitinated proteins were immunoprecipitated from HeLa cells coexpressing HA-PML and Flag-ubiquitin with or without GFP-CRAG and analyzed by immunoblot with anti-HA and anti-Flag. Bars, 20 μm.
We noticed that ubiquitin signals were accumulated in these GFP-CRAG–associated inclusions but not in GTPase mutants (Fig. 2 d). Therefore, we examined whether CRAG stimulated ubiquitin ligase activity in PML immunoprecipitates. An in vitro ubiquitin ligase assay revealed that the activity in PML immunoprecipitates was undetectable in the absence of GFP-CRAG, but GFP-CRAG coexpression induced ubiquitin ligase activity in immunoprecipitates of PML WT but not PML RING (really interesting new gene)–finger mutants (C51S/C54S; Fig. 2 e). For in vivo ubiquitination, ubiquitinated proteins were purified from cells coexpressing HA-PML and Flag ubiquitin with or without GFP-CRAG and analyzed by immunoblot with anti-HA and anti-Flag antibodies. As shown in Fig. 2 f, ubiquitinated proteins significantly increased by GFP-CRAG expression. Collectively, these results suggest that CRAG is an inducer for the association of unknown ubiquitin ligases with PML or a direct activator of PML-associated ubiquitin ligase. Because PML contains a RING-finger domain that confers on it an E3 ubiquitin ligase activity, it is possible that PML is an E3 ubiquitin ligase.
The fact that polyglutamine diseases are characterized by the presence of ubiquitinated, PML-associated NIs suggested the possible involvement of CRAG in polyglutamine diseases. To ascertain a possible involvement of CRAG in polyglutamine diseases, we examined subcellular localization of CRAG in the brains of Machado-Joseph disease (MJD) patients and detected specific CRAG inclusion (Fig. 3 a). Similar CRAG inclusions were observed in the brains of other MJD patients (unpublished data). To further confirm this phenomenon, we generated GFP-Q12 as control and GFP-Q69 as polyQ and examined whether CRAG could interact with polyQ. Subcellular distribution analysis revealed the diffuse cytoplasmic localization of Q12 and perinuclear aggregation of misfolded Q69 in HeLa cells (Fig. 3 b, left). Compared with the cytoplasmic distribution of both Q12 and CRAG, Q69 and CRAG translocated to the nucleus and formed NIs (Fig. 3 b, right). An immunoprecipitation assay also demonstrated that CRAG interacted with GFP-Q69 but not -Q12 (Fig. 3, c and e). Similarly, in hippocampal neurons expressing GFP-Q69 or -Q12, endogenous CRAG associated with GFP-Q69 but not -Q12 (Fig. 3 d).
Figure 3.
Association of CRAG with polyQ requires ROS generation. (a) Accumulation of CRAG NI in the brain of an MJD patient. A brain slice (pons) obtained from an MJD patient was immunostained with anti-CRAG antibody. The arrowhead indicates the NI of CRAG. (b) Colocalization of CRAG with polyQ in the NIs. HeLa cells expressing GFP-Q12 or -Q69 are shown as control. HeLa cells coexpressing GFP-Q12/HA-CRAG or GFP-Q69/HA-CRAG were stained with anti-HA antibody (red) and Hoechst 33258 (blue). (c) Association of CRAG with GFP-Q69 but not -Q12. HeLa cells transfected with the indicated constructs were immunoprecipitated with anti-HA antibody (left) or anti-GFP antibody (right) and immunoblotted with anti-GFP or anti-HA antibodies. (d) Association of endogenous CRAG with GFP-Q69 but not -Q12. DRG neuronal cells transfected with the indicated constructs were immunoprecipitated with anti-GFP antibody and immunoblotted with anti-CRAG antibodies and anti-GFP antibodies. (e) ROS scavenger blocked Q69-mediated CRAG activation. COS-7 cells were transfected with the indicated vectors. After 24 h of incubation with or without ROS scavenger Radicut (100 μg/ml), CRAG was immunoprecipitated with anti-HA antibody and an in vitro GTPase assay was performed. (f) ROS scavenger blocked colocalization of CRAG with Q69. COS-7 cells were cotransfected with GFP-Q69 and HA-CRAG in the presence of 100 μg/ml Radicut. After 24 h, an immunofluorescence assay was performed with anti-HA (red) and Hoechst 33258 (blue). (g) ROS scavenger blocked the interaction of CRAG with GFP-Q69. COS-7 cells were cotransfected with GFP-Q69 and HA-CRAG in the presence of the indicated concentration of Radicut. After 24 h, CRAG was immunoprecipitated with anti-HA and immunoblotted with anti-GFP and anti-HA antibodies. Bars, 20 μm.
NI formation by CRAG was induced by various stress stimuli generating reactive oxygen species (ROS) such as UV. Indeed, we found that H2O2 induced CRAG nuclear translocation and that this was blocked by ROS scavenger edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one; Radicut; Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200505079/DC1). An in vitro GTPase assay indicated that H2O2 activated CRAG GTPase (unpublished data). To understand the mechanism by which CRAG recognizes and interacts with polyQ, we focused on the role of ROS generation by polyQ in the activation of CRAG. An in vitro GTPase assay demonstrated that coexpression with Q69 but not Q12 activated CRAG GTPase, and this activation was blocked by the treatment of ROS scavenger Radicut (Fig. 3 e). Actually, Radicut inhibited Q69-mediated ROS generation (unpublished data). Furthermore, in Radicut-treated cells, CRAG failed to colocalize with Q69 and induce Q69 nuclear translocation (Fig. 3 f). A coimmunoprecipitation assay also indicated that Radicut blocked the association of CRAG with Q69 in a Radicut dose–dependent manner (Fig. 3 g). Thus, ROS may be required for CRAG activation and interaction with polyQ.
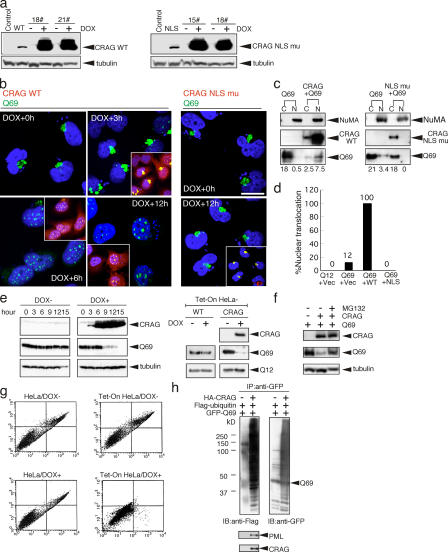
To confirm this, doxycycline (DOX)-inducible Tet-On HeLa cell lines expressing HA-CRAG WT and NLS mutants were established. Two clones each selected from WT and NLS mutants were treated with DOX, and induction of HA-CRAG expression was checked by immunoblot analysis using anti-HA antibody (Fig. 4 a). That equal amount of total protein was loaded is shown by tubulin immunoblots (Fig. 4 a, bottom). Using these cell systems, we monitored the time-dependent nuclear translocation of Q69 by CRAG. As shown in Fig. 4 b, no nuclear localization of Q69 was detected before DOX treatment, but 3 h after DOX treatment, a major part of CRAG was colocalized with Q69 aggregates at perinuclear sites and a small part of the CRAG–Q69 complex was detected in the NIs. After 6–12 h, almost all CRAG and Q69 translocated to the nucleus and formed NIs. Merged images of CRAG and Q69 (Fig. 4 b, insets, yellow) demonstrated the colocalization of CRAG and Q69 in the nucleus. As a negative control, cells treated with solvent for 12 h revealed no nuclear translocation of Q69 (unpublished data). Similar results were obtained in other Tet-On HeLa cell lines (unpublished data). On the other hand, NLS mutants of CRAG could colocalize with Q69 but not induce nuclear translocation of Q69 at all (Fig. 4 b, right). Merged images (Fig. 4 b, insets, yellow) showed the perinuclear aggregates of the CRAG–Q69 complex. Subcellular fractionation analysis also indicated CRAG-dependent nuclear translocation of Q69 and no nuclear localization of Q69 in cells coexpressing NLS mutants (Fig. 4 c). Statistical analysis also indicated that CRAG WT, but not NLS mutants, induced Q69 nuclear translocation (Fig. 4 d). These results demonstrated that CRAG promoted Q69 nuclear translocation in an NLS-dependent manner.
Figure 4.
CRAG promotes nuclear translocation and degradation of polyQ through the ubiquitin–proteasome pathway. (a) Establishment of DOX-inducible Tet-On HeLa cell lines expressing HA-CRAG WT and NLS mutants. Two clones each were treated with or without 1 μM DOX for 24 h and immunoblotted with anti-HA or anti-tubulin antibodies. Cells transiently transfected with control vector, CRAG WT, or NLS mutants were indicated in the first two lanes. (b) NLS-dependent nuclear translocation of Q69. At 24 h after transient transfection of Tet-On HeLa cells with Q69-myc (DOX + 0 h), cells were treated with DOX for the indicated times and immunostained with anti-myc (green), anti-HA antibodies (red), and Hoechst 33258 (blue). Merged images are shown in insets. (c) Biochemical analysis of nuclear translocation of Q69 by CRAG. Cytosolic (C) and nuclear (N) fractions from HeLa cells transfected with the indicated vectors were separated and immunoblotted with anti-HA, anti-myc, and anti–nuclear mitotic apparatus protein (NuMA; nuclear marker) antibodies. The optical density of Q69 bands (bottom) was analyzed by the NIH Image software. (d) Effect of CRAG WT and NLS mutants on the nuclear translocation of Q69. The percentage of cells showing nuclear translocation of Q69 was calculated from 100 HeLa cells transfected with the indicated vectors. (e) CRAG-dependent disappearance of Q69. HA-CRAG–inducible Tet-On HeLa cells were transfected with Q69-myc. At 24 h after transient transfection, cells were treated with or without 1 μM DOX for the indicated times and immunoblotted with anti-HA, anti-myc, and anti-tubulin antibodies. DOX alone did not affect the Q69 protein level in Tet-On HeLa cells without CRAG (WT; right). Note that CRAG expression did not affect the Q12 protein level. (f) MG132 blocked the CRAG-induced Q69 degradation. COS-7 cells expressing Q69-myc or Q69-myc/HA-CRAG were treated with or without 10 μM MG132 for 6 h and immunoblotted with anti-HA and anti-myc antibodies. (g) CRAG suppressed Q69-induced cell toxicity. Apoptosis in Tet-On HeLa cells without (left) or with (right) CRAG in the presence or absence of DOX was determined by FACS analysis using annexin-V/propidium iodide staining (right). Apoptotic cells were collected in the top right box. (h) GFP-CRAG enhanced in vivo ubiquitination of Q69. GFP-Q69 was immunoprecipitated from cells coexpressing GFP-Q69 and Flag-ubiquitin with or without GFP-CRAG and analyzed by immunoblot with the indicated antibodies. Bars, 20 μm.
The disappearance of polyQ at PML body has been previously reported as a pathological finding of polyglutamine diseases (Takahashi et al., 2003). We examined CRAG-dependent degradation of Q69 using a DOX-inducible Tet-On HeLa cell line expressing HA-CRAG. Upon DOX treatment, CRAG expression was up-regulated, whereas Q69 protein level was concomitantly down-regulated. That equal amount of total protein was loaded is demonstrated by tubulin immunoblots. A distinct background band sometimes appeared as a cross-reacting protein in the anti-HA immunoblots (Fig. 4 e, left). We confirmed that DOX alone did not affect Q69 protein level in Tet-On HeLa cells without CRAG (Fig. 4 e, right). As shown in Fig. 4 f, the proteasome inhibitor MG132 blocked Q69 down-regulation by CRAG WT, suggesting that CRAG may promote Q69 degradation through the ubiquitin–proteasome pathway. Furthermore, FACS analysis of cells stained with annexin-V/propidium iodide demonstrated that CRAG WT expression suppressed Q69-induced cell death (Fig. 4 g). In contrast, DOX alone could not rescue Q69-mediated cell death in Tet-On HeLa cells without CRAG (Fig. 4 g). To confirm ubiquitination of Q69 by CRAG, an in vivo ubiquitination assay was performed. As shown in Fig. 4 h, the ubiquitinated Q69 significantly increased by CRAG expression. Degradation of Q69 was not enhanced by NLS or GTPase mutants, and DNA ladder formation assays consistently revealed that the two mutants could not rescue Q69-mediated cell death (unpublished data). Because NLS and GTPase activity in CRAG are critical for its interaction with PML, CRAG-mediated activation of PML-associated ubiquitin ligase may be responsible for this ubiquitination.
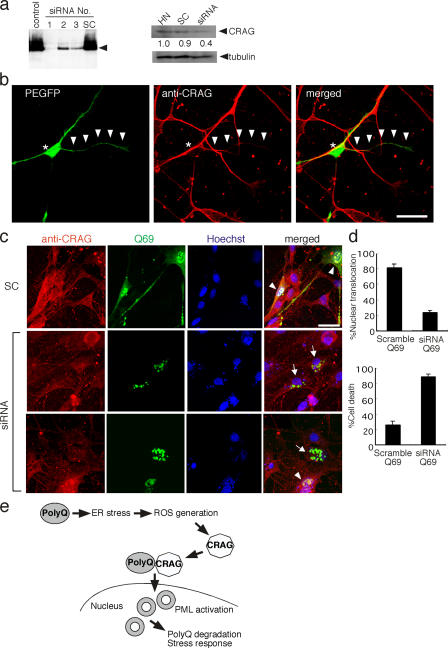
We next examined the effect of CRAG knockdown by RNA interference method on nuclear translocation of polyQ in neurons. Several small interfering RNA (siRNA) oligonucleotides specifically targeted to the CRAG sequence were generated, and their inhibitory effects were estimated using the COS-7 cell expression system. All three RNA interference oligonucleotides, but not scramble oligonucleotides, suppressed the expression of HA-CRAG (Fig. 5 a, left). Among them, one siRNA (No. 1) showing the strongest inhibitory effect on CRAG expression was used for the following experiments. This siRNA or scramble with Q69 plus pEGFP vector as a marker were introduced into cultured rat hippocampal neurons, and the inhibitory effect on endogenous CRAG expression was evaluated by CRAG immunoblotting and immunostaining. As shown in Fig. 5 (a [right] and b) this siRNA, but not scramble, was found to suppress endogenous CRAG expression, indicating that this siRNA against the CRAG gene is useful to assess CRAG function.
Figure 5.
Knockdown of CRAG in neuronal cells blocked nuclear translocation of polyQ and polyQ-mediated cell death. (a) Effect of siRNA oligonucleotides on CRAG expression in COS-7 cells and neurons. Three different siRNA oligonucleotides against CRAG sequence (411–429, 525–543, and 698–716) were synthesized and transfected with HA-CRAG into COS-7 cells. Scramble oligonucleotide (SC) was used as a negative control. (left) The arrowhead indicates the position of CRAG. SC and siRNA (No. 1) were transfected into hippocampal neurons (HN). Inhibitory effect on CRAG expression was estimated by immunoblot with the indicated antibodies. The optical density of CRAG bands was analyzed by the NIH Image software. (b) Knockdown of endogenous CRAG by siRNA. Hippocampal neurons were fixed at 24 h after transfection of siRNA (No.1) plus pEGFP vector and immunostained with anti-CRAG antibody (red). Asterisks and arrowheads indicate CRAG-deficient cell body and neurite, respectively. (c) Effect of siRNA-mediated CRAG depletion on nuclear translocation of Q69. Mouse DRG neurons were cotransfected with Q69-myc and scramble or CRAG siRNA. After 24 h of incubation, an immunofluorescence assay was performed with anti-CRAG (red), anti-myc antibodies (green), and Hoechst 33258 (blue). Arrows and arrowheads indicate CRAG-deficient and not deficient cells, respectively. (d) Knockdown of CRAG blocks nuclear translocation of Q69 and promotes Q69-induced cell death. The percentage of cells exhibiting nuclear translocation of Q69 was measured from 100 DRG neurons expressing Q69 (top). An immunofluorescence assay was performed as described in panel c and the percentage of cell death at 48 h after transfection was measured from 100 DRG neurons expressing Q69, judged by chromatin condensation with Hoechst 33258 staining (bottom). Error bars represent SD. n = 3. (e) Schematic model of CRAG action on polyQ. Bars, 20 μm.
To determine whether endogenous CRAG is involved in nuclear translocation, inclusion body formation, and decreased cell toxicity of polyQ in neuronal cells, the effects of CRAG knockdown on subcellular distribution and cell toxicity of Q69 were examined. In scramble siRNA–cotransfected DRG neurons, endogenous CRAG and Q69 translocated to the nucleus and formed NIs (Fig. 5 c, top). In contrast, perinuclear aggregations of Q69 were dominantly detected in siRNA-mediated CRAG-deficient cells (Fig. 5 c, middle). Moreover, almost all CRAG-deficient cells revealed a typical apoptotic phenotype including chromatin condensation as shown in Fig. 5 c (bottom). Statistical analysis indicated that siRNA-mediated CRAG depletion blocked the nuclear translocation of Q69 in DRG neurons (Fig. 5 d, top). In addition, analysis of a cell death assay, judging from chromatin condensation, showed that >80% of CRAG-deficient cells underwent cell death, whereas only 30% of scramble siRNA–transfected cells died by Q69 expression 48 h after transfection (Fig. 5 d, bottom). CRAG knockdown by siRNA did not cause death in cells that were not expressing Q69 (unpublished data). These results demonstrate that endogenous CRAG mediates nuclear translocation and NI formation by polyQ and confers resistance to cell death under the conditions of polyQ accumulation. A schematic model of CRAG action on polyQ is shown in Fig. 5 e.
Patients with polyglutamine diseases and transgenic mouse model carrying polyQ showed the late-onset and gradually progressive neurological pathology (Turmaine et al., 2000; Katsuno et al., 2003). Indeed, CRAG expression is very high in the developing brain and decreased thereafter in the adult brain (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200505079/DC1). This developmentally regulated expression of CRAG may be closely related to the appearance of polyglutamine disease; a decreased level of CRAG expression fails to scavenge unfolded proteins at PML bodies and permits an accumulation of polyQ aggregates in the nucleus, thereby conferring a toxic gain of function that is selectively deleterious to neurons. If so, CRAG is a rate-limiting factor in the degradation of pathological forms of polyQs and targeted expression of CRAG is a potential gene therapy for polyglutamine disease.
Materials and methods
Materials
Anti-Flag M2 monoclonal and anti–α-tubulin antibodies were obtained from Sigma-Aldrich. Anti-HA high affinity, anti-HA affinity matrix, and anti–c-myc mouse mAbs were obtained from Roche. Anti-GFP rabbit polyclonal antibody, annexin V, propidium iodide (PI), and secondary antibodies conjugated with Alexa Fluor 488, 594, and 647 were obtained from Invitrogen. Anti-6×His monoclonal, anti-GFP mouse monoclonal, and Hoechst 33258 were obtained from Nacalai Tesque. DOX and mouse MTN blot were obtained from CLONTECH Laboratories, Inc. HA-probe was purchased from Santa Cruz Biotechnology, Inc. Nuclear mitotic apparatus protein antibody 1 was purchased from Biocarta. Anti-ubiquitin antibody was obtained from Santa Cruz Biotechnology, Inc. Anti-HA mouse mAb was purchased from Covance. Anti-PML mouse mAb was purchased from Santa Cruz Biotechnology, Inc.
Identification of CRAG in CRAM immunoprecipitates from developing rat brain
CRAM (CRMP-5) has been implicated in semaphorin signaling (Inatome et al., 2000). We have searched for CRAM-interacting proteins from developing rat brain. The purification procedure was performed by using anti-CRAM antibody affinity column chromatography and specific elution with antigen peptides for anti-CRAM antibody. Among purified proteins, we focused on a 42-kD protein, and partial amino acid sequence analysis revealed several peptide sequences, including KSALVHRYLTGTYVQEESPEGGRF. Based on this information, we cloned a novel gene encoding a GTPase from the mouse brain cDNA library (available from GenBank/EMBL/DDBJ under accession no. AB078345).
Expression constructs
CRAG WT, GTPase mutants, NLS mutants, and murine PML1 isoform (available from GenBank/EMBL/DDBJ under accession no. BC020990) cDNA tagged with the HA epitope at the NH2 terminus were subcloned into pCMV5 expression vectors. Murine PML1 shows 67% homology to human PML1 at the amino acid level. CRAG GTPase mutation was generated by missense mutation S114N. CRAG NLS mutation was generated by missense mutation KR342-343EE. The plasmid encoding NH2-terminal–truncated ataxin-3 with Q69 together with the COOH-terminal myc epitope and the NH2-terminal HA epitope was described previously (Yoshizawa et al., 2000). In this plasmid, 286 amino acid residues of ataxin-3 were deleted from the NH2-terminal side. For our experiment, the HA epitope was removed.
The Q12 gene was created by PCR using the primers 5′-CCCAAGCTTGGGATGGCCTACTTTGAAAAACAG-3′ and 5′-GGGGTACCCCAGGGAATGAAGAATAATG-3′ and subcloned into pcDNA3.1- myc His. A RING-finger domain mutation of PML was generated by the missense mutation C51S/C54S using the PCR primers 5′-CCTGCACACGCTGAGCTCCGGAAGCCTGGAGGCGC-3′ and 5′- GCGCCTCCAGGCTTCCGGAGCTCAGCGTGTGCAGG-3′.
Immunoprecipitation, immunoblotting, and immunofluorescence microscope
These procedures were performed as described previously (Mitsui et al., 2002; Hotta et al., 2005). Fluorescence images were analyzed on a confocal microscope (LSM 510 META; Carl Zeiss MicroImaging, Inc.) equipped with three lasers (UV.Ar. 364, Ar. 488, and HeNe 543) using Plan-Apochromat 63× oil-immersion (NA 1.40) objective. LSM 510 META 3.0 software (Carl Zeiss MicroImaging, Inc.) was used for image acquisition from confocal microscopy. Photoshop 6.0 software (Adobe) was used for minor adjustments to contrast and overlaying.
Immunohistochemistry with pathological specimens
Postmortem brain tissues were obtained from two MJD patients and analyzed as described previously (Yoshizawa et al., 1990).
GTPase assay for CRAG
The GTPase assay was performed as described previously (Der et al., 1986).
In vitro ubiquitin ligase assay
Immunoprecipitates were washed three times with lysis buffer and once with ubiquitination buffer (50 mM Tris-HCL, pH 7.4, 10 mM MgCl2, 5 mM ATP, and 2 mM dithiothreitol) and incubated in 50 μl of the same buffer supplemented with 100 ng E1 (Honda et al., 1997), 500 ng E2ubcH5(a,b,c) (Kobirumaki et al., 2005), and 2.5 μg His-tagged ubiquitin (Calbiochem) for 30 min at 25°C. Samples were analyzed with anti-His antibody.
Online supplemental material
Fig. S1 a shows the hollow, doughnut-like nuclear bodies of CRAG in UV-irradiated hippocampal neurons, and Fig. S1 b shows colocalization of CRAG doughnut-like bodies with PML1 in a UV-irradiated DRG neuron. Fig. S2 shows NIs of CRAG in response to H2O2 stimulation in HeLa cells. Fig. S3 shows a comparison of CRAG expression in adult and developing mouse brains. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200505079/DC1.
Supplementary Material
Acknowledgments
We thank Dr. S. Jahangeer for critical reading of this manuscript.
This study was supported in part by Grants-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology and the Japan Society for the Promotion of Science.
Q. Qin and R. Inatome contributed equally to this paper.
Abbreviations used in this paper: CRAM, CRMP-associated molecule; CRMP, collapsin response mediator protein; DOX, doxycycline; DRG, dorsal root ganglion; GTPase, guanosine triphosphatase; MJD, Machado-Joseph disease; NI, nuclear inclusion; NLS, nuclear localization signal; PML, promyelocytic leukemia protein; polyQ, polyglutamine protein; RING, really interesting new gene; ROS, reactive oxygen species; siRNA, small interfering RNA; WT, wild type.
References
- Der, C.J., T. Finkel, and G.M. Cooper. 1986. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 44:167–176. [DOI] [PubMed] [Google Scholar]
- Hodges, M., C. Tissot, K. Howe, D. Grimwade, and P.S. Freemont. 1998. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am. J. Hum. Genet. 63:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25–27. [DOI] [PubMed] [Google Scholar]
- Hotta, A., R. Inatome, J. Yuasa-Kawada, Q. Qin, H. Yamamura, and S. Yanagi. 2005. Critical role of CRMP-associated molecule CRAM for filopodia and growth cone development in neurons. Mol. Biol. Cell. 16:32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inatome, R., T. Tsujimura, T. Hitomi, N. Mitsui, P. Hermann, S. Kuroda, H. Yamamura, and S. Yanagi. 2000. Identification of CRAM, a novel unc-33 gene family protein that associates with CRMP3 and protein-tyrosine kinase(s) in the developing rat brain. J. Biol. Chem. 275:27291–27302. [DOI] [PubMed] [Google Scholar]
- Katsuno, M., H. Adachi, A. Inukai, and G. Sobue. 2003. Transgenic mouse models of spinal and bulbar muscular atrophy (SBMA). Cytogenet. Genome Res. 100:243–251. [DOI] [PubMed] [Google Scholar]
- Kobirumaki, F., Y. Miyauchi, K. Fukami, and H. Tanaka. 2005. A novel UbcH10-binding protein facilitates the ubiquitinylation of cyclin B in vitro. J. Biochem. (Tokyo). 137:133–139. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and W.C. Earnshaw. 1998. Structure and function in the nucleus. Science. 280:547–553. [DOI] [PubMed] [Google Scholar]
- Mitsui, N., R. Inatome, S. Takahashi, Y. Goshima, H. Yamamura, and S. Yanagi. 2002. Involvement of Fes/Fps tyrosine kinase in semaphorin3A signaling. EMBO J. 21:3274–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H.T. 2001. Beyond the Qs in the polyglutamine diseases. Genes Dev. 15:925–932. [DOI] [PubMed] [Google Scholar]
- Ross, C.A. 1997. Intranuclear neuronal inclusions: a common pathogenic mechanism for glutamine-repeat neurodegenerative diseases? Neuron. 19:1147–1150. [DOI] [PubMed] [Google Scholar]
- Sato, A., T. Shimohata, R. Koide, H. Takano, T. Sato, M. Oyake, S. Igarashi, K. Tanaka, T. Inuzuka, H. Nawa, and S. Tsuji. 1999. Adenovirus-mediated expression of mutant DRPLA proteins with expanded polyglutamine stretches in neuronally differentiated PC12 cells. Preferential intranuclear aggregate formation and apoptosis. Hum. Mol. Genet. 8:997–1006. [DOI] [PubMed] [Google Scholar]
- Takahashi, J., H. Fujigasaki, K. Iwabuchi, A.C. Bruni, T. Uchihara, K.H. El Hachimi, G. Stevanin, A. Durr, A.S. Lebre, Y. Trottier, et al. 2003. PML nuclear bodies and neuronal intranuclear inclusion in polyglutamine diseases. Neurobiol. Dis. 13:230–237. [DOI] [PubMed] [Google Scholar]
- Turmaine, M., A. Raza, A. Mahal, L. Mangiarini, G.P. Bates, and S.W. Davies. 2000. Nonapoptotic neurodegeneration in a transgenic mouse model of Huntingtons disease. Proc. Natl. Acad. Sci. USA. 97:8093–8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, K., K.J. Hurt, F.Y. Wu, M. Fang, H.R. Luo, J.J. Hong, S. Blackshaw, C.D. Ferris, and S.H. Snyder. 2000. Pike. A nuclear GTPase that enhances PI3kinase activity and is regulated by protein 4.1N. Cell. 103:919–930. [DOI] [PubMed] [Google Scholar]
- Yoshizawa, T., O. Shinmi, A. Giaid, M. Yanagisawa, S.J. Gibson, S. Kimura, Y. Uchiyama, J.M. Polak, T. Masaki, and I. Kanazawa. 1990. Endothelin: a novel peptide in the posterior pituitary system. Science. 247:462–464. [DOI] [PubMed] [Google Scholar]
- Yoshizawa, T., Y. Yamagishi, N. Koseki, J. Goto, H. Yoshida, F. Shibasaki, S. Shoji, and I. Kanazawa. 2000. Cell cycle arrest enhances the in vitro cellular toxicity of the truncated Machado-Joseph disease gene product with an expanded polyglutamine stretch. Hum. Mol. Genet. 9:69–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.