Abstract
Upon the accumulation of unfolded proteins in the mammalian endoplasmic reticulum (ER), X-box binding protein 1 (XBP1) premessenger RNA (premRNA) is converted to mature mRNA by unconventional splicing that is mediated by the endonuclease inositol-requiring enzyme 1. The transcription factor protein (p) XBP1 spliced (S), which is translated from mature XBP1 mRNA, contains the nuclear localization signal and the transcriptional activation domain and activates the transcription of target genes, including those encoding ER chaperones in the nucleus. We show that pXBP1 unspliced (U) encoded in XBP1 pre-mRNA was constitutively expressed and markedly accumulated at the recovery phase of ER stress. pXBP1(U) contained the nuclear exclusion signal instead of the transcriptional activation domain and shuttled between the nucleus and the cytoplasm. Interestingly, pXBP1(U) formed a complex with pXBP1(S), and the pXBP1(U)–pXBP1(S) complex was sequestered from the nucleus. Moreover, the complex was rapidly degraded by proteasomes because of the degradation motif contained in pXBP1(U). Thus, pXBP1(U) is a negative feedback regulator of pXBP1(S), which shuts off the transcription of target genes during the recovery phase of ER stress.
Introduction
The folding of nascent proteins is an extremely error-prone process, and cells must deal with malfolded proteins, which tend to form aggregates, by using molecular chaperones and protein degradation machinery. The membrane of the ER in mammalian cells contains three sensors (PKR-like ER-resistant kinase [PERK], activating transcription factor 6 [ATF6], and inositol requiring enzyme 1 [IRE1]) that can monitor the accumulation of unfolded proteins in the ER (ER stress) and activate elaborate defense mechanisms known collectively as the ER stress response to alleviate the burden of unfolded proteins (Kaufman, 1999; Mori, 2000; Urano et al., 2000; Patil and Walter, 2001). The first sensor molecule, PERK, is a transmembrane kinase that is activated in response to ER stress (Harding et al., 1999) and phosphorylates the α subunit of eukaryotic translational initiation factor 2, leading to translational attenuation to avoid further accumulation of unfolded proteins in the ER (Harding et al., 2000). The second sensor, ATF6, a transmembrane transcription factor, is transported to the Golgi apparatus upon ER stress and is sequentially cleaved by site-1 and -2 proteases (Yoshida et al., 1998; Haze et al., 1999, 2001; Ye et al., 2000). The liberated cytoplasmic fragment of ATF6, containing a basic leucine zipper motif (pATF6α(N)), translocates into the nucleus, binds to the cis-acting ER stress response element (ERSE), and activates transcription of ER chaperones such as BiP, GRP94, and calreticulin (Yoshida et al., 1998, 2000, 2001b).
The third sensor, IRE1, is a transmembrane RNase (Tirasophon et al., 1998; Wang et al., 1998; Niwa et al., 1999; Iwawaki et al., 2001) involved in the splicing of XBP1 pre-mRNA (Yoshida et al., 2001a; Calfon et al., 2002). XBP1 is a basic leucine zipper–type transcription factor containing a DNA-binding domain and a transcriptional activation domain, each encoded by a separate open reading frame on the pre-mRNA. Upon ER stress, XBP1 pre-mRNA is cleaved by the activated IRE1 and ligated by an unidentified RNA ligase to form mature (spliced) XBP1 mRNA, which encodes pXBP1(S) (Yoshida et al., 2001a; Calfon et al., 2002). pXBP1(S) binds to ERSE to induce transcription of ER chaperones, and to another cis-acting element, unfolded protein response element, to induce transcription of other genes (probably genes involved in ER-associated protein degradation [ERAD]; Yoshida et al., 2003). The IRE1 signaling pathway is well conserved from yeast to mammals. In the budding yeast Saccharomyces cerevisiae, Ire1p converts HAC1 pre-mRNA to mature mRNA, which allows translation of the active transcription factor Hac1p to induce transcription of ER chaperones and ERAD components (Cox et al., 1993; Mori et al., 1993, 1996; Cox and Walter, 1996).
The splicing of HAC1 and XBP1 pre-mRNAs by IRE1 is quite unconventional (Patil and Walter, 2001; Yoshida et al., 2001a; Calfon et al., 2002). The conventional splicing involves an elaborate complex of proteins and RNAs, called the spliceosome, and occurs exclusively in the nucleus, whereas the splicing reaction of HAC1 and XBP1 pre-mRNA simply requires IRE1 and RNA ligase, which is completely independent of the spliceosome, and takes place in the cytoplasm (Ruegsegger et al., 2001). Because the removal of an intron from the HAC1 and XBP1 pre-mRNAs causes a switching of the reading frame in the COOH-terminal portion of the respective proteins, such splicing could be called “frame switch splicing” (Yoshida et al., 2003) or “cytoplasmic splicing” (Ruegsegger et al., 2001).
One of the unresolved issues regarding XBP1 is whether XBP1 pre-mRNA encodes a functional protein. In yeast, HAC1 pre-mRNA has a long (252 nt) intron that inhibits translation (Chapman and Walter, 1997; Kawahara et al., 1997; Ruegsegger et al., 2001). In contrast, unspliced (U) XBP1 pre-mRNA contains a much shorter (26 nt) intron and is actively translated to produce a protein (pXBP1(U)), although pXBP1(U) is rapidly degraded by the proteasome and not detected by immunoblotting (Yoshida et al., 2001a; Calfon et al., 2002). It remained possible, however, that pXBP1(U) expression was enhanced in certain situations and played an important physiological role. Lee et al. (2003) reported that pXBP1(U) mutants whose lysine residues were replaced with arginine residues were resistant to degradation by proteasomes and that overexpression of the mutants repressed transcriptional induction by pXBP1(S), suggesting that pXBP1(U) could modulate function of pXBP1(S) if its expression was induced.
We examined this problem and revealed an elaborate regulatory mechanism of mammalian ER stress response taking advantage of the dynamic interplay between pXBP1(U) and pXBP1(S) that appears critical for swiftly adapting to physiological changes in the ER.
Results
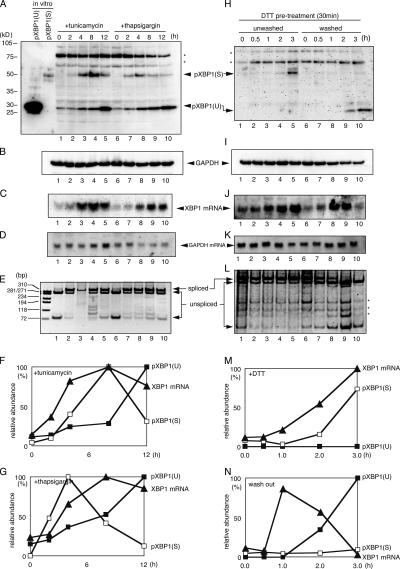
Expression of pXBP1(U) is induced during the recovery phase of ER stress
It had been postulated that XBP1 pre-mRNA is not translated into a functional protein because pXBP1(U) encoded in the pre-mRNA is rapidly degraded by the proteasome and because endogenous pXBP1(U) cannot be detected by immunoblotting (Yoshida et al., 2001a; Calfon et al., 2002). By improving the protein extraction protocol, it became possible to detect pXBP1(U) protein accumulated in the cells even without prior proteasome inhibitor treatment. In cells not treated with ER stress inducers, a very small amount of 29-kD pXBP1(U) was detected (Fig. 1 A, lanes 1 and 6). Northern blot analysis revealed a certain amount of XBP1 mRNA in these cells (Fig. 1 C, lanes 1 and 6). When RT-PCR analysis was performed to differentiate spliced XBP1 mRNA from unspliced pre-mRNA, using a pair of primers designed to encompass the intron containing a PstI site (Fig. 2 A), only the bands corresponding to unspliced XBP1 mRNA cut with PstI (63 and 297 nt) were detected in uninduced cells (Fig. 1 E, lanes 1 and 6). Upon addition of tunicamycin (an inhibitor of N-linked glycosylation) or thapsigargin (an inhibitor of calcium ATPase in the ER), the band of spliced mature mRNA (334 nt) was markedly induced (Fig. 1 E, lanes 2–4 and 7–9). This induced splicing was shortly followed by a marked expression of pXBP1(S) and XBP1 mRNA, whereas the amount of pXBP1(U) increased gradually (Fig. 1, A and C, lanes 2–4 and 7–9, respectively). At the later phase of ER stress, the amount of pXBP1(S) gradually decreased, whereas the amount of pXBP1(U) continued to increase (Fig. 1 A, lanes 5 and 10). At this stage, a large amount of XBP1 mRNA was still accumulated (Fig. 1 C, lanes 5 and 10), and the amount of unspliced XBP1 mRNA was increased (Fig. 1 E, lanes 5 and 10; see the bands of 63 nt). These results implied that XBP1 transcription is still vigorous at this stage, but an appreciable portion of newly transcribed XBP1 pre-mRNA remains unspliced, possibly because of the gradual inactivation of IRE1, leading to the enhanced synthesis of pXBP1(U). This suggested the interesting possibility that pXBP1(U) expression is induced at the recovery phase of ER stress. To exclude the possibility that induction of pXBP1(U) at the later phase is caused by stabilization of pXBP1(U), we examined the stability of pXBP1(U) during ER stress. HeLa cells expressing pXBP1(U) were treated with cycloheximide to block de novo protein synthesis, and degradation of pXBP1(U) was monitored by immunoblotting with anti–XBP1-A antiserum. Degradation of pXBP1(U) was enhanced in the later phase of ER stress (Fig. 2 L).
Figure 1.
pXBP1(U) expression is induced during the recovery phase of ER stress. (A and B) Accumulation of pXBP1(U) and pXBP1(S) upon ER stress. HeLa cells were treated with 2 μg/ml tunicamycin (Tm) or 300 nM thapsigargin (Tg) for the indicated period, and cell lysates were subjected to immunoblotting with anti–XBP1-A (A) and anti-GAPDH antisera (B). In vitro–translated pXBP1(U) and pXBP1(S) were included for comparison. (C and D) Accumulation of XBP1 mRNA upon ER stress. Total RNA from cells treated as described in A was subjected to Northern blotting with XBP1 (C) and GAPDH (D) cDNA probes. (E) Splicing of XBP1 pre-mRNA during ER stress. RT-PCR was performed using total RNA extracted from cells treated as described in A as a template and the PCR primers shown in Fig. 2 A to discriminate spliced mature XBP1 mRNA from unspliced pre-mRNA. The amplified cDNA was digested with PstI, which specifically cleaves cDNA of XBP1 pre-mRNA, and separated on polyacrylamide gel. (F and G) Quantified data for A–D. pXBP1(U) (closed box), pXBP1(S) (open box), and XBP1 mRNA (closed triangle) are shown. (H and I) Accumulation of pXBP1(U) in the recovery phase. HeLa cells treated with 1 mM DTT for 30 min were washed with PBS as indicated, and lysates were prepared and analyzed by immunoblotting with anti-XBP1-A (H) antiserum that detects both pXBP1(U) and pXBP1(S) and anti-GAPDH (I) antiserum. *, nonspecific bands. (J and K) Accumulation of XBP1 mRNA during the recovery phase. Total RNA isolated from cells treated as in H was subjected to Northern blotting with XBP1 (J) and GAPDH (K) cDNA probes. (L) Splicing of XBP1 pre-mRNA in the recovery phase. RT-PCR was performed with total RNA as in E. (M and N) Quantified data for H–K. pXBP1(U) (closed box), pXBP1(S) (open box), and XBP1 mRNA (closed triangle) are shown.
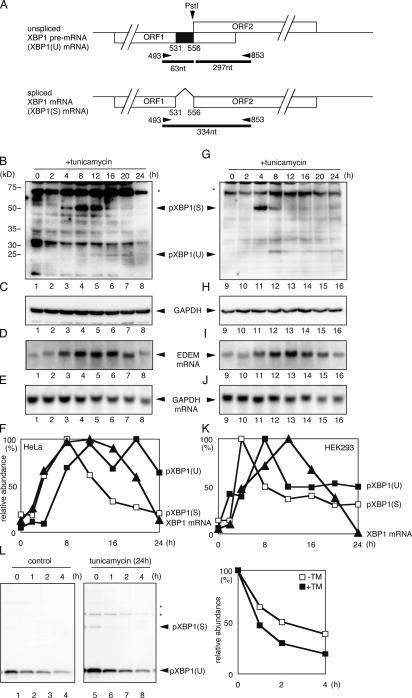
Figure 2.
Expression of the XBP1 target gene wanes during the late phase of ER stress. (A) Schematic representation of RT-PCR. (B–K) Comparison of induction time course of pXBP1(U), pXBP1(S), and EDEM mRNA. HeLa (B–F) and HEK293 cells (G–K) were analyzed as in Fig. 1. (L) Stability of pXBP1(U) during ER stress. HeLa cells transfected with pCMV-HA-pXBP1(U) were incubated in the presence or absence of 10 μg/ml tunicamycin for 24 h, and then treated with 40 μM cycloheximide for the indicated period. Whole cell lysates were extracted and subjected to immunoblotting with anti–XBP1-A antiserum.
We also examined pXBP1(U) expression in human embryonic kidney 293 (HEK293) cells and obtained results similar to those observed in HeLa cells (Fig. 2, B–K). Interestingly, the kinetics and duration of ER degradation–enhancing mannosidase-like protein (EDEM) mRNA induction, one of XBP1's targets (Yoshida et al., 2003), correlated well with the expression level of pXBP1(S) in HeLa and HEK293 cells (Fig. 2, D and I), and EDEM mRNA expression actually waned in the later phase (Fig. 2, D and I, lanes 6–8 and 14–16), though the peak of EDEM mRNA level was delayed as compared with that of pXBP1(S) level. It should be noted that the level of pXBP1(S) determines the rate of transcription of EDEM mRNA, whereas the data observed in the Northern blots reflects accumulation of EDEM mRNA.
To confirm the notion that pXBP1(U) expression is induced at the recovery phase of ER stress, the expression of pXBP1(U) was examined in cells recovering from ER stress induced with DTT, which is a potent inducer yet easily washed out to accelerate recovery. As expected, DTT markedly induced transcription and splicing of XBP1 mRNA and expression of pXBP1(S) protein (Fig. 1, H–L, lanes 1–5). In contrast, when DTT was washed out after 30 min of treatment, transcription and splicing of XBP1 mRNA gradually decreased to the basal level, whereas the accumulation of pXBP1(U) increased (Fig. 1, H–L, lanes 6–10). This clearly showed that pXBP1(U) expression is induced during recovery from ER stress, when XBP1 mRNA splicing is halted, leading to accumulation of pre-mRNA. This temporal regulation of pXBP1(U) expression suggested that pXBP1(U) may have an important regulatory role in mammalian ER stress response, especially during the recovery phase.
pXBP1(U) shuttles between the nucleus and cytoplasm
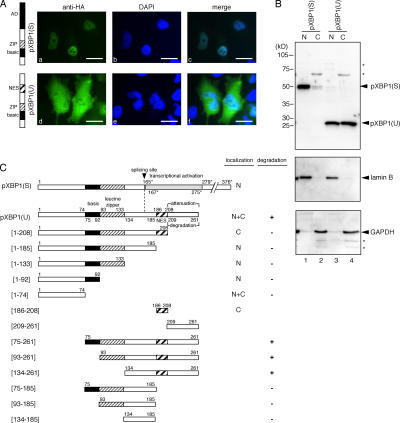
We next analyzed subcellular localization of pXBP1(U) using immunofluorescent microscopy. When HeLa cells were transfected with plasmid expressing HA-tagged pXBP1(U) or pXBP1(S), pXBP1(U) was found in the cytoplasm as well as in the nucleus (Fig. 3 A, d–f), whereas pXBP1(S) was specifically localized to the nucleus, as expected (Fig. 3 A, a–c). We confirmed this result by subcellular fractionation and immunoblotting of HeLa cells (Fig. 3 B).
Figure 3.
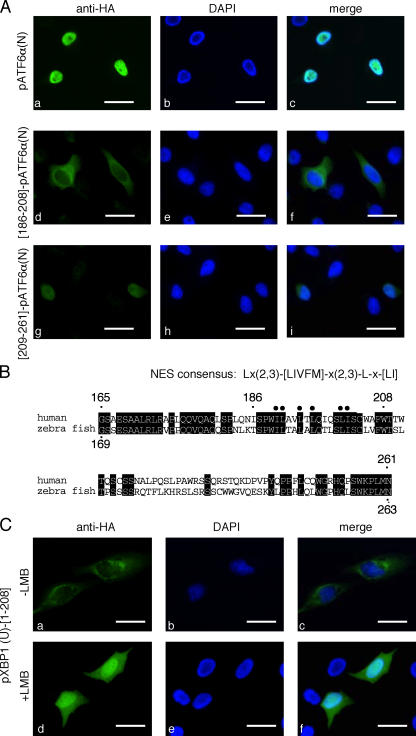
pXBP1(U) resides in both the nucleus and cytoplasm. (A) Subcellular localization of pXBP1(S) and pXBP1(U). HeLa cells transiently transfected with pCMV-HA-pXBP1(S) or pCMV-HA-pXBP1(U) were stained with anti-HA antiserum (left), and the nucleus was stained with DAPI (middle). (right) Superimposed image. (B) Subcellular fractionation of pXBP1(S) and pXBP1(U). Cells transfected as in A were separated into the nuclear (N) and postnuclear (C) fractions, and subjected to immunoblotting using anti–XBP1-A. The same blot was also probed with anti–lamin B (a nuclear marker) and anti-GAPDH (a cytoplasmic marker) antisera. (C) Deletion constructs of XBP1 used (number represents amino acid residues encoded in unspliced XBP1 mRNA, and asterisk indicates amino acid residues encoded in spliced mRNA) and a summary of results. The splicing site is indicated by a dotted line. A summary of subcellular localization (N, nuclear; C, cytosolic) and degradation by the proteasome (Fig. 6; +, rapidly degraded; −, relatively stable) is shown on the right. Bars, 10 μm.
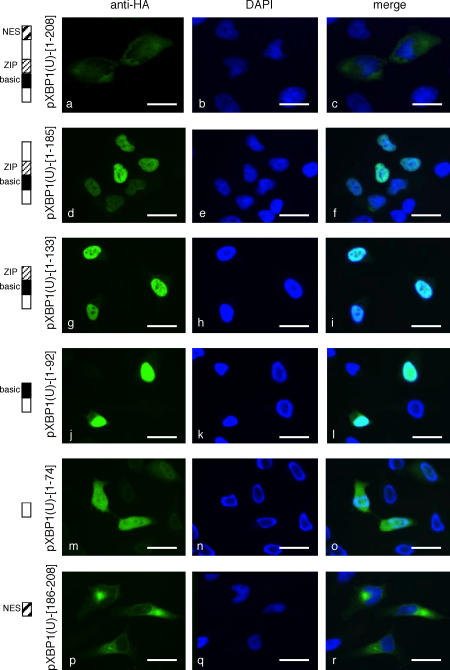
To determine the amino acid sequence responsible for this unexpected localization of pXBP1(U), we constructed and analyzed a set of deletion mutants of pXBP1(U) (Fig. 3 C). The COOH-terminal deletion mutant pXBP1(U)-[1–185] was exclusively localized to the nucleus (Fig. 4, d–f), suggesting that the anterior ([1–185]) and posterior ([186–261]) regions of pXBP1(U) contain the NLS and cytoplasmic localization signal, respectively. Analysis of further COOH-terminal deletions revealed that pXBP1(U)-[1–133] and pXBP1(U)-[1–92] containing the basic domain were localized to the nucleus (Fig. 4, g–l), whereas pXBP1(U)-[1–74], which was lacking the basic domain, resided in both the cytoplasm and nucleus (Fig. 4, m–o), indicating that the basic domain ([75–92]) functions as a NLS.
Figure 4.
Localization of deletion mutants of pXBP1(U). HeLa cells transiently transfected with plasmid expressing indicated deletion mutants were analyzed by immunofluorescence microscopy as in Fig. 3 A. Bars, 10 μm.
On the other hand, pXBP1(U)-[1–208] was predominantly localized to the cytoplasm (Fig. 4, a–c), as opposed to pXBP1(U)-[1–185], which was found only in the nucleus. This indicates that the [186–208] region contains a strong nuclear exclusion signal (NES) that overcomes the function of the NLS. Indeed, this small fragment of 22 amino acid residues was exclusively localized to the cytoplasm (Fig. 4, p–r). To further examine the function of the NES, this segment was fused to the transcription factor pATF6α(N), which contains a basic domain and is solely expressed in the nucleus (Fig. 5 A, a–c; Haze et al., 1999). When expressed in HeLa cells, the fusion protein [186–208]-pATF6α(N) was localized to the cytoplasm (Fig. 5 A, d–f), whereas [209–261]-pATF6α(N) resided in the nucleus (Fig. 5 A, g–i).
Figure 5.
pXBP1(U) contains a conventional NES. (A) NES of pXBP1(U) is functional when fused with a heterologous protein. Localization of pATF6α(N) fused with the indicated regions of pXBP1(U) was analyzed as in Fig. 3 A. (B) Comparison of amino acid sequences between human pXBP1(U)-[165–261] and the corresponding region of zebrafish pXBP1(U). Identical residues are shaded, and conserved hydrophobic residues are marked. (C) Nuclear export of pXBP1(U) is inhibited by LMB. HeLa cells transiently transfected with pCMV-HA-pXBP1(U)-[1–208] were treated with 10 nM LMB for 2 h and analyzed by immunofluorescence microscopy as in A. Bars, 10 μm.
The 22-residue NES region represents a leucine-rich sequence that is conserved between human and zebrafish pXBP1(U), and is similar to a conventional NES (Lx(2,3)-[LIVFM]-x(2,3)-L-x-[LI]; Fig. 5 B; la Cour et al., 2003). To confirm that the nuclear export of pXBP1(U) is mediated by the conventional nuclear export machinery, HeLa cells transfected with plasmid expressing pXBP1(U)-[1–208] were treated with leptomycin B (LMB), an inhibitor for the nuclear export receptor CRM1 (Fornerod et al., 1997), and the localization was analyzed. pXBP1(U)-[1–208] was expressed in the cytoplasm in the absence of LMB (Fig. 5 C, a–c), whereas it was clearly concentrated in the nucleus in the presence of LMB (Fig. 5 C, d–f). This suggested that the nuclear export of pXBP1(U) is mediated by the conventional nuclear export machinery and that pXBP1(U) shuttles between the nucleus and cytoplasm.
Finally, a comparison of pXBP1(U)-[1–208] (localized to the cytoplasm; Fig. 4, a–c) with pXBP1(U)-[1–261] (found in both the cytoplasm and nucleus; Fig. 3 A, d–f) suggested that the [209–261] region partially attenuates the NES activity of the [185–208] region, possibly by sterically hindering the NES from interacting with NES receptors.
Identification of the degradation domain in pXBP1(U)
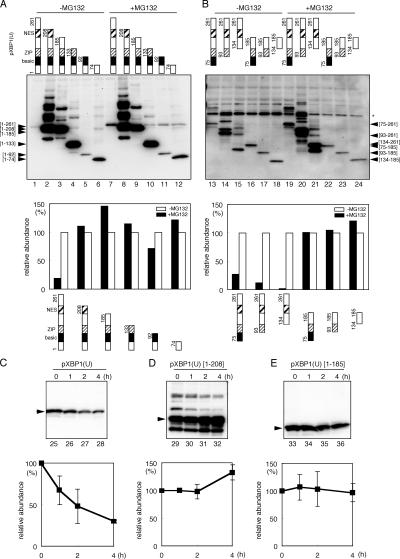
Because pXBP1(U) is very unstable and is rapidly degraded by the proteasome (Yoshida et al., 2001a; Calfon et al., 2002), we analyzed the region involved in rapid degradation. Plasmids carrying deletion derivatives of HA-tagged pXBP1(U) were transfected into HeLa cells, and degradation of each mutant protein was evaluated by comparing the protein level in the presence or absence of the proteasome inhibitor MG132. The amount of intact pXBP1(U) was much increased when cells were treated with MG132, which is consistent with rapid degradation by the proteasome (compare Fig. 6 A, lanes 1 and 7), the level of COOH-terminal deletion mutants lacking the [209–261] region was little affected (Fig. 6 A, lanes 2–6 and 8–12). This indicated that the [209–261] region is required for rapid degradation by the proteasome. The level of NH2-terminal deletion mutants retaining the [209–261] region also increased by MG132 (Fig. 6 B, lanes 13–15 and 19–21), whereas that of deletion mutants lacking this region was little affected (Fig. 6 B, lanes 16–18 and 22–24), confirming our previous conclusion.
Figure 6.
Identification of the region responsible for rapid degradation. Cells were transfected with plasmid expressing the indicated deletion mutants of HA-tagged pXBP1(U) and treated with 10 μM MG132 for 2 h as indicated. Cell lysate was analyzed by Western blotting using anti–XBP1-A antiserum (A) and anti-HA serum (B). (bottom) Data quantified by an image analyzer. (C–E) Stability of pXBP1(U) and its mutants. Cells expressing pXBP1(U) (left), pXBP1(U)-[1–208] (middle), and pXBP1(U)-[1–185] (right) were treated with 40 μM cycloheximide for the indicated period, and cell lysates were subjected to immunoblotting with anti–XBP1-A antiserum. (bottom) Averages from two independent experiments are presented with standard deviation values.
We also examined the turnover of pXBP1(U) and its deletion mutants. Cells expressing pXBP1(U) or its derivatives were treated with cycloheximide, and whole cell lysates were subjected to immunoblotting. pXBP1(U) was rapidly degraded (Fig. 6 C), whereas pXBP1(U) mutants lacking the [209–261] region were more stable (Figs. 5 E and 6 D). This confirmed the aforementioned conclusion that the [209–261] region is indispensable for rapid degradation of pXBP1(U).
pXBP1(U) binds to pXBP1(S)
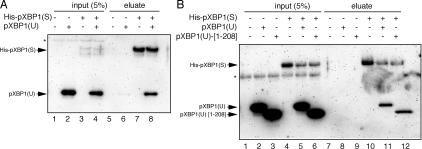
The aforementioned results prompted us to examine whether pXBP1(U) can modulate the function of pXBP1(S), possibly as a negative regulator, to shut off transcriptional induction by pXBP1(S). Thus, we investigated whether pXBP1(U) binds to pXBP1(S) when coexpressed in vivo. HeLa cells were transiently transfected with a plasmid expressing Histidine-tagged pXBP1(S), together with a plasmid expressing XBP1(U), and whole cell lysate was incubated with Ni-NTA resin to bind His-pXBP1(S). Evidently, pXBP1(U) was specifically copurified with His-pXBP1(S) in this pull-down assay (Fig. 7 A, lanes 5–8), suggesting that pXBP1(U) associates with pXBP1(S) under these conditions. We then tested whether pXBP1(U) binds to pXBP1(S) synthesized in vitro. His-pXBP1(S) and pXBP1(U) (or a deletion mutant pXBP1(U)-[1–208]) were simultaneously translated in the same reaction using rabbit reticulocyte lysates, and the resulting products were treated with Ni-NTA resin. pXBP1(U), as well as pXBP1(U)-[1–208], was specifically copurified with His-pXBP1(S) (Fig. 7 B, lanes 7–12). These results suggested that pXBP1(U) synthesized in vitro can directly bind to pXBP1(S).
Figure 7.
Specific binding of pXBP1(U) to pXBP1(S). (A) Coexpression of the two proteins in vivo. Cell lysates were prepared from HeLa cells transfected with pCMV-His-pXBP1(S) and/or pCMV-pXBP1(U), and the lysates were subjected to pull-down assays using Ni-NTA agarose. Input and bound materials were analyzed by immunoblotting with anti–XBP1-A antibody. (B) Mixing of two proteins synthesized in vitro. Indicated proteins were translated in vitro using rabbit reticulocyte lysates and analyzed essentially as in A. *, nonspecific banding.
pXBP1(U) sequesters pXBP1(S) from the nucleus and enhances degradation
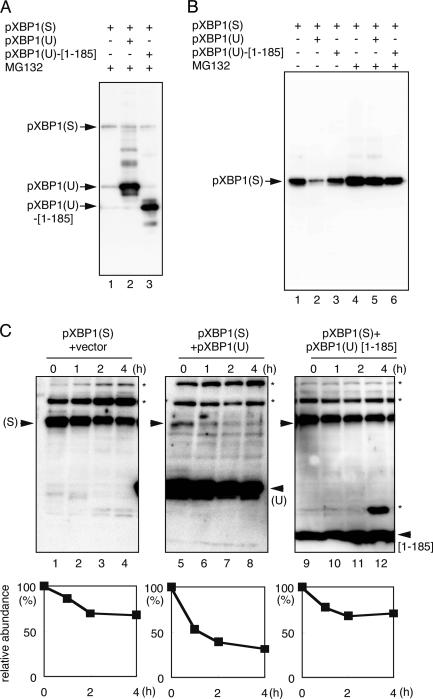
We next examined the physiological implication of the interaction between pXBP1(S) and pXBP1(U). Based on the results presented in Fig. 7, most pXBP1(S) was expected to interact with pXBP1(U) in the presence of excess pXBP1(U). To compare the stability of the pXBP1(S)–pXBP1(U) complex with pXBP1(S), HeLa cells were transfected with a plasmid expressing pXBP1(S), together with a ninefold excess of a plasmid expressing pXBP1(U) or vector alone, and the accumulation of pXBP1(S) was examined by immunoblotting using anti–XBP1-C that specifically recognizes pXBP1(S) (Yoshida et al., 2001a). Under these conditions, pXBP1(U) was expressed in great excess over pXBP1(S) (Fig. 8 A, lane 2), and the level of pXBP1(S) was markedly reduced by overexpression of pXBP1(U) as compared with cells cotransfected with vector alone (Fig. 8 B, lanes 1 and 2). This effect of pXBP1(U) overexpression was greatly compromised by the proteasome inhibitor MG132 (Fig. 8 B, lanes 4 and 5), suggesting that pXBP1(S)–pXBP1(U) is more susceptible to degradation by the proteasome than pXBP1(S). Interestingly, overexpression of pXBP1(U)-[1–185] lacking the “degradation domain” identified in Fig. 6 had little effect on the stability of pXBP1(S) (Fig. 8 B, lanes 3 and 6), though pXBP1(U)-[1–185] was expressed at a level similar to pXBP1(U) (Fig. 8 A, lane 3). This is also consistent with the notion that the degradation domain of pXBP1(U) is indispensable for rapid degradation of the pXBP1(S)–pXBP1(U) complex.
Figure 8.
Level and stability of the pXBP1(S)–pXBP1(U) complex. (A and B) HeLa cells were transiently cotransfected with pCMV-pXBP1(S) and a ninefold excess of pCMV-pXBP1(U) or pCMV-pXBP1(U)-[1–185] and were treated with 10 μM of the proteasome inhibitor MG132 for 2 h before harvest, as indicated. Whole cell lysate was subjected to Western blotting using anti–XBP1-A antiserum (A) or anti–XBP1-C antiserum (B). (C) Cells transfected as in B were treated with 40 μM cycloheximide for the indicated period, and cell lysates were subjected to immunoblotting with anti–XBP1-A antiserum. *, nonspecific banding.
We confirmed these notions by examining the turnover of pXBP1(S) in the presence or absence of abundant pXBP1 (U). Cells transfected as in Fig. 8 B were treated with cycloheximide, and whole cell lysates were subjected to immunoblotting. Coexpression of pXBP1(U) significantly accelerated pXBP1(S) degradation (Fig. 8 C, left and middle), whereas that of pXBP1(U)-[1–185] hardly effected pXBP1(S) stability (Fig. 8, right).
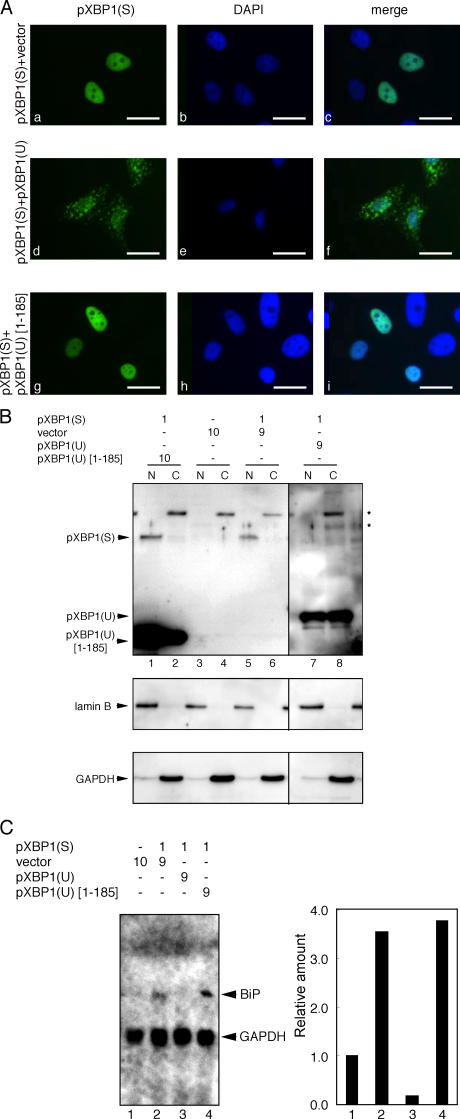
We also analyzed subcellular localization of the pXBP1(S)–pXBP1(U) complex using HeLa cells cotransfected with plasmid expressing pXBP1(S) and a ninefold excess of plasmid expressing pXBP1(U), which resulted in speckled staining in the cytoplasm and reduced staining in the nucleus (Fig. 9 A, d–f). In contrast, pXBP1(S) was located solely in the nucleus (Fig. 9 A, a–c). Interestingly, coexpression of pXBP1(U)-[1–185] lacking the NES did not affect pXBP1(S) localization (Fig. 9 A, g–i). This suggested that pXBP1(S)–pXBP1(U) was exported from the nucleus to cytoplasm and that the NES contained in pXBP1(U) is essential for this export, although the significance of the speckled staining observed remained unknown.
Figure 9.
Subcellular localization of the pXBP1(S)–pXBP1(U) complex. (A) HeLa cells transfected as in Fig. 8 C were stained with anti–XBP1-C antiserum (left) or with DAPI (middle). (B) Cells that were transfected as in A were separated into the nuclear (N) and postnuclear fractions (C), and subjected to immunoblotting as in Fig. 3 B. (C) Total RNA extracted from cells that were transfected as in A was subjected to Northern blotting with BiP and GAPDH cDNA probes. (right) Quantified data. The level of BiP mRNA observed in lane 1 is set to 1. Bars, 10 μm.
We confirmed this notion by subcellular fractionation experiments. Cells transfected as in Fig. 9 A were separated into the nuclear and postnuclear (cytoplasmic) fractions and subjected to immunoblotting with anti–XBP1-A antiserum. pXBP1(S) was localized in the nuclear fraction when it was expressed (Fig. 9 B, lanes 5 and 6). When pXBP1(U) was coexpressed, most of pXBP1(S) was actually found in the postnuclear fraction (Fig. 9 B, lanes 7 and 8), whereas coexpression of pXBP1(U)-[1–185] did not affect pXBP1(S) localization (Fig. 9 B, lanes 1 and 2).
Finally, we examined the effect of pXBP1(U) coexpression on expression of pXBP1(S) target genes. Total RNA was extracted from cells transfected as in Fig. 9 A, and subjected to Northern blotting. Accumulation of BiP mRNA was enhanced in cells expressing pXBP1(S) (Fig. 9 C, lanes 1 and 2). In contrast, this induction was abolished by coexpression of pXBP1(U) (Fig. 9 C, lane 3), whereas it was not affected by coexpression of pXBP1(U)-[1–185] (Fig. 9 C, lane 4). These results strongly suggested that pXBP1(U) functions as a negative feedback regulator of pXBP1(S).
Discussion
We revealed that pXBP1(U) translated from the unspliced pre-mRNA represents a functional protein with an important role in mammalian ER stress response. One interesting characteristic of pXBP1(U) is that it contains three distinct subcellular localization determinants, which are the NLS, NES, and NES attenuator (Fig. 3 B). The functional balance among these signals appears to determine the subcellular location of XBP1 proteins under diverse physiological and environmental conditions. pXBP1(U), which contains all of these signals, can shuttle back and forth between the nucleus and the cytoplasm (Fig. 3 A). pXBP1(S) contains only the NLS and is located exclusively in the nucleus (Fig. 3 A). Interestingly, the pXBP1(S)–pXBP1(U) complex was localized to the cytoplasm (Fig. 9). Conformational changes evoked by the formation of this complex might cancel the NES attenuator activity. Although the molecular mechanism behind the attenuation remains unknown, it is possible that the attenuator sterically prevents NES receptors such as CRM1 from approaching the NES. The XBP1(U)-[186–261] region contains two highly conserved domains, a conventional NES motif ([186–208]), and a SWKPLMN motif at the very end (Fig. 5 B), and the latter might be important for the attenuation or for degradation by the proteasome.
pXBP1(U) also contains a “degradation domain” in the [209–261] region that makes it more susceptible to degradation by the proteasome. Because pXBP1-[1–208], which lacks the degradation domain and is exclusively localized to the cytoplasm, is clearly more stable than pXBP1(U) (Fig. 6 A), the cytoplasmic localization by itself is not sufficient for rapid degradation. Although pXBP1(S) does not contain the degradation domain, it becomes less stable when it forms a complex with pXBP1(U), possibly because the degradation domain of pXBP1(U) is presented to the degradation machinery.
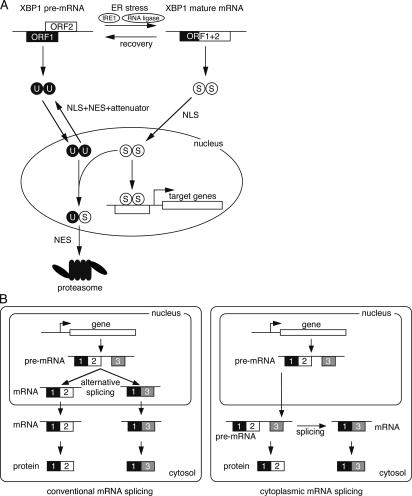
Taking our findings into account, as well as the fact that pXBP1(U) expression is significantly induced during recovery from ER stress, we propose a working model of pXBP1(U) function in mammalian ER stress response (Fig. 10 A). In the absence of ER stress, pXBP1(U) protein is translated from XBP1 pre-mRNA and shuttles between the cytoplasm and the nucleus to monitor the level of pXBP1(S), and possibly that of inopportune expression of pATF6(N) as well because pXBP1(U) can form a heterooligomer with pATF6(N) (Newman and Keating, 2003). Upon ER stress, activated IRE1 splices XBP1 pre-mRNA into mature mRNA, from which pXBP1(S) is translated. pXBP1(S) is then translocated into the nucleus, and activates transcription of targets such as ER chaperones and ERAD components. When enough ER chaperones and ERAD components are produced and ER stress has subsided, IRE1 becomes inactive, but a certain level of pXBP1(S) remains in the nucleus, leading to further transcription of XBP1 pre-mRNA and production of pXBP1(U). pXBP1(U) shuttles between the cytoplasm and the nucleus and forms a complex with pXBP1(S) left over in the nucleus. The pXBP1(S)–pXBP1(U) complex that may expose NES is efficiently exported from the nucleus to the cytoplasm and degraded by the proteasome by virtue of the degradation motif in pXBP1(U), resulting in a complete shutoff of transcription of the target genes. Thus, it seems very likely that pXBP1(U) functions as a negative feed-back regulator specific to pXBP1(S) (and possibly to pATF6(N) as well). Such a shutoff mechanism would clearly be beneficial for cell survival because constitutive activation of the ER stress response would be harmful and is known to retard cell growth in yeast (Cox and Walter, 1996; Kawahara et al., 1997).
Figure 10.
Models. (A) The model for pXBP1(U) function in mammalian ER stress response. pXBP1(U) and pXBP1(S) are depicted as U and S, respectively. (B) Comparison between conventional and cytoplasmic splicing.
To our knowledge, this study identified the first case in which a functional protein was translated from pre-mRNA. The findings suggest that cytoplasmic mRNA splicing is a very sophisticated mechanism of gene regulation as compared with conventional nuclear mRNA splicing, which is catalyzed by the spliceosome (Fig. 10 B). In conventional splicing, which occurs in the nucleus, it would be difficult to resplice mRNA if it has already been exported to the cytoplasm. Thus, it would be necessary to transcribe and splice the primary transcript de novo to accommodate possible change in amino acid sequence of product protein translated from the mRNA. In contrast, in the case of cytoplasmic splicing, pre-mRNA that is transported to the cytoplasm and used for translation can be respliced when necessary to change the structure of product protein translated from the mRNA, in response to extracellular or intracellular signals. Thus, cytoplasmic splicing would be a very rapid, versatile, and energy-efficient mechanism with minimum garbage, as compared with conventional mRNA splicing. The ER stress response may require such a sophisticated system to effectively exploit the interplay between pXBP1(U) and pXBP1(S) to deal with malicious unfolded proteins accumulated in the ER. Moreover, it is conceivable that other systems or cellular processes, such as development or antiviral response, adopt similar mechanisms in which cytoplasmic splicing plays an essential role. Identification of such systems would greatly increase our understanding of cellular response mechanisms to cope with various physiological and pathological situations.
Materials and methods
Cell culture
HeLa cells were grown in DME/glucose at 4.5 g/liter supplemented with 10% fetal calf serum, 2 mM glutamine and antibiotics, 100 U/ml penicillin/100 μg/ml streptomycin. Cells were maintained at 37°C in a humidified 5% CO2/95% air atmosphere.
Transient transfection of cultured cells
Transient transfection was performed by the standard calcium phosphate method (Sambrook et al., 1989; Yoshida et al., 2001a). In brief, HeLa cells cultured in 60-mm dishes were incubated with precipitates of calcium phosphate, including plasmids, for 6 h at 37°C. After washing with PBS to remove calcium phosphate precipitates, cells were incubated in fresh medium for 24 h and harvested for analysis. For subcellular fractionation, transfected cells were suspended in 100 μl of ice-cold PBS containing protease inhibitors and 5% NP-40 and separated into the nuclear and postnuclear fractions by centrifugation at 14,000 rpm for 1 min. For turnover analysis, transfected cells were treated with 40 μM cycloheximide for the indicated period and subjected to immunoblotting. LMB was provided by M. Yoshida (Institute of Physical and Chemical Research, Saitama, Japan).
Construction of plasmids
To construct pCMV-HA-pXBP1(U) and pCMV-HA-pXBP1(S), 1,787- and 1,761-bp fragments, respectively, of XBP1 cDNA encoding pXBP1(U) and pXBP1(S) were cloned into an XhoI site of pCMV-HA vector (CLONTECH Laboratories, Inc.). Expression plasmids for a series of pXBP1(U) deletion mutants fused with HA tag were made by ligating the PCR product of the corresponding region with pCMV-HA. pCMV-HA-pATF6α(N) was constructed by inserting a cDNA encoding the [1–373] region of human ATF6α into BglII–XhoI sites of pCMV-HA vector. For construction of pCMV-HA-pXBP1-[165–261]-pATF6α(N), cDNA encoding the [165–261] region of human pXBP1(U) was cloned into a BglII site of pCMV-HA-pATF6α(N). pcDNA-His-pXBP1(S) was made by inserting pXBP1(S) cDNA into an XhoI site of pcDNA3.1-His vector (Promega).
Immunoblotting
Cells grown in a 60-mm culture dish were harvested with a rubber policeman and pelleted by centrifugation. The pellet was suspended in 20 μl of ice-cold PBS containing protease inhibitors (100 μM 4-(2-Aminoethyl)benzenesulphonyl fluoride, 80 μM aprotinin, 1.5 μM E-64, 2 μM leupeptin, 5 μM bestatin, and 1 μM pepstatin A), mixed with 20 μl of 4× SDS sample buffer (200 mM Tris-Cl, pH 6.8, 400 mM DTT, 8% SDS, and 40% glycerol), and immediately boiled at 100°C. 10-μl portions of samples were subjected to SDS-PAGE using 4–20% gradient gels, transferred onto a Hybond-P filter (GE Healthcare), and incubated with various antisera according to the standard protocol (Sambrook et al., 1989). Anti–XBP1-A detects both pXBP1(U) and pXBP1(S), whereas anti–XBP1-C detects only pXBP1(S) (Yoshida et al., 2001a). An ECL Western blotting detection kit (GE Healthcare) and lumino-image analyzer (model LAS-3000; Fuji) were used to detect antigens.
Northern blot hybridization analysis
Total RNA extracted with guanidine-phenol was separated by electrophoresis with 2% agarose gel containing 2.2 M formaldehyde, blotted onto a Hybond-N+ filter (GE Healthcare), hybridized with alkaline phosphatase-conjugated cDNA probes, and detected with a LAS-3000 using the Gene Images AlkPhos direct labeling and detection system (GE Healthcare).
RT-PCR
RT-PCR of XBP1 mRNA was performed essentially as described previously (Yoshida et al., 2001a). In brief, 10 μg of total RNA was reverse-transcribed with M-MLV reverse transcriptase (Invitrogen) and amplified with Ex Taq polymerase (TaKaRa) using a pair of primers that correspond to nt 493–512 (CGCGGATCCGAATGAAGTGAGGCCAGTGG) and 834–853 (GGGGCTTGGTATATATGTGG) of XBP1 mRNA, respectively (Fig. 2 A). Amplified fragments covering a 26-nt intron (nt 531–556) and flanking exon fragments were digested with PstI (a unique PstI site existed at nt 556) and separated on 4–20% polyacrylamide gels. cDNA was visualized by staining with SYBR Gold (Invitrogen) and detected by a Fluor-image analyzer (model FLA-3000; Fuji).
Immunocytochemistry
HeLa cells grown on coverslips were transiently transfected with appropriate expression plasmids by the calcium phosphate method. Cells were fixed with 2% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100 for 10 min, and stained with the appropriate antisera. Coverslips were mounted with 90% glycerol/10% PBS containing 100 ng/ml DAPI. Images were acquired using a microscope (model TE2000; Nikon) and a digital charge-coupled device camera (ORCA-ER; Hamamatsu Photonics).
Pull-down assay
For in vivo pull-down assays, HeLa cells were cotransfected with expression plasmids for His-pXBP1(S) and pXBP1(U), harvested by rubber policeman, lysed in binding buffer (20 mM Hepes, pH 7.9, 100 mM KCl, 10% glycerol, 1 mM MgCl2, 1 mM mercaptoethanol, 0.1% Tween-20, and 20 mM imidazole), and centrifuged. Supernatants were mixed with Ni-NTA agarose in binding buffer containing protease inhibitors for 1 h at 4°C, washed with binding buffer three times, and then suspended in 1× SDS sample buffer. Samples were boiled at 100°C and subjected to 4–20% SDS-PAGE. pXBP1(U) coprecipitated with His-pXBP1(S) was detected by immunoblotting with anti–XBP1-A antiserum. For in vitro pull-down assays, both His-pXBP1(S) protein and pXBP1(U) protein were cotranslated using TNT Quick Coupled Transcription/Translation systems (Promega), incubated with Ni-NTA agarose for 1 h at 4°C, and processed like the HeLa cells.
Acknowledgments
We appreciate the Radioisotope Research Center of Kyoto University. We also thank Drs. Takashi Yura and Tohru Yoshihisa for critical reading of the manuscript and Ms. Kaoru Miyagawa for technical-secretarial assistance.
This work was supported by the Precursory Research for Embryonic Science and Technology-Solution Oriented Research for Science and Technology program of the Japan Science and Technology Agency and grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (17370061 and 17026022 to H. Yoshida).
Abbreviations used in this paper: ATF, activating transcription factor; EDEM, ER degradation–enhancing mannosidase-like protein; ERAD, ER-associated protein degradation; ERSE, ER stress response element; HEK, human embryonic kidney; IRE, inositol requiring enzyme; LMB, leptomycin B; NES, nuclear exclusion signal; p, protein; PERK, PKR-like ER-resistant kinase; S, spliced; U, unspliced; XBP1, X-box binding protein 1.
References
- Calfon, M., H. Zeng, F. Urano, J.H. Till, S.R. Hubbard, H.P. Harding, S.G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. [DOI] [PubMed] [Google Scholar]
- Chapman, R.E., and P. Walter. 1997. Translational attenuation mediated by an mRNA intron. Curr. Biol. 7:850–859. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 87:391–404. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., C.E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73:1197–1206. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., M. Ohno, M. Yoshida, and I.W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 90:1051–1060. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 397:271–274. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., I.I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108. [DOI] [PubMed] [Google Scholar]
- Haze, K., T. Okada, H. Yoshida, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 355:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki, T., A. Hosoda, T. Okuda, Y. Kamigori, C. Nomura-Furuwatari, Y. Kimata, A. Tsuru, and K. Kohno. 2001. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 3:158–164. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211–1233. [DOI] [PubMed] [Google Scholar]
- Kawahara, T., H. Yanagi, T. Yura, and K. Mori. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell. 8:1845–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour, T., R. Gupta, K. Rapacki, K. Skriver, F.M. Poulsen, and S. Brunak. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 31:393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A.H., N.N. Iwakoshi, K.C. Anderson, and L.H. Glimcher. 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA. 100:9946–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 101:451–454. [DOI] [PubMed] [Google Scholar]
- Mori, K., W. Ma, M.J. Gething, and J. Sambrook. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 74:743–756. [DOI] [PubMed] [Google Scholar]
- Mori, K., T. Kawahara, H. Yoshida, H. Yanagi, and T. Yura. 1996. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic- leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1:803–817. [DOI] [PubMed] [Google Scholar]
- Newman, J.R., and A.E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 300:2097–2101. [DOI] [PubMed] [Google Scholar]
- Niwa, M., C. Sidrauski, R.J. Kaufman, and P. Walter. 1999. A role for presenilin-1 in nuclear accumulation of Ire1 fragments and induction of the mammalian unfolded protein response. Cell. 99:691–702. [DOI] [PubMed] [Google Scholar]
- Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349–355. [DOI] [PubMed] [Google Scholar]
- Ruegsegger, U., J.H. Leber, and P. Walter. 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 107:103–114. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Second Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Tirasophon, W., A.A. Welihinda, and R.J. Kaufman. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12:1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano, F., A. Bertolotti, and D. Ron. 2000. IRE1 and efferent signaling from the endoplasmic reticulum. J. Cell Sci. 113:3697–3702. [DOI] [PubMed] [Google Scholar]
- Wang, X.Z., H.P. Harding, Y. Zhang, E.M. Jolicoeur, M. Kuroda, and D. Ron. 1998. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17:5708–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J., R.B. Rawson, R. Komuro, X. Chen, U.P. Dave, R. Prywes, M.S. Brown, and J.L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 6:1355–1364. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., K. Haze, H. Yanagi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273:33741–33749. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. a. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. b. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol. Cell. Biol. 21:1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., T. Matsui, N. Hosokawa, R.J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 4:265–271. [DOI] [PubMed] [Google Scholar]