Abstract
Focal adhesions are specialized regions of the cell surface where integrin receptors and associated proteins link the extracellular matrix to the actin cytoskeleton. To define the cellular role of the focal adhesion protein zyxin, we characterized the phenotype of fibroblasts in which the zyxin gene was deleted by homologous recombination. Zyxin-null fibroblasts display enhanced integrin-dependent adhesion and are more migratory than wild-type fibroblasts, displaying reduced dependence on extracellular matrix cues. We identified differences in the profiles of 75- and 80-kD tyrosine-phosphorylated proteins in the zyxin-null cells. Tandem array mass spectrometry identified both modified proteins as isoforms of the actomyosin regulator caldesmon, a protein known to influence contractility, stress fiber formation, and motility. Zyxin-null fibroblasts also show deficits in actin stress fiber remodeling and exhibit changes in the molecular composition of focal adhesions, most notably by severely reduced accumulation of Ena/VASP proteins. We postulate that zyxin cooperates with Ena/VASP proteins and caldesmon to influence integrin-dependent cell motility and actin stress fiber remodeling.
Introduction
Cell locomotion is critical for normal embryonic development, cell-mediated immunity, and wound healing, as well as for pathological conditions such as inflammation and tumor metastasis. The process of cell motility depends on the ability of a cell to extend a leading edge in a polarized fashion and to adhere to an underlying substratum. Integrin receptors for extracellular matrix are concentrated at sites of substratum attachment, providing the structural support necessary for lamellar protrusion. Recent work has revealed that substratum adhesion sites display functional heterogeneity, with the small adhesions near the leading edge of a cell representing the sites of greatest mechanical force transmission to the substratum (Beningo et al., 2001). Heterogeneity in the molecular composition of adhesion sites is likely to account for their functional diversity (Zamir et al., 1999; Katz et al., 2000).
In mammalian cells, one component of substratum adhesions that is associated with mature focal adhesions, but not with Rac-induced nascent adhesions that are concentrated at the leading edge, is zyxin (Zaidel-Bar et al., 2003). Zyxin is a LIM domain protein that has been implicated in the regulation of actin assembly, cell division control, and nuclear–cytoplasmic communication (Nix and Beckerle, 1997; Beckerle, 1998; Hirota et al., 2000). A role for zyxin in actin assembly was proposed when it was recognized that a series of proline-rich repeats in the ActA protein of the intracellular pathogen Listeria monocytogenes are also present in zyxin (Golsteyn et al., 1997; Niebuhr et al., 1997). The FPPPP sequences present in L. monocytogenes ActA stimulate actin assembly on the pathogen's surface and promote bacterial motility within the cytoplasm of a eukaryotic host. A common mechanism by which zyxin and ActA are thought to promote actin polymerization involves recruitment of Ena/VASP proteins, which are proposed to act by recruiting profilin-actin and by controlling the access of capping protein to the barbed ends of actin filaments (Fradelizi et al., 2001; Bear et al., 2002).
Imaging of zyxin in living cells has revealed that zyxin accumulation at adhesion sites is highly dynamic. Zyxin levels are low near the leading edge of a migrating cell, where high tension and propulsive forces are generated (Beningo et al., 2001). Moreover, the accumulation of zyxin at substratum adhesion sites is correlated with cessation of membrane protrusion (Zaidel-Bar et al., 2003) and with a dramatic reduction in traction forces (Beningo et al., 2001). Because of these intriguing observations that suggest a link between the presence of zyxin and the properties of substratum adhesion sites, we sought to assess the consequences of eliminating zyxin function for cell adhesion, motility, and actin organization.
Results
Isolation and characterization of zyxin-null fibroblasts
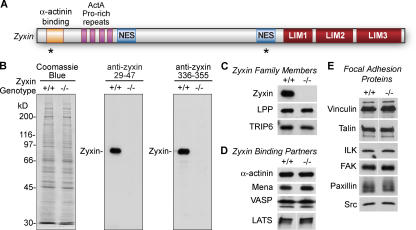
Murine zyxin is a 564-aa LIM protein (Fig. 1 A) whose binding partner repertoire had suggested a role in actin assembly, cell cycle control, cell motility, and cell signaling. To directly assess the contributions of zyxin to cell function, we have isolated fibroblasts from mice in which the zyxin gene is disrupted by homologous recombination (Hoffman et al., 2003). Neither full-length nor truncated zyxin protein is present in the zyxin−/− cells (Fig. 1 B). Loss of gene function can lead to compensatory up-regulation of other family members. Two proteins that are closely related to zyxin, the lipoma preferred partner (LPP; Petit et al., 2000) and thyroid receptor-interacting protein 6 (TRIP6; Yi and Beckerle, 1998), are also expressed in fibroblasts. We detected no alteration in the level of expression of either LPP or TRIP6 in the zyxin−/− cells (Fig. 1 C), which is consistent with the view that cells do not compensate for the elimination of zyxin by modulating the expression of these highly related proteins.
Figure 1.
Immunoblot analysis of zyxin-null fibroblasts. (A) Domains of zyxin include the α-actinin binding site, four ActA Ena/VASP binding repeats, two nuclear export signals (NES), and three LIM domains. Sites of epitopes for zyxin-specific antibodies 1D1 (zyxin aa 29–47) and B71 (zyxin aa 336–355) are identified by an asterisk. (B) Cell lysates of wild-type (+/+) and zyxin-null (−/−) fibroblasts were electrophoresed (10% SDS-PAGE) and either stained to confirm equal protein loading (Coomassie blue) or transferred to nitrocellulose for immunoblot analysis with anti-zyxin aa 29–47 (1D1) and anti-zyxin aa 336–355 (B71). (C–E) Immunoblot analysis of wild-type and zyxin−/− cells for expression of zyxin family members (C), zyxin-binding partners (D), and other focal adhesion proteins (E).
Some focal adhesion constituents depend on the presence of their binding partners for their stability (Fukuda et al., 2003). Therefore, we examined whether the levels of zyxin-binding partners were compromised when zyxin was absent and detected no differences in the levels of several zyxin-binding partners (Fig. 1 D). Likewise, other focal adhesion proteins including vinculin, talin, integrin-linked kinase (ILK), FAK, paxillin, and src were detected at similar levels in wild-type and zyxin−/− cells (Fig. 1 E).
Elimination of zyxin does not alter mitotic progression
Zyxin displays mitosis-dependent phosphorylation and has been shown to interact directly with the h-warts/LATS1 tumor suppressor, a serine/threonine kinase implicated in cell cycle control (Hirota et al., 2000). Disturbance of the LATS1–zyxin interaction by use of a dominant-interfering LATS1 truncation product (LATS1 aa 136–700) was shown to interfere with zyxin localization during mitosis and delay mitotic progression, leading to the suggestion that LATS1-dependent recruitment of zyxin to the mitotic apparatus is required for regulated exit from mitosis (Hirota et al., 2000).
With the availability of zyxin−/− cells, we were able to test directly whether zyxin is essential for normal mitotic progression. We detected no change in LATS1 protein levels in the zyxin−/− cells (Fig. 1 C); therefore, zyxin is not essential for LATS1 stability. To look for specific changes in the cell cycle caused by loss of zyxin, flow cytometric analysis of cell cycle progression in exponentially growing wild-type and zyxin−/− cells was used (Fig. 2 A). No statistically significant differences in cell cycle progression (Fig. 2 B) or the percentage of binucleate cells (not depicted) were observed. These results illustrate that, at least in fibroblasts, mitotic progression and cytokinesis can proceed normally in the absence of zyxin.
Figure 2.
The cell cycle is not altered in zyxin-null cells. (A) Flow cytometry profiles of the propidium-iodide DNA content per cell (2 and 4 n) for wild-type (+/+) and zyxin-null (−/−) fibroblasts. (B) Graph of cell cycle distribution of wild-type and zyxin−/− fibroblasts modeled as diploid populations. Data represent the mean ± SEM. n = 4.
Zyxin-null cells display enhanced migration
Many of zyxin's binding partners and biochemical activities suggest that it may play a role in integrin- and actin-linked processes. Therefore, we focused the balance of our analysis on the possible role of zyxin in motility, adhesion, and cytoarchitecture. To determine if loss of zyxin altered cell motility, the behavior of zyxin−/− cells was evaluated using a monolayer wound assay. In blind studies, zyxin−/− fibroblasts were consistently observed to reach the midline of the wound sooner than the wild-type cells (Fig. 3 A). Quantitative analysis revealed that the zyxin−/− cells displayed a mean migration velocity of 32 μm/h compared with the wild-type migration rate of 18 μm/h (Fig. 3 B). The enhanced migration of zyxin−/− fibroblasts suggests that zyxin acts as a negative regulator of cell motility.
Figure 3.
Monolayer wound assays demonstrate enhanced migration of fibroblasts lacking zyxin. A wound was scraped across monolayer wild-type and zyxin-null fibroblasts, and migration was monitored by time-lapse microscopy. (A) Time points (0, 4, 8, and 12 h) of wild-type fibroblasts (top) and zyxin−/− fibroblasts (bottom). (B) The results were examined in a blind fashion and converted into rate measurements (the distance to the midline of the wound over the time it took for cells to reach the midline; mean ± SEM; n = 4). Primary MEFs were used for wound assays. **, P < 0.005.
Zyxin-null cells appear primed for migration, independent of matrix cues
Integrin-dependent adhesion is essential for communicating intracellular contractility to the underlying extracellular matrix and is necessary to drive motility. To explore the contribution of zyxin to cell motility in greater detail, we examined the haptotactic migration of wild-type and zyxin−/− cells toward a variety of integrin ligands using a Boyden chamber transwell migration assay (Fig. 4 A). We observed a striking difference in haptotactic motility when comparing wild-type and zyxin−/− cells. Except in the cases of 20 μg/ml fibronectin or vitronectin, the zyxin-null cells displayed statistically significant enhancement of migration, relative to wild-type cells.
Figure 4.
Boyden chamber transwell assays demonstrate enhanced migration of zyxin-null fibroblasts. (A) Transwell migration (6 h) toward culture medium with 10% FBS on BSA or 20 μg/ml of collagen type I (Col I), collagen type IV (Col IV), fibronectin (Fib), laminin (Lam), superfibronectin (Sfib), or vitronectin (Vit). Data represent the mean ± SEM. n = 4. (B) Wild-type and zyxin-null cell migration with increasing fibronectin concentration (n = 4). (C) Transwell Matrigel invasion assays (24 h) of wild-type and zyxin-null fibroblasts, and zyxin-null cells expressing a GFP-zyxin transgene. (D) GFP-zyxin expressed in zyxin−/− fibroblasts localized properly to focal adhesions. (E) Immunoblot analysis of zyxin and GFP-zyxin. Immortalized cell lines were used for transwell migration assays. *, P < 0.05; **, P < 0.005.
Matrix proteins have differential, concentration-dependent capacities to stimulate cell migration. As can be seen in Fig. 4 B, migration of wild-type fibroblasts exhibited sensitivity to fibronectin concentration, whereas zyxin-null fibroblasts migrated efficiently independent of fibronectin concentration. The migration of the zyxin-null cells in the absence of matrix cues was statistically indistinguishable from the maximal migration of wild-type cells toward fibronectin. Zyxin-null cells also displayed enhanced migratory potential relative to their wild-type counterparts in a Matrigel invasion assay (Fig. 4 C). Introduction of a GFP-tagged version of zyxin into the null cells resulted in suppression of cell migration (Fig. 4 C). GFP-zyxin localized as expected at focal adhesions in the zyxin−/− cells (Fig. 4 D) and was expressed at levels approximating that of zyxin in wild-type cells (Fig. 4 E). GFP, which did not affect motility, displayed diffuse cellular localization (not depicted). The successful rescue of the null cells by reintroduction of a zyxin transgene illustrates that the migration phenotype observed for the zyxin−/− cells is directly attributable to loss of zyxin.
Zyxin-null cells display enhanced adhesion without any increase in integrin expression
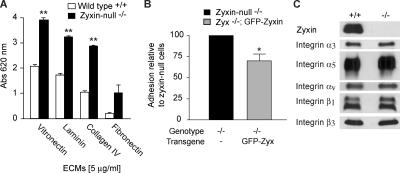
We next tested whether altered cell–substratum adhesion might account for the enhanced motility of the zyxin−/− cells. Zyxin-null fibroblasts were more adherent than wild-type fibroblasts when plated on a variety of extracellular matrix proteins (Fig. 5 A), and reexpression of GFP-zyxin in null cells resulted in reduced adhesion (Fig. 5 B). One possible explanation for the enhanced cell adhesion observed in the zyxin−/− cells could be elevated integrin expression. However, immunoblot analysis revealed no difference in integrin levels or subunit expression (Fig. 5 C).
Figure 5.
Adhesion to ECM proteins is enhanced in zyxin-null fibroblasts, but integrin expression is unchanged. (A) Wild-type and zyxin-null fibroblasts were evaluated for adhesion to vitronectin, laminin, collagen IV, and fibronectin (5 μg/ml; 25 min). After removal of unattached cells, the adherent cells were fixed, stained with toluidine blue, and quantitated by absorbance at 620 nm. Data represent the mean ± SEM. n = 4. (B) GFP-zyxin reintroduced into zyxin−/− cells decreased the adhesion to laminin, relative to zyxin−/− cells. (C) Immunoblot analysis of wild-type and zyxin−/− fibroblasts for integrins α3, α5, αV, β1 and β3. Immortalized cell lines were used for adhesion assays. *, P < 0.05; **, P < 0.005.
Distinct profile of tyrosine-phosphorylated proteins in zyxin-null fibroblasts
The behavior of zyxin-null fibroblasts on integrin ligands suggested that cells lacking zyxin might somehow be primed for integrin-dependent adhesion and migration. One molecular hallmark of the integrin activation state is enhanced tyrosine phosphorylation of proteins such as FAK, which is a tyrosine kinase that colocalizes with zyxin at focal adhesions. Therefore, we compared the subcellular distributions and molecular mass profiles of tyrosine-phosphorylated proteins in wild-type and zyxin-null cells. Both wild-type and zyxin-null cells displayed phosphotyrosine (pY) associated with focal adhesions (Fig. 6, A and B), and Western immunoblot analysis of pY-containing proteins revealed no apparent difference in the region where FAK migrates (Fig. 6 C). However, the wild-type cells displayed a 75-kD tyrosine-phosphorylated protein (pY75) that showed reduced abundance in zyxin-null cells. In contrast, we observed an 80-kD tyrosine-phosphorylated protein (pY80) in the zyxin-null cells that was not evident in the wild-type cells. Using two-dimensional (2D) gel electrophoresis of wild-type and zyxin-null cell lysates, followed by Western immunoblot with antiphosphotyrosine antibody, both pY75 and pY80 were resolved as a cluster of isoelectric point variants ranging from ∼6.5 to 8.0 (Fig. 6 D).
Figure 6.
An altered tyrosine-phosphorylated protein in zyxin-null cells is identified as the actomyosin regulator caldesmon. Indirect immunofluorescence of pY-containing proteins in wild-type (A) and zyxin-null (B) fibroblasts. (C) Immunoblot analysis of equivalent amounts of protein from wild-type and zyxin−/− cells using pY-specific antibody 4G10 detected a 75-kD signal (arrowheads) in wild-type cells, and an 80-kD signal (arrowhead) in zyxin−/− cells. (D) 2D gel electrophoresis and anti-pY immunoblot analysis identified a slower mobility series of pY spots from zyxin-null fibroblasts compared with wild-type fibroblasts. (E) Wild-type and zyxin-null pY-positive spots of interest were excised from 2D gels, partially digested with trypsin, and then subjected to LC-MS/MS for protein sequence analysis. Both sets of spots contained multiple peptide sequences for the actomyosin regulator caldesmon, which are shown aligned with l-caldesmon coding exons1′–13. Exons 3b and 4, which would be found in h-caldesmon, are shown above the l-caldesmon structure. (F) Immunoblot for caldesmon in wild-type and zyxin−/− cell lysates separated by 10% SDS-PAGE. Short exposure (left) showed nonmuscle caldesmon (l-caldesmon) migrating between 75–80 kD and longer exposure (right) identified the 140-kD smooth-muscle caldesmon (h-caldesmon). (G) Tissue extracts (lung, bladder, and spleen) from wild-type (+/+), zyxin-heterozygous (+/−), and zyxin-null (−/−) mice were electrophoresed, transferred to nitrocellulose, and probed for zyxin (B71 rabbit anti-serum; top) and caldesmon (bottom).
The actomyosin regulator caldesmon is altered in zyxin-null cells
Mass spectrometric (LC-MS/MS) sequence analysis of the protein found four pY75 2D gel spots derived from wild-type cells and four pY80 2D gel spots derived from zyxin-null cells, such as those in Fig. 6 D, and identified all eight tyrosine-phosphorylated spots as caldesmon (Fig. 6 E). Caldesmon is an actomyosin regulator that influences stress fiber formation, cell contractility, and cell motility (Wang, 2001). There are two major isoforms of caldesmon derived by alternative splicing (Fig. 6 E): nonmuscle or low molecular weight caldesmon (l-caldesmon) and smooth muscle or high molecular weight caldesmon (h-caldesmon; Wang, 2001). Our sequence results are consistent with recovery of l-caldesmon. With the exception of sequence encoded by exon 1′, which was only obtained for pY75 isolated from wild-type cells, sequence from all other classical l-caldesmon exons (Hayashi et al., 1992; Guo and Wang, 2005) were represented and were nearly identical for pY75 and pY80 (Fig. 6 E).
In wild-type mouse fibroblasts, a caldesmon-specific antibody detected a prominent 75-kD immunoreactive band, as well as a minor 80-kD immunoreactive band, which is consistent with l-caldesmon, whereas zyxin-null fibroblasts displayed only the slower migrating 80-kD species (Fig. 6 F). A similar quantitative shift to a slower mobility species occurred for the 140-kD h-caldesmon in the zyxin-null fibroblasts, as seen in a longer exposure (Fig. 6 F, right). This shift in caldesmon mobility was detected in multiple zyxin-null primary fibroblast isolates, as well as in immortalized zyxin-null fibroblast lines (unpublished data). Moreover, the caldesmon mobility shift was observed in cells derived from zyxin knock-out mice generated from two independently targeted embryonic stem cell lines (not depicted) and in a variety of tissues, including lung, bladder, and spleen, which were isolated directly from the zyxin-null mice (Fig. 6 G). The slower migrating caldesmon isoform is evident only in tissues derived from homozygous mutant animals, suggesting that complete loss of zyxin is required to promote the change in caldesmon.
Posttranslational modification does not account for the caldesmon isoform variation observed in the zyxin-null cells
Phosphorylation can affect a protein's electrophoretic mobility and, in the case of caldesmon, is known to regulate its ability to control contractility and integrin-dependent motility (Wang, 2001). We tested directly whether l-caldesmon from wild-type and zyxin-null cells was differentially phosphorylated on Ser497 and Ser527, two ERK-dependent phosphorylation sites implicated in regulation of caldesmon function (D'Angelo et al., 1999). Both the 75- and 80-kD caldesmon species were recognized with the phosphospecific antibody (Fig. 7 A). For a more global assessment of whether differential phosphorylation might be responsible for the caldesmon mobility difference we have observed, we treated cell lysates from wild-type and zyxin-null cells with phosphatase and assayed caldesmon and control paxillin protein mobility by SDS-PAGE (Fig. 7 B). Although the phosphatase treatment resulted in the collapse of paxillin bands indicative of dephosphorylation, no impact on caldesmon mobility was observed, suggesting that phosphorylation is unlikely to account for the altered caldesmon mobility seen in the zyxin-null cells. To directly test whether caldesmon is subject to differential posttranslational modification that affects its mobility, we transfected wild-type and zyxin-null cells with a his-caldesmon cDNA expression construct and monitored mobility of the expressed protein with Western immunoblot. The mobility of the his-tagged caldesmon was indistinguishable in wild-type and zyxin-null cells, whereas the endogenous caldesmon exhibited the mobility shift (Fig. 7 C). Collectively, our results suggest that differential posttranslational modification of caldesmon is not likely to be responsible for the altered caldesmon mobility we observe in the zyxin-null cells. Re-expression of zyxin does not restore caldesmon to its wild-type mobility (Fig. 7 D), which illustrates that the phenotypes observed in the zyxin-null cells are linked to loss of zyxin and are not attributable to alteration of caldesmon.
Figure 7.
Caldesmon protein in wild-type and zyxin-null fibroblasts. Fibroblasts were grown for 24 h in high (10% FBS) and low (0.5% FBS) serum, and then collected for immunoblot analysis. (A) Phospho-ser527/789-specific antibody detected caldesmon from both wild-type and zyxin-null cells, with a greater signal in high serum conditions (as expected for the phosphorylation-specific antibody). The caldesmon-specific immunoblot confirmed approximately equivalent levels of caldesmon in the lysates. (B) Incubation of fibroblast lysates with lambda-phosphatase followed by immunoblot analysis for caldesmon and paxillin. (C) His-tagged mouse l-caldesmon was expressed in wild-type and zyxin-null fibroblasts, and then evaluated by immunoblot analysis using his-specific and caldesmon-specific antibodies. (D) Immunoblot analysis of wild-type, zyxin-null, and zyxin-null cells expressing GFP-zyxin, with caldesmon- and zyxin-specific antibodies.
Depletion of Mena and VASP from focal adhesions of zyxin-null cells
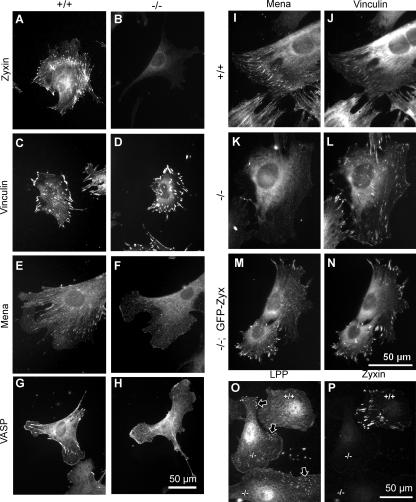
Zyxin is concentrated in the focal adhesions of wild-type fibroblasts (Fig. 8 A); no immunoreactivity is detected in zyxin-null cells (Fig. 8 B). Elimination of zyxin did not lead to changes in focal adhesion morphology or number, as assessed by qualitative inspection of vinculin localization (Fig. 8, C and D). The distributions of several other focal adhesion markers, including FAK and paxillin, also appeared unperturbed in the zyxin-null cells (unpublished data). In contrast, we detected a dramatic diminution in the localization of the zyxin-binding partners, Mena and VASP, at focal adhesions (Fig. 8, E–H; and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200512115/DC1) even though their cellular levels were unchanged (Fig. 1 C). In double-labeling experiments, it was evident that Mena colocalized with vinculin at focal adhesions of wild-type cells (Fig. 8, I and J) and was depleted from vinculin-rich focal adhesions in zyxin-null cells (Fig. 8, K and L). Thus, the altered Mena localization was not attributable to loss of focal adhesion structures. Expression of GFP-zyxin in the null cells restored localization of Mena to vinculin-rich focal adhesions (Fig. 8, M and N). Similar results were obtained for VASP (unpublished data). These observations are consistent with a major role for zyxin in localizing Ena/VASP family members at focal adhesions. Residual Mena and VASP accumulation at focal adhesions in the zyxin−/− cells likely reflects the presence of other focal adhesion proteins that can dock Ena/VASP family members. For example, both vinculin and LPP are present in focal adhesions and have the capacity to bind Ena/VASP proteins (Brindle et al., 1996; Petit et al., 2000). Interestingly, LPP was detected more prominently at focal adhesions of zyxin-null cells, as compared with cocultured wild-type cells (Fig. 8, O and P).
Figure 8.
Localization of proteins in wild-type and zyxin-null fibroblasts demonstrate the importance of zyxin for proper localization of Mena and VASP. Protein localization in wild-type (+/+) and zyxin-null (−/−) cells was evaluated by indirect immunofluorescence microscopy using antibodies specific for zyxin (A and B; rabbit polyclonal antiserum B71), vinculin (C and D), Mena (E and F), and VASP (G and H). Wild-type cells display colocalized Mena (I) and vinculin (J) at focal adhesions. Mena failed to localize normally to zyxin-null focal adhesions (K) although vinculin persisted (L). Expression of GFP-zyxin in zyxin−/− cells restored the localization of Mena to focal adhesions (M), colocalizing with vinculin (N). To show the enhanced LPP focal adhesion staining (arrows) in fibroblasts lacking zyxin, a mixed population of wild-type (+/+) and zyxin-null (−/−) cells was stained for LPP (O) and for zyxin (P; mouse monoclonal antibody 1D1).
Zyxin-null cells fail to build robust actin stress fibers when stimulated with jasplakinolide
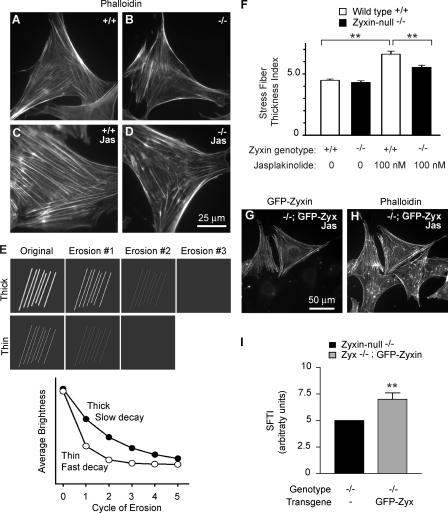
The mislocalization of Ena/VASP proteins and the alteration of caldesmon observed in the zyxin-null cells raised the possibility that actin filament assembly or organization might be disturbed when zyxin function is compromised. Likewise, previous work involving expression of a dominant-negative zyxin deletion fragment (Nix et al., 2001), RNA interference (Harborth et al., 2001), and exposure of cells to mechanical stimuli (Yoshigi et al., 2005) suggested that zyxin might be required for actin cytoskeletal assembly or maintenance. To assess this possibility, we compared the actin cytoskeletons of wild-type and zyxin-null cells using phalloidin staining (Fig. 9, A and B). The presence of actin filament arrays in the zyxin-null fibroblasts illustrated that zyxin was not absolutely essential for establishment and maintenance of actin stress fibers. To explore whether we might detect an effect of eliminating zyxin when cells were challenged to reorganize actin arrays, we compared the response of wild-type and zyxin-null fibroblasts with the membrane-permeable drug jasplakinolide which stabilizes actin filaments and decreases the critical concentration of monomers required for polymerization (Bubb et al., 2000). Jasplakinolide-treated wild-type fibroblasts developed robust actin filament arrays detected by phalloidin staining, but jasplakinolide-treated zyxin-null fibroblasts did not exhibit such robust stress fiber arrays (Fig. 9, C and D). To quantitate a stress fiber thickness index (SFTI), we used an “erosion” spatial filtering approach to calculate decay constants of brightness curves (Fig. 9 E; see Materials and methods). This analysis confirmed that there was no statistically significant difference in phalloidin-stained actin filaments between untreated wild-type and zyxin-null fibroblasts (Fig. 9 F). However, jasplakinolide-induced actin stress fibers in wild-type fibroblasts were thicker than the fibers in zyxin-null fibroblasts (Fig. 9 F). When GFP-zyxin was expressed in the zyxin-null cells, the ability of the cells to generate robust actin filament arrays in response to jasplakinolide was restored. Zyxin-null cells expressing GFP-zyxin (Fig. 9 G) showed more robust actin stress fibers (Fig. 9 H) after exposure to jasplakinolide than adjacent cells that were not expressing the transgene. Rescue of the zyxin-null cells' ability to build robust stress fibers by expression of GFP-zyxin was confirmed by quantitative SFTI analysis (Fig. 9 I).
Figure 9.
Zyxin-null cells fail to augment actin stress fibers in response to jasplakinolide. Wild-type (A) and zyxin-null (B) fibroblasts were treated with vehicle or jasplakinolide and then fixed and stained by phalloidin. Jasplakinolide induced thick actin stress fibers in wild-type cells (C), but noticeably thinner and less robust actin in zyxin−/− cells (D). A method to quantitate an SFTI was developed using a computer algorithm based on erosion filtering (E), as described in Materials and methods. 20 images of phalloidin-stained cells were randomly recorded, and the average of SFTI was compared using analysis of variance between groups. (F) No difference between wild-type and zyxin−/− actin filaments was detected in untreated controls, but a statistically significant difference between jasplakinolide-treated wild-type and zyxin−/− fibroblasts was observed. A population of jasplakinolide-treated zyxin-null fibroblasts, some of which were expressing GFP-zyxin (G), was stained with phalloidin (H). More robust actin stress fibers were observed in the null cells that expressed GFP-zyxin. (I) SFTI analysis confirmed that reintroduced zyxin enhanced the actin filaments in zyxin-null cells. Data represent the mean ± SEM of 33 measurements. **, P < 0.005.
Dynamic regulation of zyxin localization during stress fiber remodeling
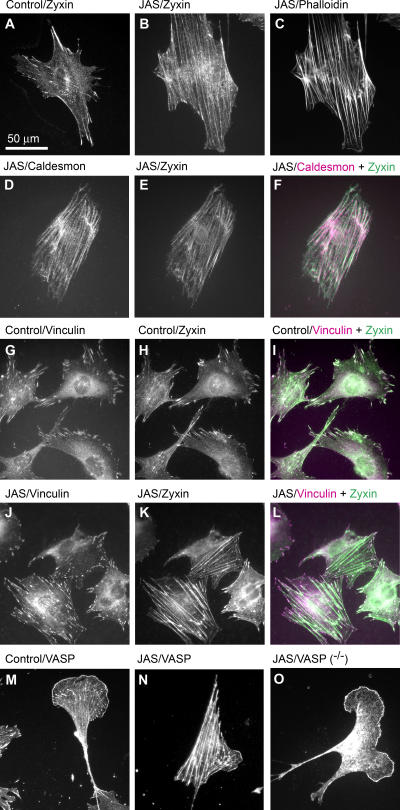
To begin to assess how zyxin might contribute to the cellular response to jasplakinolide, we examined the behavior of zyxin in jasplakinolide-treated cells. As can be seen in Fig. 10 (A and B), zyxin underwent a dramatic redistribution in response to exposure of cells to jasplakinolide. Zyxin was lost from focal adhesions and accumulated along the actin stress fibers (Fig. 10, B and C). Under those conditions, zyxin was colocalized with caldesmon along actin filaments (Fig. 10, D–F). In untreated fibroblasts, zyxin was colocalized with vinculin at focal adhesions (Fig. 10, G–I). In contrast to zyxin, which left focal adhesions in response to jasplakinolide, vinculin was retained at focal adhesions (Fig. 10, J–L), illustrating the specificity of zyxin's redistribution and also illustrating that the focal adhesions were not disassembling in response to jasplakinolide. In contrast with vinculin, which was retained at focal adhesions in cells exposed to jasplakinolide, Ena/VASP proteins redistributed from focal adhesions to actin filaments concomitant with zyxin redistribution (Fig. 10, M and N). This VASP redistribution to actin stress fibers depended on the presence of zyxin (Fig. 10 O). VASP accumulation in lamellar extensions was zyxin independent (Fig. 10 O).
Figure 10.
Jasplakinolide induces zyxin and VASP localization to actin filaments. Wild-type fibroblasts treated with jasplakinolide (JAS) or DMSO (control) were evaluated by indirect immunofluorescence microscopy. Zyxin localization in control fibroblast (A) and in jasplakinolide-treated fibroblast (B). (C) Phalloidin staining of F-actin in jasplakinolide-treated fibroblast. Jasplakinolide-treated fibroblast stained for caldesmon (D) and zyxin (E). (F) Caldesmon (pink) and zyxin (green) merge to white with colocalization. (G) Vinculin at focal adhesions in control fibroblasts colocalizes with zyxin (H) as seen in I (merge to white with colocalization). (J) Vinculin at focal adhesions in jasplakinolide-treated fibroblasts and zyxin (K) along actin filaments. (L) Merge (white) of vinculin (pink) and zyxin (green) signals shows the loss of colocalization upon jasplakinolide treatment. (M) Focal adhesion and ruffle localization of VASP in wild-type fibroblast. (N) VASP localizes along actin stress fibers in a jasplakinolide-treated wild-type fibroblast. (O) In a zyxin-null fibroblast, VASP fails to translocate to actin stress fibers in response to jasplakinolide.
Discussion
Focal adhesions serve as sites of substratum attachment, signal transduction, and actin filament anchorage. We have demonstrated that elimination of the LIM protein zyxin by targeted gene disruption affects cell adhesion, migration, and stress fiber remodeling. Our analysis identifies several molecular alterations in the zyxin-null cells that provide insight into the mechanism by which loss of zyxin affects these processes.
Elimination of zyxin by targeted gene disruption enhances fibroblast adhesion and motility
Zyxin-null cells display enhanced adhesion to integrin ligands and enhanced migration in both wound healing and haptotactic motility assays. Of particular interest, zyxin-null cells behave as if they are insensitive to contextual cues provided by extracellular matrix and, instead, appear primed for adhesion. It is interesting that studies in which zyxin localization and traction force generation in migrating cells were simultaneously analyzed demonstrated that zyxin accumulation at adhesion sites is inversely correlated with traction strength (Beningo et al., 2001). The high traction adhesions with relatively low zyxin accumulation are concentrated near the advancing edge of a cell and represent key regions for driving motility. Our findings are consistent with a model in which zyxin reduces the capacity for generating strong traction forces. In that view, when zyxin is absent, as in our knock-out cells, high affinity integrin attachments that generate strong traction forces may persist, leading to enhanced motility. As we will discuss later in the study, the loss of zyxin also affects the actin cytoskeleton, in particular actin stress fibers, and the enhanced motility of the zyxin-null cells could also result from that change.
Our finding that knockout of zyxin is associated with enhanced cell motility is consistent with studies in which stable expression of the oncoprotein associated with Ewing's sarcoma was shown to result in decreased zyxin levels and a corresponding increase in motility (Amsellem et al., 2005). However, the increased motility of the zyxin-null cells contrasts with previous findings in which zyxin function was probed using a nongenetic approach. In that case, microinjection of a peptide derived from the NH2 terminus of zyxin was used to displace zyxin from its normal subcellular location and decreased cell motility was observed (Drees et al., 1999). The peptide inhibition approach is distinct from the genetic approach in several ways. In the case in which the zyxin gene is disrupted or expression is down-regulated, cellular proteins that normally bind to zyxin would be free to circulate in the absence of zyxin. On the other hand, in the case of peptide-induced displacement of zyxin, many of these partners may still be bound to zyxin but not present in their normal location, which may have distinct functional consequences.
The fact that zyxin-null mice are viable and fertile (Hoffman et al., 2003) is somewhat surprising, given the cellular phenotypes associated with loss of zyxin function; however, this is not without precedence. There are several cases in which the loss-of-function phenotype at the whole organism level appears less severe and provides less mechanistic insight compared with cell-level analysis. For example, MyoD knock-out mice fail to display muscle defects (Rudnicki and Jaenisch, 1995), whereas cell-based assays revealed a role for MyoD in muscle differentiation. In the case of zyxin, our findings suggest that unchallenged organisms are relatively insensitive to a twofold increase in fibroblast motility or that the functional deficits observed in vitro are somehow masked in vivo. It will be interesting in future studies to challenge the zyxin-null mice to assess whether the enhanced cell motility observed in vitro might be evident as a hyperactive inflammatory response or enhanced tumor cell invasiveness.
Zyxin provides the primary docking site for Ena/VASP proteins at focal adhesions
Previous works have demonstrated that the NH2-terminal EVH1 domain of Ena/VASP interacts with the FPPPP (ActA) motif and is responsible for focal adhesion targeting (Gertler et al., 1996; Niebuhr et al., 1997; Drees et al., 2000). Several proteins that display ActA repeats are present in focal adhesions (Renfranz and Beckerle, 2002), and the relative contribution of each to Ena/VASP recruitment had not been established. Our analysis of Ena/VASP localization in cells lacking zyxin demonstrates that zyxin provides the primary focal adhesion targeting information for Ena/VASP. The observation that LPP levels appear elevated in zyxin-null focal adhesions suggests that enhanced localization of LPP may provide some compensation for the elimination of zyxin. Other focal adhesion constituents are unaffected by elimination of zyxin, thus, underscoring the specificity of the Ena/VASP deficiency and providing definitive evidence that zyxin is not essential for the assembly or maintenance of focal adhesions.
Lack of zyxin does not disturb the capacity of Ena/VASP to concentrate at the leading edge, suggesting that zyxin does not normally contribute to the recruitment of Ena/VASP to this region of a cell. This result is consistent with studies reporting that Ena/VASP proteins in cell surface projections are not extensively colocalized with zyxin (Rottner et al., 2001). Moreover, a recent study has identified another ActA repeat-containing protein, lamellipodin, as critical for targeting Ena/VASP to sites of lamellipodial protrusion (Krause et al., 2004). Thus, it appears that zyxin plays a key role in targeting Ena/VASP to focal adhesions, but not to other cell areas, where Ena/VASP accumulation depends on other factors.
Cells that lack zyxin display deficits in actin cytoskeletal remodeling
Zyxin promotes actin assembly when targeted to the plasma membrane or to mitochondria, and this effect depends on the ability of zyxin to bind Ena/VASP proteins (Golsteyn et al., 1997; Fradelizi et al., 2001). Moreover, exposure of fibroblasts to mechanical force induces the reinforcement of actin stress fibers by a mechanism that has been shown to require zyxin (Yoshigi et al., 2005). We demonstrate that treatment of wild-type fibroblasts with the actin assembly-promoting drug, jasplakinolide, results in thickening of stress fibers, and this response is abrogated in zyxin-null cells. These findings illustrate a role for zyxin in the control of stress fibers.
The importance of zyxin for the proper subcellular localization of Ena/VASP proteins suggests that zyxin could contribute to enhancement of actin stress fiber assembly by recruitment of Ena/VASP proteins to the fast growing ends of actin filaments that are anchored at focal adhesions. At those sites, Ena/VASP proteins could enhance actin assembly by recruitment of profilin-actin and/or by interference with the ability of capping protein to block filament barbed ends (Bear et al., 2002). In that regard, it is intriguing that exposure of wild-type cells to jasplakinolide (this study) or mechanical force (Yoshigi et al., 2005) is accompanied by a dramatic shift in the subcellular localization of both zyxin and Ena/VASP from focal adhesions onto actin stress fibers, whereas many other focal adhesion proteins remain associated with the substratum attachment sites.
The mechanism by which zyxin translocates from focal adhesions to actin stress fibers in response to jasplakinolide or physiological effectors that induce actin filament stabilization/assembly remains to be clarified. One possibility is that zyxin and its partners are released from the focal adhesions into the cytoplasm and can then assemble along the lengths of actin stress fibers. An alternative possibility is that zyxin and its partners are primarily associated with the barbed ends of actin filaments and stimulation of actin assembly, e.g., by jasplakinolide, causes their centripetal movement because of the addition of new monomer at focal adhesions. From this perspective, the mobilization of zyxin and Ena/VASP from focal adhesions to actin stress fibers could provide a marker for actin polymerization.
Modification of the actomyosin regulator, caldesmon, in zyxin-null cells
Caldesmon negatively regulates actomyosin-dependent contractility (Wang, 2001), and overexpression of caldesmon results in the diminishment of actin stress fibers (Helfman et al., 1999). Our findings illustrate that zyxin has an opposite, enhancing impact on stress fibers. Given this functional relationship, it is interesting that we consistently observe an altered mobility isoform of caldesmon in cells derived from mice in which the zyxin gene is ablated. A modification of caldesmon that abrogated its capacity to inhibit contractility could provide compensation for the loss of zyxin. In this view, one might expect a more severe stress fiber disruption if zyxin function were compromised in the absence of the hypothesized compensatory change in caldesmon. Indeed, it has been reported that instantaneous elimination of zyxin expression by siRNA results in loss of stress fibers (Harborth et al., 2001).
Additional work is required to elucidate the precise mechanism underlying the caldesmon isoform shift observed in zyxin-null cells; however, our results suggest that altered posttranslational modification is not responsible for the change in caldesmon mobility. Moreover, the fact that we observe the slower mobility caldesmon variant in homozygous mutant cells, but not in tissues derived from heterozygous mice, provides evidence that the change in caldesmon is not caused by an insertional mutagenesis event in the caldesmon gene that constitutively affects the size of the protein product. Expression of an alternatively spliced form of caldesmon that is regulated at either the transcriptional or translational level could account for the observed caldesmon mobility shift in the zyxin-null cells. L-caldesmon is encoded by 11 invariant exons (2, 3a, and 5–13; Hayashi et al., 1992; Payne et al., 1995), and we detected sequence encoded by each of these exons in caldesmon isolated from both wild-type and zyxin-null cells. Thus, variation in the usage of these exons is not likely to account for the mobility shift observed in the zyxin-null cells. Our sequence analysis confirmed the usage of exon 1′ in l-caldesmon isolated from wild-type cells. However, we did not detect exon 1′ encoded sequence in caldesmon derived from zyxin-null cells, suggesting that differential inclusion of the alternative exon 1 or an uncharacterized 5′ exon may account for the caldesmon variation observed in the zyxin-null cells.
Molecular heterogeneity of caldesmon has been previously noted; however, the molecular mechanism for this isoform diversity has not been explained (Okamoto et al., 2000). Usage of exon 1 is associated with tumor neovasculature for glioblastomas, oligodendrogliomas, and astrocytomas (Zheng et al., 2004). In addition, a caldesmon mobility shift similar to what we describe is also observed in mice lacking calponin, another regulator of actomyosin-dependent contractility (Matthew et al., 2000). This observation reinforces the concept that expression of alternative forms of caldesmon may provide a means for compensation when contractility regulation is compromised by mutation, and provides a link between zyxin and this function. It is interesting that reintroduction of zyxin into the null cells does not result in reversion of caldesmon to the species found in wild-type cells. We interpret this to mean that the change is genetically or epigenetically fixed in the absence of zyxin. Importantly, because reexpression of zyxin in the null cells does restore normal actin architecture, Ena/VASP localization, and motility, these phenotypes can be directly attributed to perturbation of zyxin levels.
Zyxin as a modulator of actin function and cell motility
We have shown that cells lacking zyxin display augmented adhesive properties, enhanced migration, reduced capacity to build robust actin stress fibers in response to a chemical stimulus, and disturbed focal adhesion accumulation of actin regulators in the Ena/VASP family. Reintroduction of a zyxin transgene into the null cells causes reversion of these phenotypes. We have also uncovered an unusual property of both zyxin and VASP proteins, which is their specific mobilization from focal adhesions to actin stress fibers in response to an agent that stimulates actin assembly. The critical role of zyxin in chemically triggered actin assembly is revealed by analysis of zyxin-null cells, which fail to respond efficiently to jasplakinolide. Finally, we have identified an unanticipated link between caldesmon and zyxin based on the demonstration that targeted deletion of the zyxin gene is consistently accompanied by an alteration in the contractility modulator caldesmon. Our analysis of zyxin function, coupled with previous work on caldesmon, suggests that these two proteins are functionally linked, perhaps antagonizing each other to regulate actin stress fiber assembly.
Disturbance of cytoskeletal architecture has long been recognized as a hallmark of cellular transformation. Cancer cells lack robust actin stress fiber arrays, which are associated with enhanced motility. Interestingly, zyxin levels are dramatically reduced in invasive bladder carcinoma (Sanchez-Carbayo et al., 2002) and in a Ewing's sarcoma model system (Amsellem et al., 2005). Methylation-sensitive silencing of zyxin expression has also been shown to be a characteristic of several breast cancer cell lines (Wang et al., 2005). The results reported here suggest a mechanism by which reduction in zyxin expression could contribute to tumor progression by affecting cytoarchitecture and motility.
Materials and methods
Antibodies
Commercial antibody sources included ICN Biomedicals (α-actinin), Sigma-Aldrich (talin and vinculin), BD Biosciences (caldesmon, ILK, paxillin, and integrins α3, αV, β1, β3), Upstate Cell Signaling (FAK 4–47, pY-4G10, and src), Santa Cruz Biotechnology, Inc. (FAK C-20 and pY-20), CHEMICON International, Inc. (integrin α5), Cell Signaling Technology (p-Tyr-100), and Qbiogene (VASP). Zyxin (B71, B72, and 1D1), TRIP6 (B65; Hoffman et al., 2003), and phospho-Ser527/789-caldesmon (D'Angelo et al., 1999) antibodies were previously described. Other investigators supplied anti-LPP MP2 (M. Petit and J.M. Van de Ven, University of Leuven, Leuven, Belgium; Petit et al., 2000), anti-Mena 2197 and anti-VASP 2010 (F. Gertler, Massachusetts Institute of Technology, Boston, MA; Bear et al., 2000), and anti-warts kinase/LATS C2 (T. Hirota and H. Saya, Kumamoto University, Hanjo, Japan; Hirota et al., 2000).
Mouse fibroblasts
Primary mouse embryonic fibroblasts (MEFs) were derived from day 14 embryos of wild-type or zyxin-null mice (backcrossed nine generations into the inbred C57BL/6 line [Charles River Laboratories] to ensure genetic homogeneity, except at the zyxin locus; Hoffman et al., 2003). Primary MEFs were expanded and frozen as passage 2 stocks. For experiments, the cells were thawed and used within a week (passages 4–6) as early-passage MEFs. To generate immortalized cell lines, cells were cultured from tissue (torso) explants of newborn mice. After spontaneous immortalization (generally passages 8–12), viable cell lines were isolated using cloning cylinders, expanded, and frozen for future use. All cells were grown in high glucose DME supplemented with pyruvate, glutamine, penicillin/streptomycin (Invitrogen), 10% FBS (HyClone) at 37°C, and 5% CO2. Cells were treated with 100 nM of jasplakinolide or DMSO control for 2 h in DME supplemented with 10% FBS (Bubb et al., 2000). Unless specified in the figure legend, experiments used both primary and immortalized cell lines, with similar results obtained for both. Immortalized cells were used for rescue experiments.
An enhanced GFP-mouse zyxin DNA was inserted into the retroviral vector LINXv-myc (Hoshimaru et al., 1996) and Phoenix-Eco cells (American Type Culture Collection) were used to generate retrovirus. Zyxin-null fibroblast cell lines were infected and sorted for GFP to select enhanced GFP-zyxin-myc–expressing cells. Caldesmon-6histidine DNA was inserted into pBABEpuro and transiently transfected with Lipofectamine 2000 (Invitrogen).
Cell cycle, adhesion, and migration assays
Propidium iodide staining of DNA and flow cytometry followed established procedures (Iida et al., 2004) using a FACScan instrument and CellQuest software (Becton Dickinson).
Adhesion assays were performed essentially as previously described (Xu et al., 1998), with 105 cells seeded for 25 min into ECM-coated wells, and were performed four times with two pairs of independently derived wild-type and zyxin−/− cell lines.
Monolayer wound assay was performed as previously described (Xu et al., 1998), with confluent MEFs in optical 35-mm plates on a 37°C heated stage apparatus (Bioptechs) monitored by time-lapse (every 15 min) phase-contrast microscopy with an inverted microscope (TE300; Nikon), a 10× DIC-L objective, 0.3 NA, a 5-Mhz digital camera (MicroMax; Princeton Instruments), and QED software (Media Cybernetics).
Haptotactic cell migrations on Boyden chamber transwells (24-well cell culture inserts of polyethylene terephthalate membranes with 8-μm pores; Becton Dickinson) moved from serum-free DME to DME supplemented with 10% FBS over 6 h (Xu et al., 1998). BioCoat Matrigel invasion chambers with 8-μm pores (BD Biosciences) in 24-well plates were seeded with 15,000 or 30,000 cells for 4- or 24-h migration. Migratory cells in five fields per well (20× phase objective) were counted for three individual wells per condition.
Immunofluorescence
Cell staining followed established procedures, using the primary antibodies described in Antibodies and Alexa Fluor secondary antibodies and phalloidin (Invitrogen). Cell images were captured at room temperature using a fluorescent microscope (AxioPhot; Carl Zeiss MicroImaging Inc.) with a Plan-Apochromat 63×, 1.40 NA, oil objective and 100×, 1.30 NA, and 40×, 1.30 NA, objectives, a MicroMax digital camera (Princeton Instruments), and Openlab imaging software (Improvision), and were incorporated into figures using Photoshop 8.0 and Illustrator 11.0 software (Adobe).
Protein analysis
Cell and tissue lysates were prepared, electrophoresed, and immunoblotted as previously described (Hoffman et al., 2003). 2D gel analysis used the Protean IEF Cell system with immobilized pH gradient gel strips (pH 3–10; Bio-Rad Laboratories) followed by denaturing 10% acrylamide gels. For MS analysis, proteins were stained with Colloidal blue (Invitrogen), excised from the gel, washed twice in 50% acetonitrile/H2O, and then frozen. Peptide digestion, purification, tandem array-liquid chromatography LC/MS-MS, and sequence analysis were performed at the University of Utah Mass Spectrometry Core Facility (Salt Lake City, UT) and at the Harvard Microchemistry Facility (Cambridge, MA) by microcapillary reverse-phase HPLC nanoelectrospray tandem mass spectrometry (μLC MS/MS) on a quadrupole ion trap mass spectrometer (Finnigan LCQ DECA XP; Thermo Electron Corporation).
SFTI
SFTI was quantitated based on erosion spatial filtering (Russ, 2002). An erosion filter is equal to the ninth rank filter, which sorts 9 pixels in a 3 × 3 pixel matrix by their pixel brightness and replaces the center pixel with the darkest value found in the matrix. This “erodes” contours of objects in the digital image. Decay curves of averaged brightness during filtration cycles reflect thickness of the original lines. Nonlinear curve fit was applied to compute decay constants of the brightness curves and were termed SFTI. This method was superior to other methods based on line profiles because it reflected thickness of lines in areas, rather than at points.
Statistics
Data analysis included analysis of variance between groups and unpaired t tests using GraphPad Prism software (GraphPad Software). A P value of <0.05 was considered statistically significant.
Online supplemental material
Additional examples of the Mena and VASP mislocalization in the zyxin-null fibroblasts are available in Fig. S1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200512115/DC1.
Supplementary Material
Acknowledgments
Antibodies were provided by M. Petit, W. Van de Ven, F. Gertler, T. Hirota, and H. Saya. K. Andrews made the GFP-zyxin retrovirus using DNA from F. Gage and cells from G. Nolan. W.S. Lane (Harvard Microchemistry Facility) and P. Krishna (University of Utah Mass Spectrometry Core Facility) provided advice and mass spectrometry analysis. University of Utah Core resource directors R. Schackmann, W. Green, C. Rodesch, and A. Szabo provided technical support. Comments from members of the Beckerle laboratory and graphic design by Diana Lim were invaluable.
This work was supported by the National Institutes of Health (grants P30 CA42014 and RO1GM50877) and the Huntsman Cancer Foundation.
Abbreviations used in this paper: 2D, two-dimensional; h, high molecular weight; ILK, integrin-linked kinase; l, low molecular weight; LPP, lipoma preferred partner; MEF, mouse embryonic fibroblast; pY, phosphotyrosine; SFTI, stress fiber thickness index; TRIP, thyroid receptor-interacting protein.
References
- Amsellem, V., M.H. Kryszke, M. Hervy, F. Subra, R. Athman, H. Leh, C. Brachet-Ducos, and C. Auclair. 2005. The actin cytoskeleton-associated protein zyxin acts as a tumor suppressor in Ewing tumor cells. Exp. Cell Res. 304:443–456. [DOI] [PubMed] [Google Scholar]
- Bear, J.E., J.J. Loureiro, I. Libova, R. Fassler, J. Wehland, and F.B. Gertler. 2000. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 101:717–728. [DOI] [PubMed] [Google Scholar]
- Bear, J.E., T.M. Svitkina, M. Krause, D.A. Schafer, J.J. Loureiro, G.A. Strasser, I.V. Maly, O.Y. Chaga, J.A. Cooper, G.G. Borisy, and F.B. Gertler. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521. [DOI] [PubMed] [Google Scholar]
- Beckerle, M. 1998. Spatial control of actin filament assembly: lessons from Listeria. Cell. 95:741–748. [DOI] [PubMed] [Google Scholar]
- Beningo, K.A., M. Dembo, I. Kaverina, J.V. Small, and Y.L. Wang. 2001. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle, N.P., M.R. Holt, J.E. Davies, C.J. Price, and D.R. Critchley. 1996. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 318:753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb, M.R., I. Spector, B.B. Beyer, and K.M. Fosen. 2000. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 275:5163–5170. [DOI] [PubMed] [Google Scholar]
- D'Angelo, G., P. Graceffa, C.A. Wang, J. Wrangle, and L.P. Adam. 1999. Mammal-specific, ERK-dependent, caldesmon phosphorylation in smooth muscle. Quantitation using novel anti-phosphopeptide antibodies. J. Biol. Chem. 274:30115–30121. [DOI] [PubMed] [Google Scholar]
- Drees, B.E., K.M. Andrews, and M.C. Beckerle. 1999. Molecular dissection of zyxin function reveals its involvement in cell motility. J. Cell Biol. 147:1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B., E. Friederich, J. Fradelizi, D. Louvard, M.C. Beckerle, and R.M. Golsteyn. 2000. Characterization of the interaction between zyxin and members of the Ena/Vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 275:22503–22511. [DOI] [PubMed] [Google Scholar]
- Fradelizi, J., V. Noireaux, J. Plastino, B. Menichi, D. Louvard, C. Sykes, R.M. Golsteyn, and E. Friederich. 2001. ActA and human zyxin harbour Arp2/3-independent actin polymerization activity. Nat. Cell Biol. 3:699–707. [DOI] [PubMed] [Google Scholar]
- Fukuda, T., K. Chen, X. Shi, and C. Wu. 2003. PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J. Biol. Chem. 278:51324–51333. [DOI] [PubMed] [Google Scholar]
- Gertler, F.B., K. Niebuhr, M. Reinhard, J. Wehland, and P. Soriano. 1996. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 87:227–239. [DOI] [PubMed] [Google Scholar]
- Golsteyn, R.M., M.C. Beckerle, T. Koay, and E. Friederich. 1997. Structural and functional similarities between the human cytoskeletal protein zyxin and the ActA protein of Listeria monocytogenes. J. Cell Sci. 110:1893–1906. [DOI] [PubMed] [Google Scholar]
- Guo, H., and C.L. Wang. 2005. Specific disruption of smooth muscle caldesmon expression in mice. Biochem. Biophys. Res. Commun. 330:1132–1137. [DOI] [PubMed] [Google Scholar]
- Harborth, J., S.M. Elbashir, K. Bechert, T. Tuschl, and K. Weber. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114:4557–4565. [DOI] [PubMed] [Google Scholar]
- Hayashi, K., H. Yano, T. Hashida, R. Takeuchi, O. Takeda, K. Asada, E. Takahashi, I. Kato, and K. Sobue. 1992. Genomic structure of the human caldesmon gene. Proc. Natl. Acad. Sci. USA. 89:12122–12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman, D.M., E.T. Levy, C. Berthier, M. Shtutman, D. Riveline, I. Grosheva, A. Lachish-Zalait, M. Elbaum, and A.D. Bershadsky. 1999. Caldesmon inhibits nonmuscle cell contractility and interferes with the formation of focal adhesions. Mol. Biol. Cell. 10:3097–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, T., T. Morisaki, Y. Nishiyama, T. Marumoto, K. Tada, T. Hara, N. Masuko, M. Inagaki, K. Hatakeyama, and H. Saya. 2000. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J. Cell Biol. 149:1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L.M., D.A. Nix, B. Benson, R. Boot-Hanford, E. Gustafsson, C. Jamora, A.S. Menzies, K.L. Goh, C.C. Jensen, F.B. Gertler, et al. 2003. Targeted disruption of the murine zyxin gene. Mol. Cell. Biol. 23:70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshimaru, M., J. Ray, D.W. Sah, and F.H. Gage. 1996. Differentiation of the immortalized adult neuronal progenitor cell line HC2S2 into neurons by regulatable suppression of the v-myc oncogene. Proc. Natl. Acad. Sci. USA. 93:1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, S., T. Hirota, T. Morisaki, T. Marumoto, T. Hara, S. Kuninaka, S. Honda, K. Kosai, M. Kawasuji, D.C. Pallas, and H. Saya. 2004. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 23:5266–5274. [DOI] [PubMed] [Google Scholar]
- Katz, B.Z., E. Zamir, A. Bershadsky, Z. Kam, K.M. Yamada, and B. Geiger. 2000. Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Mol. Biol. Cell. 11:1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, M., J.D. Leslie, M. Stewart, E.M. Lafuente, F. Valderrama, R. Jagannathan, G.A. Strasser, D.A. Rubinson, H. Liu, M. Way, et al. 2004. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell. 7:571–583. [DOI] [PubMed] [Google Scholar]
- Matthew, J.D., A.S. Khromov, M.J. McDuffie, A.V. Somlyo, A.P. Somlyo, S. Taniguchi, and K. Takahashi. 2000. Contractile properties and proteins of smooth muscles of a calponin knockout mouse. J. Physiol. 529:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr, K., F. Ebel, R. Frank, M. Reinhard, E. Domann, U.D. Carl, U. Walter, F.B. Gertler, J. Wehland, and T. Chakraborty. 1997. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 16:5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix, D.A., and M.C. Beckerle. 1997. Nuclear–cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 138:1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix, D.A., J. Fradelizi, S. Bockholt, B. Menichi, D. Louvard, E. Friederich, and M.C. Beckerle. 2001. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 276:34759–34767. [DOI] [PubMed] [Google Scholar]
- Okamoto, K., N. Kashihara, Y. Yamasaki, K. Kanao, Y. Maeshima, T. Sekikawa, H. Sugiyama, T. Murakami, and H. Makino. 2000. Caldesmon isoform associated with phenotypic modulation of mesangial cells. Exp. Nephrol. 8:20–27. [DOI] [PubMed] [Google Scholar]
- Payne, A.M., P. Yue, K. Pritchard, and S.B. Marston. 1995. Caldesmon mRNA splicing and isoform expression in mammalian smooth-muscle and non-muscle tissues. Biochem. J. 305:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, M.M.R., J. Fradelizi, R.M. Golsteyn, T.A.Y. Ayoubi, B. Menichi, D. Louvard, W.J.M. Van De Ven, and E. Friederich. 2000. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell. 11:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfranz, P.J., and M.C. Beckerle. 2002. Doing (F/L)PPPPs: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr. Opin. Cell Biol. 14:88–103. [DOI] [PubMed] [Google Scholar]
- Rottner, K., M. Krause, M. Gimona, J.V. Small, and J. Wehland. 2001. Zyxin is not colocalized with vasodilator-stimulated phosphoprotein (VASP) at lamellipodial tips and exhibits different dynamics to vinculin, paxillin, and VASP in focal adhesions. Mol. Biol. Cell. 12:3103–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki, M.A., and R. Jaenisch. 1995. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 17:203–209. [DOI] [PubMed] [Google Scholar]
- Russ, J.C. 2002. The Image Processing Handbook. Fourth ed. CRC Press LLC, Boca Raton, FL. 744 pp.
- Sanchez-Carbayo, M., N.D. Socci, E. Charytonowicz, M. Lu, M. Prystowsky, G. Childs, and C. Cordon-Cardo. 2002. Molecular profiling of bladder cancer using cDNA microarrays: defining histogenesis and biological phenotypes. Cancer Res. 62:6973–6980. [PubMed] [Google Scholar]
- Wang, C.L. 2001. Caldesmon and smooth-muscle regulation. Cell Biochem. Biophys. 35:275–288. [DOI] [PubMed] [Google Scholar]
- Wang, W., G. Huper, Y. Guo, S.K. Murphy, J.A. Olson, and J.R. Marks. 2005. Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene. 24:2705–2714. [DOI] [PubMed] [Google Scholar]
- Xu, W., H. Baribault, and E.D. Adamson. 1998. Vinculin knockout results in heart and brain defects during embryonic development. Development. 125:327–337. [DOI] [PubMed] [Google Scholar]
- Yi, J., and M.C. Beckerle. 1998. The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics. 49:314–316. [DOI] [PubMed] [Google Scholar]
- Yoshigi, M., L.M. Hoffman, C.C. Jensen, H.J. Yost, and M.C. Beckerle. 2005. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 171:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar, R., C. Ballestrem, Z. Kam, and B. Geiger. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613. [DOI] [PubMed] [Google Scholar]
- Zamir, E., B.Z. Katz, S. Aota, K.M. Yamada, B. Geiger, and Z. Kam. 1999. Molecular diversity of cell-matrix adhesions. J. Cell Sci. 112:1655–1669. [DOI] [PubMed] [Google Scholar]
- Zheng, P.P., A.M. Sieuwerts, T.M. Luider, M. van der Weiden, P.A. Sillevis-Smitt, and J.M. Kros. 2004. Differential expression of splicing variants of the human caldesmon gene (CALD1) in glioma neovascularization versus normal brain microvasculature. Am. J. Pathol. 164:2217–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.