Abstract
The Apaf-1 protein is essential for cytochrome c–mediated caspase-9 activation in the intrinsic mammalian pathway of apoptosis. Although Apaf-1 is the only known mammalian homologue of the Caenorhabditis elegans CED-4 protein, the deficiency of apaf-1 in cells or in mice results in a limited cell survival phenotype, suggesting that alternative mechanisms of caspase activation and apoptosis exist in mammals. In Drosophila melanogaster, the only Apaf-1/CED-4 homologue, ARK, is required for the activation of the caspase-9/CED-3–like caspase DRONC. Using specific mutants that are deficient for ark function, we demonstrate that ARK is essential for most programmed cell death (PCD) during D. melanogaster development, as well as for radiation-induced apoptosis. ark mutant embryos have extra cells, and tissues such as brain lobes and wing discs are enlarged. These tissues from ark mutant larvae lack detectable PCD. During metamorphosis, larval salivary gland removal was severely delayed in ark mutants. However, PCD occurred normally in the larval midgut, suggesting that ARK-independent cell death pathways also exist in D. melanogaster.
Introduction
Programmed cell death (PCD), which is often carried out by a morphologically distinct process called apoptosis, is essential for both proper animal development and cellular homeostasis (for review see Baehrecke, 2002). The caspase family of cysteine proteases are the main effectors of apoptosis. These proteases specifically target a number of cellular proteins for cleavage, leading to the disassembly of cells undergoing apoptosis (for reviews see Hengartner, 2000; Adams, 2003). In most cases, caspases are produced as inactive zymogens that become active in response to apoptotic stimuli. In Caenorhabditis elegans, the loss of function of a single caspase CED-3 or its adaptor CED-4, which is required for CED-3 activation, results in a complete block in developmental PCD (Hengartner, 2000). The mammalian caspase family can be divided into initiator and effector caspases. Initiator caspases resemble CED-3 and are characterized by the presence of protein–protein interaction motifs such as a caspase recruitment domain (CARD), e.g., caspase-2 and -9, or a pair of death effector domains, e.g., caspase-8 and -10 (Hengartner, 2000; Adams, 2003). Initiator caspases undergo adaptor-assisted self-activation, whereas the effector caspases, lacking a CARD or death effector domains, require proteolytic processing by an initiator caspase to become active. In mammals, the activation of the initiator caspase, caspase-9, which is mediated via its adaptor protein Apaf-1, is necessary for stress-induced cellular apoptotic responses (Zou et al., 1999; Hengartner, 2000; Adams, 2003). However, Apaf-1 and caspase-9 knockout mice, or cells derived from knockout animals, show limited phenotypic abnormalities, and caspase activity and apoptosis is still seen in many tissues from the knockout animals (Marsden et al., 2002; Ekert et al., 2004). Therefore, it is likely that although the caspase-9–Apaf-1 pathway is required for specific PCD, caspase activation and apoptosis pathways not requiring these conserved proteins also exist in mammals.
Drosophila melanogaster has one Apaf-1 homologue, ARK (DARK/dApaf-1/Hac-1), which is required for the activation of DRONC, the only CARD-containing orthologue of CED-3/caspase-9 in D. melanogaster (Dorstyn et al., 1999; Kanuka et al., 1999; Rodriguez et al., 1999; Zhou et al., 1999; for reviews see Kumar and Doumanis, 2000; Cashio et al., 2005). Apaf-1 activation has an obligate requirement for cytochrome c released from mitochondria for apoptosome assembly, whereas an ARK apoptosome-like complex can assemble in the absence of cytochrome c (Yu et al., 2006). The DIAP-1 (D. melanogaster inhibitor of apoptosis protein-1) directly binds DRONC, preventing its activation by blocking the ARK–DRONC interaction (Cashio et al., 2005). The REAPER, HID, and GRIM proteins antagonize DIAP-1 function to facilitate DRONC activation (Cashio et al., 2005). Genetic and cell culture data suggest that DRONC is required for most developmental and stress-induced cell death (Quinn et al., 2000; Chew et al., 2004; Daish et al., 2004; Waldhuber et al., 2005; Xu et al., 2005). Interestingly, in animals lacking DRONC, some embryonic PCD and larval midgut histolysis occur normally, indicating that DRONC is not essential for all PCD (Daish et al., 2004; Xu et al., 2005).
Previous studies with hypomorphic ark alleles and cell-based RNA interference analyses suggest that ARK is required for PCD (Kanuka et al., 1999; Rodriguez et al., 1999; Zhou et al., 1999; Zimmermann et al., 2002). However, the ark hypomorphs are viable and show restricted phenotypic abnormalities, making it difficult to fully assess the function of ARK in PCD. In this paper, we describe the analysis of two independent ark mutants that are strong alleles of ark and demonstrate that ARK is essential for normal development, most developmental PCD, and stress-mediated apoptosis. However, similar to DRONC, some PCD is ARK independent, suggesting that the ARK–DRONC pathway controls most, but not all, PCD in the fly.
Results and discussion
Generation of specific ark mutants
ark alleles were obtained in a screen conducted using mitotic recombination for mutations, which results in an increased relative representation of mutant over wild-type (WT) tissue. In these mutants, the mutant clones were larger than the corresponding WT twin spots. The screen of the right arm of chromosome 2 identified mutations in the hippo locus that have been previously described (Harvey et al., 2003). Four alleles of ark were also obtained from the same screen, which were all lethal at the pupal stage of development as homozygotes or in trans to each other. Sequencing revealed point mutations or deletions in the coding sequence of the ark gene in each of the mutant chromosomes (Fig. 1 A). ark1 had a G to A mutation, resulting in the truncation of the protein after residue 206; ark2 had a C to T mutation, causing protein truncation after residue 660; and ark3 had a deletion after residue 592, generating a frameshift mutation, whereas ark4 possessed a T to G mutation, causing protein truncation after residue 1,357. The mutation in ark1 is predicted to affect both of the reported alternately spliced transcripts of the ark gene (Kanuka et al., 1999). Because all ark mutants were lethal at a similar stage, only ark1 and ark2 were analyzed in our studies.
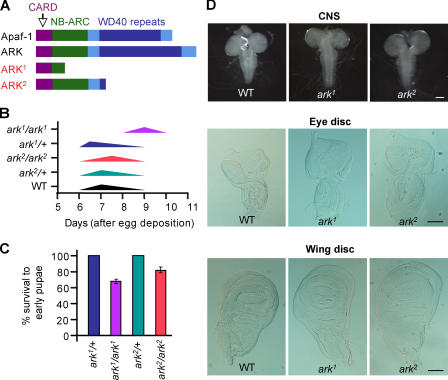
Figure 1.
Generation and analysis of ark mutants. (A) Structure of ARK and predicted ARK mutant proteins. The domain structures of ARK and its mammalian counterpart Apaf-1 are shown. Both ark alleles are truncation mutants, deleting most of the WD40 domain region. (B) ark1 homozygous animals show delayed development. Peaks indicate the point at which the majority of animals reach the wandering larval stage. (C) Survival rates of early pupae were from two experiments. Data are means ± SEM. n values for WT, ark1, and ark2 animals were 1,018, 755, and 1,065, respectively. The data for heterozygous animals were normalized to 100%. (D) CNS (top), eye discs (middle), and wing discs (bottom) from homozygous ark mutant and WT control late third instar larvae. Selected examples of tissues showing hyperplasia are shown. Bars, 100 μm.
ARK is essential for development
Similar to Apaf-1, ARK consists of a CARD, a nucleotide-binding NB-ARC domain, and multiple WD40 repeats (Fig. 1 A). ark1 mutation truncates the protein in the NB-ARC (CED-4 domain), whereas ark 2 leads to a protein lacking most of the WD40 repeats (Fig. 1 A). Both mutants are lethal and have very similar phenotypes, suggesting that they are strong loss-of-function alleles. The phenotypes also indicate that both the NB-ARC and the WD40 domains are essential for ARK function. Unlike the published hypomorphs (Kanuka et al., 1999; Rodriguez et al., 1999; Zhou et al., 1999), all homozygous ark1 and ark2 animals die as pupae. Despite the similar overall phenotypes for ark1 and ark2 alleles, development of ark1 mutants to pupation was significantly delayed when compared with WT or ark2 alleles (Fig. 1 B), suggesting that ark1 may be a stronger allele than ark2. Consequently, the survival of ark1-null animals to early pupae stage was lower than that of the heterozygotes (Fig. 1 C). Although larvae and pupae from both ark mutants appeared grossly normal externally, some larval tissues derived from late third instar animals showed hyperplasia. For example, consistent with previous observations (Rodriguez et al., 1999; Kanuka et al., 1999), the larval central nervous system (CNS) was enlarged in both ark mutants (Fig. 1 D). This was particularly evident in the ventral ganglion that appeared to be elongated and contained longer nerve fibers. In ∼40% of ark1 and most of the ark2 animals, the wing discs were enlarged (Fig. 1 D). In a small number of both mutants, the eye discs were also enlarged (Fig. 1 D).
Defective PCD in ark mutant animals
dronc mutant embryos contain extra cells, and the removal of maternal dronc abolishes most cell death during embryogenesis (Quinn et al., 2000; Xu et al., 2005). dronc-deficient embryos also show an enlargement of the CNS, which is presumably caused by reduced PCD (Xu et al., 2005). By staining embryos with anti–embryonic lethal abnormal visual protein (ELAV) antibody to visualize neurons in the CNS and peripheral nervous system, we found extra neurons in chordotonal cell clusters in ark mutant embryos (Fig. 2, A and B). There were up to three extra cells per cluster in most ark mutant embryos analyzed (Fig. 2 B). Staining of embryos with BP102 antibody, which recognizes CNS axons, showed gross abnormalities in many mutant animals, with ark2 animals often showing more dramatic features (Fig. 2 C). We consistently observed stronger staining of CNS axons in ark mutant embryos compared with WT animals, which could result from more densely packed axons. In many mutant animals, the ventral nerve cord appeared to be improperly compacted and the spacing between longitudinal axonal tracts was enlarged (Fig. 2 C). This could be attributable to additional cells in the mutants caused by reduced PCD.
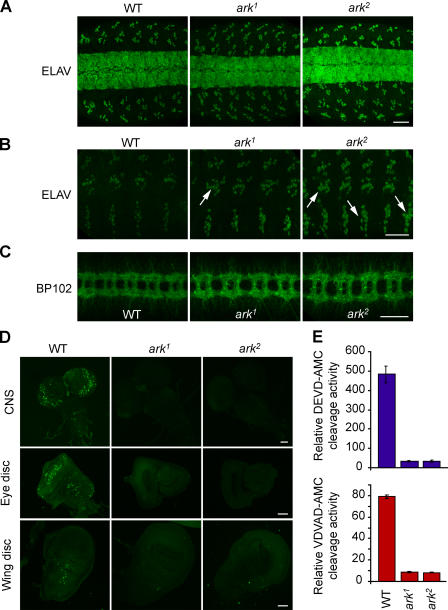
Figure 2.
ARK is required for developmental PCD in embryos and in larval tissues. (A and B) ELAV staining of stage 15 embryos. (B) Enlarged views of the anti-ELAV–stained abdominal hemisegments of the peripheral nervous system are shown. (arrows) Examples of cell clusters containing extra cells in ark1 and ark2. (C) BP102 antibody staining of WT and ark mutant embryos. Selected ark1 and ark2 animals show aberrant ventral nerve cords where commissures appear wider than in WT animals. (D) CNS, eye discs, and wing discs were dissected from late third instar larvae of ark1, ark2, and WT animals and stained with AO. (E) Caspase activity in prepupal extracts was measured on DEVD-AMC and VDVAD-AMC substrates. Data are means ± SEM. n = 6. Bars: (A–C) 50 μm; (D) 100 μm.
To investigate whether ark mutants have fewer cells undergoing PCD, we analyzed various larval organs for apoptosis using acridine orange (AO) staining. In ark1 and ark2 brain lobes, AO staining was almost absent when compared with WT, which showed many AO-positive cells (Fig. 2 D). Furthermore, AO staining in wing discs was greatly reduced in both ark mutants (Fig. 2 D). Larval eye discs also displayed a dramatic decrease in the number of dying cells when compared with WT (Fig. 2 D). As the only predicted function of ARK is to activate caspases, we analyzed caspase activity in prepupal lysates. Caspase activity, which was measured using two different substrates, was lower in both ark mutants (Fig. 2 E). Although reduced, some caspase activity was detectable in mutant animals at different stages of larval development (unpublished data), suggesting that ARK-independent caspase activation may occur in some tissues and/or cell types. We conclude that ARK is essential for most PCD in the embryo and larval tissues. These data also suggest that ARK and DRONC function in the same PCD pathway.
Delayed salivary gland PCD in ark mutants
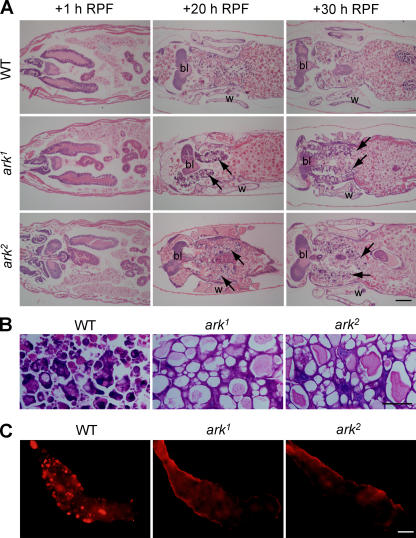
ark and dronc are transcriptionally up-regulated in salivary glands after the late prepupal ecdysone pulse, and dronc mutants show a delay in salivary gland PCD (Cakouros et al., 2002, 2004; Daish et al., 2004; Waldhuber et al., 2005; for reviews see Kumar and Cakouros, 2004; Yin and Thummel, 2005). To examine a role for ARK in larval salivary gland removal, we analyzed PCD in this tissue in ark mutants (Fig. 3 A). Larval salivary gland removal was markedly delayed in both ark1 and ark2 animals. Histological analysis indicated that both ark mutants had persistent or partially degraded salivary glands with an intact lumen at 20 h relative to puparium formation (RPF) at a time when, in the WT animals, salivary glands had been completely removed (Fig. 3 A). We could also see intact salivary glands at 30 h RPF in all ark mutant animals. The persistent salivary glands were highly vacuolated and appeared to be similar to persistent glands in dronc mutant animals and prehistolyzed WT glands (Fig. 3 B; Daish et al., 2004). Adult structures, such as wings, were forming in both ark mutants as in the WT controls, indicating continuing pupal development (Fig. 3 A). Thus, the persistence of the salivary glands cannot be attributed to a global delay in development. No TUNEL-positive nuclei were observed in ark mutant salivary glands at the time when WT glands were TUNEL positive (Fig. 3 C), indicating that DNA fragmentation, which requires caspase activation, does not occur in the absence of ARK in salivary glands. These results indicate that ARK is required for caspase-dependent removal of the larval salivary glands.
Figure 3.
ARK is required for the timely removal of larval salivary glands. (A) Larval salivary glands persist in ark mutant animals. Hematoxylin- and eosin-stained sections of WT and ark mutant animals at +1 h, +20 h, and +30 h RPF are shown. Note adult wing (w) structures are forming in both ark mutants and WT animals. (arrows) Persistent glands in ark1 and ark2 mutants. bl, brain lobes. (B) Enlarged views of hematoxylin- and eosin-stained sections of WT animals showing remnants of histolyzed salivary gland cells and ark mutants showing highly vacuolated intact glands at +20 h RPF. (C) TUNEL of WT, ark1, and ark2 salivary glands from animals staged to 14 h RPF. Bars: (A) 200 μm; (B and C) 50 μm.
Normal caspase activation and PCD in larval midguts from ark mutants
Along with dronc, ark is up-regulated in the midgut after the late third instar larval ecdysone pulse, which triggers PCD in this organ (Lee et al., 2002; Kumar and Cakouros, 2004; Yin and Thummel, 2005). However, our data demonstrate that in contrast to salivary glands, midgut removal was largely normal in both ark mutants (Fig. 4). Larval midguts are eliminated in the few hours after puparium formation. Removal of this tissue involves shortening of the gastric caecae with a concomitant reduction in midgut length, leading to the formation of the adult epithelia around the regressing larval structure (Jiang et al., 1997). By 4 h RPF, the proventriculus is markedly reduced in size, whereas the gastric caecum and midgut no longer resemble the larval form (Lee et al., 2002).
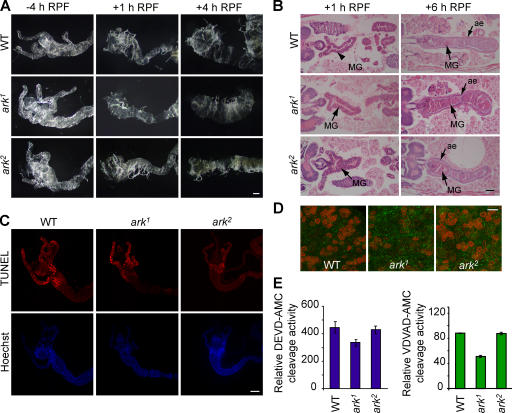
Figure 4.
Metamorphic PCD occurs normally in larval midguts from ark mutant animals. (A) Larval midguts from both ark mutants undergo deletion in concert with WT animals, as judged by gross morphology. (B) Hematoxylin- and eosin-stained sections of WT and ark mutant animals at 6 h RPF. MG, larval midgut; ae, adult epithelium. (C) TUNEL of WT and ark mutant midguts. (bottom) Nuclear (Hoechst) staining of the same midgut samples as in the top images. (D) Confocal images of WT and ark mutant midguts immunostained for caspase-3–like activity (green) and nuclear lamin (red). In control experiments, no background staining was seen with the secondary antibodies used here (not depicted). (E) Caspase activity is present in ark mutant midguts at the commencement of metamorphosis. Enzyme activities were measured on DEVD-AMC and VDVAD-AMC substrates. Data are means ± SEM and were derived from three experiments. n = 6. Bars: (A and B) 100 μm; (C) 200 μm; (D) 20 μm.
Both WT and ark mutant animals showed shortening of the gastric caecae by 1 h RPF and complete removal by 4 h RPF (Fig. 4 A). Histological analysis showed that by 6 h RPF both WT and ark mutants contained condensed larval midguts within detached adult epithelial lumen (Fig. 4 B). Within 1 h RPF, TUNEL staining, similar to that seen in WT midguts, was evident in midguts from both ark mutants (Fig. 4 C), indicating that DNA fragmentation occurs in the absence of ARK. We further analyzed caspase-3–like activity in prepupal (+1 h RPF) WT, ark 1, and ark2 midguts using the active caspase-3 monoclonal antibody. Caspase-3–like activity was present in both mutant and control prepupal midguts (Fig. 4 D). Midguts from both ark mutants showed that caspase activities were generally comparable to WT, as measured by substrate cleavage, except for ark1 animals that showed lower activity on VDVAD-AMC (Fig. 4 E). These data indicate that caspase activation occurs normally in the midgut of ark mutants and that ARK is not essential for midgut PCD. These results are consistent with our observations of dronc mutants (Daish et al., 2004) and indicate that the ARK–DRONC pathway of caspase activation is not essential for midgut PCD and that an alternative mechanism of caspase activation is likely to be functioning in this tissue.
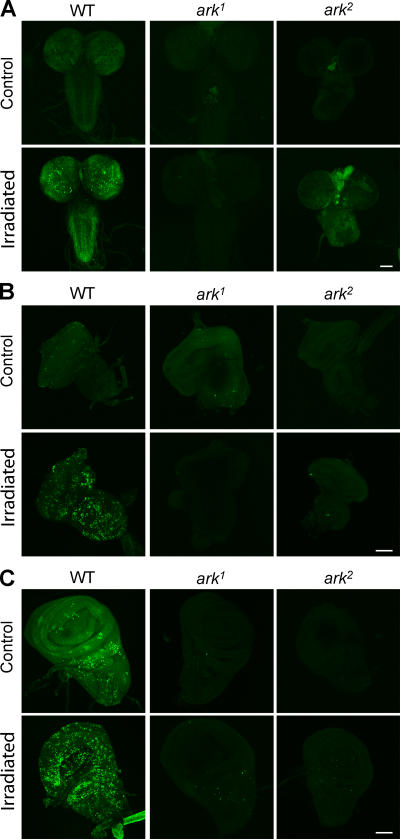
ARK is essential for radiation-induced apoptosis
A previous study suggests that ark transcription is induced by irradiation (Zhou et al., 1999). In dronc mutants, stress-mediated cell death, including radiation-induced apoptosis, is completely abolished (Chew et al., 2004; Daish et al., 2004; Waldhuber et al., 2005). To investigate the role of ARK in DNA damage-induced PCD, we irradiated larvae with γ rays and analyzed the effect on apoptosis. As expected, the basal number of AO-positive cells in tissues from both ark mutants were very low (or absent; Fig. 5). After irradiation, WT larvae showed large increases in apoptosis in brain lobes, eye discs, and wing discs, as observed by increased AO staining (Fig. 5, A–C). However, we did not see any increase in AO staining in any tissue from ark1 and ark2 animals (Fig. 5), suggesting that these organs in ark mutants were resistant to apoptosis that was induced by γ irradiation. These data, combined with previous studies (Zhou et al., 1999; Chew et al., 2004; Daish et al., 2004; Waldhuber et al., 2005), suggest that the ARK–DRONC pathway is essential for mediating radiation-induced apoptosis.
Figure 5.
Radiation-induced cell death is inhibited in ark mutants. AO staining of WT and ark third instar larval CNS (A), eye discs (B), and wing discs (C) from untreated controls or after treatment with 12 Gy γ radiation. No significant increase in AO staining in response to irradiation is observed in ark1 and ark 2 brain lobes, eye discs, and wing discs compared with WT. Untreated ark1 and ark 2 tissues have little or no AO-positive cells compared with untreated WT eye discs. Bars, 100 μm.
Conclusions
The in vivo data presented using two independent ark mutant alleles suggest that ARK plays an essential function in most developmental PCD in flies and that removal of ark results in profound developmental defects and lethality. Our results, combined with previously published data (Kanuka et al., 1999; Rodriguez et al., 1999; Zhou et al., 1999; Zimmermann et al., 2002), indicate that ARK is required for most PCD that occurs during embryonic and larval development, for the efficient removal of larval salivary glands during larval/pupal metamorphosis, and for stress-mediated apoptosis. Thus, it seems that ARK has a more global function in PCD than Apaf-1, its mammalian counterpart. However, consistent with Apaf-1 knockout data (Marsden et al., 2002), the results also suggest that some caspase activation and cell death can still occur in the absence of ARK.
As ark mutants essentially phenocopy the loss-of-dronc function, our data argue that these proteins act in a common pathway. Previous experiments using RNA interference have shown that ARK is required for DRONC activation (Muro et al., 2004). These results suggest that the primary function of ARK is to facilitate DRONC activation. The observation that metamorphic midgut cell death occurs normally, whereas salivary gland PCD is significantly delayed, suggests that the midgut may provide a model system for studying novel caspase activation and cell death pathways that are independent of the evolutionarily conserved canonical pathway.
Materials and methods
Fly genetics
Ethylmethanesulfonate-generated ark alleles were identified in a genetic screen for genes that confer a survival advantage to mutant tissue (Harvey et al., 2003). Meiotic recombination mapping was used to initially map the mutant locus to a 1-Mb interval around 53C and 54B. Lethal complementation analysis was then performed with small deficiencies from this region. The mutant locus failed to complement y; Df(2R)P803-Delta15, cn1, which is a deletion spanning 14 annotated genes, but complemented w; Df(2R)ED1, which contains intact coding sequence for ark, but lacks the remaining 13 genes of the y; Df(2R)P803-Delta15, cn1 deletion. This revealed that mutations in the ark gene were likely causing the lethality of the mutant alleles we isolated. For experimental analysis we used stocks balanced over Cyo Kr-GFP to allow identification of homozygous animals. Lethality tests were performed at 25°C. Embryos deposited over 4 h were counted, and development to early pupae was monitored for 12 d. Developmental delay was analyzed by scoring the emergence of early pupae over the indicated times. Survival rates of homozygous animals were calculated following Mendelian principles using the observed number of heterozygous animals at early pupal stage to determine the expected homozygous complement. Larvae were staged by gut clearance after feeding on food supplemented with 5% Bromophenol blue.
Cell death detection
TUNEL of embryos was performed essentially as previously described (Quinn et al., 2000). Dissected larval tissues were fixed for 20 min in 4% formaldehyde in PBS/Tween-20, washed in PBS/Tween-20, permeabilized by incubation in 100 mM sodium citrate/0.1% Triton X-100 at 65°C for 30 min, and then TUNEL assayed using a kit (Roche). Tissues were mounted in 80% glycerol with 4 μg/ml Hoechst for confocal analysis. For AO staining, larval tissues were dissected in 1.6 μM AO/PBS, incubated for 10 min, washed in PBS, and analyzed by confocal microscopy (see Microscopy and image capture).
Immunohistochemistry
Active caspase-3 staining was performed essentially as previously described (Daish et al., 2004). Anti-active caspase-3 (Cell Signaling Technology) and anti-lamin DmO ADL67.10 (Developmental Studies Hybridoma Bank) antibodies were used at 1:50 and 1:400, respectively. For ELAV and BP102 staining, embryos were fixed in 4% formaldehyde and blocked in 10% goat/sheep sera. Anti-ELAV and BP102 antibodies (both from Developmental Studies Hybridoma Bank) were used at 2 μg/ml. Alexa Fluor 488– and 568–coupled secondary antibodies (Invitrogen) were used at 1:500.
Histology
Staged animals were fixed in 85% ethanol/4% formaldehyde/5% acetic acid/1% glutaraldehyde; then they were paraffin embedded, sectioned, stained, and analyzed by light microscopy (see next section).
Microscopy and image capture
Images in Figs. 1 D (top), 3 A, 4 A, and 4 B were obtained using a microscope (model SZ40; Olympus) with a 110AL 2× objective and captured using a camera (model DP11; Olympus). Images in Figs. 1 D (middle and bottom) and 3 B were obtained using a microscope (model BX51; Olympus) with UPlanApo objectives, fitted with a camera (model DP70; Olympus) and processed with Olysia Bioreport software (Olympus). Images in Figs. 2 (A–D), 3 C, 4 (C and D), and 5 were captured using a confocal microscope (Radiance 2100; Bio-Rad Laboratories) equipped with three lasers, an Argon ion 488 nm (14 mW); a Green HeNe 543 nm (1.5 mW); and a Red Diode 637 nm (5 mW), and an inverted microscope (model IX70; Olympus) with UApo objectives. The dual-labeled cells/tissues were imaged with two separate channels (photomultiplier tubes) in a sequential setting. All image acquisitions were performed at room temperature. Images were compiled using Photoshop 6.0 (Adobe).
Caspase assays
20–50 μg of animal or tissue lysates were used for caspase assays following established protocols (Dorstyn et al., 2002, 2004). Cleavage of the caspase substrates VDVAD-AMC (a preferred DRONC substrate) and DEVD-AMC (a substrate for effector caspases such as DRICE and DCP-1) was used for determining enzyme activities.
Acknowledgments
We thank T. Shandala for helpful discussions, G. Sarvestani for assistance with confocal microscopy, the Bloomington Stock Center for fly strains, and the Developmental Studies Hybridoma Bank for the supply of antibodies.
This work was supported by the National Health and Medical Research Council (S. Kumar) and also in part by the National Institutes of Health (I.K. Hariharan).
Abbreviations used in this paper: AO, acridine orange; CARD, caspase recruitment domain; CNS, central nervous system; ELAV, embryonic lethal abnormal visual protein; PCD, programmed cell death; RPF, relative to puparium formation; WT, wild-type.
References
- Adams, J.M. 2003. Ways of dying: multiple pathways to apoptosis. Genes Dev. 17:2481–2495. [DOI] [PubMed] [Google Scholar]
- Baehrecke, E.H. 2002. How death shapes life during development. Nat. Rev. Mol. Cell Biol. 3:779–787. [DOI] [PubMed] [Google Scholar]
- Cakouros, D., T. Daish, D. Martin, E.H. Baehrecke, and S. Kumar. 2002. Ecdysone-induced expression of the caspase DRONC during hormone- dependent programmed cell death in Drosophila is regulated by Broad-Complex. J. Cell Biol. 157:985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakouros, D., T.J. Daish, and S. Kumar. 2004. Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc, regulating its expression in specific tissues. J. Cell Biol. 165:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashio, P., T.V. Lee, and A. Bergmann. 2005. Genetic control of programmed cell death in Drosophila melanogaster. Semin. Cell Dev. Biol. 16:225–235. [DOI] [PubMed] [Google Scholar]
- Chew, S.K., F. Akdemir, P. Chen, W.-J. Lu, K. Mills, T. Daish, S. Kumar, A. Rodriguez, and J.M. Abrams. 2004. The apical caspase, dronc, governs programmed and unprogrammed cell death in Drosophila. Dev. Cell. 7:897–907. [DOI] [PubMed] [Google Scholar]
- Daish, T.J., K. Mills, and S. Kumar. 2004. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell. 7:909–915. [DOI] [PubMed] [Google Scholar]
- Dorstyn, L., P.A. Colussi, L.M. Quinn, H. Richardson, and S. Kumar. 1999. Dronc, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. USA. 96:4307–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn, L., S.H. Read, D. Cakouros, J.R. Huh, B.A. Hay, and S. Kumar. 2002. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 156:1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn, L., K. Mills, Y. Lazebnik, and S. Kumar. 2004. The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J. Cell Biol. 167:405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekert, P.G., S.H. Read, J. Silke, V. Marsden, H. Kaufmann, C.J. Hawkins, R. Gerl, S. Kumar, and D.L. Vaux. 2004. Apaf-1 and caspase-9 accelerate apoptosis, but do not determine whether factor-deprived or drug-treated cells die. J. Cell Biol. 165:835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, K.F., C.M. Pfleger, and I.K. Hariharan. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 114:457–467. [DOI] [PubMed] [Google Scholar]
- Hengartner, M.O. 2000. The biochemistry of apoptosis. Nature. 407:770–776. [DOI] [PubMed] [Google Scholar]
- Jiang, C., E.H. Baehrecke, and C.S. Thummel. 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 124:4673–4683. [DOI] [PubMed] [Google Scholar]
- Kanuka, H., K. Sawamoto, N. Inohara, K. Matsuno, H. Okano, and M. Miura. 1999. Control of the cell death pathway by Dapaf-1, a Drosophila APAF-1/CED-4 related caspase activator. Mol. Cell. 4:757–769. [DOI] [PubMed] [Google Scholar]
- Kumar, S., and J. Doumanis. 2000. The fly caspases. Cell Death Differ. 7:1039–1044. [DOI] [PubMed] [Google Scholar]
- Kumar, S., and D. Cakouros. 2004. Transcriptional control of the core cell-death machinery. Trends Biochem. Sci. 29:193–199. [DOI] [PubMed] [Google Scholar]
- Lee, C.Y., B.A. Cooksey, and E.H. Baehrecke. 2002. Steroid regulation of midgut cell death during Drosophila development. Dev. Biol. 250:101–111. [DOI] [PubMed] [Google Scholar]
- Marsden, V.S., L. O'Connor, L.A. O'Reilly, J. Silke, D. Metcalf, P.G. Ekert, et al. 2002. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature. 419:634–637. [DOI] [PubMed] [Google Scholar]
- Muro, I., K. Monser, and R.J. Clem. 2004. Mechanism of Dronc activation in Drosophila cells. J. Cell Sci. 117:5035–5041. [DOI] [PubMed] [Google Scholar]
- Quinn, L.M., L. Dorstyn, K. Mills, P.A. Colussi, P. Chen, M. Coombe, J. Abrams, S. Kumar, and H. Richardson. 2000. An essential role for the caspase Dronc in developmentally programmed cell death in Drosophila. J. Biol. Chem. 275:40416–40424. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A., H. Oliver, H. Zou, P. Chen, X. Wang, and J.M. Abrams. 1999. DARK is a Drosophila homologue of APAF-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat. Cell Biol. 1:272–279. [DOI] [PubMed] [Google Scholar]
- Waldhuber, M., K. Emoto, and C. Petritsch. 2005. The Drosophila caspase DRONC is required for metamorphosis and cell death in response to irradiation and developmental signals. Mech. Dev. 122:914–927. [DOI] [PubMed] [Google Scholar]
- Xu, D., Y. Li, M. Arcaro, M. Lackey, and A. Bergmann. 2005. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 132:2125–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, V.P., and C.S. Thummel. 2005. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin. Cell Dev. Biol. 16:237–243. [DOI] [PubMed] [Google Scholar]
- Yu, X., L. Wang, D. Acehan, X. Wang, and C.W. Akey. 2006. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 355:577–589. [DOI] [PubMed] [Google Scholar]
- Zhou, L., Z. Song, J. Tittel, and H. Stellar. 1999. HAC-1 a Drosophila homolog of Apaf-1 and CED-4, functions in developmental and radiation-induced apoptosis. Mol. Cell. 4:745–755. [DOI] [PubMed] [Google Scholar]
- Zimmermann, K.C., J.-E. Ricci, N.M. Droin, and D. Green. 2002. The role of ARK in stress-induced apoptosis in Drosophila cells. J. Cell Biol. 156:1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H., Y. Li, X. Liu, and X. Wang. 1999. An APAF-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274:11549–11556. [DOI] [PubMed] [Google Scholar]