Abstract
Separase is an evolutionarily conserved protease that is essential for chromosome segregation and cleaves cohesin Scc1/Rad21, which joins the sister chromatids together. Although mammalian separase also functions in chromosome segregation, our understanding of this process in mammals is still incomplete. We generated separase knockout mice, reporting an essential function for mammalian separase. Separase-deficient mouse embryonic fibroblasts exhibited severely restrained increases in cell number, polyploid chromosomes, and amplified centrosomes. Chromosome spreads demonstrated that multiple chromosomes connected to a centromeric region. Live observation demonstrated that the chromosomes of separase-deficient cells condensed, but failed to segregate, although subsequent cytokinesis and chromosome decondensation proceeded normally. These results establish that mammalian separase is essential for the separation of centromeres, but not of the arm regions of chromosomes. Other cell cycle events, such as mitotic exit, DNA replication, and centrosome duplication appear to occur normally. We also demonstrated that heterozygous separase-deficient cells exhibited severely restrained increases in cell number with apparently normal mitosis in the absence of securin, which is an inhibitory partner of separase.
Introduction
Equal delivery of replicated genetic information to daughter cells is essential for dividing cells during mitosis. Chromosome segregation, which occurs at the metaphase/anaphase transition, is the critical event in this delivery process. The securin–separase complex is responsible for sister chromatid separation (Yanagida, 2000; Amon, 2001; Nasmyth, 2002). Separase is a protease that cleaves cohesin, which joins the sister chromatids together. Securin, which is an inhibitor of separase, is degraded by the proteasome after anaphase-promoting complex/cyclosome–mediated polyubiquitination at the metaphase/anaphase transition; the destruction of securin activates separase in anaphase. Although these key proteins are functionally conserved from fungi to vertebrates, some of the additional properties of these proteins differ between species. Deletion of securin is lethal to fission yeast and flies (Hirano et al., 1986; Uzawa et al., 1990; Stratmann and Lehner, 1996), but does not affect the viability of mice (Mei et al., 2001; Wang et al., 2001). In vertebrates, cyclin-dependent kinases potently down-regulate separase activity (Stemmann et al., 2001). To determine the mechanisms by which separase and securin are involved in human development and disease, more direct studies in mammalian systems are required. Using mouse reverse genetics, we demonstrate that separase is essential for the early development of mice. In mouse embryonic fibroblast (MEF) cells, this protein is essential for the elimination of centromeric chromosomal cohesion during mitosis.
Results
Separase is essential for mouse development
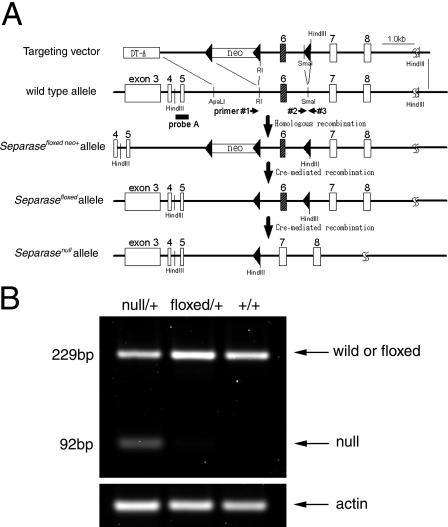
To examine mammalian separase function in vivo, we performed targeted inactivation of this gene (Shibata et al., 1997) in mice. Using homologous recombination followed by loxP-mediated deletion (Sternberg and Hamilton, 1981), we deleted a 1.4-kb genomic DNA fragment containing exon 6 of the separase gene from embryonic stem (ES) cells.
RT-PCR analysis of separase gene transcripts detected a reduction in wild-type separase expression to approximately half of normal levels. This analysis also discovered an additional transcript specifically in ES cells bearing the exon-deleted mutant allele (Fig. 1 B). Sequencing of the amplified fragment identified that this aberrant transcript was generated by the splicing of exons 5–7, which encoded a frame-shift mutation at codon 452. This transcript was present at ∼30% of the levels seen for the wild-type transcript, likely because of non-sense–mediated decay. Therefore, we concluded that functional separase expression was inactivated in cells bearing the exon-deleted mutant allele, which was designated Separasenull (Fig. 1).
Figure 1.
Generation of Separasefloxed and Separasenull alleles. (A) The exon–intron structures of the NH2-terminal region of the wild-type and mutant mouse Separase genes are shown. The linearized targeting vector (top line) contained a pGK-neo sequence inserted into intron 5 flanked by a pair of loxP sequences and an additional loxP sequence in intron 6. This construct was electroporated into mouse ES cells to generate the Separasefloxed neo+ allele by homologous recombination. After Cre-PAC electroporation, transient expression of Cre recombinase of ES cells containing the Separasefloxed neo+ allele generated the Separasefloxed or Separasenull loci by Cre-mediated recombination of either the pair of loxP sequences flanking pGK-neo or the outermost loxP sequences, respectively. The Separasefloxed allele retained a pair of loxP sequences in the introns flanking exon 6. In the Separasenull allele, the 1.5-kb fragment containing exon 6 was deleted, leaving a single loxP sequence. The position of the HindIII sites and the Southern blotting probe (probe A) used to confirm the structures of these loci are indicated. The positions of the primers used to confirm the loci structure by PCR analysis are also shown (#1, 2, and 3). (B) RT-PCR analysis of mRNA derived from Separasefloxed/+, Separase +/+, and Separasenull/+ ES cells. The levels of wild-type separase mRNA expressed were quantified from the amount of the 229-bp amplified cDNA fragment. The levels of separase expression in Separasefloxed/+ cells were comparable to those seen in Separase +/+ cells, whereas that in Separasenull/+ cells was reduced approximately to the half the levels observed in Separase +/+ or Separasefloxed/+ cells. In Separasenull/+ cells, an additional cDNA fragment of 92 bp was also detected; this fragment was confirmed by sequencing to be the region of exon 6 containing the deletion (not depicted). The levels of aberrant fragment amplified from the mutant transcript were ∼30% of the levels of the wild-type fragment seen in Separase +/+ cells. The relative amounts of the 229-bp band in Separasefloxed/+ and Separasenull/+ cells were 1.08 and 0.53, respectively, normalized to the amount seen in Separase +/+ cells, whereas the relative amount of the 92-bp band in Separasenull/+ cells was 0.16.
Mutant mice generated from these ES cells that were heterozygous for the Separasenull allele were intact and fertile. Homozygous mutants, however, could not be obtained by the breeding of heterozygotes, indicating that separase deficiency was embryonically lethal (Table I). In utero analyses discovered the death of homozygous mutant embryos before embryonic day (E) 8.5, prompting us to investigate early development in separase-deficient animals. Separasenull/+ female mice were crossed to Separasenull/+ male mice, and blastocysts were obtained by uterine washes at E3.5. Genotype analysis identified homozygous blastocysts present at Mendelian ratios. Homozygous blastocysts could be identified by gross appearance under a dissection microscope (Fig. 2, B and D); the mutants were smaller than heterozygous or wild-type blastocysts (Fig. 2, A and C) that were obtained from the same female.
Table I.
Genotype of offspring from breeding of Separasenull mutant heterozygotes
| Stage | Separase +/+ | Separasenull/+ | Separasenull/null | Not identified | Total |
|---|---|---|---|---|---|
| E3.5 | 19 | 47 | 13 | 16 | 95 |
| E3.5 + 1-d culture | 9 | 26 | 10 | 13 | 58 |
| E3.5 + 3-d culture | 17 | 31 | 5 | 7 | 60 |
| E8.5 | 10 | 23 | 0 | 33 | |
| E10.5 | 6 | 15 | 0 | 21 | |
| Term | 40 | 71 | 0 | 111 |
No Separasenull homozygous mice could be observed in a total of 111 live births from Separasenull heterozygous intercrosses. Although Separasenull homozygous blastocysts could be obtained at E3.5, no Separasenull homozygous embryos were observed after E8.5.
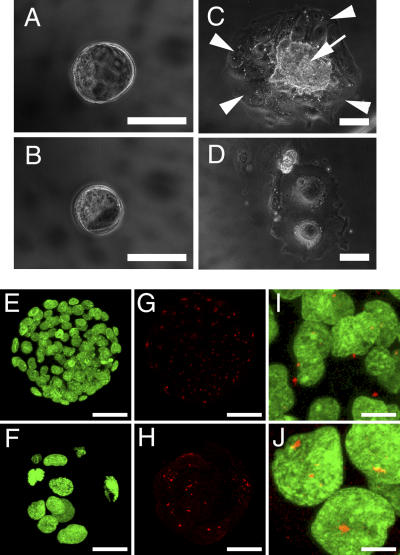
Figure 2.
Abnormal features of Separasenull/null blastocysts. Blastocysts heterozygous (A, C, E, G, and I) and homozygous (B, D, F, H, and J) for the Separasenull mutation were obtained from pregnant female mice at E3.5 (A and B) and cultured in Terasaki plates for 3 d (C and D). Expansion of the inner cell mass (arrow) and spreading of the growing trophoblasts (arrowheads) were observed in heterozygous, but not homozygous, Separasenull blastocysts. After 1 d of culture, immunostaining of blastocysts with both Cytox green to detect DNA (E, F, I, and J, green) and an anti-pericentrin antibody to visualize centrosomes (G–J, red) identified a smaller number of cells with enlarged nuclei and multiple centrosomes in the blastocysts homozygous for Separasenull. Bars: (A–D) 100 μm; (E–H) 50 μm; (I and J) 12.5 μm.
Culture of blastocysts in vitro for 1 d revealed that blastocysts homozygous for Separasenull (Fig. 2, F, H, and J) were easily distinguishable from wild-type or heterozygous blastocysts by their smaller numbers of cells with abnormally large nuclei (Fig. 2, E, G, and I). The total number of cells in blastocysts homozygous for Separasenull was only 10% of those seen in heterozygotes. In contrast, the mean diameter of mutant cell nuclei, detected by Cytox green staining, was approximately twice that of heterozygotes (Fig. 2, compare F with E; quantitative data not depicted). Immunostaining of homozygous blastocysts with antibodies against pericentrin, which is a component of centrosomes (Fig. 2, G–J, red), revealed an increased number of centrosomes per cell (red pericentrin signals of the merged images in Fig. 2 J).
After an additional 3-d culture, neither expansion of the inner cell mass nor spreading of trophoblasts on the dish surface could be observed for homozygous Separasenull blastocysts (Fig. 2 D). Heterozygous blastocysts cultured for 3 d (Fig. 2 C) exhibited a normal inner cell mass (arrow) with observable spreading of the growing trophobalsts (arrowheads). DNA staining of homozygous embryos with Cytox green could not detect an increased incidence of apoptotic cell death (unpublished data), indicating that separase-deficient embryos suffered from cell cycle arrest or retarded growth at the blastocyst stage that resulted in death at an early embryonic stage.
Separase-deficient MEFs show growth retardation and increased ploidy
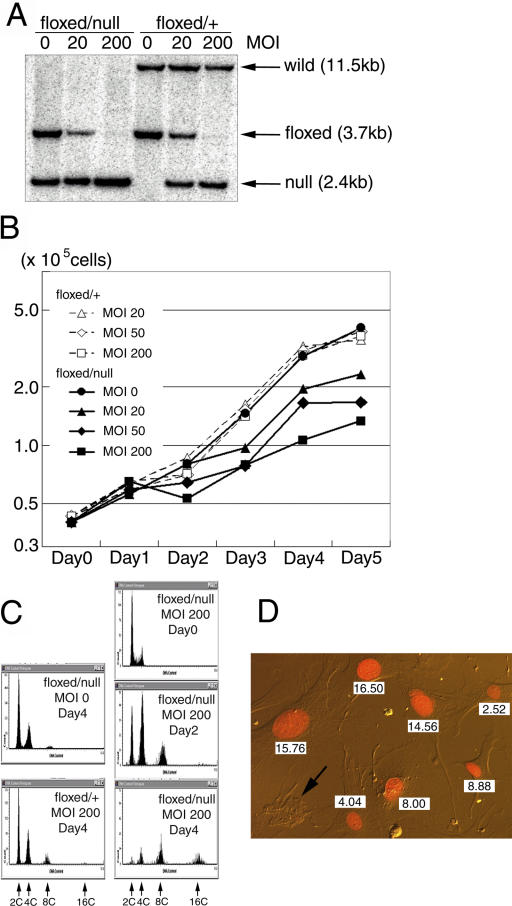
To investigate the growth defects of separase-deficient cells, we established mutant mice carrying a conditional allele of mutant separase (Separasefloxed). These animals were generated by the insertion of a pair of loxP sequences into introns 5 and 6 (Fig. 1 A). RT-PCR analysis of separase gene transcripts detected separase expression in Separasefloxed/+ ES cells that was equal to the expression observed in wild-type cells, suggesting that the Separasefloxed allele is functionally intact (Fig. 1 B). Mutant animals homozygous for Separasefloxed, which were healthy and fertile, were crossed to Separasenull/+ mice. MEFs, which were prepared from the resulting embryos at E14.5, were infected with a recombinant adenovirus bearing the Cre gene (AxCre; Shibata et al., 1997) to inactivate separase expression. We analyzed the growth profile of separase-deficient MEFs by quantitating cell numbers at the specified time points after infection (Fig. 3 B).
Figure 3.
Reduced cell number increases and high ploidy in separase-deficient MEF. MEFs were obtained at E14.5 from embryos heterozygous for Separasefloxed or heterozygous for both the Separasenull and Separasefloxed alleles. These cells were infected with AxCre at varying MOI to inactivate the separase gene by converting the Separasefloxed allele to the Separasenull allele. (A) Conversion to the Separasenull allele after AxCre infection was confirmed by Southern blot analysis. Genomic DNA was analyzed by Southern blotting after harvest of cells 3 d after infection with AxCre at MOI of 0, 20, or 200. The Separase +, Separasefloxed, and Separasenull alleles were detected as bands of 11.3, 3.7, and 2.4 kb, respectively. The Separasefloxed allele was completely inactivated by conversion to the Separasenull allele after infection at an MOI of 200 in both Separasefloxed/null and Separasefloxed/+ MEF cells. This conversion was incomplete in cells infected at an MOI of 20. (B) The growth characteristics of cells after inactivation of separase were analyzed by quantitation of cell numbers. After infection with AxCre at MOIs of 0 (closed circles), 20 (closed triangles), 50 (closed diamonds), or 200 (closed squares), cells were cultured for 5 d. Quantitation of cell numbers each day after Cre expression revealed growth retardation of MEFs heterozygous for both Separasenull and Separasefloxed (floxed/null; continuous lines), but not those heterozygous for Separasefloxed alone (floxed/+; dashed lines). (C) DNA content in MEF after infection was analyzed by LSC. 4 d after infection, Separasefloxed/+ MEF infected with AxCre at an MOI of 200 and mock-infected Separasefloxed/null MEF exhibited normal DNA content (2C and 4C). Although Separasefloxed/null MEF infected with AxCre at an MOI of 200 contained normal DNA content (2C and 4C) at day 0 (AxCre infection), cells with an abnormally high DNA content accumulated progressively, with the appearance of 8C cells at 2 d and 8C and 16C cells 4 d after infection. (D) Separasefloxed/null MEF were infected with AxCre at an MOI of 200. 4 d after infection, cellular DNA content and nuclear morphology were analyzed by LSC. Nuclei were stained with propidium iodide; relative DNA content is indicated as integers (2C = 2.00). Each MEF with high DNA ploidy contained a single abnormally large nucleus. Anuclear cell fragments were also observed (arrow).
Separasefloxed/+ MEFs exhibited a growth capacity identical to that of wild-type MEFs after infection with AxCre at a high multiplicity of infection (MOI; Fig. 3 B, dashed lines). AxCre infection, however, significantly retarded the growth of Separasefloxed/null MEFs (Fig. 3 B, continuous lines). This effect was not observed after mock infection. This growth inhibitory effect was amplified by infections at higher MOIs, suggesting that the observed growth retardation resulted from separase deficiency. As we could not detect an increased incidence of apoptosis in the growth-retarded cells (unpublished data), we hypothesized that separase deficiency inhibited the observed increases in MEF cell numbers.
To understand the mechanisms underlying this inhibition, we performed a cell cycle analysis of cultures using laser scanning cytometry (LSC; Fig. 3 C). After infection with AxCre, Separasefloxed/+ MEFs maintained normal ploidy (2C and 4C; Fig. 3 C, bottom left), whereas cultures of Separasefloxed/null MEFs revealed an accumulation of cells with an abnormally high ploidy, usually 8C and 16C after 2 and 4 d, respectively (Fig. 3 C, right). The increase in DNA content was not observed in mock-infected cells (Fig. 3 C, top left). As the total DNA content in Separasefloxed/null MOI 200 cells was similar to that in the mock-infected Separasefloxed/null control cells (unpublished data), additional rounds of DNA replication appeared to have occurred in the separase-defective MEF cells. Interestingly, the actual DNA content of each Separasefloxed/null MOI 200 cell correlated roughly with the size of the nuclei (Fig. 3 D), suggesting that enlargement of the nuclei in Separasefloxed/null MEFs infected with AxCre may follow the additional rounds of DNA replication. 4 d after AxCre infection, the numbers of centrosomes per cell also increased in Separasefloxed/null MOI 200 MEFs (6.4 ± 4.3) from the numbers observed in mock-infected Separasefloxed/null MEFs (2.1 ± 1.2). Therefore, we concluded that separase deficiency suppressed nuclear division and centrosome separation accompanied by cytokinesis in MEFs, resulting in the accumulation of cells with a high DNA content.
Heterozygous mutation of separase causes embryonic lethality on a securin-deficient background
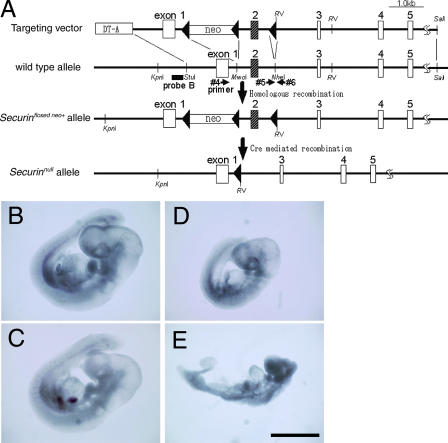
We also generated a mutant mouse line deficient in Pttg, a mammalian homologue of securin. We used homologous recombination followed by Cre-loxP–mediated recombination in ES cells to generate a securin/Pttg mutant (Securinnull) with the deletion of exon 2 (Fig. 4 A). As reported, homozygous mutants for Securinnull were viable and fertile (Mei et al., 2001; Wang et al., 2001). We crossed Securinnull heterozygotes with Separasenull heterozygotes. Double heterozygotes (Separasenull/+ Securinnull/+) were obtained at Mendelian ratios (unpublished data). These mice were subsequently crossed with Securinnull homozygotes. Genotyping of offspring at birth revealed the absence of Separasenull/+ Securinnull/null mutants, suggesting that this genotype is also embryonically lethal (Table II). Further analysis demonstrated that Separasenull/+ Securinnull/null mutant embryos died by E11.5. These embryos were easily distinguishable from their littermates as early as E9.5 by severely retarded growth (Fig. 4, B–E). These results strongly suggest that heterozygosity for separase function in the absence of Securin function exhibits haploinsufficiency. A more detailed analysis of mutant embryos will be required to elucidate the molecular mechanisms responsible for this phenotype.
Figure 4.
Separasenull/+Securinnull/null embryos exhibit developmental defects. (A) Generation of the Securinnull allele. The exon–intron structures of the wild-type and mutant mouse Pttg/Securin genes are shown schematically. The linearized targeting vector contained the pMC1-neo sequence inserted into the MwoI site within intron 1 flanked by a pair of loxP sequences and an additional loxP sequence at the NheI site within intron 2. The Securinfloxed neo+ locus was generated by homologous recombination after introduction of this vector into mouse J1 ES cells by electroporation. Cre recombinase was transiently expressed after Cre-PAC electroporation in ES cells carrying the Securinfloxed neo+ allele. Cre-mediated recombination between the outermost loxP sequences generated the Securinnull allele, which lacked exon 2 of the Securin gene. The positions of the restriction sites (KpnI and EcoRV) and the Southern blotting probe (probe B) used to confirm the structures of these loci are indicated. The positions of the primers used to confirm the locus structure by PCR analysis are also indicated (#4, 5, and 6). (B–E) Separasenull/+ Securinnull/+ female mice were crossed to Separase +/+ Securinnull/null male mice. The resulting embryos were examined at each embryonic stage. At E9.5, Separasenull/+ Securinnull/null embryos (D and E) were obtained at the expected Mendelian rate. These embryos, however, were small and less developed in comparison to the either Separasenull/+ Securinnull/+ embryos (B) or Separase +/+ Securinnull/null embryos (C). Bar, 1 mm.
Table II.
Genotype of offspring resulting from crosses of Separasenull/+Securinnull/+ females and Separase+/+Securinnull/null males
| Stage | Separase+/+ Securinnull/+ | Separase +/+ Securinnull/null | Separasenull/+ Securinnull/+ | Separasenull/+ Securinnull/null | Total |
|---|---|---|---|---|---|
| E9.5 | 6 | 4 | 11 | 6 (1) | 27 (1) |
| E10.5 | 11 | 11 (1) | 12 | 10 (5) | 44 (6) |
| E11.5 | 6 | 1 | 3 | 6 (6) | 16 (6) |
| E12.5 | 5 | 3 | 4 | 0 | 12 |
| E14.5 | 1 | 5 | 5 | 0 | 11 |
| Term | 14 | 13 | 15 | 0 | 42 |
Separasenull/+ Securinnull/+ female mice were crossed to Separase +/+ Securinnull/null male mice. No Separasenull/+ Securinnull/null mice could be observed in a total of 42 live births. Living Separasenull/+ Securinnull/null embryos could be obtained at E9.5 and E10.5. After E11.5, however, no living Separasenull/+ Securinnull/null embryos could be observed. The number of dead embryos is indicated in parentheses.
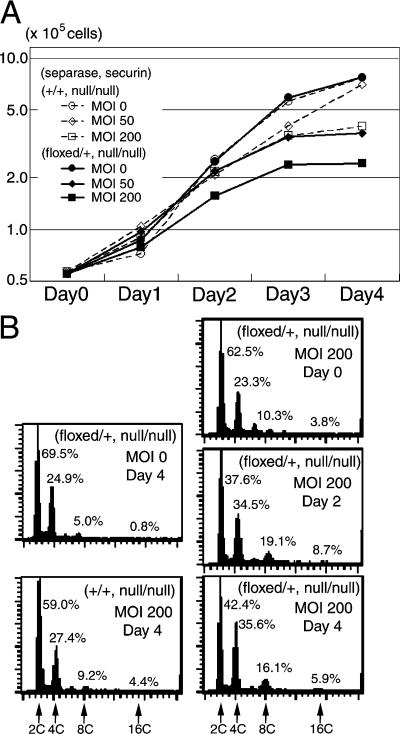
Heterozygous separase mutation causes growth retardation of MEF cells on a securin-deficient background
The death of Separasenull/+ Securinnull/null embryos by E11.5 suggested a genetic interaction between these genes during embryonic development. To elucidate the molecular nature of this interaction, we analyzed separase and securin functions in MEF cells. Separasefloxed/+ Securinnull/+ mice were crossed to Securinnull homozygotes; MEFs prepared from the resulting Separasefloxed/+ Securinnull/null and Separase +/+ Securinnull/null embryos at E14.5 were infected with AxCre to inactivate separase. We observed only modest growth retardation of Separase +/+ Securinnull/null MEFs, which did not carry any floxed alleles, after AxCre infection. This growth suppression was increasingly evident in cells infected at higher MOIs (Fig. 5 A). As AxCre infection did not alter the growth profile of wild-type or Separasefloxed/+ MEF cells (Fig. 3 B and not depicted), this result suggests that Cre expression alone by recombinant adenovirus may suppress the growth of MEFs lacking a functional securin gene. In comparison to this modest effect, however, Separasefloxed/+ Securinnull/null MEFs exhibited severely restrained increases in cell number after infection with AxCre (Fig. 5 A), suggesting that a single functional Separase allele is not sufficient to support normal growth in securin-deficient MEF cells. To examine this defect in detail, we analyzed the ploidy profiles of these cells by FACS analysis (Fig. 5 B). The majority of Separasefloxed/+ Securinnull/null MEF cells possessed 2C or 4C DNA, even 4 d after infection with AxCre, indicating that the additional DNA replication resulting in polyploidy that was observed in Separasefloxed/null MEF cells did not occur in Separasefloxed/+ Securinnull/null MEF cells. We also did not observe any changes in the incidence of mitotic cells, suggesting that mitosis proceeded normally in the presence of one intact separase gene, despite the absence of securin. We also could not detect any increased incidence of cell death in these cells. In contrast, the proportion of 4C cells significantly increased; 27.4% of Separase +/+ Securinnull/null and 35.6% of Separasefloxed/+ Securinnull/null MEF cells exhibited 4C DNA at 4 d after infection with AxCre. These results suggest that cell cycle progression may be restrained during G2 phase in Separasefloxed/+ Securinnull/null MEF cells.
Figure 5.
Heterozygous deletion of separase causes growth retardation and abnormal DNA content of MEF on a securin-deficient background. MEF were obtained from Separasefloxed/+ Securinnull/null (floxed/+, null/null) or Separase +/+ Scurinnull/null (+/+, null/null) embryos at E14.5. (A) Cells were cultured for 4 d after infection with AxCre at an MOI of 0 (circles), 50 (diamonds), or 200 (squares). Analysis of the increases in cell number indicated growth retardation in both Separasefloxed/+ Securinnull/null and Separase +/+ Scurinnull/null MEFs after AxCre infection. The extent of growth retardation, however, was more severe in Separasefloxed/+ Securinnull/null cells (continuous lines) than in Separase +/+ Scurinnull/null cells (dashed lines) at the same MOI. (B) DNA content in MEF after AxCre infection was analyzed by flow cytometry. Separase +/+ Scurinnull/null cells infected with AxCre at an MOI of 200 and mock-infected Separasefloxed/+ Securinnull/null cells exhibited normal DNA content after 4 d of culture. Although Separasefloxed/+ Securinnull/null MEFs demonstrated normal DNA content at day 0 (AxCre infection), they displayed abnormal DNA content 2 and 4 d after infection with increased numbers of 4C cells and decreased 2C cells.
Separase is essential for the separation of centromeric regions of sister chromatids
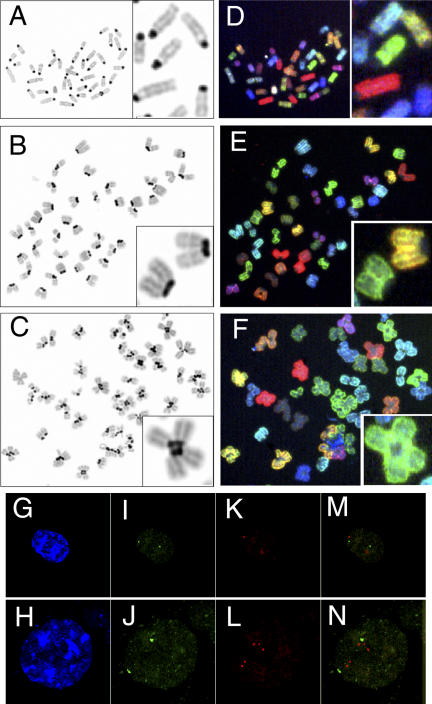
To examine the chromosomal structure of MEF cells with abnormally high ploidy, we performed karyotype analysis. 3 d after infection with AxCre, MEF cells were mitotically arrested using colcemid; prepared chromosomal spreads were then observed. Wild-type cells arrested at prometaphase each contained 40 chromosomes, including a pair of acrocentric chromatids (Fig. 6 A). In separase-deficient cells, the majority of the spreads contained chromosomal clusters (Fig. 6, B and C, inset). The total number of chromosomal clusters per cell was ∼40; each chromosomal cluster contained two or four pairs of chromatids. These pairs were attached at their centromeric regions to form diploid or quadruple chromosomes, respectively (Fig. 6, B and C).
Figure 6.
Aberrant structure and localization of chromosomes in separase-deficient MEF. Karyotype analysis of Separase +/+ (A and D) and Separasefloxed/null (B, C, E, and F) MEF was performed after arrest by colcemid treatment 3 d after AxCre infection. Chromosome spreads were stained by DAPI (A–C) or processed for SKY analysis (D–F). The typical images of the chromosomes were enlarged and shown in the insets of A–F. Separase +/+ MEF contained diploid chromosomes; each chromosome exhibited typical acrocentric figures (A, inset). Tetraploid (B and E) and octaploid (C and F) cells, however, were frequently observed in Separasefloxed/null MEF. Two or four chromosomes were connected at their centromeric regions, forming diploid (B, inset) or quadruple chromosomes (C, inset), respectively. SKY analysis demonstrated that these diploid or quadruple chromosomes consisted of multiples of the same sister chromatid (E and F). The interphase nuclei of AxCre-infected Separasefloxed/+ (G, I, K, and M) and Separasefloxed/null (H, J, L, and N) MEFs were analyzed by FISH with probes for the centromeric (I, J, M, and N, green) and telomeric (K, L, M, and N, red) regions of chromosome 5. Nuclei were also stained with DAPI (G and H, blue). After AxCre infection, the majority of Separasefloxed/+ MEF exhibited two centromeric signals and two telomeric signals per nucleus (G, I, K, and M), whereas Separasefloxed/null MEF frequently displayed multiple telomeric signals scattered throughout the nucleus, and the centromeric signals formed two large clusters (H, J, L, and N).
To determine if these abnormal chromosome clusters contained multiple copies of the same chromatid, we applied the spectral karyotyping (SKY) method to these chromosome spreads. All chromosomes within clusters stained with a single color, demonstrating that they were copies of the same sister chromatid (Fig. 6, E and F; D was control). In separase-deficient cells, these abnormal connections of chromosomal pairs and tetramers at their centromeric regions likely resulted from defects in chromosome segregation during previous rounds of mitosis. It is also possible that in the absence of separase cohesin is abnormally abundant at the centromeric regions of chromosomes, resulting in aberrant DNA replication that creates abnormal chromosome connections.
We next analyzed the localization of genomic DNA in the nuclei of interphase cells by FISH (Fig. 6, G–N). In control MEF cells expressing normal separase, all of the probes detecting specific chromosomal regions (chromosome 5–specific centromeric and telomeric probes are shown in green and red, respectively) detected two spots within each nucleus (Fig. 6, I, K, and M). This result likely reflects normal diploidy; sister chromatids were located in such close vicinity that resolution of the two was not possible, even after DNA replication. In separase-deficient cells, telomeric probes detected several scattered spots reflective of the high ploidy. The centromeric probes, however, detected either two spots or clusters (Fig. 6, H, J, L, and N). These results indicated that abnormal connections of chromatids at their centromeric regions existed in separase-deficient cells, even during interphase. Separase thus plays an essential role in the separation of the centromeric regions of sister chromatids in mouse cells; separase deficiency resulted in the formation of aberrant centromeric connections between chromosomal pairs or tetramers.
Because FACS analysis also detected a slight increase of the proportion of cells with high DNA contents, such as 8C and 16C, in Separasefloxed/+ Securinnull/null MEF cells after infection with AxCre (Fig. 5 B), we also analyzed chromosomal structures of these cells by karyotype analysis. From the chromosomal spreads of Separasefloxed/+ Securinnull/null MEF cells infected with AxCre, 14 spreads, each of them containing 80 chromosomes (8C), were picked up and analyzed. In these spreads, the centromeres of most chromosomes (78.4%) were apart from each other and localized free from other chromosomes. This incidence was almost identical to that observed in Separase +/+ Securinnull/null MEF cells infected with AxCre (79.6%). These results suggested that Separase expressed from a single allele is enough to separate centromeric connections of chromatids in Separasenull/+ Securinnull/null MEF cells suffering from the arrest or severe delay of cell cycle.
Separase is essential for chromosome segregation, but not for exit from mitosis
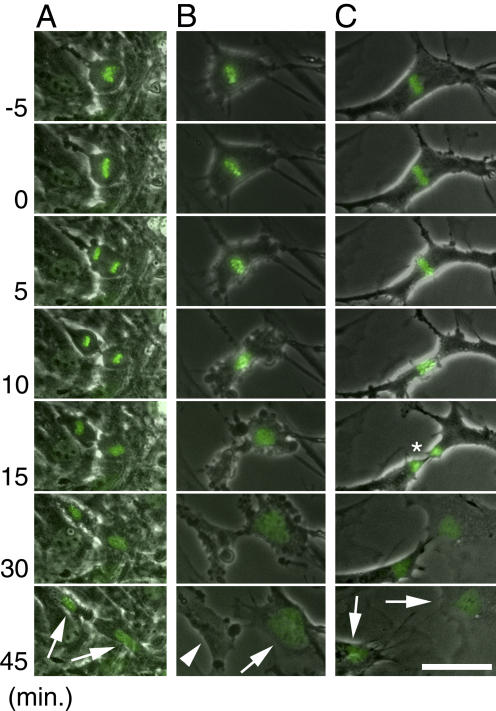
These results suggested that the cell cycle progresses in separase-deficient cells, despite the persistence of connections between multiple chromosomes at their centromeric regions. To analyze the defect in cell division observed in the absence of separase, we infected Separasefloxed/null and Separasefloxed/+ MEF with the recombinant adenoviruses AxCre and AxH-GFP, which encode GFP-tagged histone H2B. Using this technique, we monitored chromosomal dynamics during the cell cycle by time-lapse microscopy (Fig. 7). In Separasefloxed/+ cell cultures infected with AxCre, the majority of cells that had initiated mitosis during the observation period underwent normal mitotic division into two daughter cells (Fig. 7 A). In contrast, a normal pattern of chromosomal segregation was rarely observed in separase-deficient cells. The proportion of cells undergoing chromosomal condensation, however, was similar in both separase-deficient and control cell populations, indicating that the defect in segregation occurred after condensation. In the majority of separase-deficient cells (62.8%), the condensed chromosomes aligned to form the metaphase plate, but never segregated. As the cells could not enter anaphase (Fig. 7, B and C), the nonsegregated chromosomes then decondensed, reforming a single nucleus (Fig. 7 B). Nuclear reformation was accompanied by a cytokinesis-like cytoplasmic division, resulting in the production of a subset of anuclear cells (Fig. 7 B, arrowhead). In 9.3% of all mitoses in separase-deficient cells (4/43), cytokinesis divided the decondensing chromosomes (Fig. 7 C, asterisk), separating the nuclear chromatin into two blocks (arrows). This type of abnormal mitosis (cytokinesis in the absence of sister chromatid separation) is reminiscent of the “cut” (cell untimely torn) phenotype, which is observed in cut1/separase-deficient fission yeast (Hirano et al., 1986; Uzawa et al., 1990).
Figure 7.
Defect in chromosomal segregation in separase-deficient MEF. MEFs, which were obtained as in Fig. 3, were infected with AxCre at an MOI of 50 and AxH2B-GFP at an MOI of 2 and analyzed by time-lapse microscopy for 2 d. Representative patterns of chromosomal segregation in Separasefloxed/+ (A) and Separasefloxed/null (B and C) MEFs after infection with AxCre are shown in micrographs representing the 50–min period surrounding metaphase. Metaphase was defined by the condensation of chromosomes aligned at the metaphase plate (0 min). In Separasefloxed/+ MEFs at metaphase, condensed chromosomes separated into two clusters; the cytokinesis that accompanied chromosome decondensation produced two daughter cells (A, arrows). In the majority of Separasefloxed/null MEF, however, condensed chromosomes at metaphase initiated decondensation in the absence of segregation. Decondensation was accompanied by aberrant cytokinesis (B and C), which resulted in the production of one daughter cell with a single large nucleus and another anuclear daughter cell (arrow and arrowhead in B, respectively). At low frequency, cytokinesis tore apart the nonsegregated chromosomes in Separasefloxed/null MEF into two pieces (C, asterisk), resulting in the production of two cells with broken nuclei (C, arrows). Bar, 50 μm.
We also performed live-cell analysis on Separasefloxed/+ Securinnull/null MEF cells infected with both AxCre and AxH-GFP. In these cultures, cells undergoing chromosome condensation were identified and the mitotic process was then analyzed by time-lapse microscopy. The majority of these cells (87.0% or 20/23) segregated their chromosomes normally. No mitotic defects could be detected.
Discussion
In this study, we reported the construction and characterization of separase mutant mice. Using a conditional inactivation system, we also characterized separase-deficient MEF cells, which showed severely restrained increases in cell numbers. Our analysis demonstrated that separase is essential for chromosome segregation in mammalian cells. This result correlates well with those of previous studies in lower eukaryotes, including yeast, flies, and worms (Uzawa et al., 1990; Jager et al., 2001; Siomos et al., 2001).
Although chromosome segregation was significantly impaired, time-lapse analysis of separase-deficient MEF could not identify any apparent defect or delay in either the condensation or metaphase alignment of chromosomes. Chromosome decondensation and cytokinesis also proceeded in the absence of chromosome segregation in separase-deficient cells. This abnormal form of cell division typically produced one cell containing a single large nucleus and a second anuclear cell-like structure, explaining the accumulation of cells with an abnormally large nucleus of high DNA content containing amplified centrosomes. We also observed this phenotype in separase-deficient blastocysts in vivo. Therefore, the presence of nonsegregated chromosomes in separase-deficient cells does not appear to cause additional defects in cell cycle progression events, such as DNA replication or centrosome duplication. In fission yeast, the cut1/separase mutation results in cell death after the appearance of unsegregated chromosomes torn apart by cytokinesis (the cut phenotype; Hirano et al., 1986; Uzawa et al., 1990). Inhibition of cytokinesis in cut1/separase mutant cells, however, prevents cell death and allows cells to enter the cell cycle (Creanor and Mitchison, 1990; Uzawa et al., 1990), suggesting that failure of chromosomal segregation is not the cause of cell death in separase-deficient cells. From yeast to mammals, accumulating evidence suggests that although separase inactivation may prevent chromosome segregation, it does not interfere with other events in cell cycle progression, such as chromosome condensation and decondensation, metaphase chromosome alignment, cytokinesis, DNA replication, and centrosome duplication.
All of the chromosomes in our chromosome spreads from separase-deficient MEFs exhibited abnormal chromosomes, which were connected only at the centromeres. This chromosome structure suggested that separase was required for the separation of sister centromeres, but not of the arm regions. Indeed, SKY analysis demonstrated that these abnormal chromosomes were multiples of identical sister chromatids. Retention of these centromeric connections was confirmed by FISH analysis of interphase cells. As cell cycle progression occurred normally in separase-deficient MEF, these abnormal chromosomes may be generated by extra rounds of DNA replication of nonsegregated chromosomes, suggesting that selective centromeric linkages are maintained throughout the cell cycle in the absence of mammalian separase. In this model, the sister chromatid pairs that had failed to segregate in separase-deficient cells would replicate again during the next S phase, then condense normally at the next mitosis, retaining their abnormal centromeric connections. Although this type of aberrant chromosome was not observed in separase-deficient HeLa cells generated with knockdown technology (Waizenegger et al., 2002), inactivation of separase expression may not have been complete. Our data strongly suggest that mammalian separase is essential for centromere separation, but not for chromosome arm separation. There does not appear to be a checkpoint system capable of detecting and/or repairing the abnormal centromeric connections anywhere throughout the cell cycle in mammals.
Although homozygous mutant mice deficient in separase underwent embryonic lethality, heterozygous mutants were viable and apparently normal. On a securin-deficient background, however, heterozygous separase mutants also exhibited embryonic lethality. Heterozygous separase-deficient MEF on a securin-deficient background also exhibited severely restrained increases in cell numbers, as seen in homozygous separase-deficient MEF. These results suggest that securin may play a positive role in promoting separase function. Securin has been reported to function as a chaperone to stabilize separase in human cells and fission yeast (Jallepalli et al., 2001; Nagao et al., 2004). Therefore, separase heterozygosity would be insufficient to support increases in cell number on a securin-deficient background.
In contrast to the phenotype of separase-deficient MEF, we could not identify any apparent mitotic abnormalities in heterozygous cells on securin-deficient background by time-lapse analysis of living cells. Karyotype analysis also failed to detect abnormal chromosomes, such as diploid or quadruple chromosomes, indicating that separase heterozygosity is sufficient for sister chromatid separation, even on a securin-deficient background. Instead, we observed an accumulation of 4C cells, suggesting a possible defect in interphase. Although we are not able to exclude the possibility that additional mitotic defects might be concealed by the limited number of mitosis observed on a securin-deficient background, these results suggest that the cell cycle was significantly delayed in G2 phase. A function for separase in interphase has recently been reported. In fission yeast, separase-mediated cleavage of cohesin during interphase was essential for DNA repair (Nagao et al., 2004). Autocleavage of human separase also plays a role at the G2/M transition (Papi et al., 2005). Our results also suggest an interphase function for separase in cell cycle progression that is independent of its role in mitosis. DNA damage that occurs spontaneously in these cells may not be efficiently repaired, causing the cell cycle to be delayed in G2 by activation of the damage checkpoint.
Our analyses of mutant mice established that mammalian separase is essential for the separation of sister chromatid centromeres, probably through the separase-mediated proteolytic cleavage of cohesin in the centromeric regions. Cohesin complexes in mammalian cells are released from the chromosome arm regions without the requirement of separase-mediated cleavage (Losada and Hirano, 2005). If, however, a small amount of separase was present in the heterozygous cells, mitosis progressed normally; no polyploid chromosomes could then be observed, even in the absence of securin, whereas the cell number increase was severely retarded. These results suggest that a small amount of separase may be sufficient for the removal of centromeric cohesin during mitosis, even in the absence of securin, but more separase is required in the absence of securin for progression through interphase, a phase in which separase performs a function that remains to be identified.
Materials and methods
Generation of mutant animals
We obtained an ∼20-kb mouse genomic DNA fragment containing the NH2-terminal separase sequence by screening a 129SVJ mouse genomic DNA phage library with an NH2-terminal human separase cDNA fragment as a probe. To construct a targeting vector, we inserted an 11.5-kb mouse genomic fragment between the ApaLI site in separase intron 5 and the NotI site within the cloning site of a phage clone. A pGK-neo-polyA fragment flanked by a pair of loxP sequences was inserted into the EcoRI site of intron 5. An additional loxP sequence was inserted into the SmaI site of intron 6. A DT-A fragment was ligated to the 5′ end of the targeting vector to facilitate negative selection. After linearization by digestion with SacII, the targeting vector was electroporated into J1 ES cells as previously described (Nakai et al., 1995). After selection in G418, homologous recombinants were identified by Southern blot analysis using a 375-bp HindIII–EcoRI fragment containing separase exon 5 as a probe. Positive clones were electroporated with pCre-PAC (Taniguchi et al., 1998), which transiently expresses the Cre recombinase. Clones containing the Separasefloxed or Separasenull loci were identified by Southern blot hybridization. After HindIII digestion, hybridizing fragments of 11.3, 5.4, 3.7, or 2.4 kb should correspond to the wild-type, Separasefloxed neo+, Separasefloxed, or Separasenull alleles, respectively. We injected the mutant clones into C57BL/6J blastocysts to create chimeric mice. These animals were crossed to C57BL/6J mice, and germline transmission was confirmed by either genomic Southern blotting or PCR analysis of mouse tail DNA. For PCR, the combination of primer #2 (5′-CAGATCCTTGCCCTAGATCTCAGGC-3′) and primer #3 (5′-CTACCCAGGCTAGTGCCCTCTACTG-3′) detected a 272-bp fragment derived from the wild-type allele and a 414-bp fragment derived from the Separasefloxed allele. The combination of primer #1 (5′-TCCTGGCACTTGGGAACCAGAGGTG-3′) and primer #3 detected a 356-bp fragment derived from the Separasenull allele. The use of animals in this research study complied with all relevant guidelines for the ethical treatment of animals of the Japanese government and the Japanese Foundation for Cancer Research Cancer Institute.
RT-PCR analysis and sequencing of separase cDNA
Total mRNA of ES cell clones was isolated using Micro-Fast Track (Invitrogen) and reverse transcribed using random primers. A Separase cDNA fragment, including the region surrounding exon 6, was amplified using primers #12 (5′-TGTTGGAGGCCTTAGAGGGCCTGTC 3′) and #13 (5′-CTCTCCACATGCAGCCTGAAGCACC-3′), which correspond to sequences within exons 5 and 7 of the separase gene, respectively. The amplified fragments were separated by electrophoresis and analyzed by sequencing after subcloning into a plasmid.
Blastocyst analysis
Blastocysts, obtained at E3.5 from Separasenull/+ female mice crossed with Separasenull/+ male mice, were cultured on Terasaki plates at 37°C. Bright field images were acquired with an inverted microscope (DM IRE2; Leica). For immunofluorescence microscopy, blastocysts were fixed in 4% PFA and permeabilized with Triton X-100. Cytox green (Invitrogen) and an anti-pericentrin polyclonal antibody (Covance) were used to stain DNA and centrosomes, respectively. Immunofluorescence images were taken through a microscope (DM RE; Leica) with a confocal microscopy system (TCS SP2; Leica). Each blastocyst was carefully recovered and genotyped by PCR, as described in the previous section.
Cell culture and adenovirus infection
MEFs were obtained from embryos at E14.5, as previously described (Todaro and Green 1963), and were maintained in DME containing 10% fetal bovine serum at 37°C. Cells were used for analyses within three passages. Exponentially growing cells were plated at 5 × 104 cells per well in 6-well dishes. After a 12-h incubation, cells were infected with AxCre (3.3 × 109 plaque-forming units) at an MOI of 20 for chromosome analysis or 200 for FISH analysis. Cre-mediated recombination was confirmed by both genomic Southern blot and PCR analysis. To count centrosome numbers, cells were fixed with cold methanol and stained with anti–γ-tubulin antibody (Sigma-Aldrich). For flow cytometric analysis, cells were fixed in 70% ethanol and stained with 100 μg/ml propidium iodide solution after treatment with 2.5 mg/ml RNase A for 30 min. Cellular DNA content was also analyzed by laser scanning cytometry (LSC2 system; Olympus). For time-lapse imaging, cells were plated in 35-mm dishes before coinfection with AxH-GFP (MOI of 2), which encodes GFP-tagged histone-H2B, and AxCre (MOI of 50). Time-lapse images were taken at 1-min intervals though an inverted microscope (Leica) with a time-lapse system (AS MDW; Leica) at 37°C.
Chromosome analysis
To obtain chromosome spreads, MEF were exposed to 0.1 μg/ml colcemid for 2 h, treated with a hypotonic 0.075 M KCl solution for 15 min, and fixed in ice-cold Carnoy's fixative. For SKY analysis, chromosome spreads were treated with a 0.003% pepsin solution (0.01 M HCl) for 15 min and stained with a SkyPaint kit (Applied Spectral Imaging). Chromosomes were also counterstained with DAPI. SKY images were acquired through a fluorescence light microscope (BX50; Olympus) with a spectral imaging system (SpectraView SD-300; Applied Spectral Imaging).
FISH analysis
FISH analysis was performed as previously described (Inazawa et al., 1992; Imoto et al., 2000) using bacterial artificial chromosomes as probes. Centromeric (RP23-315O5) and telomeric (RP23-159N17) probes specific for chromosome 5 were labeled with biotin-16-dUTP and digoxigenin-11-dUTP (Roche), respectively, by nick-translation. These labels were detected with FITC-avidin and anti–digoxigenin-rhodamine, respectively. FISH images were acquired through a microscope (Axioplan2; Carl Zeiss MicroImaging, Inc.) with a confocal microscopy system (LSM 510; Carl Zeiss MicroImaging, Inc.).
Acknowledgments
We would like to thank T. Kobayashi, T. Hirota, N. Kudo, K. Nagao, and Y. Toyoda for their helpful discussion and advice and H. Yamanaka, S. Ito, M. Fukuda, and N. Ito for their excellent technical assistance.
This work was supported by a Grant-Aid for Scientific Research from the Ministry on Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used in this paper: E, embryonic day; ES, embryonic stem; LSC, laser scanning cytometry; MEF, mouse embryonic fibroblast; MOI, multiplicity of infection; SKY, spectral karyotyping.
References
- Amon, A. 2001. Together until separin do us part. Nat. Cell Biol. 3:E12–E14. [DOI] [PubMed] [Google Scholar]
- Creanor, J., and J.M. Mitchison. 1990. Continued DNA synthesis after a mitotic block in the double mutant cut1 cdc11 of the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 96:435–438. [DOI] [PubMed] [Google Scholar]
- Hirano, T., S. Funahashi, T. Uemura, and M. Yanagida. 1986. Isolation and characterization of Schizosaccharomyces pombe cut mutants that brock nuclear division but not cytokinesis. EMBO J. 5:2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inazawa, J., T. Ariyama, and T. Abe. 1992. Physical ordering of three polymorphic DNA markers spanning the regions containing a tumor suppressor gene of renal cell carcinoma by three-color fluorescent in situ hybridization. Jpn. J. Cancer Res. 83:1248–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto, I., A. Pimkhaokham, T. Watanabe, F. Saito-Ohara, E. Soeda, and J. Inazawa. 2000. Amplification and overexpression of TGIF2, a novel homeobox gene of the TALE superclass, in ovarian cancer cell lines. Biochem. Biophys. Res. Commun. 276:264–270. [DOI] [PubMed] [Google Scholar]
- Jager, H., A. Herzig, C.F. Lehner, and S. Heidmann. 2001. Drosophila separase is required for sister chromatid separation and binds to PIM and THR.Genes Dev. 15:2572–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli, P.V., I.C. Waizenegger, F. Bunz, S. Langer, M.R. Speicher, J.M. Peters, K.W. Kinzler, B. Vogelstein, and C. Lengauer. 2001. Securin is required for chromosomal stability in human cells. Cell. 105:445–457. [DOI] [PubMed] [Google Scholar]
- Losada, A., and T. Hirano. 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19:1269–1287. [DOI] [PubMed] [Google Scholar]
- Mei, J., X. Huang, and P. Zhang. 2001. Securin is not required for cellular viability, but is required for normal growth of mouse embryonic fibroblasts. Curr. Biol. 11:1197–1201. [DOI] [PubMed] [Google Scholar]
- Nagao, K., Y. Adachi, and M. Yanagida. 2004. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 430:1044–1048. [DOI] [PubMed] [Google Scholar]
- Nakai, S., H. Kawano, T. Yudate, M. Nishi, J. Kuno, A. Nagata, K. Jishage, H. Hamada, H. Fujii, K. Kawamura, et al. 1995. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 9:3109–3121. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science. 297:559–565. [DOI] [PubMed] [Google Scholar]
- Papi, M., E. Berdougo, C.L. Randall, S. Ganguly, and P.V. Jallepalli. 2005. Multiple roles for separase auto-cleavage during the G2/M transition. Nat. Cell Biol. 7:1029–1035. [DOI] [PubMed] [Google Scholar]
- Shibata, H., K. Toyama, H. Shioya, M. Ito, M. Hirota, S. Hasegawa, H. Matsumoto, H. Takano, T. Akiyama, K. Toyoshima, et al. 1997. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 278:120–123. [DOI] [PubMed] [Google Scholar]
- Siomos, M.F., A. Badrinath, P. Pasierbek, D. Livingstone, J. White, M. Glotzer, and K. Nasmyth. 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11:1825–1835. [DOI] [PubMed] [Google Scholar]
- Stemmann, O., H. Zou, S.A. Gerber, S.P. Gygi, and M.W. Kirschner. 2001. Dual inhibition of sister chromatid separation at metaphase. Cell. 107:715–726. [DOI] [PubMed] [Google Scholar]
- Sternberg, N., and D. Hamilton. 1981. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150:467–486. [DOI] [PubMed] [Google Scholar]
- Stratmann, R., and C.F. Lehner. 1996. Separation of sister chromatids in mitosis requires the Drosophila pimples product, a protein degraded after the metaphase/anaphase transition. Cell. 84:25–35. [DOI] [PubMed] [Google Scholar]
- Taniguchi, M., M. Sanbo, S. Watanabe, I. Naruse, M. Mishina, and T. Yagi. 1998. Efficient production of Cre-mediated site-directed recombinants through the utilization of the puromycin resistance gene, pac: a transient gene-integration marker for ES cells. Nucleic Acids Res. 26:679–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro, G.J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa, S., I. Samejima, T. Hirano, K. Tanaka, and M. Yanagida. 1990. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 62:913–925. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I., J.F. Gimenez-Abian, D. Wernic, and J.M. Peters. 2002. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 12:1368–1378. [DOI] [PubMed] [Google Scholar]
- Wang, Z., R. Yu, and S. Melmed. 2001. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 15:1870–1879. [DOI] [PubMed] [Google Scholar]
- Yanagida, M. 2000. Cell cycle mechanisms of sister chromatid separation; roles of Cut1/separin and Cut2/securin. Genes Cells. 5:1–8. [DOI] [PubMed] [Google Scholar]