Abstract
Separase is a protease whose liberation from its inhibitory chaperone Securin triggers sister chromatid disjunction at anaphase onset in yeast by cleaving cohesin's kleisin subunit. We have created conditional knockout alleles of the mouse Separase and Securin genes. Deletion of both copies of Separase but not Securin causes embryonic lethality. Loss of Securin reduces Separase activity because deletion of just one copy of the Separase gene is lethal to embryos lacking Securin. In embryonic fibroblasts, Separase depletion blocks sister chromatid separation but does not prevent other aspects of mitosis, cytokinesis, or chromosome replication. Thus, fibroblasts lacking Separase become highly polyploid. Hepatocytes stimulated to proliferate in vivo by hepatectomy also become unusually large and polyploid in the absence of Separase but are able to regenerate functional livers. Separase depletion in bone marrow causes aplasia and the presumed death of hematopoietic cells other than erythrocytes. Destruction of sister chromatid cohesion by Separase may be a universal feature of mitosis in eukaryotic cells.
Introduction
Cohesion between sister chromatids is essential for the biorientation of chromosomes on mitotic spindles (Tanaka et al., 2000). By resisting the tendency of microtubules to pull sister chromatids apart, cohesion creates the tension needed to stabilize the attachment of microtubules to kinetochores (Nicklas and Ward, 1994). Chromosome segregation cannot, however, actually take place until the links holding bioriented sister chromatids together are broken, a process that occurs simultaneously on all chromosomes a few minutes after the last chromosome has bioriented (Rieder et al., 1994). Thus, loss of sister chromatid cohesion triggers what is possibly one of the most dramatic events in the life of any eukaryotic cell—the sudden migration of sister chromatids to opposite poles, an event known as the metaphase–anaphase transition.
Sister chromatid cohesion is mediated by a complex called cohesin (Nasmyth and Haering, 2005) whose two structural maintenance of chromosomes proteins (Smc1 and -3) and a single α kleisin (Scc1/Rad21) subunit join together to create a tripartite ring within which, it has been proposed, sister DNAs are topologically entrapped (Gruber et al., 2003). Crucially, sister chromatid cohesion is suddenly destroyed at the onset of anaphase by the cleavage of cohesin's α kleisin subunit by a protease called Separase (Uhlmann et al., 1999), which opens the cohesin ring and causes it to dissociate from chromosomes.
Because loss of sister chromatid cohesion before chromosome biorientation is disastrous for chromosome segregation, cleavage of cohesin by Separase is tightly controlled. For most of the cell cycle, Separase is bound by a chaperone called Securin, which inhibits its proteolytic activity (Ciosk et al., 1998; Uhlmann et al., 1999; Hornig et al., 2002; Waizenegger et al., 2002). Once all chromosomes have been bioriented, Securin is targeted for proteasomal destruction by a ubiquitin ligase called the anaphase promoting complex or cyclosome (APC/C; Cohen-Fix et al., 1996; Funabiki et al., 1996b; Zou et al., 1999), resulting in Separase activation. In vertebrate cells, Separase is inhibited not only by Securin but also by phosphorylation at the hands of Cdk1 (Stemmann et al., 2001). In these cells, therefore, APC/C triggers Separase activation through the simultaneous destruction of Securin and of Cdk1's activating subunit cyclin B.
In most, if not all, organisms, Securins have both positive and negative effects on Separase activity. Thus, in Schizosaccharomyces pombe and Drosophila melanogaster, inactivation of the Securins cut2 (Funabiki et al., 1996a) and pimples (Stratmann and Lehner, 1996), respectively, is lethal and causes phenotypes very similar to inactivating Separase. Though not lethal, deletion of the Securin genes in mice (Mei et al., 2001), human tissue culture cells (Jallepalli et al., 2001), or Saccharomyces cerevisiae (Ciosk et al., 1998) also has adverse effects on sister chromatid separation. In the yeasts S. cerevisiae and S. pombe, either inactivation of Separase or expression of noncleavable α kleisin subunits prevents sister chromatid separation (Uhlmann et al., 1999; Tomonaga et al., 2000), and in S. cerevisiae, α kleisin cleavage is even sufficient for triggering anaphase (Uhlmann et al., 2000).
Given the importance of the metaphase–anaphase transition and the degree of control that is exerted over this process, it is essential to know whether the chemistry of sister chromatid separation unearthed in yeast is shared by all eukaryotes, including humans. However, little is known about the functions of both α kleisin cleavage and Separase in organisms other than yeast. Thus, in D. melanogaster (Jager et al., 2001) and Caenorhabditis elegans (Siomos et al., 2001), Separase is known to be required for sister chromatid separation, but whether it triggers anaphase by cleaving α kleisins is not known. In mammals, α kleisin can be cleaved by Separase purified from tissue culture cells, a small fraction is indeed cleaved at the metaphase–anaphase transition (Waizenegger et al., 2000), and expression of a noncleavable version interferes with chromatid segregation at anaphase (Hauf et al., 2001). Investigation of Separase's in vivo function has hitherto been confined to the use of RNA interference to deplete it from tissue culture cells, which interferes with chromosome segregation and causes the production of highly abnormal (polyploid) nuclei (Waizenegger et al., 2002; Chestukhin et al., 2003). However, it has so far not been possible to directly observe the entry into and passage through mitosis of cells known to lack Separase. It is therefore not yet known for certain whether Separase is essential for sister chromatid separation in mammalian cells.
It is in fact not a forgone conclusion that Separase is essential for sister chromatid separation in mammals because most cohesin dissociates from chromosome arms (but not centromeres) during prophase and prometaphase (Losada et al., 1998; Sumara et al., 2000; Waizenegger et al., 2000). This process is called the prophase pathway and is at least partly dependent on phosphorylation of cohesin's Scc3-SA2 subunit (Hauf et al., 2005) but not, apparently, by cleavage of its α kleisin subunit. A similar process could conceivably also contribute to sister chromatid separation at anaphase, when cohesin persisting at centromeres disappears from chromosomes.
To address as rigorously as possible the role of Separase during the chromosome cycle of mammalian cells, especially when they are growing in the context of real tissues within animals, we used homologous recombination in embryonic stem (ES) cells to replace the wild-type Separase gene by a version in which the eight COOH-terminal exons encoding part of its conserved protease domain are flanked by loxP sites and can therefore be deleted from the genome by Cre-recombinase expression. We chose this approach because manipulation of the genome has proven to be a more reliable method of altering gene function than methods that merely alter the abundance or activity of gene products. We find that deletion of Separase specifically blocks sister chromatid separation but not other aspects of mitosis, mitotic exit, cytokinesis, or even chromosome rereplication.
Results
Generation of conditional Separase and Securin alleles
To generate a conditional Separase allele, we created a targeting vector in which the most COOH-terminal eights exons of the Separase locus, which encode part of the conserved COOH terminus, including its catalytic dyad, were flanked by loxP sites. An identical strategy was used to create a targeting vector for the Securin gene in which its three COOH-terminal exons were flanked by loxP sites (Fig. 1, A and C).
Figure 1.
Generation of Separase and Securin floxed and Δ alleles. (A) Targeting strategy for Separase. Shown are the Separase genomic locus, the targeting vector, and the targeted allele. Exons are indicated as black boxes, the conserved peptidase domain starting at exon 18 (nucleotide 3980 of the mRNA) as red boxes, and the exon containing the conserved histidine and cysteine as a green box. For Separase the eight COOH-terminal exons were flanked by loxP sites (triangles). The selection cassette Neo-Tk is represented as red and green boxes and the DTA-cassette as a blue box. Cre-mediated recombination (dashed lines) was used to obtain Separase floxed or Δ alleles. (B) Southern blot to confirm germ-line transmission of Separase flox and Δ alleles with EcoRV-digested DNA and the internal probe c1. (C) Targeting strategy for generating Securin floxed and Δ alleles. (D) Southern blot to confirm germ-line transmission of Securin Δ alleles with BamHI-digested DNA using the internal probe c1.
HM1 ES cells were transfected separately with the Separase and Securin targeting vectors, and G418-resistant HM1 ES cell clones in which a single Separase or Securin locus (allele) had been replaced by the targeting construct were identified by Southern blotting. Transient transfection of these clones with a plasmid expressing Cre recombinase created floxed or deletion alleles. Three independent ES cell clones carrying floxed or deletion alleles of Separase or Securin were injected into C57BL/6 blastocytes. Chimeras were crossed with C57BL/6 mice to obtain germ-line transmission (Fig. 1, B and D).
Separase is essential for embryonic development
To determine whether Separase is essential for embryonic development, heterozygous Separase Δ/+ mice were intercrossed. Of 60 21-d-old progeny, 18 were Separase +/+, 42 were Separase Δ/+, and none were Separase Δ/Δ (Table I). We detected no obvious difference between the development, health, or behavior of +/+ and Δ/+ mice. These data imply that a single copy of the Separase gene is both necessary and sufficient for embryonic development. We also determined the genotypes of 54 embryos from Separase Δ/+ intercrosses at 6.5, 7.5, and 8.5 days post coitus (dpc). 14 were +/+, 40 were Δ/+, and none were Δ/Δ, which implies that Separase is required for early embryogenesis (Table I). We have no explanation for the slight but significant excess of Δ/+ embryos. Other crosses yielded at roughly expected frequencies mice homozygous for Separase floxed alleles (Separaseflox/flox) as well as mice with one deleted and one floxed allele (Separase Δ/flox). The normal appearance of such mice implies that the floxed allele of Separase is possibly as functional as wild type.
Table I.
Deletion of Separase is embryonic lethal
| Day of embryonic development |
Total | Embryos | Resorptions | Embryos analyzed by PCR |
Separase +/+ | Separase Δ/+ | Separase Δ/Δ |
|---|---|---|---|---|---|---|---|
| 6.5 dpc | 49 | 41 | 8 | 38 | 13 | 25 | 0 |
| 7.5 dpc | 10 | 8 | 2 | 8 | 1 | 7 | 0 |
| 8.5 dpc | 9 | 8 | 1 | 8 | 0 | 8 | 0 |
| p21 | 60 | 60 | — | 60 | 18 | 42 | 0 |
Deletion of both Separase alleles is embryonic lethal. Intercrosses between Separase Δ/+ mice. No Separase Δ/Δ mice were born in a total of 60 live births. At 6.5, 7.5, and 8.5 dpc, all embryos were Separase +/+ or heterozygous for the Separase Δ allele. p, postnatal day.
Securin is important but not essential for Separase activity in vivo
Mice homozygous for the Securin deletion were obtained from intercrosses between Securin Δ/+ mice at roughly the expected frequencies. These mice were fertile, both as males and females, albeit less so than wild type (unpublished data). This confirms previous reports that the mouse Securin gene is not essential for either mitosis or meiosis (Mei et al., 2001; Wang et al., 2001).
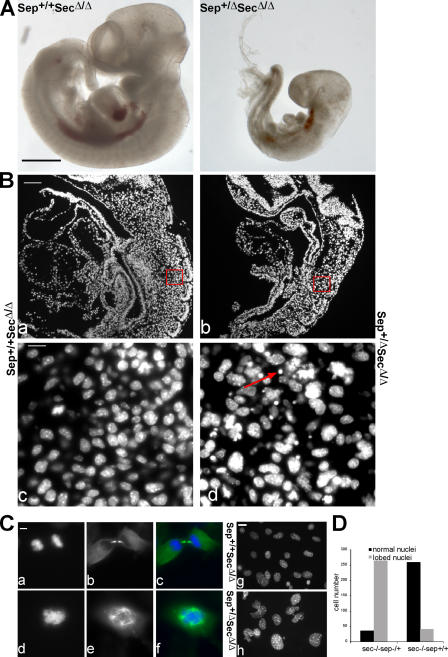
To determine whether Separase is less active in mice lacking Securin, we crossed Securin Δ/Δ Separase +/+ and Securin Δ/+ Separase Δ/+ mice. Out of 69 progeny, 22 were Securin Δ/+ Separase +/+, 22 were Securin Δ/+ Separase Δ/+, 25 were Securin Δ/Δ Separase +/+, and none were Securin Δ/Δ Separase Δ/+ (Table II). Mice with the Securin Δ/Δ Separase Δ/+ genotype should have been as frequent as the other three classes, and their absence suggests that embryonic development in the absence of Securin requires both copies of the Separase gene. Securin Δ/Δ Separase Δ/+ embryos are smaller than Securin Δ/Δ Separase +/+ embryos at 10.5 dpc, they have irregular somites and abnormal neurale tubes (Fig. 2 A), and they die at 11.5 dpc. Hoechst staining of longitudinal paraffin sections from 9.5 dpc embryos revealed extensive cell death and larger lobed nuclei in several organs (somites, heart, and brain) of Securin Δ/Δ Separase Δ/+ but not Securin Δ/Δ Separase +/+ embryos (Fig. 2 B, c and d). To determine whether such nuclei arise from mitotic defects, we cultured mouse embryonic fibroblasts (MEFs) from 10.5 dpc embryos. Despite a much lower plating efficiency, it proved possible to analyze the DNA and α-tubulin distribution of Securin Δ/Δ Separase Δ/+ embryonic cells after 10 d in culture. Securin Δ/Δ Separase Δ/+ MEFs possessed abnormally large lobed nuclei (Fig. 2, C and D). Out of 24 mitotic cells, 16 had multipolar spindles and 8 were undergoing anaphase with lagging chromosomes. Out of 24 mitotic cells from Securin Δ/Δ Separase +/+ embryos, only one was undergoing anaphase with lagging chromosomes and none contained multipolar spindles. However, chromosome spreads from Securin Δ/Δ Separase Δ/+ MEFs did not reveal the diplochromosomes characteristic of MEFs completely lacking Separase (see Loss of Separase causes polyploidy). These data suggest that Separase function is so compromised in Securin Δ/Δ embryos that a further twofold reduction in its activity leads to lethal chromosome missegregation.
Table II.
Deletion of Securin and one Separase allele is embryonic lethal
| Sec Δ/+ Sep +/+ | Sec Δ/+ Sep Δ/+ | Sec Δ/Δ Sep +/+ | Sec Δ/Δ Sep Δ/+ | Total | |
|---|---|---|---|---|---|
| SecΔ/Δ m × SecΔ/+SepΔ/+ f | |||||
| p10 | 9 | 7 | 13 | 0 | 29 |
| SecΔ/Δ f × SecΔ/+SepΔ/+ m | |||||
| p10 | 13 | 15 | 12 | 0 | 40 |
Securin Δ/Δ Separase +/+ and Securin Δ/+ Separase Δ/+ mice were intercrossed. No Securin Δ/Δ Separase Δ/+ mice were born in a total of 69 live births. m, male; f, female; p, postnatal day.
Figure 2.
SecurinΔ/ΔSeparaseΔ/+ embryos show developmental defects. (A) Securin Δ/+ Separase Δ/+ female mice were crossed with Securin Δ/+ Separase +/+ male mice. At 10.5 dpc, Securin Δ/Δ Separase Δ/+ embryos were obtained at the expected Mendelian ratio. However, Securin Δ/Δ Separase Δ/+ embryos were smaller and less developed than Securin Δ/+ Separase +/+ embryos. Bar, 1 mm. (B) Paraffin longitudinal sections of Securin Δ/+ Separase +/+ (a) and Securin Δ/Δ Separase Δ/+ (b) embryos 9.5 dpc were stained with Hoechst to visualize the nuclei. Squared regions in panels a and b are shown enlarged in panels c and d. Bars: (a and b) 100 μm; (c and d) 10 μm. Arrow points to a dead cell. (C) MEFs from Securin Δ/+ Separase +/+ and Securin Δ/Δ Separase Δ/+ embryos (10.5 dpc) were cultured and analyzed by immunofluorescence microscopy. Cells were stained with DAPI (a, d, g, and h) and α-tubulin (b and e). Bars, 10 μm. (D) Number of lobed nuclei in Securin Δ/+ Separase +/+ and Securin Δ/Δ Separase Δ/+ MEFs.
Separase is dispensable in quiescent hepatocytes
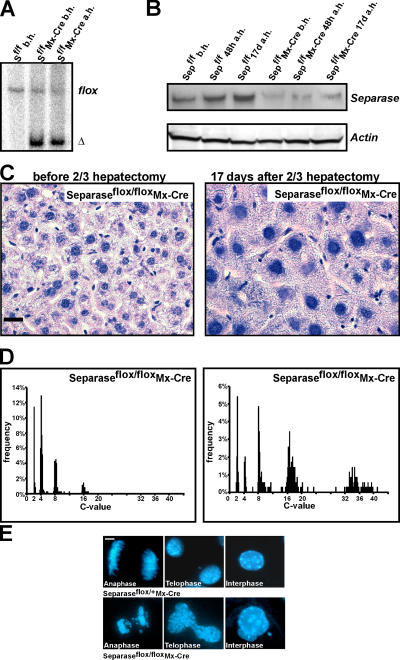
Separase is mainly expressed in proliferating cells. However, modest amounts can be detected by Western blotting in liver extracts from adult mice (Fig. 3 B; unpublished data). To determine whether Separase has a function in resting hepatocytes, we used an Mx-Cre transgene to delete both copies of Separase from liver cells of Separaseflox/flox mice. Despite efficient deletion of the Separase gene (Fig. 3 A), which leads to a reduction in the level of Separase in their livers (Fig. 3 B), 10 out of 10 Separaseflox/floxMx-Cre mice survived >6 mo without any overt pathology (not depicted).
Figure 3.
Hepatocytes after two thirds hepatectomy are highly polyploid. (A) Southern blot analysis of Separaseflox/flox and Separaseflox/floxMx-Cre livers before and 17 d after the two thirds hepatectomy. The Separase flox band is efficiently deleted in Separaseflox/floxMx-Cre livers. (B) Western blot from livers before, 48 h after, and 17 d after the two thirds hepatectomy. The Separase protein is detected in Separaseflox/flox hepatocytes, whereas in Separaseflox/floxMx-Cre hepatocytes the Separase protein is down-regulated. b.h., before the two thirds hepatectomy; a.h., after the two thirds hepatectomy. (C) Hematoxylin/ eosin staining of livers from Separaseflox/floxMx-Cre mice before and 17 d after the two thirds hepatectomy. Separaseflox/floxMx-Cre hepatocytes increase in ploidy and in cell size after the two thirds hepatectomy. Bar, 10 μm. (D) Single-cell DNA measurement of Feulgen-stained hepatocyte nuclei demonstrating the increase in DNA content in Separaseflox/floxMx-Cre hepatocytes at 17 d after the two thirds hepatectomy. (E) Hoechst staining of hepatocytes in culture. Compared with Separase flox/+ Mx-Cre hepatocytes, Separaseflox/floxMx-Cre hepatocytes demonstrate anaphases with sister chromatid missegregation, telophases with DNA bridges between daughter cells and multinucleated hepatocytes in interphase. Bar, 1 μm.
Liver regeneration in the absence of Separase involves polyploidization
To address the function of Separase in proliferating hepatocytes, we induced entry into the cell cycle of resting hepatocytes by surgical removal (hepatectomy) of two thirds of the liver. Mice can survive with the reduced liver mass for several days, but regrowth is required for long-term survival. 10 Separaseflox/flox Mx-Cre mice, 10 Separaseflox/flox, and 10 Separase flox/+ Mx-Cre mice were first injected with 400 μl poly(I)poly(C) (pI/C), and hepatectomy was performed 3 d later. Remarkably, all 30 mice, including all those with a Separaseflox/floxMx-Cre genotype, survived for several months after the hepatectomy. Livers from all three sets of mice reached their original size ∼3 wk after hepatectomy. Southern and Western blotting confirmed that Separase had been efficiently deleted in the Separaseflox/floxMx-Cre livers, both before and 17 d after the two thirds hepatectomy (Fig. 3, A and B). This result was surprising because it implies that liver regeneration does not require Separase.
To further investigate the process of liver regeneration in the absence of Separase, 10 Separaseflox/floxMx-Cre, 10 Separase flox/+ Mx-Cre, and 10 Separaseflox/flox mice were analyzed at different time points after pI/C injection and two thirds hepatectomy. Histological analysis at 3, 5, and 17 d after hepatectomy revealed that the size of cells and their nuclei was greatly increased after regeneration in livers lacking Separase (Fig. 3 C), whereas that of control livers was unaltered (not depicted). Feulgen-staining revealed that in the Separaseflox/flox livers, some hepatocytes had a 2- or 8C DNA content but most had a 4C DNA content both before and 17 d after the two thirds hepatectomy (unpublished data). The DNA contents of hepatocytes from Separaseflox/flox Mx-Cre mice (n = 3) resembled that of their controls before hepatectomy, but their DNA contents had increased to 8C, 16C, 32C, or even higher 17 d after hepatectomy (Fig. 3 D). These data indicate that liver regeneration without Separase is accompanied by several rounds of genome rereplication in the absence of cell proliferation, which leads to the production of highly polyploid, albeit apparently functional, hepatocytes.
Separase is required for anaphase in hepatocytes
To investigate the mechanism that gives rise to polyploidy in regenerating hepatocytes, we cultured hepatocytes from Separaseflox/floxMx-Cre and Separase flox/+ Mx-Cre mice (after collagenase perfusion) 3 d after injection with pI/C and analyzed them by live cell video microscopy (Fig. S1, available at http:/www.jcb.org/cgi/content/full/jcb.200506119/DC1). Under these conditions, hepatocytes divide just once, and a maximum mitotic index of 5–10% is reached 48 h after cultivation. Mitosis in Separase flox/+ Mx-Cre hepatocytes invariably produced two identically sized nuclei, but it was only sometimes accompanied by cell division. This means that mitosis usually creates a binucleate hepatocyte (Guidotti et al., 2003). Separaseflox/flox Mx-Cre hepatocytes entered mitosis and aligned their chromosomes on metaphase plates, but in 80% of anaphase cells, sister chromatids failed to disjoin properly at the onset of anaphase. Cells nevertheless exited from mitosis and produced progeny containing micronuclei as well as abnormally large nuclei (Fig. 3 E and Fig. S1; n = 100 mitotic cells per experiment). These data suggest that the abnormal polyploidy of Separase-deficient hepatocytes arises not because of their failure to enter mitosis but because of their failure to undergo anaphase.
Rapid cell death of Separase-deficient hematopoietic cells
pI/C causes efficient expression of Mx-Cre in bone marrow cells as well as in hepatocytes (Kuhn et al., 1995). To investigate the consequences of Separase depletion in this compartment, we isolated hematopoietic cells from bone marrow at 2 and 3 d after pI/C injection of 10 Separaseflox/floxMx-Cre, 10 Separase flox/+ Mx-Cre, and 10 Separaseflox/flox mice. This revealed severe bone marrow aplasia in Separaseflox/floxMx-Cre mice but few if any abnormalities in Separase flox/+ Mx-Cre and Separaseflox/flox mice. By day 3, bone marrow from Separaseflox/flox Mx-Cre mice contained only erythrocytes (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200506119/DC1). Separaseflox/floxMx-Cre mice survived despite their severe bone marrow aplasia and fully reconstituted their bone marrow by day 31 with Separaseflox/flox or Separase flox/Δ hematopoietic cells (Fig. S2 A) whose nuclear divisions and cell sizes appeared normal. Unlike hepatocytes, diploid hematopoietic cells appear to undergo rapid cell death in the absence of Separase.
Loss of Separase causes polyploidy in immortalized fibroblasts
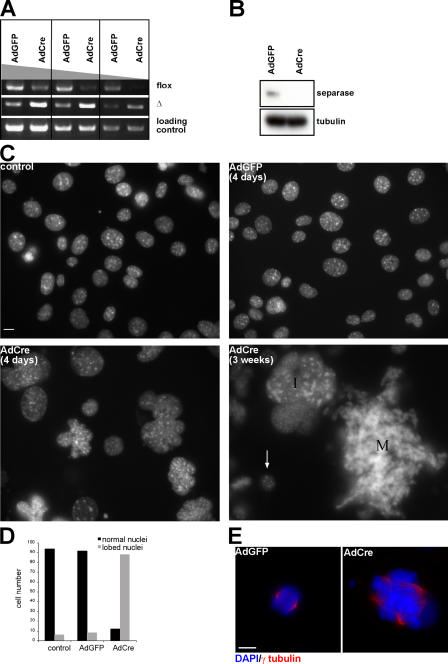
To analyze the consequences of Separase inactivation in greater detail, we isolated embryonic fibroblasts from Separase Δ/flox embryos and used the 3T3 protocol to create an immortalized MEF (iMEF). To inactivate the floxed allele, we used adenovirus expressing Cre recombinase (AdCre). Infection with AdCre but not with adenovirus expressing GFP (AdGFP) within 4 d caused deletion of most floxed alleles (Fig. 4 A), which was accompanied by a reduction in the level of Separase as measured by Western blotting (Fig. 4 B) and the accumulation of cells with large and multilobed nuclei (Fig. 4, C and D). These abnormal nuclei were not observed in iMEFs carrying a wild-type Separase locus infected with AdCre (unpublished data) or in Separase Δ/flox iMEFs infected with AdGFP (Fig. 4 C) and must therefore be caused by deletion of the Separase locus and not by infection with adenovirus or expression of Cre by itself.
Figure 4.
iMEFs lacking Separase become polyploid with multipolar spindle. (A–E) Separase Δ/flox iMEFs were infected with AdCre and -GFP, respectively. (A) Genomic PCR was performed to reveal the deletion of the last eight exons in the Separase genomic locus 4 d after virus infection. PCR primers were used to amplify the flox and the deletion allele. The region located in the Separase NH2-terminal coding sequence was amplified for loading control. (B) Western blot analysis of Separase protein level in the cells 4 d after viral infection. Tubulin was used as a loading control. (C) Nuclear morphology revealed by DAPI staining. Control represents Separase Δ/flox iMEFs not infected with the virus. iMEFs were infected with AdGFP and -Cre, respectively, and DAPI staining was performed 4 d later. Note that the size of the cells 3 wk after AdCre transduction can be directly compared with small nuclei from cells that were most likely not infected with the virus (arrow). I, interphase cell; M, mitotic cell. (D) Number of lobed nuclei 4 d after viral infection counted in 100 interphase cells. (E) Immunofluorescence staining of the cells 3 d after viral infection. γ-tubulin was used to stain the spindle. Bars, 10 μm.
Remarkably, cells with even larger multilobed nuclei accumulated 3 wk after infection, and such cells entered mitosis with huge numbers of chromosomes (Fig. 4 C). These observations suggest that, like hepatocytes, iMEFs lacking Separase undergo multiple rounds of DNA replication despite failing to segregate their chromosomes at mitosis. At early stages, rereplication of chromosomes was accompanied by centrosome reduplication. Thus, 90% of Separase Δ/flox iMEFs infected with AdCre, but 5% or fewer of those infected with AdGFP, contained multipolar spindles (Fig. 4 E).
Separase is required for sister chromatid separation but not for chromosome reduplication
To measure DNA replication in cells lacking Separase, Separase Δ/flox iMEFs were first grown to 100% confluency, which led to contact inhibition, and then infected with AdCre or -GFP, and 48 h later the cultures were split to stimulate their entry into the cell cycle (Fig. 5 A). FACS sorting revealed an increase in 4- and 8C cells relative to 2C cells 48 h after splitting of iMEFs infected with AdCre (Fig. 5 B) and an appreciable number of cells with DNA contents of 16C or more after 72 and 96 h, whereas cultures infected with AdGFP showed no increase in DNA content. AdCre but not -GFP also caused a large increase in the number of cells with <2C DNA contents, and more floating cells were observed in these tissue culture plates, which indicates the accumulation of apoptotic cells.
Figure 5.
iMEFs lacking Separase reveal higher ordered chromosomes. (A) Schematic overview of viral infection and harvesting procedure. (B) A portion of cells harvested for the analysis in C was methanol fixed, and their DNA content was analyzed by flow cytometry. Arrows indicate peaks showing <2C and >16C that were not present in the cells infected with AdGFP. (C) Chromosome spreads of cells harvested at the different time points. Before harvesting, cells were treated with nocodazole for 5 h to enrich mitotic cells and collected by mitotic shakeoff. (D) Higher magnification of the high-ordered chromosomes resulting from cells infected with AdCre and harvested at different time points. Number of chromatids is indicated. Control cells that underwent the same procedure except that they were not infected had a FACS profile very similar to that of the cells infected with AdGFP (not depicted). Bars, 10 μm.
To analyze the state of chromosomes, cells at each time point after splitting were incubated for 5 h in the presence of nocodazole to enrich mitotic cells, which were then collected by shakeoff, spread on glass slides, and stained with Giemsa. In samples collected 48 h after splitting, 89% of mitotic cells from Separase Δ/flox iMEFs infected with AdCre (but none infected with AdGFP) contained diplochromosomes in which two sets of sister chromatids were either closely aligned in parallel or remained attached at their centromeres (Fig. 5, C and D). Samples collected at 72 h frequently contained quadrupled chromosomes, that is, four sets of sister chromatids associated with each other in the region of their centromeres. In cells that had undergone yet another round of DNA replication, we sometimes observed, albeit rarely, karyotypes in which eight sets of sister chromatids remained associated (Fig. 5 D). Spreads from mitotic cells sampled 96 h after splitting had even higher numbers of chromosomes, but most were single chromosomes containing a single pair of sister chromatids. These observations imply that cells lacking Separase fail to separate sister chromatids when they enter mitosis but nevertheless subsequently reduplicate their chromosomes. Remarkably, reduplication gives rise to chromosomes in which the two sets of sister chromatids produced by reduplication frequently remain associated with each other. We do not know whether the association between pairs of sister chromatids after reduplication in the absence of Separase is, like that between sister chromatids themselves, mediated by cohesin.
Separase is required for chromosome segregation at anaphase but not for cytokinesis
To address which aspects of mitosis fail to take place without Separase, mitotic cells were collected 24 h after splitting of Separase Δ/flox iMEFs infected with AdCre or -GFP or with no virus. Cells were cultured as in Fig. 5 A and either collected by mitotic shakeoff and stained with Giemsa (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200506119/DC1) or processed for immunofluorescence microscopy (Fig. 6). Infection with AdCre caused a reduction in the frequency of anaphase and telophase cells but not that of prometaphase or metaphase cells (Fig. S3). AdCre also caused the appearance of abnormal metaphase-like cells that contained partly decondensed chromosomes (Fig. S3 B, a–c) and three types of highly abnormal telophases (Fig. S3 B), namely, cells whose chromosomes were untimely torn by cell cleavage (28% from abnormal telophases), cells with chromatin bridges connecting highly asymmetric chromosome masses (31%), and cells containing a single nucleus on one side of their cleavage furrow (40%).
Figure 6.
Cytokinesis occurs in some iMEFs lacking Separase. The infection and harvesting procedure was the same as in Fig. 5 A, except that a single time point 24 h after splitting was taken. (A) Immunostaining was performed using anti–Aurora B (red) and anti-MkLp1 (green) antibodies. DNA was stained with DAPI (blue). Prometaphase or metaphase (a), anaphase (b), and telophase (c) are shown. Arrows indicate unequal distribution of the DNA in cells infected with AdCre. (B) Different mitotic stages were scored in cells not infected with any virus (control) and cells infected with AdCre and -GFP, respectively. Mitotic stages were defined according to the Aurora B, MkLp1, and localization. n = 100 per cell type. (C) Immunostaining was performed using anti–cyclin B. DNA was stained with DAPI. (D) Cyclin B–positive and –negative metaphase plates were scored in cells infected with AdCre and -GFP. n = 100 per cell type. Bars, 10 μm.
A strong reduction in normal anaphases and telophases was also observed by immunofluorescence microscopy when cells infected with AdCre were stained with DAPI and antibodies to the spindle midzone protein MkLp1 and the mitotic kinase Aurora B (Fig. 6, A and B). Aurora B normally relocates from centromeres to the midspindle in anaphase and accumulates at the midbody in telophase. In yeast the association of Aurora B/Ipl1 with the spindle depends on Separase (Pereira and Schiebel, 2003). Our experiments revealed that Aurora B is located at centromeres in prometaphase cells that are lacking Separase, but in many metaphase-like cells, Aurora B was distributed throughout the cytoplasm (Fig. 6 A). These observations imply that Aurora B can dissociate from chromosomes in the absence of Separase but then fails to associate with microtubules. It is unclear whether cells lacking Separase can form a proper midspindle. The inability of Aurora B to associate with microtubules could thus be either a direct or an indirect consequence of Separase depletion. These cells were nevertheless able to undergo cytokinesis, and during this process Aurora B became enriched in the cortical region of the ingressing cleavage furrow and later in the bridge that connects daughter cells (Fig. 6 A). Separase activity is therefore not essential for cytokinesis in mouse fibroblasts.
To determine whether the lack of anaphase or telophase cells could be caused by a failure to activate the APC/C, we measured cyclin B levels by immunofluorescence. In cells infected with AdGFP, most prometaphase and metaphase cells were cyclin B positive, whereas anaphase and telophase cells were negative. In cells infected with AdCre, 66% of metaphase cells were cyclin B negative (Fig. 6, C and D). These cells have presumably activated the APC/C but despite this failed to separate their sister chromatids.
To visualize chromosome segregation, we generated Separase Δ/flox cells that stably express an mRFP-tagged version of histone H2B and filmed cells as they formed metaphase plates. All eight cells infected with AdGFP underwent anaphase and cytokinesis, yielding daughter cells whose nuclei had equal amounts of mRFP fluorescence (Fig. 7 A). In contrast, none of the 16 cells infected with AdCre managed to segregate their chromosomes, despite forming apparently normal metaphase plates. All 16 cells nevertheless formed cleavage furrows even though their chromosomes remained at the spindle equator. In two cells, the furrows attempted to bisect the chromosomes as they decondensed, causing constriction of the chromosomes. These furrows later regressed, leading to the formation of binucleate cells. In two other cells, the cleavage furrow constricted the chromosomes in an asymmetric fashion. These cells completed cytokinesis, producing one interphase cell and another that did not attach to the plate. In the remaining 12 cells, cleavage produced one cell that contained all or most chromosomes and another that contained few if any and did not attach to the plate (Fig. 7 B). Irrespective of their precise fate, all 16 cells decondensed their chromosomes, indicating that they exited from a mitotic state. We conclude that Separase is necessary for segregating chromosomes at anaphase but not for mitotic exit or cytokinesis.
Figure 7.
Live cell imaging of cells lacking Separase. (A and B) Separase Δ/flox iMEF cells expressing mRFP-tagged histone H2B were grown to 100% confluency, infected with the virus, and split 48 h later. After 24 h, live cell imaging was performed using a fluorescence microscope. Stacks of six different z plane images were obtained every 10 min, and projected images for several time points are shown. Bars, 50 μm.
Separase is required to remove cohesin from chromosomes
To address whether Separase is needed to remove cohesin from chromosomes, we generated a stable Separase Δ/flox cell line expressing a myc-tagged version of the cohesin subunit Scc1 (Fig. 8). This coimmunoprecipitated with Smc1 and -3 (Fig. 8, A and B) and was concentrated between pairs of CREST (calcinosis, Raynaud's phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasia) dots at centromeres of mitotic cells (treated with nocodazole) after extraction of bulk soluble cohesin (Fig. 8 C). On the diplochromosomes produced by infection with AdCre, Scc1-myc staining was enriched at both centromeres but not between them (Fig. 8 F). Faint staining was also observed on chromosome arms, exclusively between the sister chromatids.
Figure 8.
Cohesin stays on the chromosomes in mitotic cells lacking Separase. A Separase Δ/flox stable cell line expressing Scc1-myc was generated. (A–C) Characterization of the cell line. (A) Silver stain of the IP products from cells expressing Scc1-myc and control cells using an anti-myc antibody. (B) Western blot of the same IP reaction as in A using anti-myc and anti-Smc3 antibodies. E, eluate; S, supernatant; ce, cell extract. (C) Immunostaining using anti-myc antibody (green) and CREST serum (red). Cells were treated with nocodazole for 30 min and spun on glass slides. Note that Scc1-myc staining was observed in the centromeric region between two CREST dots. (D) Cells were infected with the virus as in Fig. 5 A, and 24 h after splitting cells were processed for immunofluorescence microscopy. Mitotic cells were spun on glass slides and analyzed for cohesin with an antibody to the myc epitope and P-H3. (E) Cells positive for myc and P-H3 as well as negative for myc but positive for P-H3 were scored. n = 200 per cell type. (F) Cells were infected in the same way as in D and collected 48 h after splitting. Before harvesting, cells were treated with nocodazole for 30 min to disrupt the spindle, leading to better spreading of chromosomes. Bars, 10 μm.
In cells infected with AdGFP, Scc1-myc staining on chromosomes was detectable in two thirds of mitotic cells positive for phosphorylated histone H3 (P-H3; Fig. 8, D [top] and E). Thus, cells that have initiated anaphase remain P-H3 positive for a considerable period after Scc1-myc has disappeared from chromosomes. In contrast, in cells infected with AdCre, Scc1-myc staining was detectable on chromosomes in nearly 99% of P-H3–positive cells (Fig. 8, D [bottom] and E). We also observed a small portion (∼5%) of cells positive for Scc1-myc on chromosomes but negative for P-H3 that we never observed in cells infected with AdGFP. These data suggest that cohesin persists at mitotic centromeres longer than in wild-type cells. The failure of cells lacking Separase to segregate their chromosomes may therefore be caused by their failure to remove cohesin.
Discussion
In the yeast S. cerevisiae, separation of sister chromatids is triggered by cleavage of cohesin's Scc1 subunit by a site-specific protease called Separase. Previous work using RNA interference is consistent with the notion that Separase has a similar function in mammalian tissue culture cells (Waizenegger et al., 2002). However, this method has two important limitations. First, it can produce off-target or nonspecific phenotypes; second, gene product depletion is rarely complete, often yielding hypomorphic phenotypes. We therefore introduced into the mouse germ-line a floxed Separase allele that permitted us to induce deletion of Separase's conserved protease domain upon induction of the Cre recombinase. We also created a floxed allele of Securin, Separase's inhibitory chaperone. By these means, we have shown that Separase is essential for mammalian embryonic development, that its activity is severely compromised in mice lacking Securin, and that Separase is essential for chromosome segregation at the onset of anaphase, both in hepatocytes in vivo and in iMEFs in vitro. It has been recently suggested that Separase is required for timely entry into mitosis (Papi et al., 2005). Our experiments would not have detected a modest delay in the G2–M phase transition. However, our finding that cells lacking Separase repeatedly enter mitosis after reduplicating their chromosomes is inconsistent with the notion that Separase has a major role in promoting M phase entry. It is important to point out that our Separase deletion could in principle lead to the continued synthesis of a truncated polypeptide that, though lacking any protease activity, might nevertheless have other cell cycle functions.
iMEFs lacking Separase enter mitosis and align chromosomes on metaphase plates but fail to completely segregate their chromosomes. This catastrophic failure is not detected by any surveillance mechanism (checkpoint) capable of arresting cell cycle progression. Thus, cells lacking Separase form cleavage furrows. These furrows sometimes bisect the chromosomes but more often leave them on one side of the dividing cell, producing one daughter cell with most chromosomes and another with few, if any. The former invariably exit from mitosis and with high frequency reenter the cell cycle, reduplicate their chromosomes, and fail again to undergo anaphase after entering mitosis. The polyploidization caused by inactivation of Separase in mammalian cells is broadly similar to that observed in fungi (Baum et al., 1988; Uzawa et al., 1990; May et al., 1992; McGrew et al., 1992). It also resembles the phenotype caused by inactivating Separase in D. melanogaster (Gatti and Baker, 1989; Jager et al., 2001; Pandey et al., 2005). Remarkably, both iMEFs and larval brain cells in D. melanogaster can undergo many rounds of chromosome reduplication in the absence of Separase, creating huge cells containing hundreds of chromosomes. A very similar sequence of events appears to occur in hepatocytes in vivo when they are stimulated by hepatectomy to undergo cell division in the absence of Separase. In this case, the polyploid cells produced in the absence of Separase appear fully capable of sustaining liver function.
Several lines of evidence suggest that the lack of chromosome segregation in cells lacking Separase might be caused by a failure to destroy sister chromatid cohesion. First, cells lacking Separase clearly biorient their chromosomes on metaphase plates. If we assume that the spindle forces during biorientation are similar to those that segregate chromosomes during anaphase, then the largely successful biorientation of chromosomes in Separase-deficient cells indicates that their spindles should also be capable of pulling chromatids to opposite poles of the cell during anaphase were it not for some other defect, for instance, in sister chromatid separation. The lack of chromosome segregation cannot be caused by a failure to activate the APC/C because cells destroy cyclin B, exit from mitosis, and usually undergo cytokinesis. Second, centromeric cohesin fails to disappear in mitotic cells lacking Separase, which indicates that sister chromatid cohesion may never be destroyed. Third, the finding of diplochromosomes after one round of rereplication in the absence of Separase and quadruplochromosomes after two rounds implies that sister chromatids that should have been separated at anaphase remain in sufficient proximity to each other during and after the next round of DNA replication that the two pairs of sister chromatids so produced remain closely associated. Fourth, the spectrum of phenotypes caused by inactivation of Separase in iMEFs resembles in many regards the spectrum of phenotypes caused by expression of noncleavable versions of cohesin's Scc1 subunit (Hauf et al., 2001). This includes the production of polyploid cells, cells that attempt to cleave through a central mass of decondensing chromosomes, and the production of diplochromosomes.
It should be noted that most of these phenotypes are much more penetrant in Separase-deficient iMEFs than they are in HeLa cells expressing noncleavable Scc1. There is little or no chromosome segregation in iMEFs lacking Separase, and diplochromosomes are generated in virtually all cells that reduplicate their chromosomes. In contrast, many HeLa cells in which noncleavable Scc1 expression has been induced still undergo chromosome segregation, and it is difficult to know whether this is due to insufficient expression of noncleavable Scc1. Furthermore, diplochromosomes accumulated in only 5.4% of cells. There are two possible explanations for this discrepancy. Either the supposedly noncleavable Scc1 alleles are still partly cleavable in vivo or Separase has functions besides Scc1 cleavage that are necessary for efficient chromosome segregation at anaphase. To distinguish between these possibilities, it may be necessary to test whether artificial cleavage of Scc1 is sufficient to trigger anaphase, as has been performed in yeast (Uhlmann et al., 2000).
Our filming of iMEFs suggests that they exit from mitosis and undergo cytokinesis with high efficiency in the absence of Separase or sister chromatid separation. Cells in D. melanogaster embryos behave likewise in the absence of the Securin-like protein Pimples, which appears to be essential for Separase activity (Pandey et al., 2005). There is therefore no evidence so far that Separase has an essential role in promoting mitotic exit in somatic animal cells.
The formation of diplo-, quadruplo-, and even octuplochromosomes in cells lacking Separase has also been observed in D. melanogaster mutants (Jager et al., 2001). In iMEFs, cohesin is clearly associated with the sister centromere pairs of diplochromosomes, but it is unclear whether appreciable amounts also connect the two sets of sister chromatids. This raises an interesting question as to the fate of sister chromatid cohesion inherited from the previous cycle in Separase mutants. Do cohesin-mediated bridges survive the next round of DNA replication and thereby somehow hold sister chromatid pairs together as well as sister chromatids, or do these bridges merely survive until the next round of DNA replication, and does the resulting close proximity of sister chromatids during replication cause the de novo production of connections, albeit abnormal ones, between the two sets of sister chromatids? Such abnormal connections could be mediated either by cohesin or by DNA catenation.
In summary, our data show that Separase is essential for sister chromatid separation in mammalian cells as well as in yeast (Ciosk et al., 1998), flies (Jager et al., 2001), and worms (Siomos et al., 2001). The so-called prophase pathway that involves phosphorylation of cohesin's Scc3-SA2 subunit is capable of removing cohesin from chromosome arms in mammalian cells (Hauf et al., 2005) but is prevented from removing cohesin from centromeres by the presence of shugoshins at this location of the chromosome (McGuinness et al., 2005). Separase alone can resolve centromeric cohesion in mammalian cells. This dependence on Separase is a crucial aspect of mitosis because Separase (via control of the APC/C) and not the prophase pathway is subject to the highly sophisticated regulation needed to ensure that sister chromatid separation does not commence until all chromosomes have bioriented on mitotic spindles.
Materials and methods
Targeting strategy for the Separase and Securin allele
Mouse Separase and Securin genomic DNA were isolated from a 129/Sv bacterial artificial chromosome (BAC) library (Research Genetics) by using a cDNA probe derived from dbEST AA165880 for Separase and from AA790273 for Securin. BAC clone 297K4 was used for construction of the Separase targeting vector and BAC clone 435D15 for the Securin targeting vector. The construction of the targeting vector and the gene targeting in HM1 ES cells was performed as described in Wirth et al. (2004).
Correct integration of the targeting construct at the Separase genomic locus was analyzed on EcoRV-digested ES cell DNA by using an external probe. The presence of the COOH-terminal loxP site was confirmed by using an Asp718 digest and an internal probe. Floxed or Δ alleles were obtained by electroporation with 25 μg pMC-Cre and confirmed by Southern blot analysis of an EcoRV digest with an internal probe.
Correct integration of the Securin targeting vector at the genomic locus was analyzed by an Asp718 digest using an external probe. With an internal probe and an Asp718 digest, the presence of the third loxP site was confirmed. Floxed and Δ alleles were identified on Southern blot using a BamHI and Asp718 digest and an internal probe. Chimeric mice were created as described in Wirth et al. (2004).
Induction of Cre in vivo
Cre-recombinase expression was induced as described in Wirth et al. (2004). In hepatocytes, pI/C was injected 2×, and in hematopoietic cells 1×, within an interval of 72 h.
BAC library high-density filter hybridization and Southern blot analysis
BAC library high-density filter hybridization and Southern blot analysis were done as described previously (Wirth et al., 2004).
Two thirds hepatectomy and single-cell DNA measurement
Two thirds hepatectomy and single-cell DNA measurement were done as described previously (Oppedal et al., 1988; Wirth et al., 2004).
Live cell video microscopy and immunofluorescence microscopy of hepatocytes
Hepatocyte culture, live cell video microscopy, and immunofluorescence microscopy of hepatocytes were performed as described previously (Guidotti et al., 2003).
Paraffin sections
For paraffin sections, embryos were fixed overnight in 4% PFA/PBS, dehydrated to 100% EtOH, embedded in paraffin, and sectioned at 5 μm. After the deparaffinization procedure, sections were treated with Proteinase K, stained with Hoechst, and mounted.
Primers for genomic DNA amplification in iMEFs
The following primers were used: 5′-ACATGACTCTGGGTGTGTCTTCTC-3′ and 5′-TTCATCACCCAAGCTCCAGCAG-3′ for the deletion allele; 5′-ACTGACCGTGACATTGACCGTTAC-3′ and 5′-TTCATCACCCAAGCTCCAGCAG-3′ for the flox allele; and 5′-ATGAGGAACTTCAAAGGAGTCAACTTC-3′ and 5′-GCGCAAGCCTTTAATCCCAG-3′ for the loading control.
Antibodies
To detect Separase on Western blot, we used mouse monoclonal (7A6) antibody (Waizenegger et al., 2000). Other antibodies used in this study were as follows: mouse anti–Aurora B antibody (anti–AIM-1; BD Biosciences), CREST serum (a gift from A. Kromminga, Univiersity of Mainz, Mainz, Germany), mouse anti-myc antibody (clone 4A6; Upstate), antibody to phosphohistone H3 (Ser10[P-H3], a mouse monoclonal antibody that detects histone H3 when phosphorylated at serine 10; Cell Signaling Technology), anti-Topoisomerase IIα (Chemicon), mouse anti–cyclin B1 antibody (Santa Cruz Biotechnology, Inc.), and anti–γ-tubulin (Sigma-Aldrich).
Cell culture and infection
iMEFs were cultured in DME supplemented with 10% FCS, 0.2 mM l-glutamine, 100 U/ml penicillin, 100 μm/ml streptomycin, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol, and nonessential amino acids. For infection, cells were grown to 100% confluency, washed with PBS, and infected with the virus (7,000 particles/cell) in DME supplemented with 2% FCS. After 24 h, cells were transferred to fresh medium. Cells were split after 24 h, and samples were harvested at different time points. Nocodazole was used at a final concentration of 100 ng/ml.
AdCre and adenovirus expressing EGFP (Ad5 CMV Cre and EGFP) were purchased from The University of Iowa (Iowa City, IA). For generation of the H2B-mRFP stably expressing line, cells were infected with plasmid H2B-mRFP (a gift from J. Ellenberg, European Molecular Biology Laboratory, Heidelberg, Germany) using lipofectamine reagent (Invitrogen). Stable expressants were selected in a complete medium containing 800 μg/ml G418 and were screened by fluorescence microscopy for expression of H2B-mRFP.
For the generation of the Separase Δ/flox iMEF cell line stably expressing Scc1, murine Scc1 was COOH-terminally tagged with nine myc epitopes and inserted into pREVTRE vector (CLONTECH Laboratories, Inc.). The resulting plasmid was infected into φNX-Eco cells (Stanford University Medical Center, Stanford, CA) using lipofectamine reagent. Supernatant containing the virus was used to infect Separase Δ/flox cells containing Tet off transactivator. Selection with 100 μg/ml hygromycin B was started 48 h later. After 2 wk, cell lines arising from single cells were picked and tested for Scc1-myc expression by immunofluorescence microscopy.
For flow cytometric analysis, cells fixed in 70% methanol were washed with PBS and subsequently stained in PI buffer (10 μm/ml propidium iodide, 10 mM Tris-Hcl, pH 7.5, 5 mM MgCl2, and 200 μm/ml RNase A) for 20 min at 37°C.
Preparation of hepatocytes and iMEF cell extracts
Preparation of hepatocytes and iMEF cell extracts for Western blotting was done as described previously (McGuinness et al., 2005).
Immunofluorescence microscopy and chromosome spreads
Immunofluorescence microscopy and chromosome spreads were done as described previously (McGuinness et al., 2005).
Online supplemental material
Fig. S1 shows that Separaseflox/floxMx-Cre hepatocytes undergo abnormal mitosis. Fig. S2 shows aplastic bone marrow in Separase-deficient mice. Fig. S3 shows that there is no anaphase but abnormal telophases in iMEFs lacking Separase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200506119/DC1.
Acknowledgments
We thank Hartmut Beug, Meinrad Busslinger, Annette Neubüser, Anton Wutz, Erwin Wagner, and their groups for advice and reagents; Dónal O'Carroll for the introduction to ES cell work; and Hans Christian Theussl for blastocyte injections. We also thank Christine Hartmann for help with embryo isolation and paraffin sections, Alexander Schleiffer for bioinformatic searches, Irene Waizenegger for Separase antibodies, Olivier Bregerie for help with the hepatocyte culture and time-lapse microscopy, and Sigrun Kirste for technical assistance.
The Institute of Molecular Pathology is funded by Boehringer Ingelheim, and this work was partly supported by a Marie Curie fellowship from the European Community for R. Ricci (HPFM-CT 2001-01133), a Research Fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft) for K. Wirth (WI 1679/1-1), and a Vienna Science and Technology Fund Life Science grant to J.-M. Peters.
K.G. Wirth and G. Wutz contributed equally to this paper.
K. Nasmyth's present address is University of Oxford, Oxford OX1 2JD, England.
Abbreviations used in this paper: AdCre, adenovirus expressing Cre recombinase; AdGFP, adenovirus expressing GFP; APC/C, anaphase promoting complex or cyclosome; BAC, bacterial artificial chromosome; CREST, calcinosis, Raynaud's phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasia; dpc, days post coitus; ES, embryonic stem; iMEF: immortalized MEF; MEF, mouse embryonic fibroblast; P-H3, phosphorylated histone H3; pI/C, poly(I)poly(C).
References
- Baum, P., C. Yip, L. Goetsch, and B. Byers. 1988. A yeast gene essential for regulation of spindle pole duplication. Mol. Cell. Biol. 8:5386–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chestukhin, A., C. Pfeffer, S. Milligan, J.A. DeCaprio, and D. Pellman. 2003. Processing, localization, and requirement of human separase for normal anaphase progression. Proc. Natl. Acad. Sci. USA. 100:4574–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An Esp1/Pds1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 93:1067–1076. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., J.-M. Peters, M.W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081–3093. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., K. Kumada, and M. Yanagida. 1996. a. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., H. Yamano, K. Kumada, K. Nagao, T. Hunt, and M. Yanagida. 1996. b. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 381:438–441. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and B.S. Baker. 1989. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 3:438–453. [DOI] [PubMed] [Google Scholar]
- Gruber, S., C.H. Haering, and K. Nasmyth. 2003. Chromosomal cohesin forms a ring. Cell. 112:765–777. [DOI] [PubMed] [Google Scholar]
- Guidotti, J.E., O. Bregerie, A. Robert, P. Debey, C. Brechot, and C. Desdouets. 2003. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J. Biol. Chem. 278:19095–19101. [DOI] [PubMed] [Google Scholar]
- Hauf, S., I. Waizenegger, and J.-M. Peters. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 293:1320–1323. [DOI] [PubMed] [Google Scholar]
- Hauf, S., E. Vorlaufer, B. Koch, C. Dittrich, K. Mechtler, and J.-M. Peters. 2005. Dissociation of cohesin from chromosome arms and loss of arm cohesion during prophase depends on phosphorylation of SA2. PLoS Biol. 3:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig, N.C.D., P.P. Knowles, N.Q. McDonald, and F. Uhlmann. 2002. The dual mechanism of separase regulation by securin. Curr. Biol. 12:973–982. [DOI] [PubMed] [Google Scholar]
- Jager, H., A. Herzig, C.F. Lehner, and S. Heidmann. 2001. Drosophila separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev. 15:2572–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, R., F. Schwenk, M. Aguet, and K. Rajewsky. 1995. Inducible gene targeting in mice. Science. 269:1427–1429. [DOI] [PubMed] [Google Scholar]
- Jallepalli, P.V., I. Waizenegger, F. Bunz, S. Langer, M.R. Speicher, J.-M. Peters, K.W. Kinzler, B. Vogelstein, and C. Lengauer. 2001. Securin is required for chromosomal stability in human cells. Cell. 105:445–457. [DOI] [PubMed] [Google Scholar]
- Losada, A., M. Hirano, and T. Hirano. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, G.S., C.A. McGoldrick, C.L. Holt, and S.H. Denison. 1992. The bimB3 mutation of Aspergillus nidulans uncouples DNA replication from the completion of mitosis. J. Biol. Chem. 267:15737–15743. [PubMed] [Google Scholar]
- McGrew, J.T., L. Goetsch, B. Byers, and P. Baum. 1992. Requirement for ESP1 in the nuclear division of S. cerevisiae. Mol. Biol. Cell. 3:1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness, B.E., T. Hirota, N.R. Kudo, J.M. Peters, and K. Nasmyth. 2005. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, J., X. Huang, and P. Zhang. 2001. Securin is not required for cellular viability, but is required for normal growth of mouse embryonic fibroblasts. Curr. Biol. 11:1197–1201. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and C.H. Haering. 2005. The structure and function of smc and kleisin complexes. Annu. Rev. Biochem. 74:595–648. [DOI] [PubMed] [Google Scholar]
- Nicklas, R.B., and S.C. Ward. 1994. Elements of error correction in mitosis: microtubule capture, release, and tension. J. Cell Biol. 126:1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppedal, B.R., A. Glomstein, and A. Zetterberg. 1988. Feulgen DNA values in Wilms' tumour in relation to prognosis. Pathol. Res. Pract. 183:756–760. [DOI] [PubMed] [Google Scholar]
- Pandey, R., S. Heidmann, and C.F. Lehner. 2005. Epithelial re-organization and dynamics of progression through mitosis in Drosophila separase complex mutants. J. Cell Sci. 118:733–742. [DOI] [PubMed] [Google Scholar]
- Papi, M., E. Berdougo, C.L. Randall, S. Ganguly, and P.V. Jallepalli. 2005. Multiple roles for separase auto-cleavage during the G2/M transition. Nat. Cell Biol. 7:1029–1035. [DOI] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel. 2003. Separase regulates INCENP-Aurora B anapahse spindle function through Cdc14. Science. 302:2120–2124. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., A. Schultz, R. Cole, and G. Sluder. 1994. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127:1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomos, M.F., A. Badrinath, P. Pasierbek, D. Livingstone, J. White, M. Glotzer, and K. Nasmyth. 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11:1825–1835. [DOI] [PubMed] [Google Scholar]
- Stemmann, O., H. Zou, S.A. Gerber, S.P. Gygi, and M.W. Kirschner. 2001. Dual inhibition of sister chromatid separation at metaphase. Cell. 107:715–726. [DOI] [PubMed] [Google Scholar]
- Stratmann, R., and C.F. Lehner. 1996. Separation of sister chromatids in mitosis requires the Drosophila pimples product, a protein degraded after the metaphase/anaphase transition. Cell. 84:25–35. [DOI] [PubMed] [Google Scholar]
- Sumara, I., E. Vorlaufer, C. Gieffers, B.H. Peters, and J.-M. Peters. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151:749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., J. Fuchs, J. Loidl, and K. Nasmyth. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2:492–499. [DOI] [PubMed] [Google Scholar]
- Tomonaga, T., K. Nagao, Y. Kawasaki, K. Furuya, A. Murakami, J. Morishita, T. Yuasa, T. Sutani, S.E. Kearsey, F. Uhlmann, et al. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14:2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1p. Nature. 400:37–42. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., D. Wernic, M.A. Poupart, E. Koonin, and K. Nasmyth. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 103:375–386. [DOI] [PubMed] [Google Scholar]
- Uzawa, S., I. Samejima, T. Hirano, K. Tanaka, and M. Yanagida. 1990. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 62:913–925. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I., S. Hauf, A. Meinke, and J.-M. Peters. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 103:399–410. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I., J.F. Gimenez-Abian, D. Wernic, and J.-M. Peters. 2002. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 12:1368–1378. [DOI] [PubMed] [Google Scholar]
- Wang, Z., R. Yu, and S. Melmed. 2001. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 15:1870–1879. [DOI] [PubMed] [Google Scholar]
- Wirth, K.G., R. Ricci, J.F. Gimenez-Abian, S. Taghybeeglu, N.R. Kudo, W. Jochum, M. Vasseur-Cognet, and K. Nasmyth. 2004. Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation. Genes Dev. 18:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H., T.J. McGarry, T. Bernal, and M.W. Kirschner. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 285:418–422. [DOI] [PubMed] [Google Scholar]