Abstract
Mdm2 is required to negatively regulate p53 activity at the peri-implantation stage of early mouse development. However, the absolute requirement for Mdm2 throughout embryogenesis and in organogenesis is unknown. To explore Mdm2–p53 signaling in osteogenesis, Mdm2-conditional mice were bred with Col3.6-Cre–transgenic mice that express Cre recombinase in osteoblast lineage cells. Mdm2-conditional Col3.6-Cre mice die at birth and display multiple skeletal defects. Osteoblast progenitor cells deleted for Mdm2 have elevated p53 activity, reduced proliferation, reduced levels of the master osteoblast transcriptional regulator Runx2, and reduced differentiation. In contrast, p53-null osteoprogenitor cells have increased proliferation, increased expression of Runx2, increased osteoblast maturation, and increased tumorigenic potential, as mice specifically deleted for p53 in osteoblasts develop osteosarcomas. These results demonstrate that p53 plays a critical role in bone organogenesis and homeostasis by negatively regulating bone development and growth and by suppressing bone neoplasia and that Mdm2-mediated inhibition of p53 function is a prerequisite for Runx2 activation, osteoblast differentiation, and proper skeletal formation.
Introduction
The p53 transcription factor is activated by inappropriate cell growth stimulation or by certain types of DNA damage and regulates the expression of other genes involved in cell growth arrest, DNA repair, and apoptosis (Vousden, 2000). These various p53-mediated effects suppress tumorigenesis, and mutation of the p53 gene or of the p53 signaling pathway is commonly found in most human cancers (Soussi and Beroud, 2001). Although the ability of p53 to regulate cell growth after exposure to stress has been well established, the role of p53 in regulating normal (nondamaged) cell growth and in tissue homeostasis is uncertain. Mice deleted for p53 will form tumors with 100% penetrance yet undergo normal development (Donehower et al., 1992), albeit with a reduction from the expected numbers of female births and a small percentage of embryos presenting at midgestation with exencephaly (Armstrong et al., 1995; Sah et al., 1995). In addition, transgenic mice bearing a reporter gene under transcriptional control of a p53 response element reveal little or no postnatal p53 activity in the absence of DNA damage (Gottlieb et al., 1997). These findings suggest that p53 is important in suppressing tumorigenesis but is largely dispensable for normal cell growth, cell differentiation, and development.
In contrast, a recently generated mouse model bearing a mutated p53 allele (m allele) that increased p53 activity in vitro displayed early aging-like phenotypes, including reduced mass of various internal organs, thinning of the dermis, hair loss, and osteoporosis (Tyner et al., 2002). Interestingly, the p53 m/+ mice also had reduced incidence of cancer relative to p53 heterozygous (−/+) mice, suggesting that the mutant allele increased p53 activity encoded by the wild-type p53 gene in the m/+ mice. Although this increase in basal levels of p53 activity offered further protection from neoplasia, the authors hypothesized that the slight increase in p53 activity also reduced stem cell proliferation in affected tissues leading to reduced tissue cellularity. These results suggest that negative regulation of p53-induced apoptosis or inhibition of cell growth might be important to maintain proper tissue homeostasis in adult mice.
Mdm2 is a key negative regulator of p53 activity in the cell. Mdm2 complexes with p53 and negatively regulates p53-induced transcription of target genes, including the Mdm2 gene (for review see Iwakuma and Lozano, 2003). During times of cellular insult, p53 activates Mdm2 gene expression by binding to a p53 response element within the first intron of the Mdm2 gene (Juven et al., 1993). Induction of Mdm2 protein levels leads to an increase in Mdm2–p53 complex formation that interferes with the ability of p53 to transactivate Mdm2. Thus, Mdm2 expression is autoregulated because of the ability of Mdm2 to negatively regulate p53 (Wu et al., 1993). Mdm2 has been shown to interfere with the ability of p53 to transactivate target genes by binding and stearically hindering the NH2-terminal activation domain of the p53 protein (Momand et al., 1992; Chen et al., 1995) or by altering p53 protein modifications that regulate p53 transcriptional activation (Xirodimas et al., 2004). In addition, Mdm2 can function as an E3 ligase to coordinate the ubiquitination of p53 (Honda et al., 1997) and can induce the degradation of p53 by the 26S proteasome (Haupt et al., 1997; Kubbutat et al., 1997; Li et al., 2003). Mdm2 can also assist in shuttling p53 from the nucleus into the cytoplasm (Freedman and Levine, 1998; Geyer et al., 2000). The importance of Mdm2 in negatively regulating p53 activity is perhaps best illustrated by the finding that the early (embryonic day [E] 4–5) lethal phenotype of Mdm2-null mice can be fully rescued by the concomitant deletion of p53 (Jones et al., 1995; Montes de Oca Luna et al., 1995).
Although the requirement for Mdm2-mediated inhibition of p53 activity during early development has been well established, the role of Mdm2 in regulating p53 functions in later stages of embryogenesis or in adult tissues is unclear. However, several lines of evidence suggest that Mdm2 does function to regulate p53 activity in postnatal tissue. EuMyc transgenic mice display a delayed onset of B cell lymphoma when haploinsufficient for Mdm2, suggesting that a reduction in Mdm2-mediated suppression of p53 can reduce tumorigenesis (Alt et al., 2003). In addition, mice bearing a hypomorphic allele of Mdm2 that have ∼30% of the normal endogenous levels of Mdm2 are smaller in size, have reduced numbers of hematopoietic cells, and display excess apoptosis in the lymphoid compartment (Mendrysa et al., 2003). Crossing the Mdm2 hypomorphic allele onto a p53-deficient background reversed the various phenotypes observed in these mice, demonstrating that the phenotypic effects caused by Mdm2 reduction in this model were induced by p53. These data suggest that Mdm2 is capable of negatively regulating p53 activity in hematopoietic tissues.
To determine the absolute requirement for Mdm2 during development and in adult tissues, we have recently used Cre-loxP technology to generate Mdm2-conditional mice. Gene targeting experiments in embryonic stem cells flanked the last two exons of the Mdm2 gene encoding the zinc RING (really interesting new gene) finger domains and polyadenylation signals with loxP sites. Cre-mediated recombination of the loxP sites in the conditional allele destabilizes Mdm2 transcripts and results in loss of Mdm2 message. (Steinman and Jones, 2002). Because studies of p53 mutant mice suggest that excess p53 activity might have a deleterious effect on normal bone homeostasis, we sought to determine whether Mdm2 regulates p53 activity during osteogenesis. To this end, Mdm2- or p53-conditional mice were bred with transgenic mice in which the Cre-recombinase gene has been placed under transcriptional control of a 3.6-kb fragment of the Coll1a1 promoter. These Col3.6-Cre–transgenic mice have been previously reported to express Cre in cells of the osteoblast lineage (Liu et al., 2004). Mdm2-conditional mice bearing the Col3.6 transgene have multiple skeletal defects, including fused or otherwise altered lumbar vertebrae, reduced mineralized bone, and reduced bone length. Osteoblasts deleted for Mdm2 do not undergo apoptosis but do have elevated p53 activity, increased transactivation of p53 target genes, reduced cell proliferation, and reduced levels of the osteoblast transcriptional regulator Runx2, which is essential for osteoblast differentiation (Banerjee et al., 1997; Ducy et al., 1997; Komori et al., 1997). In contrast, osteoblasts deleted for p53 display elevated Runx2 levels, enhanced cell proliferation, and increased maturation and mineralization. Furthermore, mice specifically deleted for p53 in osteoblast progenitor cells develop osteosarcomas. These results demonstrate that p53 is an important negative regulator of osteogenesis and that Mdm2-mediated inhibition of p53 function is a critical requirement for Runx2 activation and proper osteoblast differentiation and skeletal formation.
Results
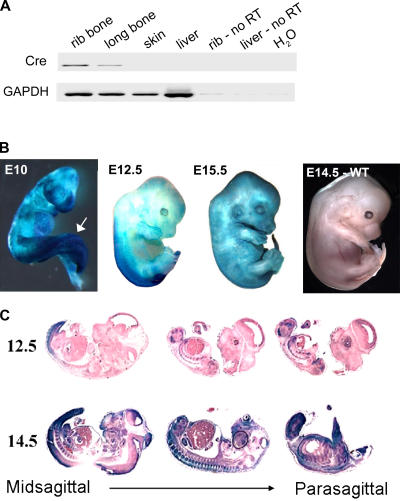
Expression of Cre recombinase in Col3.6-Cre–transgenic mice
To understand the contribution of Mdm2 and p53 signaling to bone development, we used Mdm2-conditional mice bearing Mdm2 genes (Mdm2sjcnd1) with loxP recombination sites flanking exons 11 and 12 (Steinman and Jones, 2002). Deletion of these exons will result in loss of Mdm2 in cells expressing the P1 bacteriophage Cre-recombinase transgene under transcriptional control of a rat 3.6-kb type I collagen promoter fragment (Liu et al., 2004). This transgene has been reported to induce Cre-mediated gene excision in postnatal calvarial bone and long bone osteoblasts, as well as in skin and tendons. We initially verified the expression of Cre in skeletal elements of adult mice. RT-PCR reactions using RNA isolated from the rib and femur of adult Col3.6-Cre mice demonstrated Cre expression in these tissues, whereas Cre transcripts were not detected in other tested organs (Fig. 1 A). To identify regions in Col3.6-Cre developing embryos that express Cre and induce Cre-mediated excision, we mated the Col3.6-Cre–transgenic mice to ROSA26 (R26R) reporter mice that express β-galactosidase upon Cre-induced deletion of a floxed cassette that inhibits the reporter gene expression (Soriano, 1999). Our findings demonstrate that this promoter becomes activated at two distinct points during development. Expression of the Col3.6-Cre transgene as determined by β-galactosidase activity is first detected in whole-mount stainings of E8–19 embryos and is localized initially to the caudal portion of the embryo. Reporter gene expression is strongest in this region at E10 (Fig. 1 B). This region of the developing embryo contains both neural and mesenchymal progenitor cells that give rise to the caudal axial skeleton and spinal cord. Later in development (∼E14), the Col3.6-Cre transgene undergoes robust activation in relation to the formation of connective tissue. Staining of sagital sections of R26R/Col3.6-Cre embryos for β-galactosidase activity indicates that Cre-mediated excision occurs in both the skin and developing skeletal elements (Fig. 1 C). These findings indicate that Cre transgene expression will induce Mdm2 deletion in Col3.6-Cre × Mdm2-conditional mice in the surface ectoderm and developing tail bud of E9 mice as well as in developing bone and connective tissue of these mice during midgestation.
Figure 1.
Expression of Cre recombinase in Col3.6-Cre–transgenic mice. (A) PCR performed using Cre primers on reverse-transcribed RNA isolated from adult Col3.6-Cre–transgenic mice. Primers to GAPDH were used as a positive control in the RT-PCR, whereas non–reverse-transcribed (No RT) rib bone RNA, liver RNA, and water (H20) were used as negative controls for the PCR. Cre expression is readily detected in the rib and femur of these mice. (B) Whole-mount staining of lacZ expression in Col3.6-Cre–transgenic R26R embryos at E10, -12.5, and -15.5 during development. Expression of the Cre recombinase as detected by β-galactosidase activity is detected in surface ectoderm and in the caudal portion of the embryo at E10, with the highest levels found in the tail bud (arrow). Between E12.5 and -15.5, the Col3.6-Cre transgene undergoes robust activation with the formation of connective tissue. (C) Staining for β-galactosidase activity in sagital sections of R26R/Col3.6-Cre embryos indicates that Cre-mediated excision occurs in both the skin and developing skeletal elements at E12.5–14.5.
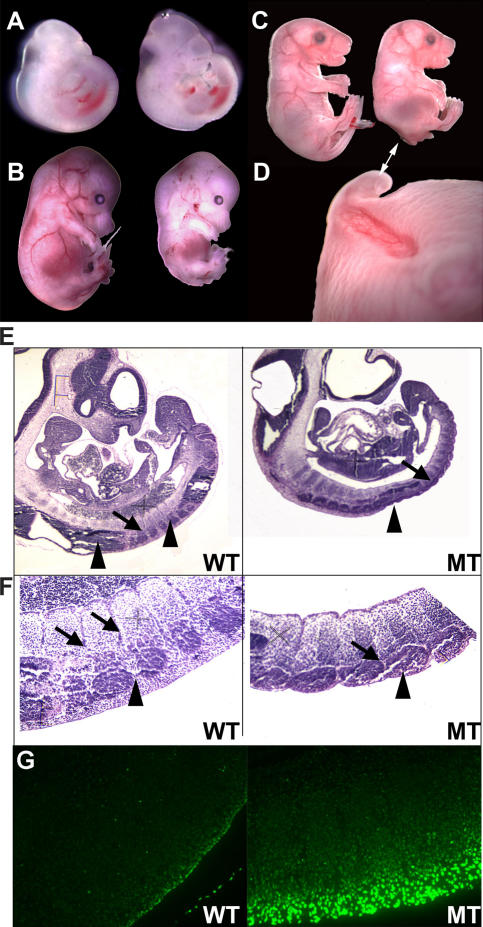
Caudal defects in Mdm2-conditional Col3.6-Cre mice
We crossed Mdm2-conditional mice (Mdm2sjcnd1/+ or Mdm2sjcnd1/sjcnd1) to Col3.6-Cre–transgenic mice containing one conditional Mdm2 allele (Mdm2sjcnd1/+,Col3.6-Cre). The resulting litters contained Mdm2sjcnd1/sjcnd1,Col3.6-Cre mice (designated as mutant), as well as Mdm2+/+,Col3.6-Cre mice and Mdm2sjcnd1/+,Col3.6-Cre mice. These last two groups of mice were indistinguishable from wild-type mice, which was expected, given that Mdm2 heterozygous mice were previously found to display no morphologic developmental defects (Jones et al., 1995).
To examine the effects of Mdm2 loss during development, timed matings of Mdm2sjcnd1/+,Col3.6-Cre–transgenic mice were performed and embryos harvested and genotyped at various times after coitum. Expected numbers of mutant Mdm2 embryos were recovered between E8.5 and -12.5 (Table I). However, reduced numbers of mutant embryos were found at E13.5 and throughout later stages of development, and no viable Mdm2sjcnd1/sjcnd1,Col3.6-Cre mutant mice were recovered at weaning or at birth, demonstrating that deletion of Mdm2 in Col3.6-Cre–transgenic mice induces lethality during later stages of embryogenesis.
Table I.
Embryonic lethality of Mdm2-conditional Col3.6-Cre mice
| Developmental stage | Mdm2 mutant/total embryos |
Percentage of total |
|---|---|---|
| E8.5–10.5 | 9/36 | 25 |
| E12.5 | 8/33 | 25 |
| E13.5 | 2/18 | 11 |
| E14.5 | 4/38 | 11 |
| E15.5–17.5 | 6/58 | 10 |
| E18.5–19.5 | 11/100 | 11 |
| Postnatal weaning | 0/100 | 0 |
Defects in the developing tail bud region of mutant mice were apparent as early as E10.5, and caudal runting was seen in all mutant mice throughout later stages of development (Fig. 2, A–C). Col3.6-Cre–induced deletion of Mdm2 expression in the caudal region of mutant mice resulted in a complete absence of tissue surrounding the somites at E10.5, as well as exposure of somitic mesenchyme at the surface of the embryo, absence of a tail, and a severe invagination in the posterior dorsal region encompassing the lumbar vertebrae (Fig. 2 D). Histologic analysis of mutant and wild-type embryos harvested at E10 was performed. Wild-type embryos had clearly segmented somites that will give rise to axial skeletal components, including vertebrae and ribs (Fig. 2 E, arrows). These somites are surrounded by primitive neural tissue and by developing dorsal root ganglia (Fig. 2 E, arrowheads). In contrast, mutant embryos lack developing neural tissue and posterior dorsal root ganglia, leaving caudal somites externalized (Fig. 2 F). To understand the mechanism underlying this tissue loss, we performed TUNEL staining on serial sections. Our results show that there is a dramatic increase in TUNEL-positive apoptotic cells in the caudal somites and surrounding tissue of mutant E10 embryos in comparison to wild-type littermates (Fig. 2 G).
Figure 2.
Caudal defects in Mdm2-conditional Col3.6-Cre mice. (A–D) Homozygous Mdm2-conditional embryos lacking (left) or containing (right) the Col3.6-Cre transgene were harvested from timed matings at E10 (A), E15 (B), and E17.5 (C). Embryos mutated for Mdm2 were generally smaller and displayed obvious runting of the caudal portion of the embryo, absence of a tail, and a severe invagination in the posterior dorsal region encompassing the lumbar vertebrae (D, double-headed arrow). (E and F) Hematoxylin and eosin staining of sagital sections of wild-type (WT) and Mdm2-conditional Col3.6-Cre (MT) embryos (E). Arrows identify somites that are surrounded by primitive neural tissue and by developing dorsal root ganglia. Mutant embryos lack developing neural tissue and posterior dorsal root ganglia (arrowheads), leaving caudal somites externalized (F). (G) TUNEL staining performed on serial sections shown in F demonstrates increased TUNEL-positive apoptotic cells in the caudal somites and surrounding tissue of mutant embryos in comparison to wild-type littermates.
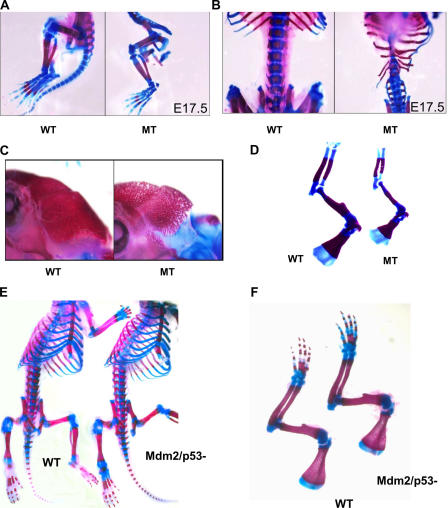
Excision of Mdm2 in skeletal tissues results in impaired bone formation
Skeletal preparations of wild-type and Mdm2sjcnd1/sjcnd1,Col3.6-Cre embryos revealed that the apoptosis observed in the E10 caudal somatic cells resulted in highly dysplastic axial skeletal elements in the mutant embryo with fused cartilaginous lumbar vertebrae (Fig. 3, A and B). However, the appendicular skeleton and bones of the skull were unaffected by the initial Cre activation. Activation of Col3.6-Cre transgene expression in the limbs and skull did not occur until the latter half of gestation (beginning at E14.5). These skeletal elements were more porous in mutant animals (Fig. 3 C), and measurements of the long bones (tibia, radius, ulna, humerus, and femur) revealed that the total long bone length was 12.1 ± 3.9% shorter in the mutant embryos and the length of mineralized portion of long bones was 12.3 ± 4.4% shorter in the mutant embryos than in wild-type littermate embryos at E17.5 (Fig. 3 D). In contrast, skeletal preparations of embryos deleted for both Mdm2 and p53 (Jones et al., 1995) revealed no vertebral dysplasia or reduction in bone length (Fig. 3, E and F). These results reveal that the deleterious effects of Mdm2 loss on skeletal formation during development are p53 dependent.
Figure 3.
Excision of Mdm2 in skeletal tissues results in impaired bone formation. Alcian blue and alizarin red staining of skeletal preparations of wild-type (WT) and Mdm2sjcnd1/sjcnd1, Col3.6-Cre (MT) embryos. (A and B) Mutant embryos harvested at E17.5 revealed highly dysplastic axial skeletal elements with fused cartilaginous lumbar vertebrae. (C) The skulls of Mdm2sjcnd1/sjcnd1,Col3.6-Cre E17.5 embryos are highly porous, and (D) appendicular bones of the forelimb were shorter in length in comparison to wild-type littermates. Skeletal preparations of wild-type neonatal mice and neonates deleted for both Mdm2 and p53 revealed no vertebral dysplasia (E) or reduction in bone length (F), indicating that the deleterious effects of Mdm2 loss on skeletal formation during development are p53 dependent.
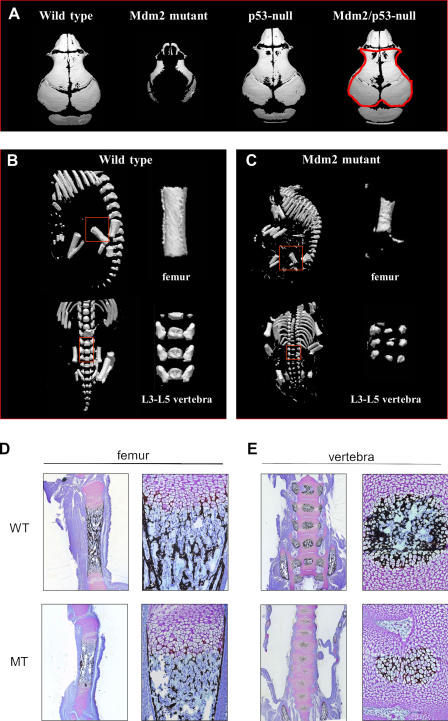
Appendicular and skull bones from E18.5 Mdm2sjcnd1/sjcnd1, Col3.6-Cre mice showed significantly less mineral deposition than wild-type littermates, as assessed by microcomputed tomography (micro-CT) scan (Fig. 4, A–C) and by silver nitrate (Von Kossa) staining (Figs. 4, D and E). Decreased bone mineralization was observed in the skull (Fig. 4 A), femur, and vertebra (Fig. 4, B and C) of Mdm2 mutant mice. Histomorphometric analysis of micro-CT scans performed on litter-matched, E18.5 embryos revealed reduced bone density in the calvaria, femur, and vertebra of Mdm2 mutant mice (Table II). However, these defects were not observed in embryos lacking both Mdm2 and p53. These results demonstrate that loss of Mdm2 in developing skeletal tissues negatively affects multiple parameters of bone quality in a p53-dependent manner. Furthermore, Von Kossa and toluidine blue staining of femur and vertebra sections (Fig. 4, D and E) revealed that the vascularized marrow cavity and other normal compartments of bone are present in the mutant mice and that Mdm2 mutant bone has a morphologically normal growth plate. These data suggest that Mdm2 mutant mice do not have a chondrogenic defect but rather a defect in ossification. However, unlike the proapoptotic effects of Mdm2 deletion in undifferentiated progenitor cells in E10 mutant mice, TUNEL assays performed on E14.5 skeletal rib elements from mutant embryos revealed no increase in the number of apoptotic cells (unpublished data).
Figure 4.
Altered mineralization and morphology observed in Mdm2 mutant mice. (A) Micro-CT analysis of skulls of wild-type; Mdm2sjcnd1/sjcnd1,Col3.6-Cre (Mdm2 mutant); p53-null; and Mdm2/p53 double-null E18.5 embryos reveals reduced bone mineralization in Mdm2 mutant mice. Histomorphometric measurements of bone volume versus total volume are provided in Table II for each skull, with a representative area analyzed for the Mdm2/p53-null sample outlined in red. Micro-CT scan of femur and vertebra (L3–5 region) of Mdm2 of wild-type E18.5 embryo (B) and Mdm2 mutant embryo (C). Regions of bones analyzed in Table II are outlined in red and shown in expanded view. Von Kossa (silver nitrate) and toluidine blue staining of sagital sections of femur (D) from Mdm2-conditional Col3.6-Cre (MT) mice showed a relatively normal growth plate region but had significantly less mineral deposition than Mdm2-conditional mice lacking the Cre transgene (WT). Similarly, less mineralization was also detected in Von Kossa and toluidine blue staining (E) of Mdm2-conditional Col3.6-Cre vertebra compared with wild-type vertebra.
Table II.
Morphometric analysis and quantitative analysis of bones
| Genotype | Skull width | Skull length | L3-5 spine | Calvaria | Femur | Vertebra |
|---|---|---|---|---|---|---|
| mm | mm | mm | % | % | % | |
| Wild type | 8.1 | 10.1 | 2.2 | 93.0 | 28.4 | 51.7 |
| Mutant | 5.2 | 7.7 | 1.3 | 31.5 | 12.7 | BT |
| p53 null | 7.8 | 10.3 | 2.2 | 88.0 | 16.4 | 40.8 |
| Mdm2/p53 null | 8.0 | 9.9 | 2.4 | 90.3 | 18.1 | 48.4 |
Data obtained from representative (E18.5) littermate embryos. Measurements are given in millimeters. Quantitative analysis is expressed as percentage of bone density as determined by the amount of bone per tissue volume (bone volume over total volume). Below threshold (BT) means bone volume was below detectable levels.
Negative regulation of p53 by Mdm2 is required for proper osteoblast differentiation
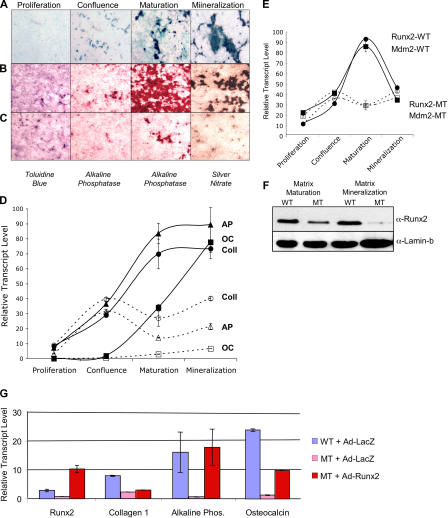
To better understand the underlying cause of the decrease in bone quality in Mdm2-conditional Col3.6-Cre mice, calvarial osteoprogenitor cells were isolated from E19 wild-type and mutant embryos as well as from R26R/Col3.6-Cre embryos to visualize the pattern of Cre-mediated excision in these cultures. Calvarial osteoprogenitors were induced to undergo osteogenic differentiation ex vivo by allowing cultures to proliferate for several days followed by the addition of ascorbic acid and inorganic phosphate to the media postconfluence. The addition of ascorbic acid to the cultured osteoprogenitors stimulates postconfluent proliferation, resulting in the formation multilayered nodules that later become mineralized (Owen et al., 1990). Examination R26R/Col3.6-Cre osteoprogenitor cells reveals that few osteoprogenitor cells had activated the transgene before isolation from the embryo and that Cre expression is induced in the osteoblast cultures during differentiation (Fig. 5 A). Because Cre-mediated recombination occurs in these cultures in maturing osteogenic nodules after reaching confluence, Mdm2 expression in Mdm2-conditional Col3.6-Cre cultures should be lost in multilayering nodules of maturing osteoblasts.
Figure 5.
Mdm2 is required for proper osteoblast differentiation. Calvarial osteoprogenitor cells were isolated and cultured ex vivo from E19 Col3.6-Cre–transgenic embryos bearing the R26R reporter gene, from E19 wild-type and Mdm2-conditional Col3.6-Cre–transgenic embryos. Calvarial osteoprogenitors were induced to undergo osteogenic differentiation ex vivo, resulting in proliferation (toluidine blue staining for total cell number), the formation of multilayered nodules and activation of alkaline phosphatase, and mineralization as detected by silver nitrate staining. (A) Examination R26R/Col3.6-Cre osteoprogenitor cells stained for β-galactosidase activity reveals that Mdm2 expression in Mdm2-conditional Col3.6-Cre cultures will be lost in multilayering nodules of maturing osteoblasts. (B) Upon reaching confluence, wild- type osteoprogenitor cell cultures underwent robust nodule formation and mineralization. (C) Mdm2-conditional Col3.6-Cre–transgenic cultures were unable to form a significant number of multilayered nodules, and subsequently only a small fraction of these cells were able to undergo differentiation, activate alkaline phosphatase activity, or induce mineralization. (D) Quantitative analysis by real-time PCR of the expression of early osteogenic genes type I collagen (Coll) and alkaline phosphatase (AP) and of osteocalcin (OC) at various stages of osteoblast maturation. Solid lines depict the relative transcript levels of genes in wild-type cell cultures, and dashed lines represent the levels of expression of the various genes in Mdm2-conditional Col3.6-Cre cell cultures. Expression levels of early and late osteogenic genes are reduced in Mdm2-conditional Col3.6-Cre–transgenic cells. (E) Real-time PCR analysis of Mdm2 and Runx2 expression in wild-type (WT) and Mdm2-conditional Col3.6-Cre–transgenic (MT) culture osteoblast progenitor cells during maturation. Solid and dashed lines depict the relative transcript levels of Mdm2 and Runx2 in wild-type or Mdm2-conditional Col3.6-Cre–transgenic cultures, respectively. As expected, Mdm2 levels do not increase in Mdm2-conditional Col3.6-Cre–transgenic cells during maturation. Similar induction of Runx2 and Mdm2 expression is observed in wild-type cells during maturation. (F) Western blot analysis of Runx2 protein levels in wild-type or Mdm2-conditional Col3.6-Cre–transgenic culture osteoblast progenitor cells during maturation and mineralization. Lamin-b is shown as a loading control. Decreased amounts of Runx2 protein are observed in Mdm2-conditional Col3.6-Cre–transgenic cells. (G) Quantitative PCR was performed on reverse-transcribed RNA isolated from osteoblast progenitor cells transduced with Runx2 or lacZ (negative control). Exogenous Runx2 up-regulated the expression of osteogenic maturation genes in Mdm2 mutant cells, including collagen 1, alkaline phosphatase, and osteocalcin. Error bars indicate SD.
Toluidine blue staining of calvarial osteoprogenitor cells cultured from wild-type and Mdm2-conditional Col3.6-Cre mice revealed that both wild-type and Mdm2 mutant–derived osteoprogenitor cultures achieved confluence simultaneously (Fig. 5, B and C, left), and BrdU incorporation indicated no difference in the rates of cell proliferation in these cultures before confluence (not depicted). However, upon reaching confluence and upon robust induction of Cre expression, wild-type osteoprogenitor cell cultures underwent robust nodule formation (Fig. 5 B), whereas Mdm2sjcnd1/sjcnd1,Col3.6-Cre cultures were unable to form a significant number of multilayered nodules. Subsequently, only a small fraction of the mutant cells were able to undergo osteoblast differentiation as reflected by alkaline phosphatase activity (Fig. 5 C). Consistent with our in vivo findings of reduced mineralization in Mdm2 mutant bone, wild-type osteoblasts ultimately formed heavily mineralized nodules in culture, whereas mutant cells failed to deposit significant mineral in the extracellular matrix (Fig. 5, B and C) as determined by silver nitrate staining (right).
Quantitative analysis by real-time PCR of osteogenic gene expression revealed that both wild-type and mutant cultures began to activate early osteogenic genes type I collagen and alkaline phosphatase upon reaching confluence. However, osteogenic gene expression was abrogated in Mdm2 mutant cultures shortly after achieving confluence (Fig. 5 D, dashed lines). Osteocalcin, a marker of late osteoblast differentiation, was not activated in mutant cultures at any time during the culture period (Fig. 5 D). Because osteoblast differentiation and expression of these osteoblast phenotypic genes is dependent on the activity of the Runt-related transcription factor Runx2 (Ducy et al., 1997; Komori et al., 1997; Otto et al., 1997), we next examined the impact of Mdm2 loss on the activity of this gene. Real-time PCR analysis revealed that Mdm2 and Runx2 are expressed at low levels in wild-type cultures during the period of proliferation but are strongly activated in postconfluent cultures during multilayering and differentiation. However, as expected, Mdm2 up-regulation is abrogated in osteoprogenitor cultures derived from Mdm2-conditional Col3.6-Cre mice during the course of differentiation (Fig. 5 E). Interestingly, both Runx2 protein and message levels were lost in mutant cultures concomitantly with the loss of Mdm2 expression (Fig. 5, E and F). Furthermore, although the G1/S ratio of preconfluent osteoprogenitor cells was unchanged in Mdm2-conditional Col3.6-Cre cells (before induction of Cre expression), postconfluent cycling of mutant cells deleted for Mdm2 was strongly inhibited as determined by BrdU uptake assays, with a G1/S ratio of 7.5:4.2 for nonmutant cells. In addition, no differences in apoptotic cell numbers were detected by TUNEL assays during differentiation of cultured osteoblast progenitor cells after Cre-mediated deletion of Mdm2 (unpublished data). Collectively, our results indicate that Mdm2 activity is required for postconfluent cell proliferation and nodule formation in osteoblast cultures and the subsequent activation of the master osteoblast transcriptional regulator Runx2. Failure of cultures lacking Mdm2 to activate the Runx2 gene ultimately results in inhibition of osteoblast differentiation and inactivity of osteoblast phenotypic genes. To confirm that reduced Runx2 expression in the Mdm2 mutant cells is the underlying cause of the maturation defect, recombinant adenovirus vectors were generated to transduce either lacZ (control) or Runx2 cDNA into the osteoblast progenitor cultures. Addition of exogenous Runx2 into Mdm2 mutant cells induced maturation of these progenitor cells and partially or fully restored the expression of mature osteogenic genes such as collagen type 1, alkaline phosphatase, and osteocalcin (Fig. 5 G).
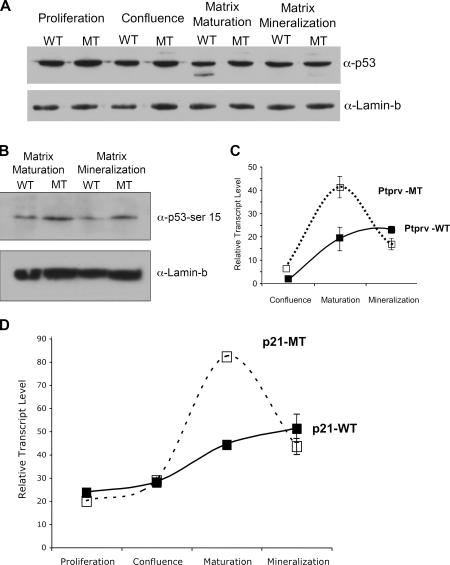
Examination of p53 levels in ex vivo osteoblast cultures revealed no change in total p53 protein levels in cultures undergoing deletion of Mdm2 (Fig. 6 A). However, a difference in the amount of activated p53 transcription factor present in the cultures was detected using a phospho-Ser15 specific antibody, with a marked induction in P-Ser15 p53 levels observed in mutant cultures relative to levels in wild-type cells (Fig. 6 B), suggesting that Mdm2 negatively regulates p53 activity but not overall p53 protein levels in differentiating osteoblasts. Furthermore, real-time PCR analysis revealed up-regulation of Ptprv and p21 gene expression (Fig. 6, C and D) in postconfluent osteoblast cultures derived from Mdm2-conditional Col3.6-Cre mice (dashed lines) relative to expression levels observed in Col3.6-Cre cells containing wild-type Mdm2 alleles (solid lines). These genes are targets of p53 transactivation known to be involved in regulating the progression of primary cells from G1 into S phase of the cell cycle (Deng et al., 1995; Doumont et al., 2005). This increase in Ptprv and p21 expression is consistent with the inhibition of postconfluent osteoprogenitor cell growth observed in the Mdm2-conditional Col3.6-Cre cultures. Interestingly, the expression levels of Bax and Puma, two proapoptotic p53 response genes, were not significantly altered in osteoblast progenitor cells after deletion of Mdm2 (unpublished data), consistent with the unaltered level of apoptosis in Mdm2 mutant and wild-type cells.
Figure 6.
Deletion of Mdm2 alters p53 activity in osteoblast cultures. (A) Western analysis of p53 levels in wild-type (WT) and Mdm2-conditional Col3.6-Cre–transgenic (MT) osteoprogenitor cells during differentiation reveals no change in total p53 protein levels in cultures undergoing deletion of Mdm2. (B) Western analysis of phosphorylated p53 in wild-type and Mdm2-conditional Col3.6-Cre–transgenic cells during maturation and mineralization reveals increased amounts of activated p53 in MT cells. Lamin-b is shown as a loading control. (C and D) Real-time PCR analysis of RNA isolated from wild-type or Mdm2-conditional Col3.6-Cre–transgenic cells during differentiation reveals up-regulation of Pptrv or p21 gene expression in postconfluent osteoblast cultures derived from Mdm2-conditional Col3.6-Cre mice (dashed line) relative to expression levels in cells containing wild-type Mdm2 alleles (solid line). Error bars indicate SD.
Regulation of osteoprogenitor differentiation and osteosarcoma formation by p53
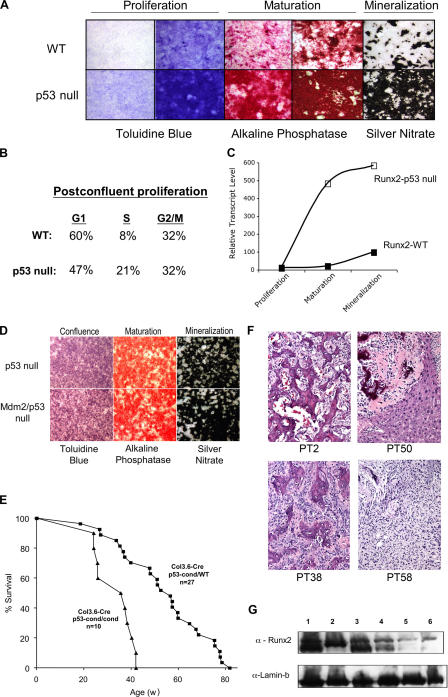
Our findings indicate that loss of Mdm2 results in a block in osteoblast differentiation that is due to excess p53 activity. To confirm that p53 negatively regulates bone maturation and mineralization, we examined the differentiation of osteoprogenitor cells harvested from the calvaria of p53-null mice. Calvarial osteoprogenitor cells were isolated from E19 wild-type or p53-null mice and induced to undergo osteogenic differentiation. The addition of ascorbic acid to the cultured osteoprogenitors greatly increased postconfluent proliferation in the p53-null cultures relative to wild-type cultures, resulting in increased formation of multilayered nodules and excess mineralized bone formation (Fig. 7 A). BrdU staining and FACS analysis of postconfluent osteoprogenitor cells revealed fewer p53-null cells in G1 and more in S phase (Fig. 7 B). Furthermore, real-time PCR analysis of Runx2 transcripts in the ex vivo cultures revealed a dramatic increase in the Runx2 expression levels during maturation and mineralization when p53 was absent (Fig. 7 C). These data confirm that p53 negatively regulates osteoblast proliferation and differentiation. Notably, there was a difference in the ex vivo maturation of p53-null and Mdm2/p53 double-null osteoblast progenitor cells (Fig. 7 D), underscoring our previous in vivo findings (Fig. 3 E and Fig. 4 A) of a p53-dependent role for Mdm2 in regulating skeletal development.
Figure 7.
Regulation of osteoblast progenitor differentiation and osteosarcoma formation by p53. Calvarial osteoprogenitor cells were cultured ex vivo from E19 wild-type (WT) and p53-null embryos. (A) Calvarial osteoprogenitors were induced to undergo osteogenic differentiation, resulting in proliferation (toluidine blue), maturation and the formation multilayered nodules and activation of alkaline phosphatase, and mineralization as detected by silver nitrate staining. Osteoprogenitor cells deleted for p53 displayed far more proliferation than wild-type cells cultured at the same initial density, increased alkaline phosphatase, and increased mineralization. (B) Proliferation was documented by BrdU uptake in postconfluent cultures of wild-type and p53-null progenitor cells just before cell maturation. The percentage of cells in each phase of the cell cycle was determined by FACS analysis after propidium iodide staining of the harvested cells. Postconfluent p53-null cells had more cells in S phase than wild-type cells. (C) Real-time PCR analysis of Runx2 expression in wild-type and p53-null cultures of osteoblast progenitor cells during maturation. Runx2 transcript levels are strongly up-regulated in p53-null cells during maturation. (D) No difference was observed in the robust osteoprogenitor cell differentiation in p53-null cells that contained (top) or lacked (bottom) Mdm2. (E) Tumorigenesis in mice deleted for p53 in osteoblasts. Col3.6-Cre–transgenic mice heterozygous for a conditional p53 allele (p53-cond/wild type) or homozygous for the p53-conditional allele (p53-cond/cond) were assayed for spontaneous tumor development. A majority of the mice presented with osteosarcomas, with a mean time to tumorigenesis of 40 wk for Col3.6-Cre, p53-cond/cond mice and 57 wk for Col3.6-Cre, p53-cond/wild-type mice. (F) Hematoxylin and eosin stains of osteosarcomas harvested from Col3.6-Cre, p53-cond/cond mice. Samples PT2, -50 (showing invasion into the liver), and -38 are all more differentiated than sample PT58, which displays a spindle-like morphology in addition to some osteoid cells. (G) Analysis of Runx2 protein levels in representative primary tumor samples from p53-conditional Col3.6-Cre mice. Runx2 levels were readily detected in representative osteosarcomas (lanes 1–3: samples PT2, -38, and -58, respectively) but reduced in fibrosarcoma (lane 4), lymphoma (lane 5), or hemangiosarcoma (lane 6) samples.
Increased proliferation of osteoprogenitor cells in the calvarial cultures suggests that negative regulation of osteoblast progenitor cell proliferation and differentiation by p53 may be an important component of p53-mediated suppression of bone tumorigenesis. To explore a link between negative regulation of osteoblast growth and neoplasia, we bred the Col3.6-Cre mice with p53-conditional mice (Marino et al., 2000). Cohorts of transgenic Col3.6-Cre mice that were either homozygous or heterozygous for the p53-conditional allele were used to perform a tumor assay. Col3.6-Cre mice heterozygous for the p53-conditional allele developed mostly bone masses starting at 20 wk of age, and all mice in the colony presented with cancer by 82 wk, whereas Col3.6-Cre mice homozygous for the p53-conditional allele all presented with tumors by 42 wk (Fig. 7 E). Histopathologic analysis of p53-conditional Col3.6-Cre mouse tumors identified 60% of these cancers as osteosarcomas, with a 20% incidence of lymphoma or fibrosarcoma. Most of the osteosarcomas were classified as high grade and were of intermediate differentiation, producing osteoid but not mature lamellar bone (Fig. 7 F, samples PT2, -50, and -38). However, a few of the osteosarcoma tumors were especially aggressive and very poorly differentiated (Fig. 7 F, sample PT58). Western blot analysis of representative primary tumor samples harvested from these mice indicated that Runx2 expression was greatly elevated specifically in osteosarcomas, regardless of their differentiation (Fig. 7 G, lanes 1–3). These results confirm that p53 not only down-regulates osteoblast cell growth and differentiation during development but also plays a critical role in suppressing osteosarcoma formation in adult bone tissue.
Discussion
The rescue of Mdm2-null mice from peri-implantation lethality by deletion of p53 demonstrates that Mdm2 plays an important role in early development by negatively regulating p53 activity (Jones et al., 1995; Montes de Oca Luna et al., 1995), but little is known about the requirement for Mdm2 throughout embryogenesis and in postembryonic tissues. We previously documented that the growth characteristics of p53-null primary fibroblasts and the tumorigenic potential of p53-null mice are indistinguishable in the presence or absence of Mdm2 (Jones et al., 1996). Thus, if Mdm2 was important in the latter stages of development, in organogenesis, or in postnatal cell growth control, it is likely due to the ability of Mdm2 to down-regulate p53 activity and not to p53-independent effects of Mdm2. In support of a role for Mdm2 in regulating p53 during the latter stages of development, recent studies of mice that contain reduced amounts of Mdm2 relative to wild-type levels indicate that Mdm2 regulates p53 activity in hematopoietic development and in B cell tumorigenesis (Alt et al., 2003; Mendrysa et al., 2003). However, the effect of complete ablation of Mdm2 activity on cellular differentiation and in organogenesis is unknown.
Coordinated proliferation and differentiation of bone-forming osteoblast progenitor cells and the deposition of extracellular matrix proteins by osteoblasts and subsequent deposition of crystalline salts for mineralization of the skeleton are two critical steps in bone modeling both during development and in the adult skeleton (Harada and Rodan, 2003). Although little is known regarding the role of p53 in this process, previous studies of mice bearing a hypermorphic p53 mutation revealed that mice with increased amounts of p53 activity exhibit symptoms of rapid aging, including osteoporosis (Tyner et al., 2002). Furthermore, a subset of mice haploinsufficient for functional p53 in all tissues develop osteosarcomas (Harvey et al., 1993b). These data suggest that p53 may regulate normal bone growth and that alterations in the levels of p53 activity can contribute to abnormal bone phenotypes.
To further explore a role for p53 in osteogenesis and to determine whether Mdm2–p53 signaling is important in bone growth and development, we bred Mdm2-conditional mice with Col3.6-Cre–transgenic mice. Deletion of Mdm2 upon expression of the Col3.6-Cre transgene resulted in midgestational caudal defects, including loss of tissue surrounding the somites, exposure of somitic mesenchyme at the surface of the embryo, absence of a tail, and a severe caudal invagination. Furthermore, TUNEL staining of serial sections of mutant embryos revealed increased apoptosis in the caudal somites and surrounding tissue of mutant embryos, suggesting that these defects arose through unregulated p53 apoptosis. Harvests of embryos from timed matings of Mdm2-conditional Col3.6-Cre–transgenic mice revealed a marked decrease in the recovery of Mdm2sjcnd1/sjcnd1,Col3.6-Cre embryos at E13.5, coincident with the robust activation of the Cre transgene in developing skeletal elements. Skeletal preparations of wild-type and of Mdm2sjcnd1/sjcnd1,Col3.6-Cre embryos documented numerous skeletal defects during the latter stages of development, including a reduction in mineralized bone and in length of appendicular bone, abnormal bone architecture, and an increase in bone porosity. However, analysis of skeletal preparations of embryos deleted for both Mdm2 and p53 revealed no skeletal defects, and no difference was observed in the growth and maturation of cultured calvarial cells deficient for p53 or for both Mdm2 and p53, indicating that the effects of Mdm2 loss on skeletal formation and osteoblast maturation are p53 dependent. In contrast to what was observed in the caudal mesoderm of E10 mutant embryos, TUNEL assays performed on E14.5 bone isolated from Mdm2-conditional Col3.6-Cre embryos and on osteoprogenitor cells cultured from E19 Mdm2-conditional Col3.6-Cre embryos revealed no increase in the number of apoptotic cells. This finding indicates that deletion of Mdm2 does not induce p53-mediated apoptosis in these cells but rather induces p53-mediated effects that block osteoblast proliferation or differentiation. To confirm that p53 plays a role in regulating osteoblast differentiation, we harvested osteoblast progenitor cells from the calvarial of p53-null mice just before birth (E19). Analysis of the growth and development of these cultured cells revealed that p53-null osteoprogenitor cells proliferated far faster than wild-type progenitor cells and underwent more robust differentiation, confirming that p53 functions to negatively regulate osteoblast maturation and mineralization.
Surprisingly, the overall level of p53 protein did not change in osteoblast cells during differentiation in the presence or absence of Mdm2; however, deletion of Mdm2 did result in an increase in the level of activated p53 as judged by the increased levels of phosphorylated p53. Furthermore, the message levels of Pptrv and the cyclin-dependent kinase inhibitor p21, two p53 target genes involved in regulating cell cycle progression from G1 to S phase, were increased in osteoblast cells after Mdm2 deletion. These results indicate that Mdm2 regulates p53 activity during osteoblast differentiation not by altering p53 stability but by inhibiting p53-mediated transactivation of genes involved in regulating osteoblast growth and differentiation.
Runx2 is a critical inducer of osteoblast differentiation in vitro and in vivo (Stein et al., 2004). Interestingly, levels of Runx2 message and protein were reduced in cells deleted for Mdm2, as were the message levels of Runx2 target genes type I collagen and alkaline phosphatase. In addition, expression of osteocalcin, a marker of late osteoblast differentiation, was not activated in cultures deleted for Mdm2 during differentiation, providing further molecular evidence for a block in osteoblast development upon deletion of Mdm2. In contrast to the reduction in Runx2 levels observed in osteoblasts deleted for Mdm2, Runx2 message levels were found to be greatly elevated in maturing osteoblasts deleted for p53. As Runx2 is a well-established master regulator of osteoblast differentiation, it is possible that p53 directly controls osteoblast maturation by negatively regulating Runx2 expression. However, analysis of the Runx2 promoter sequences failed to identify any p53 canonical binding sites, and there is no evidence present in the literature to suggest that Runx2 expression is directly regulated by p53. Therefore, we hypothesize that proper osteoblast differentiation and bone development require Mdm2 to inhibit a p53-mediated block on osteoprogenitor cell division. By permitting the postconfluent proliferation of osteoblasts through the down-regulation of p53 activity, Mdm2 indirectly facilitates Runx2 induction and osteoblast maturation. In support of this hypothesis, expression levels of mature osteogenic genes were found to be elevated in Mdm2 mutant osteoblasts after restoration of Runx2 expression.
Our results indicate that up-regulation of p53 activity due to Mdm2 deletion induces a block in bone differentiation and mineralization and causes profound skeletal defects in the developing embryo. Furthermore, deletion of p53 in osteoblasts induces hyperproliferation, greatly elevated levels of Runx2 expression, and increased bone maturation in vitro. These findings indicate that p53 is an important negative regulator of bone growth and development. Interestingly, loss of cell differentiation and reduced expression of mature osteogenic genes such as Osteocalcin are prognostic indicators in human osteosarcomas, with poorly differentiated or dedifferentiated tumors usually associated with the high-grade category (Hopyan et al., 1999). In addition, Runx2 is down-regulated in various human osteosarcoma cell lines, suggesting a link between loss of Runx2 expression, dedifferentiation, and cancer (Thomas et al., 2004). However, we observed increased osteoblast differentiation and elevated Runx2 expression in osteoblast progenitor cells derived from p53-null mice. Therefore, we examined the ability of p53 to suppress tumorigenesis in osteoprogenitor-derived cells by crossing Col3.6-Cre–transgenic mice with p53-conditional mice (Marino et al., 2000). Our results indicate that loss of p53 in osteoblasts induces a fairly rapid tumorigenesis in mice. Interestingly, Col3.6-Cre–transgenic mice heterozygous or homozygous for the p53-conditional allele display kinetics of tumor onset similar to those that have been previously documented for p53 knockout heterozygous or homozygous mice, though the tissue specificity of tumorigenesis was greatly altered. Mice deleted for p53 in all tissues die predominantly from lymphomas, chiefly of the thymus, and only occasionally will present with bone tumors (Harvey et al., 1993b). Depending on the genetic background of the mice, between 3 and 8% of p53-null mice develop osteosarcomas (Harvey et al., 1993a; Jones et al., 1996). However, this tumor spectrum may reflect the critical importance of p53 in suppressing thymic lymphomas in relatively young mice and not a reduced role for p53 in suppressing bone cancer, as osteosarcomas do constitute approximately one third of all tumor types observed in the longer lived p53 heterozygous mice (Harvey et al., 1993b). A majority of the Col3.6-Cre–transgenic, p53-conditional heterozygous mice or p53-conditional homozygous mice in our study developed osteosarcomas. Furthermore, Runx2 expression was elevated in primary osteosarcoma samples harvested from these mice, in agreement with our finding of increased Runx2 expression in the p53-null calvarial cell cultures.
The results of our p53-conditional Col3.6-Cre–transgenic mouse cross confirm that p53 is a critical tumor suppressor in bone tissue and indicate that osteosarcoma formation does not require loss of Runx2 expression. Instead, we propose that p53 inhibition of osteoblast cell proliferation is the mechanistic basis for suppression of bone osteosarcomas in this model. As we have demonstrated that Mdm2 is a key regulator of p53 activity in osteoblasts, disrupting the ability of Mdm2 to down-regulate p53 activity in these cells may prove to be a useful therapeutic strategy in treating osteosarcomas.
Materials and methods
Mouse lines
Mdm2-conditional mice were developed in our laboratory using standard gene targeting techniques in embryonic stem cells. The resulting allele contains two loxP sites flanking exons 11 and 12. Upon Cre-mediated recombination, exons 11 and 12 are excised, rendering the allele inactive (described in Steinman and Jones [2002]). Mdm2 excision was mediated by crossing Mdm2-conditional (Mdm2sjcnd1) mice to transgenic mice in which the 3.6-kb type I collagen promoter governs the expression of the Cre-recombinase enzyme (Col3.6-Cre). Visualization of tissues in which the Cre-recombinase activity has recombined target alleles was facilitated by mating Col3.6-Cre–transgenic mice to R26R reporter mice (The Jackson Laboratory) in which Cre expression results in the removal of a loxP-flanked DNA segment that prevents expression of a lacZ gene. The p53-conditional mouse model (Marino et al., 2000) was obtained from A. Berns (Netherlands Cancer Institute, Amsterdam, Netherlands). All animals were maintained and used in accordance with the University of Massachusetts Animal Care and Use Committee.
Whole-mount and histological analysis
Embryos were harvested from timed pregnant mothers at various time points during gestation followed by fixation in 4% paraformaldehyde. Tissues destined for histological sectioning were dehydrated in a graded series of ethanol and xylene, followed by infiltration with paraffin wax. Tissues from R26R/Col3.6-Cre crosses were fixed in 4% paraformaldehyde, equilibrated overnight in 30% sucrose, and embedded in optimal cutting temperature for cryosectioning. Some embryos were fixed in 4% paraformaldehyde followed by whole-mount staining for β-galactosidase activity. Parrafin sections were cut at 7 μm and counterstained with eosin. Frozen sections were cut at 10–12 μm, stained for β-galactosidase activity, and counterstained with eosin. β-Galactosidase activity was visualized by staining whole embryos, cryosections, or calvarial cultures in a solution containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, 0.2% NP-40, 0.01% sodium deoxycholate, and 1 mg/ml X-Gal in PBS, pH 7.4, at 37°C for 1–6 h. Calvarial cultures were stained for mineral content using the method of Von Kossa. In brief, sections were exposed to a solution of 3% silver nitrite under direct sunlight for 15 min, after which mineral deposits were visualized as black precipitate under brightfield microscopy. Alkaline phosphatase activity in calvarial cultures was visualized by colorimetric enzymatic reaction to a solution containing 0.5 mg/ml napthol as MX phosphate disodium salt (Sigma-Aldrich), 2.8% N,N dimethylformamide (Sigma-Aldrich), 0.1 M Tris-maleate buffer, pH 8.4, and 1 mg/ml fast red salt (Sigma-Aldrich). The reaction was performed at 37°C for 10 min. Decalcified femora and vertebrae were embedded in a mixture of methyl methacrylate: glycol methacrylate, sectioned at 2 μm, stained with Von Kossa, and counterstained with toluidine blue. TUNEL staining was performed on dewaxed paraffin sections using a fluorescein-conjugated in situ cell death detection kit (Roche) according to the manufacturer's protocol.
Skeletal preparations
Skeletons were prepared for visualization of cartilage and bone using alcian blue and alizarin red stains, respectively. Embryos were eviscerated followed by overnight fixation in 100% ethanol. The next day, cartilaginous elements were stained overnight in a solution containing 4 parts ethanol, 1 part glacial acetic acid, and 0.3 mg/ml alcian blue 8GX (Sigma-Aldrich). The next day, soft tissues were dissolved for 6 h in a 2% KOH solution followed by an overnight staining in a 1% KOH solution containing 75 μg/ml alizarin red S (Sigma-Aldrich). Skeletons were then destained in 20% glycerol and 1% KOH for several days and stored in 50% glycerol and 50% ethanol.
Bone micro-CT
Embryos were fixed overnight in 4% paraformaldehyde, rinsed three times with PBS, and scanned on a Skyscan 1072 instrument (Skyscan). Image acquisition of the head was performed using 25× magnification (50× for the limb) at 45 kV and 222 μA, with a 0.45° rotation between frames to obtain two-dimensional images. Three-dimensional reconstruction and quantitative analyses were performed on a computer (Dell) using the NRecon, ANT, and CTAn software supplied with the Skyscan instrument.
Calvarial osteoblast preparations
Calvarial osteoblasts were isolated from E19 embryos by enzymatic digestion of calvarial bones. In brief, calvaria were minced and subjected to three sequential digestions (8, 10, and 26 min) with collagenase P (Roche) at 37°C. Osteoblasts in the second and third digest were collected and resuspended in α-MEM supplemented with 10% FBS (HyClone). Cells were plated at a density of 106 cells/6-well plate (Owen et al., 1990). Differentiation was initiated after confluence by the addition of ascorbic acid and β-glycerol phosphate. Cultures were harvested at various time points and stained for β-galactosidase activity, mineral content, alkaline phosphatase activity, or total cellularity using toluidine blue. All osteoblast differentiation experiments were performed a total of three times, and each experiment used embryos of different genotypes harvested on the same day (E19) from the same litter. The proliferation, confluence, maturation, and mineralization stages of differentiation are defined as days 5, 10, 14, and 20 in culture, respectively, except for Fig. 6 C, where confluence, maturation, and mineralization stages of differentiation were reached on days 8, 12, and 17 of culture, respectively.
Image acquisition
Whole-mount photographic images of embryos (Fig. 1 B and Fig. 2, A–D), histologic sections of embryos (Fig. 1 C and Fig. 2, E and F), skeletal preps (Fig. 3), and stained osteoblast cultures (Fig. 5, A–C; and Fig. 7, A and D) were obtained using a stereoscope (MZ8; Leica) with either a 1× or 0.63× reduction lens and a digital camera (3008 Prog/Res; JenOptik) coupled to a computer (G4; Macintosh), using Photoshop 4 software (Adobe). Bone histology images (Fig. 4, D and E) were captured using an Axioskop 40 (Carl Zeiss MicroImaging, Inc.) equipped with a camera (AxioCamMRc; Carl Zeiss MicroImaging, Inc.), a Dell computer, and MRGrab software. Magnifications at source are 2.5× (low power) and 20× (high power). TUNEL photographic images (Fig. 2 G) were obtained using an inverted microscope (model 405; Carl Zeiss MicroImaging, Inc.) with a Fluor 10 (10×) plan (Nikon) and a digital camera (SPOT; Diagnostic Instruments) connected to a Dell computer using Photoshop 4 Imaging. Tumor hematoxylin and eosin–stained sections (Fig. 7 F) were imaged using a microscope (Eclipse E400; Nikon) with a 20× plan (Nikon) coupled to a SPOT digital camera and a computer (IBM) with Spot acquisition software (4.0.1) and Photoshop 7 software.
Transduction of Runx2 into calvarial cultures
Adenoviral infection of primary mouse osteoblasts was performed at day 10 of culture (confluence) with either a vector expressing Xpress-tagged mouse Runx2 under transcriptional control of the cytomegalovirus CMV5 promoter (pAd/CMV5/Xpress-Runx2/IRES/GFP) or a control vector expressing LacZ (pAd/CMV5/LacZ/IRES/GFP). Infections were performed at a multiplicity of infection of ∼100 in α-MEM containing 5% FBS (HyClone). 12 h after infection, the media was replaced with α-MEM containing 10% FBS and ascorbic acid to initiate osteogenic differentiation. Cultures were harvested 7 d after infection, and quantitative RT-PCR was performed on total RNA isolated from each sample.
RNA isolation and analysis
RNA was isolated from tissue or cell cultures using Trizol reagent (Invitrogen) according to the manufacturer's protocol. After purification, 5 μg of total RNA was DNase treated using a DNA-free RNA column purification kit (Zymo Research). 1 μg RNA was then reverse transcribed using Oligo-dT primers and a first-strand synthesis kit (SuperScript; Invitrogen) according the manufacturer's protocol. Gene expression was assessed by semiquantitative (Cre, Mdm2, and glyseraldehyde-3-phosphate dehydrogenase [GAPDH]; 25 cycles) and quantitative real-time PCR (Runx2, alkaline phosphatase, osteocalcin, collagen type I, and GAPDH). Quantitative PCR was performed using SYBR green 2× master mix (Eurogentec) and a two-step cycling protocol (anneal and elongate at 60°C and denature at 94°C). Specificity of primers was verified by dissociation temperature of amplicons. Results are representative of two or more independent experiments.
Protein analysis
Total protein was isolated from calvarial cultures or from primary tumor samples in the presence of direct lysis buffer (0.1 M Tris-HCl, pH 7.5, 10% glycerol, 0.01 M DTT, 12% urea, and 2% SDS) followed by heating for 5 min at 100°C. 40 μg of total protein was then electrophoresed through a 10% acrylamide gel followed by transfer onto a polyvinylidene fluoride (Immobilon) membrane. Membranes were blocked in PBS Tween 20 (PBST) containing 2% nonfat dry milk (Bio-Rad Laboratories) before incubation with antibodies. Antibodies were incubated with membranes in the presence of PBST containing 2% nonfat dry milk for 1 h at room temperature. Excess primary antibody was removed with three 10-min washes of PBST. Secondary antibodies were incubated with membranes for 1 h at room temperature followed by three 10-min washes with PBST to remove excess antibody. Proteins were visualized on the membrane by exposure to Western lightning chemiluminescent reagent. The Runx2 antibody was a gift from Y. Ito (National University of Singapore, Singapore). Total p53 was detected using a 50:50 mix of ab-1 and -3 (Oncogene Research Products). Activated p53 was detected using an anti-pSer15 p53 antibody (Cell Signaling Technology). All antibodies were used at a concentration of 50 ng/ml.
Acknowledgments
We thank Dr. Anton Berns for the p53-conditional mouse model; Dr. Sadiq Hussain, Kathy Hoover, and Judith Gallant for technical assistance; and Charlene Baron for assistance with manuscript preparation.
This work was supported by grants from the National Institutes of Health to G.S. Stein (AR39588), B.E. Kream (AR38933), and S.N. Jones (CA77735).
C.J. Lengner and H.A. Steinman contributed equally to this work.
C.J. Lengner's present address is Whitehead Institute for Biomedical Research, Cambridge, MA 02142.
J. Gagnon's present address is Department of Molecular Biology, Cell Biology, and Biochemistry, Brown University, Providence, RI 02912.
Abbreviations used in this paper: E, embryonic day; GAPDH, glyseraldehyde-3-phosphate dehydrogenase; micro-CT, microcomputed tomography.
References
- Alt, J.R., T.C. Greiner, J.L. Cleveland, and C.M. Eischen. 2003. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 22:1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, J.F., M.H. Kaufman, D.J. Harrison, and A.R. Clarke. 1995. High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 5:931–936. [DOI] [PubMed] [Google Scholar]
- Banerjee, C., L.R. McCabe, J.-Y. Choi, S.W. Hiebert, J.L. Stein, G.S. Stein, and J.B. Lian. 1997. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J. Cell. Biochem. 66:1–8. [DOI] [PubMed] [Google Scholar]
- Chen, J., J. Lin, and A.J. Levine. 1995. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol. Med. 1:142–152. [PMC free article] [PubMed] [Google Scholar]
- Deng, C., P. Zhang, J.W. Harper, S.J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 82:675–684. [DOI] [PubMed] [Google Scholar]
- Donehower, L.A., M. Harvey, B.L. Slagle, M.J. McArthur, C.A. Montgomery Jr., J.S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 356:215–221. [DOI] [PubMed] [Google Scholar]
- Doumont, G., A. Martoriati, C. Beekman, S. Bogaerts, P.J. Mee, F. Bureau, E. Colombo, M. Alcalay, E. Bellefroid, F. Marchesi, et al. 2005. G1 checkpoint failure and increased tumor susceptibility in mice lacking the novel p53 target Ptprv. EMBO J. 24:3093–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy, P., R. Zhang, V. Geoffroy, A.L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 89:747–754. [DOI] [PubMed] [Google Scholar]
- Freedman, D.A., and A.J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, R.K., Z.K. Yu, and C.G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569–573. [DOI] [PubMed] [Google Scholar]
- Gottlieb, E., R. Haffner, A. King, G. Asher, P. Gruss, P. Lonai, and M. Oren. 1997. Transgenic mouse model for studying the transcriptional activity of the p53 protein: age- and tissue-dependent changes in radiation-induced activation during embryogenesis. EMBO J. 16:1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, S., and G.A. Rodan. 2003. Control of osteoblast function and regulation of bone mass. Nature. 423:349–355. [DOI] [PubMed] [Google Scholar]
- Harvey, M., M.J. McArthur, C.A. Montgomery Jr., A. Bradley, and L.A. Donehower. 1993. a. Genetic background alters the spectrum of tumors that develop in p53-deficient mice. FASEB J. 7:938–943. [DOI] [PubMed] [Google Scholar]
- Harvey, M., M.J. McArthur, C.A. Montgomery Jr., J.S. Butel, A. Bradley, and L.A. Donehower. 1993. b. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat. Genet. 5:225–229. [DOI] [PubMed] [Google Scholar]
- Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature. 387:296–299. [DOI] [PubMed] [Google Scholar]
- Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25–27. [DOI] [PubMed] [Google Scholar]
- Hopyan, S., N. Gokgoz, R.S. Bell, I.L. Andrulis, B.A. Alman, and J.S. Wunder. 1999. Expression of osteocalcin and its transcriptional regulators core-binding factor alpha 1 and MSX2 in osteoid-forming tumours. J. Orthop. Res. 17:633–638. [DOI] [PubMed] [Google Scholar]
- Iwakuma, T., and G. Lozano. 2003. MDM2, an introduction. Mol. Cancer Res. 1:993–1000. [PubMed] [Google Scholar]
- Jones, S.N., A.E. Roe, L.A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 378:206–208. [DOI] [PubMed] [Google Scholar]
- Jones, S.N., A.T. Sands, A.R. Hancock, H. Vogel, L.A. Donehower, S.P. Linke, G.M. Wahl, and A. Bradley. 1996. The tumorigenic potential and cell growth characteristics of p53-deficient cells are equivalent in the presence or absence of Mdm2. Proc. Natl. Acad. Sci. USA. 93:14106–14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven, T., Y. Barak, A. Zauberman, D.L. George, and M. Oren. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 8:3411–3416. [PubMed] [Google Scholar]
- Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R.T. Bronson, Y.H. Gao, M. Inada, et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 89:755–764. [DOI] [PubMed] [Google Scholar]
- Kubbutat, M.H., S.N. Jones, and K.H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature. 387:299–303. [DOI] [PubMed] [Google Scholar]
- Li, M., C.L. Brooks, F. Wu-Baer, D. Chen, R. Baer, and W. Gu. 2003. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 302:1972–1975. [DOI] [PubMed] [Google Scholar]
- Liu, F., H.W. Woitge, A. Braut, M.S. Kronenberg, A.C. Lichtler, M. Mina, and B.E. Kream. 2004. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int. J. Dev. Biol. 48:645–653. [DOI] [PubMed] [Google Scholar]
- Marino, S., M. Vooijs, H. van Der Gulden, J. Jonkers, and A. Berns. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Mendrysa, S.M., M.K. McElwee, J. Michalowski, K.A. O'Leary, K.M. Young, and M.E. Perry. 2003. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol. Cell. Biol. 23:462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand, J., G.P. Zambetti, D.C. Olson, D. George, and A.J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 69:1237–1245. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna, R., D.S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 378:203–206. [DOI] [PubMed] [Google Scholar]
- Otto, F., A.P. Thornell, T. Crompton, A. Denzel, K.C. Gilmour, I.R. Rosewell, G.W. Stamp, R.S. Beddington, S. Mundlos, B.R. Olsen, et al. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 89:765–771. [DOI] [PubMed] [Google Scholar]
- Owen, T.A., J. Holthuis, E. Markose, A.J. van Wijnen, S.A. Wolfe, S.R. Grimes, J.B. Lian, and G.S. Stein. 1990. Modifications of protein-DNA interactions in the proximal promoter of a cell-growth-regulated histone gene during onset and progression of osteoblast differentiation. Proc. Natl. Acad. Sci. USA. 87:5129–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah, V.P., L.D. Attardi, G.J. Mulligan, B.O. Williams, R.T. Bronson, and T. Jacks. 1995. A subset of p53-deficient embryos exhibit exencephaly. Nat. Genet. 10:175–180. [DOI] [PubMed] [Google Scholar]
- Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70–71. [DOI] [PubMed] [Google Scholar]
- Soussi, T., and C. Beroud. 2001. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer. 1:233–240. [DOI] [PubMed] [Google Scholar]
- Stein, G.S., J.B. Lian, A.J. van Wijnen, J.L. Stein, M. Montecino, A. Javed, S.K. Zaidi, D.W. Young, J.-Y. Choi, and S.M. Pockinwise. 2004. Runx2 control of organization and activity of the regulatory machinery for skeletal gene expression. Oncogene. 23:4315–4329. [DOI] [PubMed] [Google Scholar]
- Steinman, H.A., and S.N. Jones. 2002. Generation of an Mdm2 conditional allele in mice. Genesis. 32:142–144. [DOI] [PubMed] [Google Scholar]
- Thomas, D.M., S.A. Johnson, N.A. Sims, M.K. Trivett, J.L. Slavin, B.P. Rubin, P. Waring, G.A. McArthur, C.R. Walkley, A.J. Holloway, et al. 2004. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J. Cell Biol. 167:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner, S.D., S. Venkatachalam, J. Choi, S. Jones, N. Ghebranious, H. Igelmann, X. Lu, G. Soron, B. Cooper, C. Brayton, et al. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature. 415:45–53. [DOI] [PubMed] [Google Scholar]
- Vousden, K.H. 2000. p53: death star. Cell. 103:691–694. [DOI] [PubMed] [Google Scholar]
- Xirodimas, D.P., M.K. Saville, J.C. Bourdon, R.T. Hay, and D.P. Lane. 2004. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 118:83–97. [DOI] [PubMed] [Google Scholar]
- Wu, X., J.H. Bayle, D. Olson, and A.J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126–1132. [DOI] [PubMed] [Google Scholar]