Abstract
Exosomes are secreted, single membrane organelles of ∼100 nm diameter. Their biogenesis is typically thought to occur in a two-step process involving (1) outward vesicle budding at limiting membranes of endosomes (outward = away from the cytoplasm), which generates intralumenal vesicles, followed by (2) endosome–plasma membrane fusion, which releases these internal vesicles into the extracellular milieu as exosomes. In this study, we present evidence that certain cells, including Jurkat T cells, possess discrete domains of plasma membrane that are enriched for exosomal and endosomal proteins, retain the endosomal property of outward vesicle budding, and serve as sites of immediate exosome biogenesis. It has been hypothesized that retroviruses utilize the exosome biogenesis pathway for the formation of infectious particles. In support of this, we find that Jurkat T cells direct the key budding factor of HIV, HIV Gag, to these endosome-like domains of plasma membrane and secrete HIV Gag from the cell in exosomes.
Introduction
The classic model of retrovirology describes virion budding and virion–cell fusion as virus-mediated processes that do not follow any normal cell biological pathway (Coffin et al., 1997; Goff, 2001). However, animal cells are capable of secreting proteins and lipids in exosomes. Exosomes are single membrane organelles that are secreted from the cell and have a similar size and the same topology as retroviral particles (Arienti et al., 1997, 2001, 2002; Denzer et al., 2000b; Carlin et al., 2001; Fritzsching et al., 2002; Saez et al., 2003). Exosomes share many additional properties with retroviral particles, including similar lipid and protein compositions. These similarities have raised the possibility that retroviruses are “Trojan exosomes” that use the exosome biogenesis pathway for virion biogenesis and can also use an exosome uptake pathway as an alternative, Env-independent pathway for infecting neighboring cells (Gould et al., 2003).
One approach to studying the possible relationship between exosomes and retrovirions is to directly compare the composition of exosomes with retrovirions released by a given cell type. The only study that directly compares exosomes with retrovirions was performed with human macrophages, and it identified several similarities between the host cell proteins that were included in or excluded from both exosomes and HIV particles (Nguyen et al., 2003). Another approach is to determine whether cells bud exosomes and retrovirions from similar or distinct locations of the cell. Several studies in macrophages demonstrated that exosome-like vesicles and HIV particles are formed in a two-step process. First, outward vesicle budding (outward = away from the cytoplasm) at the limiting membrane of endosomes generates discrete vesicles, and second, fusion of vesicle-laden endosomes with the plasma membrane secretes vesicles into the extracellular milieu (Raposo et al., 2002; Nydegger et al., 2003; Pelchen-Matthews et al., 2003; Ono and Freed, 2004).
These macrophage studies fit the common conception that exosome biogenesis occurs only by outward budding at endosomal membranes followed by fusion of vesicle-laden endosomes with the plasma membrane (Denzer et al., 2000a; Stoorvogel et al., 2002; Thery et al., 2002; Pelchen-Matthews et al., 2004). In fact, the dearth of evidence for exosome budding at the plasma membrane and the budding of HIV from the plasma membrane of T cells (Freed and Martin, 2001; Morita and Sundquist, 2004) have been cited as reasons to doubt that retroviruses are a variant form of exosomes (Pelchen-Matthews et al., 2004). In this study, we examined the cellular distribution of exosomal proteins and lipids in the human CD4+ T cell line Jurkat and found that exosomal markers colocalized primarily at discrete domains of the plasma membrane. We also observed presumptive outward budding intermediates at the plasma membrane of Jurkat T cells and found that these structures were associated with exosomal markers. Moreover, Jurkat T cells appear to treat HIV Gag as an exosomal cargo molecule, sorting it to domains of plasma membrane that are enriched in exosomal markers and secreting it from the cell in exosomes.
Results
T cell exosomes are enriched for CD81 and CD63 but depleted of CD45
Human CD4+ T cells and certain T cell lines, such as Jurkat T cells, are permissive for HIV infection and bud HIV and HIV Gag from their plasma membrane (Freed and Martin, 2001). To determine whether human T cells also bud exosomes from their plasma membrane, we first tested for the presence of known exosomal marker proteins on exosomes that are secreted from Jurkat T cells, which were grown under our laboratory conditions. Jurkat T cells were adapted to growth in serum-free medium, and exosomes were collected from the medium by filtration and differential centrifugation. The exosome-containing pellet was resuspended, subjected to sucrose density gradient centrifugation, and fractions across the gradient were assayed for density by refractometry and for specific proteins by immunoblotting.
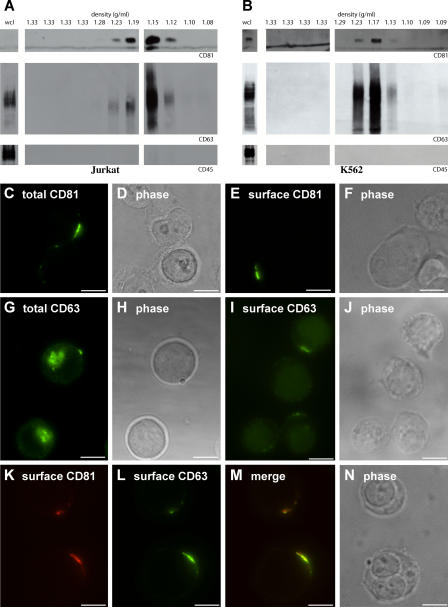
CD81 has previously been detected on the limiting membrane of endosomes (Fritzsching et al., 2002), is particularly enriched in exosomal membranes (Escola et al., 1998; Fritzsching et al., 2002), and was enriched in exosomes secreted by Jurkat T cells grown in serum-free medium (Fig. 1 A, top). CD63 has also been detected on the limiting membrane of endosomes (Metzelaar et al., 1991) and exosomal membranes (Escola et al., 1998; Heijnen et al., 1999; Clayton et al., 2001; van Niel et al., 2001; Blanchard et al., 2002; Raposo et al., 2002; van Niel and Heyman, 2002). CD63 was abundant in the peak fractions of Jurkat-derived exosomes, although perhaps not as enriched as CD81 (Fig. 1 A). In contrast to CD81 and CD63, the plasma membrane protein CD45 is excluded from T cell–derived exosomes (Blanchard et al., 2002) and was also excluded from exosomes secreted by Jurkat T cells (Fig. 1 A, bottom). Interestingly, CD45 is also excluded from T cell–derived HIV virions (Esser et al., 2001; Trubey et al., 2003). K562 cells also secreted exosomes that were enriched in CD81 and CD63 but lacked CD45 (Fig. 1 B).
Figure 1.
Jurkat T cells sort exosomal proteins to discrete domains of the plasma membrane. (A and B) Sucrose density gradient analysis of exosomes from Jurkat T cells (A) and K562 cells (B). Densities of the different fractions are listed at the top in grams/milliliters. Note the significant enrichment of CD81 and CD63 in exosome fractions relative to the levels of the proteins in the whole cell lysate (wcl; CD81 could not be detected in the whole cell lysate sample). The same amount of whole cell lysate and gradient fractions were used for all three blots. (C–J) Jurkat T cells were processed for fluorescence microscopy using antibodies specific for CD81 (C–F) or CD63 (G–J) under permeabilizing (C, D, G, and H) or nonpermeabilizing (E, F, I, and J) conditions. Shown are fluorescence (C, E, G, and I) and phase-contrast (D, F, H, and J) images of representative cells. (K–N) Unpermeabilized Jurkat T cells were fixed and processed for fluorescence microscopy using antibodies specific for CD81 (K) and CD63 (L). (M) Merge of K and L. (N) Phase-contrast image of the cell. Bars, 10 μm.
Exosomal proteins are enriched at domains of the T cell plasma membrane
We next examined the subcellular distribution of CD81 and CD63. Immunofluorescence analysis of fixed and permeabilized Jurkat cells revealed that CD81 was located predominantly at the plasma membrane where it was enriched at discrete domains (Fig. 1, C and D), a distribution that was consistent with what we observed for CD81 at the surface of fixed, unpermeabilized Jurkat cells (Fig. 1, E and F). The subcellular distribution of CD63 in permeabilized cells was a bit different, with much of the protein residing in internal organelles, although some CD63 appeared to be at the cell periphery (Fig. 1, G and H). Immunofluorescence analysis of fixed and unpermeabilized Jurkat cells confirmed that the CD63 present at the cell periphery was exposed to the extracellular milieu (Fig. 1, I and J). Moreover, double-label immunofluorescence analysis of fixed, unpermeabilized Jurkat cells revealed that CD81 and CD63 colocalized at discrete domains of the T cell surface (Fig. 1, K–N). In contrast, CD45 and 10 other plasma membrane proteins were distributed evenly over the surface of these cells (supplemental material and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200508014/DC1). Similar results were observed in K562 cells (not depicted).
Colocalization of exosomal proteins and an exosomal lipid
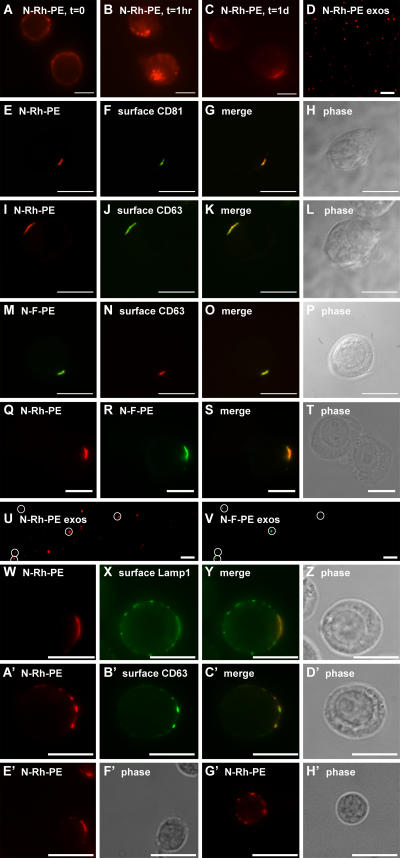
Willem et al. (1990) and Vidal et al. (1997) previously established that N-Rh-PE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[lissamine rhodamine B sulfonyl]), a fluorescent phosphatidyl ethanolamine (PE) analogue, is taken up at the plasma membrane of cells, sorted to endosomes, and secreted in exosomes. When added to human T cells, N-Rh-PE was taken up at the plasma membrane (Fig. 2 A), and, by 1 h, much of this lipid had been endocytosed (Fig. 2 B). However, by 3–4 h, a significant portion of the cell-associated N-Rh-PE was present at discrete domains of the plasma membrane. This distribution could be detected up to a day (Fig. 2 C) or more after labeling. In addition, N-Rh-PE was secreted from the cells on exosomes, which could be captured, bound to coverglasses, and visualized by fluorescence microscopy (Fig. 2 D). Double-label fluorescence microscopy experiments revealed that the plasma membrane domains enriched for N-Rh-PE were the same as those enriched for CD81 (Fig. 2, E–H) and CD63 (Fig. 2, I–L). Jurkat T cells sorted a fluorescein-labeled PE derivative, N-F-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[carboxyfluorescein]), in much the same way, as shown by its colocalization with surface CD63 (Fig. 2, M–P). When Jurkat T cells were pulse labeled with N-Rh-PE, incubated for 1 d, pulse labeled with N-F-PE, and examined 16 h later, one could detect the colocalization of these exosomal lipids at discrete domains of the plasma membrane (Fig. 2, Q–T) and their secretion on overlapping populations of exosomes (Fig. 2, U and V). Fixed, unpermeabilized, N-Rh-PE–labeled Jurkat T cells were also decorated with antibodies specific for the extracellular domain of Lamp1. This protein also colocalized with N-Rh-PE, although not completely, as Lamp1 was also detected at additional points along the plasma membrane (Fig. 2, W–Z). Similar results were obtained in K562 cells (not depicted).
Figure 2.
N-Rh-PE is sorted to discrete domains of the T cell plasma membrane. (A–C) Jurkat cells pulse labeled with N-Rh-PE at 4°C for 1 h were washed, returned to growth medium, and examined by fluorescence microscopy at 0 (A), 1 (B), and 24 (C) h after labeling. (D) 24 h after labeling, exosomes were collected, bound to coverglass, and examined by fluorescence microscopy. (E–L) Unpermeabilized Jurkat cells that had been pulse labeled with N-Rh-PE and incubated at 37°C overnight were fixed and processed for fluorescence microscopy using antibodies specific for CD81 (E–H) or CD63 (I–L). (M–P) Unpermeabilized Jurkat cells that had been pulse labeled with N-F-PE and incubated at 37°C overnight were fixed and processed for fluorescence microscopy using antibodies specific for CD63. (Q–T) Unpermeabilized Jurkat cells were pulse labeled with N-Rh-PE for 1 h, incubated for 16 h at 37°C, pulse labeled with N-F-PE for 1 h, incubated for an additional 16 h at 37°C, and processed for fluorescence microscopy. (U and V) Exosomes were collected from the second 16-h incubation, bound to coverglass, and examined by fluorescence microscopy. (U) N-Rh-PE–containing exosomes; (V) N-F-PE–containing exosomes. N-F-PE–containing exosomes are marked by white circles. (W–Z) Unpermeabilized Jurkat T cells that had been pulse labeled with N-Rh-PE and incubated at 37°C overnight were fixed and processed for fluorescence microscopy using antibodies specific for Lamp1. (A′–D′) Unpermeabilized Jurkat T cells that had been pulse labeled with N-Rh-PE were fixed, stained with antibodies specific for CD63, and examined by fluorescence microscopy. Primary T cells that had been pulse labeled with N-Rh-PE were also examined by fluorescence microscopy and found to possess single (E′ and F′) or multiple (G′ and H′) domains enriched for this lipid. Bars, 10 μm.
Approximately one fourth of Jurkat T cells possess a single, large domain enriched for these exosomal markers. An even greater proportion of the cell population, approximately two thirds, possess multiple plasma membrane domains enriched for exosomal proteins. The presence of multiple domains was evident in cells stained for CD81 and CD63 (such as the middle cell in Fig. 1 [I and J] and the upper cell in Fig. 1 [K–N]) and is shown here for N-Rh-PE–labeled cells stained for surface CD63 (Fig. 2, A′–D′). We failed to detect such domains in ∼10% of the cells (for quantitation, see supplemental material). Similar results were observed in primary human T cells (Fig. 2, E′–H′).
Domain dynamics
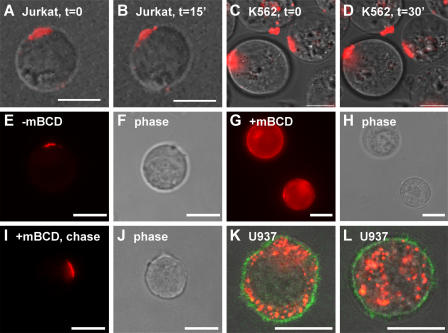
To determine whether these domains remain for just a few seconds or persist for longer periods of time, we examined the distribution of N-Rh-PE in live cells by time-lapse microscopy. The plasma membrane domains marked by this lipid appeared to be semistable, with many of them persisting for periods of 15 min or more (Fig. 3, A and B). Similar results were observed in K562 cells (Fig. 3, C and D). However, these domains were not static entities. Some domains moved laterally in the plasma membrane, whereas others formed or dispersed over time, typically by the movement of smaller domains toward or away from one another but also by movement of intracellular organelles to the cell surface.
Figure 3.
Domain dynamics. (A–D) Jurkat T cells (A and B) or K562 cells (C and D) were pulse labeled with N-Rh-PE for 1 h, incubated overnight, and examined by live cell video microscopy. (E–J) Jurkat T cells were pulse labeled with N-Rh-PE for 1 h, incubated for 16 h, incubated with 15 mM β-cyclodextrin for 30 min, washed, and incubated for various lengths of time before examination by fluorescence microscopy. (E and F) Mock-treated cells examined at t = 0 after treatment. (G and H) β-cyclodextrin–treated cells examined at t = 0 after treatment. (I and J) Methyl-β-cyclodextrin–treated cells examined at t = 8 h after treatment. (K and L) The monocyte cell line U937 was labeled with N-Rh-PE for 1 h, grown overnight, fixed, and processed for immunofluorescence microscopy using FITC-conjugated antibodies specific for CD43. Shown are two representative cells in which the N-Rh-PE (red), surface CD43 (green), and phase-contrast images have been merged. Bars, 10 μm.
The presence of discrete domains in the T cell plasma membrane raises the question of how these domains are organized and how the cell retains lipids such as N-Rh-PE and N-F-PE at these domains. To determine whether membrane lipid composition plays a role in retaining N-Rh-PE at these domains, we pulse labeled Jurkat T cells with N-Rh-PE for 1 h, incubated them for 16 h, exposed them to methyl-β-cyclodextrin for 30 min to deplete sterols and glycosphingolipids from the plasma membrane, removed the methyl-β-cyclodextrin, and followed N-Rh-PE distribution over time. Before the addition of methyl-β-cyclodextrin and after mock treatment, N-Rh-PE is concentrated at discrete domains of the plasma membrane (Fig. 3, E and F). Incubation with methyl-β-cyclodextrin for 30 min released a significant amount of N-Rh-PE from these domains (Fig. 3, G and H). This phenotype was reversible, as removal of the methyl-β-cyclodextrin allowed Jurkat T cells to redirect N-Rh-PE to discrete domains of the plasma membrane (Fig. 3, I and J). Thus, it appears that lipid composition is important for the retention of N-Rh-PE at discrete domains of the plasma membrane.
Although most Jurkat T cells possess one larger or several small discrete domains, there is no reason to presume they are present in the plasma membrane of all cells. Monocytic cell types, including macrophages and dendritic cells, are one group of cells that might be expected to lack such domains. These cells appear to use the delayed mode of exosome biogenesis, in which vesicles bud into endosomes, accumulate, and are secreted as exosomes when the endosomes fuse with the plasma membrane. To test for the presence of discrete domains on the plasma membrane of monocytic cells, we pulse labeled the human monocytic cell line U937 with N-Rh-PE and examined N-Rh-PE distribution 1 d later, counterstaining the cells with antibodies specific for the plasma membrane protein CD43. Unlike Jurkat T cells, which direct N-Rh-PE to discrete domains of the plasma membrane, U937 monocytes retain N-Rh-PE in discrete endosomes (Fig. 3, K and L).
Outward vesicle budding at discrete domains of the plasma membrane
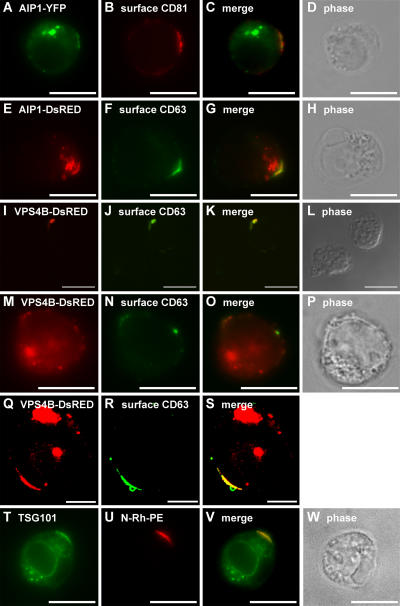
To determine whether these domains of the T cell plasma membrane share any functional similarities with bona fide endosomal membranes, we examined the subcellular distribution of a class E vacuolar protein sorting (VPS) protein, AIP1/VPS31. Class E VPS proteins are thought to mediate outward vesicle budding from the limiting membrane of endosomes, and AIP1/VPS31 has been localized to endosomes and implicated in a late stage of protein sorting to outward budding vesicles in yeasts and in animal cells (Katzmann et al., 2002; Gruenberg and Stenmark, 2004). We observed that a human AIP1/VPS31-YFP fusion protein was present at both internal organelles and at discrete domains of the T cell plasma membrane (Fig. 4, A–D). A similar distribution was observed for an AIP1/VPS31-DsRed fusion protein (Fig. 4, E–H). Another class E VPS protein, VPS4, is an ATPase that is also required at a late stage in protein sorting to outward budding vesicles. Human VPS4B has previously been described as largely cytoplasmic (Bishop and Woodman, 2000), and we found that a VPS4B-DsRed fusion protein was localized to the cytoplasm of Jurkat T cells (not depicted). However, ATPase-defective forms of VPS4 have been reported to concentrate on endosomal membranes (Babst et al., 1997; Bishop and Woodman, 2000; Piper and Luzio, 2001; Katzmann et al., 2002), and we detected human VPS4B/K180Q-DsRed at the same domains of the plasma membrane as CD63 (Fig. 4, I–L) as well as at internal organelles, presumably endosomes (Fig. 4, M–P). Similar results were observed in K562 cells (Fig. 4, Q–S). An antibody to yet a third class E VPS protein, TSG101/VPS23, also decorated these domains of plasma membrane in some N-Rh-PE–labeled Jurkat T cells (Fig. 4, T–W). However, we were unable to detect TSG101-GFP fusion proteins at such domains (not depicted).
Figure 4.
Class E VPS proteins at discrete domains of the Jurkat T cell plasma membrane. (A–D) Unpermeabilized Jurkat T cells expressing AIP1/VPS31-YFP were processed for fluorescence microscopy using antibodies specific for CD81. (A) AIP1/VPS31-YFP. (E–H) Unpermeabilized Jurkat T cells expressing AIP1/VPS31-DsRed were processed for fluorescence microscopy using antibodies specific for CD63. (E) AIP1/VPS31-DsRed. (I–S) Unpermeabilized Jurkat T cells (I–P) or K562 cells (Q–S) expressing VPS4B/K180Q-DsRed were processed for fluorescence microscopy using antibodies specific for CD63. (I, M, and Q) VPS4B/K180Q-DsRed. (T–W) N-Rh-PE–labeled Jurkat T cells were fixed, permeabilized, and processed for fluorescence microscopy using antibodies specific for TSG101. Bars (I–P and T–W), 10 μm; (Q–S) 5 μm.
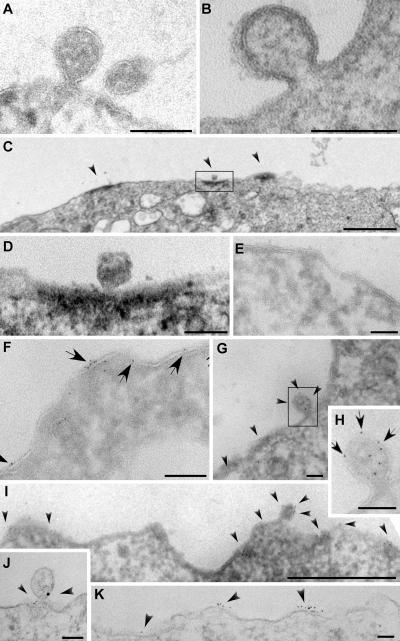
The colocalization of outward vesicle budding factors and exosomal proteins at discrete domains of the plasma membrane indicates that these domains might be sites of outward vesicle budding (i.e., sites of exosome biogenesis). To explore this possibility, we examined Jurkat T cells and K562 cells by electron microscopy. Most thin sections of these cells lacked presumptive outward vesicle budding intermediates. However, we did occasionally detect such structures (Fig. 5, A and B). Using HRP-conjugated secondary antibodies and 3,3′-DAB–based cytochemistry for HRP activity (which deposits an electron-dense reaction product in the vicinity of the antigen), we observed CD63 enrichment at discrete domains of the plasma membrane (Fig. 5 C), one of which contained a presumptive outward budding intermediate (Fig. 5 D). Such domains were more common than presumptive outward vesicle budding intermediates but were still fairly rare. Similar results were obtained using anti-CD81 antibodies and 6-nm gold-conjugated secondary antibodies. Most regions of the plasma membrane were devoid of CD81 label (Fig. 5 E), but clusters of CD81 spanning 1–3 μm of the plasma membrane (Fig. 5, F–I) could be detected in ∼5% of sections (for quantitation of immunogold labeling, see supplemental material). For those rare cell sections that possessed presumptive outward vesicle budding intermediates (∼1%), CD81 labeling was detected on or near ∼50% or more of the buds (Fig. 5, G and H). N-Rh-PE–labeled Jurkat T cells were also examined, this time using rabbit polyclonal antibodies specific for rhodamine and mouse monoclonal antibodies specific for CD81 followed by 18-nm gold-conjugated anti–mouse IgG secondary antibodies and 6-nm gold-conjugated anti–rabbit IgG secondary antibodies. Both markers colocalized on presumptive outward budding intermediates (Fig. 5 J). Like CD81, N-Rh-PE was detected at discrete domains of the plasma membrane (Fig. 5 K). In contrast to what we observed for the exosomal markers CD81 and N-Rh-PE, labeling Jurkat T cells for the nonexosomal protein CD45 decorated the entirety of the plasma membrane (not depicted).
Figure 5.
Exosomes appear to bud from discrete domains of the Jurkat T cell plasma membrane. (A and B) A presumptive outward budding intermediate (nascent exosome) at the plasma membrane of a K562 cell (B) or Jurkat T cells (A, left) adjacent to either another outward budding vesicle or a completely budded vesicle (A, right). (C and D) HRP/DAB detection of CD63 at the Jurkat T cell surface. (C) A cluster of three CD63-positive regions (arrowheads). (D) A higher magnification image of the boxed region, possibly with a presumptive outward budding intermediate. (E–I). Immunogold labeling of Jurkat T cell cryosections. (E) Most of the Jurkat T cell was devoid of label. (F) When CD81 (arrows) could be detected, it was typically clustered. (G) CD81 (arrowheads) decorated presumptive outward vesicle budding intermediates. (H) A higher magnification view of the budding intermediate (boxed area) from G. Arrows denote the positions of immunogold labeling for CD81. (I) CD81 (arrowheads) can be detected in domains that span micrometers of the plasma membrane. (J and K) Immunogold labeling (arrowheads) of Jurkat T cells for N-Rh-PE (6 nm gold) and CD81 (18 nm gold). (J) CD81 and N-Rh-PE colocalized at presumptive outward budding intermediates. (K) N-Rh-PE was enriched at discrete domains of the plasma membrane. Bars (A, B, D–H, J, and K), 100 nm; (C and I) 1 μm.
Collectively, these results indicate that there are discrete domains of the T cell plasma membrane that are enriched for a variety of endosomal and exosomal proteins and are competent for outward vesicle budding. Because these properties are normally associated with endosomes, we propose the term “endosome-like domain” to describe these regions of the plasma membrane.
HIV Gag buds from endosome-like domains of the plasma membrane
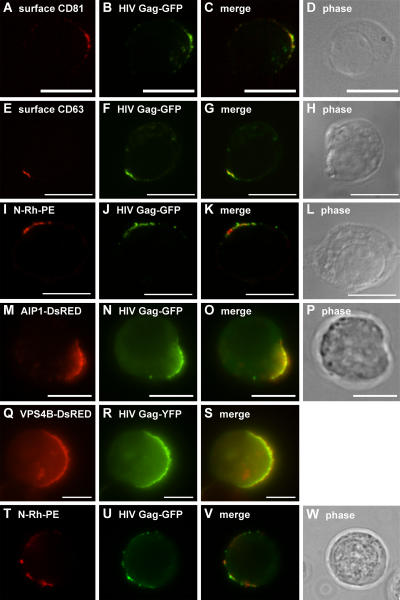
We next compared the sorting of HIV Gag in Jurkat T cells to that of exosomal proteins and lipids. HIV Gag mimics the budding of the intact retrovirus (Coffin et al., 1997; Freed, 1998; Freed and Martin, 2001; Garrus et al., 2001; von Schwedler et al., 2003; Ono and Freed, 2004), and GFP-tagged forms of HIV Gag can be used to follow its biogenesis (Sandefur et al., 1998, 2000). When expressed in human T cells, HIV Gag-GFP, like HIV Gag, is detected at discrete domains of the plasma membrane (Freed, 1998; Freed and Martin, 2001; Ono and Freed, 2004). This curiously focal distribution appears to reflect the trafficking of HIV Gag to endosome-like domains of the plasma membrane, as demonstrated by its colocalization with surface CD81 (Fig. 6, A–D), surface CD63 (Fig. 6, E–H), N-Rh-PE (Fig. 6, I–L), and AIP1/VPS31 (Fig. 6, M–P). Similar results were obtained in K562 cells, which also colocalized VPS4B/K180Q-DsRed with HIV Gag-GFP (Fig. 6, Q–S). It should be noted that HIV Gag was also present at flanking regions of the plasma membrane, raising the possibility that HIV Gag might reach endosome-like domains by lateral sorting within the plasma membrane. Moreover, those Jurkat T cells that contained multiple endosome-like domains of plasma membrane sorted HIV Gag to each of these domains (Fig. 6, T–W).
Figure 6.
HIV Gag is sorted to endosome-like domains of T cells and K562 cells. (A–H) Unpermeabilized Jurkat cells expressing HIV Gag-GFP were processed for fluorescence microscopy using antibodies specific for CD81 (A–D) or CD63 (E–H). (I–L) Unpermeabilized Jurkat cells pulse labeled with N-Rh-PE and expressing HIV Gag-GFP were processed for fluorescence microscopy. (M–P) Unpermeabilized Jurkat T cells expressing AIP1/VPS31-DsRed and HIV Gag-GFP were processed for fluorescence microscopy. (Q–S) K562 cells expressing VPS4BK180Q-DsRed and HIV Gag-YFP were processed for fluorescence microscopy. (Q) VPS4B-K180Q-DsRED. (T–W) Unpermeabilized, N-Rh-PE–labeled Jurkat T cells expressing HIV Gag-GFP were processed for fluorescence microscopy. Bars, 10 μm.
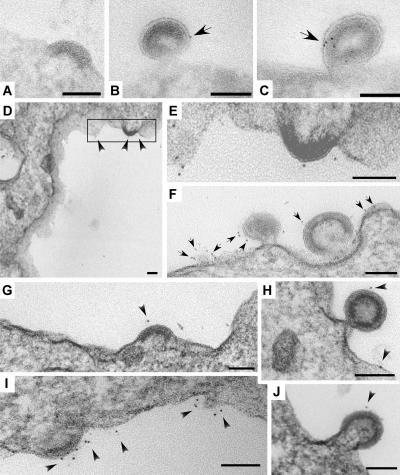
The trafficking of HIV Gag-GFP to endosome-like domains of the T cell plasma membrane indicates that it buds from these domains just as CD81, CD63, and N-Rh-PE bud from these domains. To test this directly, we examined Gag-expressing T cells and K562 cells by immunoelectron microscopy. During budding, HIV Gag oligomerizes to form an electron-dense protein core, which marks sites of Gag budding. Exosomal markers were detected using mouse monoclonal primary antibodies and gold-conjugated secondary antibodies. Although most of the Jurkat T cell plasma membrane was devoid of budding intermediates, we could occasionally detect nascent HIV Gag buds at the surface of Jurkat T cells (Fig. 7 A) in the vicinity of the exosomal marker CD81 (Fig. 7, B and C). Surface labeling of K562 cells revealed that the exosomal marker CD63 was also present on or near nascent HIV Gag budding intermediates (Fig. 7, D–J). Similar results were observed in 293T cells expressing a proviral clone of HIV (Fig. 8).
Figure 7.
HIV Gag buds from endosome-like domains of Jurkat T cells and K562 cells. (A) Jurkat T cells processed for transmission electron microscopy of cryosections possess nascent HIV Gag buds at their plasma membrane. (B and C) Jurkat T cells processed for transmission electron microscopy of cryosections using antibodies specific for CD81 and gold-conjugated secondary antibodies. Arrows highlight immunogold labeling for surface CD81. (D–J) K562 cells expressing HIV Gag were fixed, surface labeled with monoclonal antibodies specific for CD63 and gold-conjugated secondary antibodies, and processed for electron microscopy. Boxed area in D is shown at a higher magnification in E. Arrows and arrowheads highlight immunogold labeling for surface CD63. Bars, 100 nm.
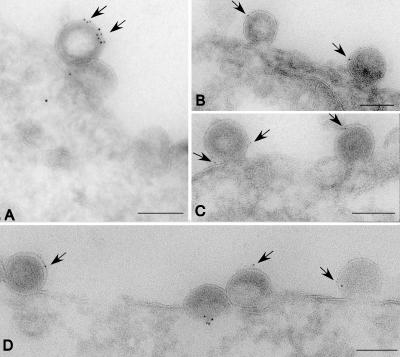
Figure 8.
HIV virions bud from the plasma membrane in association with CD81. 293T cells expressing a proviral clone of HIV were processed for cryoimmunoelectron microscopy using antibodies specific for CD81 and gold-conjugated secondary antibodies. Arrows denote position of gold particles. Bars, 100 nm.
HIV Gag is secreted in exosomes
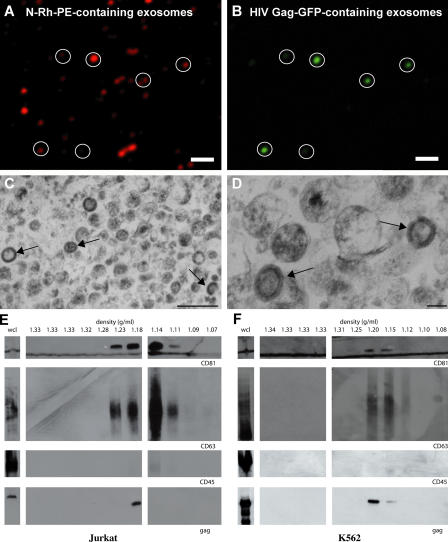
Another way to probe the relationship between exosomes and retroviruses is to collect exosomes secreted by Gag-expressing cells and test them for the presence of Gag. HIV Gag-GFP was expressed in Jurkat T cells that had been pulse labeled with N-Rh-PE, and exosomes were collected from the medium of these cells, bound to poly-l-lysine–coated coverglasses, and examined by fluorescencemicroscopy. Exosomes were detected on the basis of N-Rh-PE fluorescence (Fig. 9 A) and HIV Gag-GFP fluorescence colocalized with this exosomal marker (Fig. 9 B). A parallel exosome preparation from HIV Gag–expressing cells contained discrete, single membrane organelles of ∼100-nm diameter, which is typical of exosomes, and a subset of these exosomes contained prominent electron-dense cores of oligomerized HIV Gag (Fig. 9, C and D). Further purification of a similarexosome preparation by sucrose density gradient centrifugation revealed that Jurkat T cells secreted HIV Gag in vesicles that cofractionated with the exosomal markers CD81 and CD63 (Fig. 9 E). Immunoblot analysis for CD45 demonstrated that these exosome fractions were significantly depleted of contaminating plasma membrane microvesicles, which can potentially contaminate exosome preparations (Esser et al., 2001; Blanchard et al., 2002; Trubey et al., 2003). HIV Gag also copurified with exosomes that were secreted by K562 cells (Fig. 9 F), providing additional evidence that human cells secrete HIV Gag from the cell in exosomes.
Figure 9.
HIV Gag is secreted from cells in exosomes. (A and B) Exosomes were collected from Jurkat cells pulse labeled with N-Rh-PE and expressing HIV Gag-GFP and were examined by fluorescence microscopy. Circles mark secreted vesicles that contain HIV Gag-GFP. (C and D) Thin-section electron micrographs of exosomes collected from Jurkat cells expressing HIV Gag. Arrows denote the position of exosomes that contain the electron-dense HIV Gag core. (E and F) Immunoblot analysis of whole cell lysates (wcl) and sucrose density gradient fractions from conditioned media prepared from HIV Gag–expressing (E) Jurkat cells and (F) K562 cells. Bars (A and B), 20 μm; (C) 500 nm; (D) 100 nm.
Discussion
Most models of exosome biogenesis emphasize a delayed mode of synthesis in which vesicle budding occurs at the limiting membrane of discrete endosomes, and vesicle secretion occurs only later upon endosome–plasma membrane fusion (Denzer et al., 2000a; Stoorvogel et al., 2002; Thery et al., 2002; Pelchen-Matthews et al., 2004). However, there is no a priori basis for assuming that exosome budding and exosome secretion must be separated in time and space. In this study, we explored the possibility that human T cells possess endosome-like domains of plasma membrane that retain at least one endosomal function: outward vesicle budding. We observed that Jurkat T cells do indeed possess endosome-like domains of plasma membrane and bud exosomes from these domains. Moreover, we found evidence that these cells treat HIV Gag as exosomal cargo, sorting it to endosome-like domains of plasma membrane and secreting it from the cell in exosomes.
Endosome-like domains in the T cell plasma membrane
The enrichment of known endosomal and exosomal proteins at discrete domains of the T cell plasma membrane was unexpected from most models of exosome biology (Denzer et al., 2000a; Stoorvogel et al., 2002; Pelchen-Matthews et al., 2004). These endosome-like domains are not associated with any obvious morphological characteristics save for the rare and transient outward vesicle budding intermediate. The enrichment of exosomal proteins and lipids at these sites raises many interesting questions regarding the mechanisms that direct and/or retain cargoes at these domains. A contribution of membrane lipid organization can be inferred from the observation that methyl-β-cyclodextrin treatment releases N-Rh-PE from these domains. It remains to be determined whether the effects are results of a general effect on membrane structure or lipid raft involvement (Edidin, 2003). Another relevant feature of these domains is that they are neither highly transient nor highly stable. Rather, they appear to be dynamic entities that persist long enough to serve as platforms for exosome biogenesis and perhaps other functions but are not so static as to be permanent features on the cell surface. This dynamic instability appears to contribute to the variable numbers of endosome-like domains in the plasma membrane, their variable size and distribution on the cell surface, and even their absence or low abundance in the plasma membrane of certain cell types. Clearly, there is a need for a more sophisticated analysis of the structure and dynamics of these domains than we have presented here.
Several lines of evidence indicate that endosome-like domains of plasma membrane are sites of exosome budding. First, the colocalization of so many exosomal and endosomal molecules at these domains of the T cell plasma membrane is a strong indication that they are sites of exosome biogenesis. Second, the presence of AIP1/VPS31, VPS4B/K180Q, and TSG101 indicates that these domains have at least some of the cellular factors that have been implicated in outward vesicle budding. Third, electron microscopy analyses of Jurkat T cells revealed the presence of exosomal molecules on or near presumptive outward vesicle budding intermediates at the plasma membrane. Fourth, live cell imaging experiments demonstrated that these domains persist for minutes to hours, a time that seems sufficient to allow vesicle budding. Collectively, these observations present a compelling argument that exosomes can, in certain cells and at certain locations, bud directly from the plasma membrane.
Immediate versus delayed modes of exosome biogenesis
We have referred to the budding of exosomes directly from the plasma membrane as the “immediate mode” of exosome biogenesis (Gould et al., 2003). This differs from the more accepted view of exosome biogenesis as a delayed process, in which vesicles bud into discrete endosomes and are subsequently released upon endosome–plasma membrane fusion (Denzer et al., 2000a; Stoorvogel et al., 2002; Thery et al., 2002). Although the morphological differences between the immediate and delayed modes of exosome biogenesis are significant, there may be only slight mechanistic and/or kinetic differences between the two. After all, both modes of exosome biogenesis involve outward vesicle budding at endosome-like membranes, ferry the same cargo molecules from the cell, and are associated with the same protein sorting machinery. Moreover, even the delayed mode of exosome biogenesis requires the formation of endosome-like domains in the plasma membrane (Pelchen-Matthews et al., 2004).
The immediate mode of exosome biogenesis has not previously attracted much attention, and we believe that sampling bias has been a contributing factor. The delayed mode of exosome biogenesis involves the formation and retention of an intermediate structure, the multivesicular endosome (MVE) or multivesicular body (MVB), which is large, relatively long lived, and, therefore, easy to detect by electron microscopy and light microscopy (Denzer et al., 2000a; Stoorvogel et al., 2002; Thery et al., 2002). In the immediate mode of exosome biogenesis, there is no MVE/MVB-like intermediate, and, thus, the only morphological evidence for immediate exosome biogenesis is the occasional outward vesicle budding intermediate at the plasma membrane. These intermediates are much smaller than MVEs/MVBs, shorter lived than MVEs/MVBs, and occur only at discrete, small, and otherwise unremarkable domains of the plasma membrane. In this context, it is understandable that the delayed mode of exosome biogenesis has been easier to observe than the immediate mode of exosome biogenesis. This bias can, however, be minimized by recognizing the existence of both modes of exosome biogenesis, by identifying molecular markers of exosome biogenesis, and by using these markers to identify sites of exosome biogenesis in future studies.
Retroviral budding
The observation that human T cells bud exosomes from endosome-like domains of plasma membrane allowed us to test a key prediction of the Trojan exosome hypothesis (Gould et al., 2003). This hypothesis posits that retroviruses are at their most fundamental level a variant form of exosome. Thus, it predicts that cells will treat the key budding factors of retroviruses (typically their Gag protein) as cargo of the exosome biogenesis pathway. In the specific case of Jurkat T cells, it predicts that HIV Gag will be sorted to endosome-like domains and secreted from the cell in exosomes. Several lines of evidence confirm this prediction. First, we observed that HIV Gag colocalizes with surface CD81, surface CD63, N-Rh-PE, and AIP1/VPS31, demonstrating that Jurkat T cells do indeed sort HIV Gag to endosome-like domains of the plasma membrane. Second, electron microscopy studies revealed that HIV Gag appears to bud from endosome-like domains of the plasma membrane, as evident from the presence of exosomal markers on or near presumptive HIV Gag budding intermediates at the plasma membrane. Third, fluorescence microscopy and electron microscopy data indicated that HIV Gag is secreted from the cell in exosomes. Fourth, biochemical studies demonstrated that HIV Gag is secreted from the cell in vesicles that copurify with exosomes. Fifth, others have shown that human T cells exclude CD45 from HIV particles (Esser et al., 2001; Trubey et al., 2003), and we observed that Jurkat T cells fail to enrich CD45 at endosome-like domains of the plasma membrane and exclude CD45 from exosomes.
These observations have implications for general hypotheses of retroviral biology. The prevailing hypothesis is that HIV Gag recruits the outward vesicle budding machinery to the plasma membrane, deforms the membrane as it oligomerizes to form its core, and, by these and other mechanisms, drives the formation of free HIV virions (Freed, 1998; Freed and Martin, 2001; Morita and Sundquist, 2004; Ono and Freed, 2004). This hypothesis, however, seems overly complex because Jurkat T cells possess endosome-like domains of plasma membrane and bud exosomes from these domains before Gag expression. In contrast, the Trojan exosome hypothesis predicts these observations (Gould et al., 2003), for its core tenet is that cells will bud HIV and other retroviruses where they happen to make their exosomes. Such a mechanism also explains why monocytes bud HIV into discrete endosomes and secrete them only later after endosome–plasma membrane fusion (Raposo et al., 2002; Nguyen et al., 2003; Nydegger et al., 2003; Pelchen-Matthews et al., 2003; Ono and Freed, 2004). Although many predictions of the Trojan exosome hypothesis remain to be tested, the available data indicate that it is at least a viable, parsimonious hypothesis of retroviral biogenesis.
Materials and methods
Cells, plasmids, and antibodies
Jurkat T cells were maintained in a serum-free medium (AIM-V; Invitrogen). K562 and primary human T cells were grown/maintained in RPMI 1640 supplemented with 10% FCS. N-Rh-PE and N-F-PE were obtained from Avanti Polar Lipids, Inc. The plasmid that expresses VPS4B(K180Q)-DsRed was provided by W. Sundquist (University of Utah, Salt Lake City, UT). The HIV Gag expression vector p96ZM651gag-opt was obtained from Y. Li, F. Gao, and B.H. Hahn through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. The Gag-GFP expression vector was generated by amplifying the optimized HIV Gag ORF from p96ZM651gag-opt using the primers 5′-GAAAGGTACCATGGGCGCCCGCGCCAGCATC-3′ and 5′-CCCGGATCCCTGGCTCAGGGGGTCGCTGCC-3′. The resulting PCR products were digested with Asp718 and BamH1 and inserted between the Asp718 and BamH1 sites of pcDNA3-GFP. A sequence-confirmed clone (pcDNA3-HIVGag-GFP) with no errors was used for the experiments described in this paper. The AIP1/VPS31-YFP and -DsRed expression vectors were generated by amplifying a fragment of the AIP1 cDNA (synthesized by PCR from a human muscle cDNA library using the primers 5′-CAAGGTACCATGGCGACATTCATCTCGGTG-3′ and 5′-CAAGGATCCCTGCTGTGGATAGTAAGACTG-3′), cleaving the fragment with Asp718 and BamH1, and inserting it between the Asp718 and BamH1 sites of pcDNA3-YFP and pcDNA3-DsRed, respectively. Only sequence-confirmed clones lacking undesired mutations were used for experiments. FITC-conjugated anti–human CD63, FITC-conjugated anti–human CD81, phycoerythrin-conjugated anti–human CD81, and unlabeled monoclonal antibody directed against human CD30 (BerH8) and CD81 (JS-81) were obtained from BD Biosciences. Unlabeled mouse monoclonal antibodies directed against human Lamp1 (H4A3), CD7 (3A1E-12HT), CD43 (DF-T1), CD45 (2D-1), and CD63 (H5C6) and polyclonal rabbit antibodies specific for CD81 were obtained from Santa Cruz Biotechnology, Inc. Rabbit anti-rhodamine antibodies were obtained from Invitrogen. Polyclonal antibodies directed against HIV Gag were generated against purified p24. Secondary antibodies were obtained from Jackson ImmunoResearch Laboratories. Directly labeled anti-CD63 and anti-CD81 antibodies were used in Figs. 1 (E–F and I–N), 2 (E–P), 4 (I–P), and 6 (A–L). Unlabeled primary antibodies were used in all other experiments.
Transfections, cell manipulations, and light microscopy
5 × 106 cells were electroporated with 2 μg of plasmid using a Nucleofactor apparatus (Amaxa) and Cell Line Nucleofector Kit V (Amaxa) or by using an electroporator (ECM600; BTX) and standard protocols. Pulse labeling of cells with N-Rh-PE, N-F-PE, or N-biotin-PE involved solubilizing the phospholipid in absolute ethanol, injecting it into cold AIM-V media with a Hamilton syringe to a final concentration of 5 μM, incubating cells in this solution for 1 h at 4°C, washing the cell extensively with cold AIM-V media, and resuspending the cells in AIM-V medium at 37°C. Labeling of transfected cells with N-Rh-PE was performed ∼14 h after transfection. Cells labeled with N-Rh-PE or N-F-PE were fixed ∼20 h after labeling unless otherwise stated. Treatment with 15 mM β-cyclodextrin was for 30 min at 37°C.
For fluorescence microscopy, cells were fixed with 3% PFA in 1× PBS, pH 7.2, for 15 min and washed twice in PBS, all at 20–25°C. For the experiments with permeabilized cells (Fig. 1, C, D, G, and H; and Fig. 4, T–W), fixed cells were incubated in 1% Triton X-100 in 1× PBS, pH 7.2, for 5 min followed by two washes in 1× PBS, pH 7.2. After fixation or fixation and permeabilization, cells were incubated with either labeled primary antibodies (experiments in Fig. 1) or sequentially with unlabeled primary and labeled secondary antibodies, each diluted in 1× PBS, pH 7.2. After the antibody incubations, samples were washed extensively in PBS. Before viewing, samples were placed in mounting medium (90% glycerol, 100 mM Tris, pH 8.5, and 0.1% paraphenylenediamine). Unless otherwise noted, cells were examined at 20–25°C, and images were obtained using a fluorescence microscope (BH-2; Olympus) equipped with a 60× NA 1.4 lens and captured with a CCD camera (Sensicam QE; Cooke) linked to a MacG4 computer using IPlab software (Scanalytics). Exceptions are listed as follows. Images for Figs. 3 (A–D) and 4 (Q–S) were obtained using a confocal fluorescence microscope (LSM510 Meta; Carl Zeiss MicroImaging, Inc.) with a 63× NA 1.4 lens and dedicated imaging software (LSM image browser; Carl Zeiss MicroImaging, Inc.). Images for Figs. 2 (E–P), 4 (I–L), 6 (A–L), and 9 (A and B) were obtained using an UltraView Live Cell Imaging system (PerkinElmer) with a 63× NA 1.4 lens (Nikon), a spinning Nipkow disk with microlenses, and UltraView software (PerkinElmer). For live cell imaging (Fig. 3, A–D), cells were incubated in growth medium supplemented with 100 mM Hepes on a peltier modulated stage warmer (Instec) maintained at 37°C. Digital images were false colored, converted to Adobe Photoshop TIFF files, adjusted for brightness and contrast, and assembled in Adobe Illustrator. Fluorochromes for labeled primary antibodies are described above. For all experiments using secondary antibodies, FITC and Texas red conjugates were used, and staining patterns were dependent on prior incubation with primary antibodies. False color images correspond to idealized hues for these fluorochromes. For experiments using fusion proteins to fluorescent proteins, images correspond to direct fluorescence of the fusion protein. Images shown are representative of results from a minimum of four replicates of each experiment.
Electron microscopy
Immunogold surface labeling of cells was performed essentially as described previously (McCaffery and Farquhar, 1995). In brief, cells were fixed for 1 h at RT in 2% PFA in 100 mM PO4, pH 7.4. Cells were washed, blocked in 10% FCS in 100 mM PO4, pH 7.4, for 20 min, and incubated overnight with anti-CD63 antibodies in 100 mM PO4, pH 7.4. After washing, cells were incubated for 2 h at RT with donkey anti–mouse 5-nm Au conjugate, washed, and again fixed for 1 h in 2% glutaraldehyde, 2.5% sucrose (wt/vol), and 100 mM cacodylate, pH 7.4. The cells were spun into a tight pellet, fixed in Palade's osmium for 1 h, en bloc stained overnight in Kellenberger's uranyl acetate, dehydrated through a graded series of ethanol, and embedded in epon. 80-nm ultrathin sections were cut on an ultramicrotome (model UCT; Leica) collected onto 400 mesh high transmission nickel grids, post-stained in uranyl acetate and lead citrate, and observed on a transmission electron microscope (EM 410; Philips). Immunoperoxidase labeling was performed essentially as described previously (McCaffery and Farquhar, 1995). In brief, cells were fixed in 4% PFA and contained in 0.1 M phosphate buffer, pH 7.4, for 1 h at RT. Fixed cells were sequentially incubated with primary antibodies, washed, and incubated with HRP-conjugated secondary antibodies, all diluted in 1× PBS. Cells were washed in 0.1 M cacodylate-HCl, pH 7.4, and 7.5% sucrose followed by postfixation with 4% glutaraldehyde in 0.1 M cacodylate- HCl, pH 7.4, for 1 h. Cells were then incubated in DAB reaction buffer (3 mg/ml DAB, 0.01% H2O2, and 0.1 M Tris-HCl, pH 4) and postfixed in 0.1 M cacodylate-HCl, pH 7.4, 1% reduced OsO4, and 1% KFeCN. Cells were dehydrated through a graded series of ethanol and embedded in epon. 80-nm sections were prepared on an ultramicrotome (model UCT; Reichert), collected onto nickel grids, and stained with lead citrate. For cryoelectron microscopy, cells were fixed at 4°C in 100 mM PO4 containing 2.5% sucrose, pH 7.4, and 4% PFA overnight, washed, and harvested. Cell pellets were cryoprotected in 2.3 M sucrose containing 30% polyvinyl pyrollidone, mounted on aluminum cryopins, and frozen in liquid nitrogen. Ultrathin cryosections were then cut on an ultramicrotome (Reichert) equipped with an FCS cryostage, and sections were collected onto 300 mesh formvar/carbon-coated nickel grids. Grids were washed, blocked in 10% FCS, and incubated overnight with 10 μg/ml primary antibodies; in the case of double indirect labeling, antibodies were mixed. After washing, grids were incubated with gold-conjugated secondary antibodies for 2 h, washed, and embedded in a mixture containing 3.2% polyvinyl alcohol (103 molecular mass), 0.2% methyl cellulose (400 centiposes), and 0.2% uranyl acetate. Sections were analyzed on a transmission electron microscope (Philips), and images were collected with a digital camera (Megaview III; Soft Imaging System). Digital images were adjusted for brightness and contrast and assembled into figures using Adobe Illustrator or Canvas. Control experiments in which primary antibodies were omitted showed no labeling, and primary antibodies used for immunoelectron microscopy were also tested for labeling specificity by immunofluorescence microscopy.
Exosome purification and immunoblotting
Cells were removed from the culture supernatant by centrifugation at 2,000 g for 5 min followed by passage of the supernatant through a 0.22-μm filter. The supernatant was then subjected to two to three successive spins at 10,000 g for 30 min to remove cell debris and at 70,000 g for 1 h to pellet exosomes. Further purification of exosomes was accomplished by resuspending the 70,000 g pellet fraction and centrifuging the exosome sample for 16 h at 270,000 g through a continuous sucrose gradient. 11 fractions were collected, assayed for density by refractometry, and particulate material was collected by spinning each sample for 1 h at 350,000 g. Pellets were resuspended in 1× SDS-PAGE loading buffer, separated by SDS- PAGE, and processed for immunoblotting using specific antibodies (mouse monoclonal antibodies to CD45 and CD63 and rabbit polyclonal antibodies for CD81 and HIV Gag together with HRP-conjugated secondary antibodies to mouse or rabbit, respectively) followed by chemiluminescent detection of HRP-conjugated secondary antibodies. For fluorescence microscopy of exosomes, 70,000 g exosome pellet fractions were resuspended in 1× PBS, pH 7.2, applied to poly-l-lysine–coated coverslips, and imaged on a fluorescence microscope (BH-2; Olympus) at 20–25°C equipped with a 60× NA 1.4 lens, and images were captured using a CCD camera (Cooke) linked to a MacG4 computer using IPlab software.
Online supplemental material
Fig. S1 shows the distribution of other plasma membrane proteins on Jurkat T cells. Supplemental material provides details about the quantitation of immunofluorescence data and immunogold labeling. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200508014/DC1.
Supplementary Material
Acknowledgments
We are deeply indebted to J. Michael McCaffery and the Integrated Imaging Center of Johns Hopkins University for outstanding advice and assistance with electron microscopy and live cell imaging. We also thank James Morrell and Wanhua Yan for technical assistance and David Graham for providing 293T cells expressing an HIV provirus.
This work was supported by grants from the NIH (DK45787 and AI62479) and Johns Hopkins University (Fund for Medical Discovery) to S.J. Gould.
A.M. Booth and Y. Fang contributed equally to this article.
Abbreviations used in this paper: MVB, multivesicular body; MVE, multivesicular endosome; PE, phosphatidyl ethanolamine; VPS, vacuolar protein sorting.
References
- Arienti, G., E. Carlini, and C.A. Palmerini. 1997. Fusion of human sperm to prostasomes at acidic pH. J. Membr. Biol. 155:89–94. [DOI] [PubMed] [Google Scholar]
- Arienti, G., A. Nicolucci, F. Santi, E. Carlini, and C.A. Palmerini. 2001. Progesterone-induced increase of sperm cytosolic calcium is enhanced by previous fusion of spermatozoa to prostasomes. Cell Calcium. 30:222–227. [DOI] [PubMed] [Google Scholar]
- Arienti, G., E. Carlini, C. Saccardi, and C.A. Palmerini. 2002. Nitric oxide and fusion with prostasomes increase cytosolic calcium in progesterone- stimulated sperm. Arch. Biochem. Biophys. 402:255–258. [DOI] [PubMed] [Google Scholar]
- Babst, M., T.K. Sato, L.M. Banta, and S.D. Emr. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, N., and P. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 11:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard, N., D. Lankar, F. Faure, A. Regnault, C. Dumont, G. Raposo, and C. Hivroz. 2002. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J. Immunol. 168:3235–3241. [DOI] [PubMed] [Google Scholar]
- Carlin, L.M., K. Eleme, F.E. McCann, and D.M. Davis. 2001. Intercellular transfer and supramolecular organization of human leukocyte antigen C at inhibitory natural killer cell immune synapses. J. Exp. Med. 194:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, A., J. Court, H. Navabi, M. Adams, M.D. Mason, J.A. Hobot, G.R. Newman, and B. Jasani. 2001. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods. 247:163–174. [DOI] [PubMed] [Google Scholar]
- Coffin, J.M., S.H. Hughes, and H.E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 843 pp. [PubMed]
- Denzer, K., M.J. Kleijmeer, H.F. Heijnen, W. Stoorvogel, and H.J. Geuze. 2000. a. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113:3365–3374. [DOI] [PubMed] [Google Scholar]
- Denzer, K., M. van Eijk, M.J. Kleijmeer, E. Jakobson, C. de Groot, and H.J. Geuze. 2000. b. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J. Immunol. 165:1259–1265. [DOI] [PubMed] [Google Scholar]
- Edidin, M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257–283. [DOI] [PubMed] [Google Scholar]
- Escola, J.M., M.J. Kleijmeer, W. Stoorvogel, J.M. Griffith, O. Yoshie, and H.J. Geuze. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273:20121–20127. [DOI] [PubMed] [Google Scholar]
- Esser, M.T., D.R. Graham, L.V. Coren, C.M. Trubey, J.W. Bess Jr., L.O. Arthur, D.E. Ott, and J.D. Lifson. 2001. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J. Virol. 75:6173–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed, E.O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 251:1–15. [DOI] [PubMed] [Google Scholar]
- Freed, E.O., and M.A. Martin. 2001. HIVs and their replication. In Fields Virology. Vol. 2. D.M. Knipe and P.M. Howley, editors. Lippincott Williams & Wilkins, Philadelphia. 1971–2041.
- Fritzsching, B., B. Schwer, J. Kartenbeck, A. Pedal, V. Horejsi, and M. Ott. 2002. Release and intercellular transfer of cell surface CD81 via microparticles. J. Immunol. 169:5531–5537. [DOI] [PubMed] [Google Scholar]
- Garrus, J.E., U.K. von Schwedler, O.W. Pornillos, S.G. Morham, K.H. Zavitz, H.E. Wang, D.A. Wettstein, K.M. Stray, M. Cote, R.L. Rich, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 107:55–65. [DOI] [PubMed] [Google Scholar]
- Goff, S.P. 2001. Retroviridae: the retroviruses and their replication. In Fields Virology. Vol. 2. D.M. Knipe and P.M. Howley, editors. Lippincott Williams & Wilkins, Philadelphia. 1871–1939.
- Gould, S.J., A.M. Booth, and J.E.K. Hildreth. 2003. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA. 100:10592–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317–323. [DOI] [PubMed] [Google Scholar]
- Heijnen, H.F., A.E. Schiel, R. Fijnheer, H.J. Geuze, and J.J. Sixma. 1999. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 94:3791–3799. [PubMed] [Google Scholar]
- Katzmann, D.J., G. Odorizzi, and S.D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905. [DOI] [PubMed] [Google Scholar]
- McCaffery, J.M., and M.G. Farquhar. 1995. Localization of GTPases by indirect immunofluorescence and immunoelectron microscopy. Methods Enzymol. 257:259–279. [DOI] [PubMed] [Google Scholar]
- Metzelaar, M.J., P.L. Wijngaard, P.J. Peters, J.J. Sixma, H.K. Nieuwenhuis, and H.C. Clevers. 1991. CD63 antigen. A novel lysosomal membrane glycoprotein, cloned by a screening procedure for intracellular antigens in eukaryotic cells. J. Biol. Chem. 266:3239–3245. [PubMed] [Google Scholar]
- Morita, E., and W.I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425. [DOI] [PubMed] [Google Scholar]
- Nguyen, D.G., A. Booth, S.J. Gould, and J.E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278:52347–52354. [DOI] [PubMed] [Google Scholar]
- Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic. 4:902–910. [DOI] [PubMed] [Google Scholar]
- Ono, A., and E.O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 78:1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews, A., G. Raposo, and M. Marsh. 2004. Endosome, exosomes, and Trojan viruses. Trends Microbiol. 12:310–316. [DOI] [PubMed] [Google Scholar]
- Piper, R.C., and J.P. Luzio. 2001. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic. 2:612–621. [DOI] [PubMed] [Google Scholar]
- Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 3:718–729. [DOI] [PubMed] [Google Scholar]
- Saez, F., G. Frenette, and R. Sullivan. 2003. Epididymosomes and prostasomes: their roles in posttesticular maturation of the sperm cells. J. Androl. 24:149–154. [DOI] [PubMed] [Google Scholar]
- Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandefur, S., R.M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag). J. Virol. 74:7238–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel, W., M.J. Kleijmeer, H.J. Geuze, and G. Raposo. 2002. The biogenesis and functions of exosomes. Traffic. 3:321–330. [DOI] [PubMed] [Google Scholar]
- Thery, C., L. Zitvogel, and S. Amigorena. 2002. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2:569–579. [DOI] [PubMed] [Google Scholar]
- Trubey, C.M., E. Chertova, L.V. Coren, J.M. Hilburn, C.V. Hixson, K. Nagashima, J.D. Lifson, and D.E. Ott. 2003. Quantitation of HLA class II protein incorporated into human immunodeficiency type 1 virions purified by anti-CD45 immunoaffinity depletion of microvesicles. J. Virol. 77:12699–12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel, G., and M. Heyman. 2002. II. Intestinal epithelial cell exosomes: perspectives on their structure and function. Am. J. Physiol. Gastrointest. Liver Physiol. 283:G251–G255. [DOI] [PubMed] [Google Scholar]
- van Niel, G., G. Raposo, C. Candalh, M. Boussac, R. Hershberg, N. Cerf-Bensussan, and M. Heyman. 2001. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 121:337–349. [DOI] [PubMed] [Google Scholar]
- Vidal, M., P. Mangeat, and D. Hoekstra. 1997. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J. Cell Sci. 110:1867–1877. [DOI] [PubMed] [Google Scholar]
- von Schwedler, U.K., M. Stuchell, B. Muller, D.M. Ward, H.Y. Chung, E. Morita, H.E. Wang, T. Davis, G.P. He, D.M. Cimbora, et al. 2003. The protein network of HIV budding. Cell. 114:701–713. [DOI] [PubMed] [Google Scholar]
- Willem, J., M. ter Beest, G. Scherphof, and D. Hoekstra. 1990. A non-exchangeable fluorescent lipid analog as a membrane traffic marker of the endocytic pathway. Eur. J. Cell Biol. 53:173–184. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.