Abstract
Actin is not only a major cytoskeletal component in all eukaryotic cells but also a nuclear protein that plays a role in gene transcription. We put together data from in vitro and in vivo experiments that begin to provide insights into the molecular mechanisms by which actin functions in transcription. Recent studies performed in vitro have suggested that actin, in direct contact with the transcription apparatus, is required in an early step of transcription that is common to all three eukaryotic RNA polymerases. In addition, there is evidence from in vivo studies that actin is involved in the transcription elongation of class II genes. In this case, actin is bound to a specific subset of premessenger RNA binding proteins, and the actin–messenger RNP complex may constitute a molecular platform for recruitment of histone-modifying enzymes. We discuss a general model for actin in RNA polymerase II transcription whereby actin works as a conformational switch in conjunction with specific adaptors to facilitate the remodeling of large macromolecular assemblies at the promoter and along the active gene.
Introduction
The idea that nuclear actin plays a role in gene transcription is gaining strong support. Two milestone studies published in the 1980s presented circumstantial evidence that actin is implicated in the transcription of protein-coding genes (Egly et al., 1984; Scheer et al., 1984). However, biochemical observations that supported the idea encountered criticism, and the findings were immediately dismissed as resulting from contamination or being artifacts, primarily because of the notorious abundance of actin. Moreover, “traditional” filamentous actin structures that are commonly observed in the cytoplasm, made visible by certain drugs, such as phalloidin, were not revealed in the cell nucleus even by advanced light microscopy methods. Skepticism in the field remained, and the question of whether actin is present in the nucleus was left to drift as an “untouchable” topic (for review see Pederson and Aebi, 2002). 20 yr later, we now agree that β-actin plays a role in gene transcription associated with three different entities: components of ATP-dependent chromatin remodeling complexes (for reviews see Olave et al., 2002; Bettinger et al., 2004), RNP particles (Percipalle et al., 2001, 2002), and the three RNA polymerases in the eukaryotic cell nucleus (Fomproix and Percipalle, 2004; Hofmann et al., 2004; Hu et al., 2004; Philimonenko et al., 2004; Kukalev et al., 2005).
Mass spectrometry (Jonsson et al., 2001) and immunoreactivity criteria (Hofmann et al., 2004) have suggested that the actin in the nucleus is β-actin. However, the studies that have investigated the function of actin in gene transcription (see the following paragraphs) have not revealed whether the actin molecules involved are in monomeric or polymeric form (for review see Pederson and Aebi, 2005). F-actin was detected in the nuclei of neuronal cells by heavy meromyosin labeling (Amankwah and de Boni, 1994). Nuclear actin filaments have also been observed attached to the nuclear pore complexes in amphibian oocytes (Kiseleva et al., 2004). Recent immunochemical studies have detected several types of actin structures in the nucleus (Schoenenberger et al., 2005). Furthermore, a pool of nuclear polymeric actin has been detected by FRAP in cell lines that express GFP-tagged actin and in cells microinjected with fluorescent actin binding proteins (McDonald et al., 2006). However, we do not know whether the actin that is engaged in transcription is monomeric, oligomeric, or filamentous.
Much work is currently being performed to reveal the molecular mechanisms that underlie the role of nuclear actin in transcription. Recent investigations have examined different systems and reached somewhat different conclusions. We summarize and discuss these recent studies and describe a general model for the function of nuclear actin in transcription.
Actin and nuclear myosin 1 (NM1) in ribosomal DNA (rDNA) transcription
NM1 was found in the cell nucleus (Pestic-Dragovich et al., 2000), which suggested that nuclear actin and myosin may function in a concerted manner. In fact, both proteins have been found in mammalian nucleoli (Andersen et al., 2002; Fomproix and Percipalle, 2004; Andersen et al., 2005). Chromatin immunoprecipitation has been used to study the association of actin and NM1 with rDNA promoters. In spite of technical limitations due to epitope accessibility, the chromatin immunoprecipitation data strongly supports the view that both actin and NM1 are present on actively transcribing ribosomal genes (Philimonenko et al., 2004). Actin is associated with both active and inactive pol-I, whereas NM1 binds to the transcription machinery through the pol-I–specific transcription initiation factor IA (Philimonenko et al., 2004). Based on these data, one could speculate that recruitment of the actin–pol-I complex to the rDNA promoter brings actin and NM1 close to each other. Actin and NM1 would then be able to interact and presumably activate rDNA transcription (Philimonenko et al., 2004). Indeed, posttranscriptional NM1 gene silencing down-regulates preribosomal RNA synthesis, and both actin and NM1 are necessary for in vitro rDNA transcription (Philimonenko et al., 2004). These results suggest that the actin–NM1 complex plays a role in the assembly of a transcription-competent pol-I.
An intriguing question is whether NM1 plays a role also in transcription by pol-II. Pol-II transcription is inhibited by a peptide-specific anti-NM1 antibody (Pestic-Dragovich et al., 2000), but there is no evidence that NM1 sits on the promoter of protein-coding genes. Recent immunoelectron microscopy studies have shown that NM1 is located at nucleoplasmic transcription sites (Kysela et al., 2005), but there is evidence that NM1 is physically connected to transcribed loci via RNA, not via TFII factors, as RNase treatment of living HeLa cells specifically depletes cellular NM1 (Fomproix and Percipalle, 2004).
Actomyosin motors in transcription?
It has been suggested that actin and NM1 act as a motor that facilitates the movement of the transcription machinery relative to the DNA template during transcription elongation (for review see de Lanerolle et al., 2005). This suggestion is based on several observations: the ATPase activity of myosin depends on actin, actomyosin motors function in the cytoplasm, and both actin and myosin are present in the cell nucleus (Pestic-Dragovich et al., 2000). It is an attractive idea that actin functions by the same mechanism in both the nucleus and cytoplasm. The motor hypothesis, however, has not been experimentally validated, and several considerations argue against this hypothesis. RNA polymerase by itself is a molecular motor (for review see Gelles and Landick, 1998), and therefore the requirement for an additional motor coupled to the transcriptional machinery seems redundant. There is no evidence that NM1 binds to DNA. Actin associates with chromatin remodeling factors and histone-modifying enzymes (see the following paragraph) without requiring myosin. Current knowledge does not allow us to formally exclude any molecular mechanism, but these observations suggest that the role, or at least one of the roles, of actin in transcription does not require NM1 and does not depend on motor activity.
Nuclear actin in chromatin remodeling complexes
Important clues concerning the role of nuclear actin came when nuclear proteins associated with actin were identified. Studies of chromatin regulation induced by antigen-receptor signaling in lymphocytes led to the identification of β-actin and BAF53, an actin-related protein (Arp), as components of the BAF (BRG-associated factor) chromatin remodeling complex (Zhao et al., 1998). After this initial observation, many other Swi/Snf-like complexes and histone-modifying factors in different organisms have been found to be associated with actin and/or Arps, and numerous investigations have reinforced the idea that there is a functional link between actin and the regulation of chromatin structure (for reviews see Olave et al., 2002; Bettinger et al., 2004).
The molecular mechanisms by which actin and Arps contribute to chromatin remodeling are not fully understood. The ubiquity of actin and Arps in a large variety of chromatin remodeling complexes suggests that these proteins may be involved, directly or indirectly, in many different nuclear processes, including transcription (for review see Rando et al., 2000). In the case of the mammalian BAF complex, which participates in transcription activation in response to external stimuli, actin is directly associated with the ATPase subunit BRG1. In this context, actin is necessary for the association of BRG1 with chromatin and is required for the full activation of the BRG1's ATPase activity (Zhao et al., 1998).
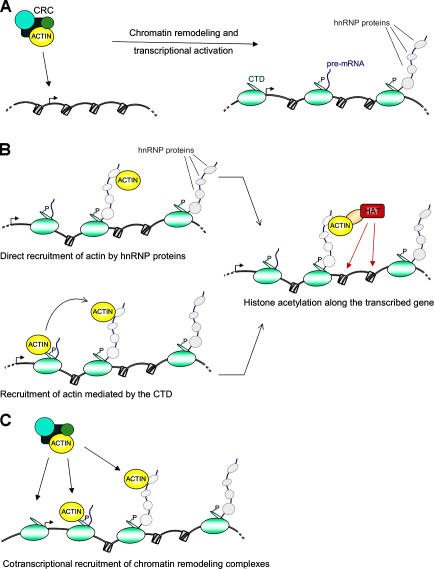
As summarized in Table I, actin can potentially be recruited to transcription complexes through different types of interactions. The presence of actin in some chromatin remodeling complexes suggests a possible mechanism for the recruitment of such complexes to active genes (Fig. 1).
Table I.
Binding partners for β-actin in the eukaryotic transcription complexes
| Actin binding protein | RNA polymerase | Reference |
|---|---|---|
| RPABC2 and -3 | pol-I, pol-II, pol-III | Hu et al., 2004 |
| RPC3 | pol-III | Hu et al., 2004 |
| Phosphorylated CTD | Elongating pol-II | Kukalev et al., 2005 |
| Pre-mRNA binding proteins | Elongating pol-II | |
| Human hnRNP A/B | Percipalle et al., 2002 | |
| Human hnRNP U | Kukalev et al., 2005 | |
| C. tentans HRP36 | Percipalle et al., 2001 | |
| C. tentans HRP65-2 | Percipalle et al., 2003 |
Figure 1.
Models for the function of actin in chromatin regulation at class II genes. (A) Actin is a component of ATP-dependent chromatin remodeling complexes (CRCs) involved in transcriptional activation. (B) Actin becomes incorporated into the growing pre-mRNP while transcription is taking place. Actin may be recruited directly by hnRNP proteins. An alternative mechanism, supported by the observed binding of actin to the CTD, is that actin is recruited to the transcription machinery via binding to the CTD and subsequently transferred from pol-II to the nascent transcript. Adaptor proteins, such as HRP65-2 in C. tentans and hnRNP U in mammals (see the text for details), help actin recruit HATs that acetylate histones along the gene and maintain the chromatin template in a transcription-competent state. (C) We speculate that actin mediates the cotranscriptional recruitment of chromatin remodeling complexes to active genes through interactions with pol-II and/or with the nascent pre-mRNP.
Actin recruits histone modifiers to class II genes
Studies in the dipteran Chironomus tentans suggest that actin can also influence chromatin structure through the recruitment of histone-modifying enzymes. Actin binds directly to the nuclear protein HRP65-2 in C. tentans. A synthetic peptide that can disrupt the interaction between actin and HRP65-2 inhibits pol-II transcription in living cells, which suggests that the actin–HRP65-2 interaction is required for transcription in vivo (Percipalle et al., 2003). A recent study has shown that the inhibitory effect of the peptide can be counteracted by trichostatin A, which is a general inhibitor of histone deacetylases. This suggests that the actin–HRP65-2 interaction is involved in acetylation/deacetylation events. This suggestion is supported by the observation that HRP65-2 binds directly to a histone acetyltransferase (HAT) called p2D10, and the interaction between actin and HRP65-2 is required for p2D10 to associate with the transcribed chromatin (Sjölinder et al., 2005). Moreover, the association of p2D10, actin, and HRP65-2 with chromatin is sensitive to ribonuclease digestion (Percipalle et al., 2001; Kiesler et al., 2005; Sjölinder et al., 2005), which indicates that these proteins are tethered to the transcribed genes by binding to the nascent transcript. In summary, these findings suggest that actin, HRP65-2, and p2D10 become assembled into nascent pre-mRNPs during transcription. Based on these observations, it has been proposed that the actin–HRP65-2–p2D10 complex is part of the nascent pre-mRNP, and it can travel along the transcribed gene, allowing p2D10 to acetylate histone H3 (Sjölinder et al., 2005). According to this proposal, the actin–HRP65-2–p2D10 complex maintains the chromatin in a transcription-competent conformation (Fig. 1 B). This model is supported by the observations that H3 acetylation is reduced (Sjölinder et al., 2005) and transcription is inhibited (Percipalle et al., 2003) when the interaction between actin and HRP65-2 is disrupted.
A recent study by Kukalev et al. (2005) suggests that the cotranscriptional binding of actin to mRNA binding proteins with the concomitant recruitment of chromatin modifiers has been conserved throughout evolution. DNase I affinity chromatography has shown that in human cells there is a specific association between actin and heterogeneous nuclear RNP (hnRNP) U. In vitro reconstitution experiments with purified proteins have shown that the interaction between actin and hnRNP U is direct and that actin binds a short amino acid sequence that is similar to the actin binding motif of C. tentans HRP65-2. Moreover, actin must interact with hnRNP U for pol-II transcription to take place (Kukalev et al., 2005). Interestingly, hnRNP U associates with the transcription activator p300/CBP, a potent HAT (Martens et al., 2002). One hypothesis is that the actin–hnRNP U complex recruits HAT activity. The HRP65-2–p2D10 case in C. tentans is thus similar to the hnRNP U–p300/CBP case in human cells, and we suggest that actin–hnRNP complexes serve as molecular platforms for the recruitment and tethering of chromatin-modifying enzymes in both insect and mammalian cells.
The investigations described in this section support the view that actin modulates the structure of the chromatin template during the transcription of class II genes. In this context, actin functions as a platform for protein–protein interactions through a mechanism that requires ongoing RNA synthesis. This mechanism is coupled to transcription elongation and does not seem to require myosin or motor activity.
Chromatin-independent roles for actin in transcription
The role of actin in the modulation of chromatin structure does not, however, exclude more direct actions of actin on the basal transcription apparatus. For both pol-I and -II, anti-actin antibodies inhibit the transcription of naked DNA templates in transcription systems containing partially purified polymerases and the necessary transcription factors (Hofmann et al., 2004; Philimonenko et al., 2004). Further, β-actin is needed for the full activation of partially purified pol-III preparations from HeLa cells, again when a pure DNA template is used (Hu et al., 2004). These results demonstrate that actin plays a role in transcription even when chromatin is not present. This conclusion is supported by the findings that actin is associated with pol-I (Fomproix and Percipalle, 2004; Philimonenko et al., 2004), pol-II (Hofmann et al., 2004; Kukalev et al., 2005), and pol-III (Hu et al., 2004). The identities of the actin binding partners in each transcription machinery are not known, but coimmuno precipitation experiments in HeLa cells have identified three pol-III subunits associated with actin (Hu et al., 2004). Two of them, RPABC2 and -3, are present in all three RNA polymerases, and the solution of the crystal structure of pol-II shows that these two subunits are located close to each other at the surface of the polymerase (Cramer et al., 2001). Hu et al. (2004) suggested that RPABC2 and -3 form an actin binding patch that is common to all three RNA polymerases. On the other hand, coimmunoprecipitation experiments have also shown that actin binds to the COOH-terminal domain of the largest subunit of pol-II (CTD; Kukalev et al., 2005), a domain that is absent in pol-I and -III. These findings lead us to propose that actin can interact with the eukaryotic RNA polymerases in two different ways. The first way is general and is probably mediated by RPABC2 and -3, whereas the second way is specific to pol-II. The latter interaction takes place via the CTD, presumably in its phosphorylated state (Table I).
We do not yet know how actin acts at a molecular level when it associates with the RNA polymerases. Actin binds to the three RNA polymerases, as described in the previous paragraph, and anti-actin antibodies reduce pol-I, -II, and -III activities to a similar extent in transcription assays in vitro (Philimonenko et al., 2004). The roles of actin may thus be similar in all three systems. Abortive transcription-initiation assays in a pol-I system have demonstrated that anti-actin antibodies do not affect the synthesis of a 3-nt-long abortive transcript, whereas they do inhibit the subsequent elongation of the 3-nt product. This suggests that actin is not involved in initiation per se but in a process that occurs shortly after transcription initiation. This process may be promoter clearance or elongation (Philimonenko et al., 2004). Similar experiments performed in a pol-II system at a lower resolution could not determine the exact stage of transcription at which actin acts, but these experiments confirmed that actin plays an early role in the transcription of class II genes (Hofmann et al., 2004). Copurification and colocalization studies have led researchers to propose that actin plays a role in the assembly of pol-II preinitiation complexes (Hofmann et al., 2004).
Actin function: conformational switch in different scenarios?
Many questions about the mechanisms by which actin functions in transcription remain unanswered. The simplest model that is compatible with our current knowledge of nuclear actin places actin in three different scenarios. First, actin is a component of some chromatin remodeling factors. Second, actin appears to be necessary in an early step of the transcription process, a step in which actin interacts directly with the transcription apparatus. This step is independent of chromatin structure and may be common for all three RNA polymerases. Third, actin becomes incorporated into the nascent mRNPs, where it is involved in the recruitment of factors that regulate chromatin structure. The molecular function of actin may be the same in all the scenarios. The structure of actin is complex (for review see Pederson and Aebi, 2002), and actin can adopt several molecular conformations in a reversible and regulated manner (Page et al., 1998). It has been proposed that actin works as a conformational switch to control the assembly or the activity of chromatin remodeling machines (Boyer and Peterson, 2000). In agreement with this idea, it is tempting to suggest that the fundamental function of actin in transcription is to mediate dynamic protein–protein interactions and to act as an allosteric factor in the remodeling of large multimolecular complexes, such as the transcriptional apparatus or the nascent mRNP.
The transcription apparatus undergoes major conformational changes after assembly of the preinitiation complex, and these changes lead to transition into the elongation phase and to promoter clearance (for review see Dvir, 2002). We speculate that actin acts in association with the eukaryotic RNA polymerases as a molecular switch in these structural transitions.
The involvement of actin in the recruitment of chromatin-modifying factors while it is part of the nascent mRNP is a more specialized function of actin in pol-II transcription. We suggest that actin has the potential to trigger the release of chromatin modifiers from the mRNP concomitantly with the termination of transcription. The newly synthesized mRNP must undergo remodeling while the pre-mRNA is processed, surveyed, and exported, and it will be interesting to determine whether actin plays a role in these events.
Finally, we note that actin is involved in signal transduction pathways in the cytoplasm, and it is therefore interesting to consider that actin-based mechanisms of transcription regulation may sense extracellular signals via cytoplasmic changes in the actin pools. The idea that actin plays a central role in the coordination of signal transduction is not new (DeMali et al., 2003; Hall, 2005). What is new is the suggestion that actin can modulate the overall transcriptional activity of the cell in response to extracellular signals.
Acknowledgments
We thank A.-K. Östlund Farrants for critical comments on the manuscript, M. Hendzel for communicating unpublished results, and G. Farrants for correcting the English.
Our research is financed by the Swedish Research Council, the Åke Wiberg Foundation, and the Carl Tryggers Foundation.
Abbreviations used in this paper: Arp, actin-related protein; CTD, COOH-terminal domain of the largest subunit of RNA polymerase II; HAT, histone acetyltransferase; hnRNP, heterogeneous nuclear RNP; NM1, nuclear myosin 1; rDNA, ribosomal DNA.
References
- Amankwah, K.S., and U. de Boni. 1994. Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp. Cell Res. 210:315–325. [DOI] [PubMed] [Google Scholar]
- Andersen, J.S., C.E. Lyon, A.H. Fox, A.K. Leung, Y.W. Lam, H. Steen, M. Mann, and A.I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1–11. [DOI] [PubMed] [Google Scholar]
- Andersen, J.S., Y.W. Lam, A.K. Leung, S.E. Ong, C.E. Lyon, A.I. Lamond, and M. Mann. 2005. Nucleolar proteome dynamics. Nature. 433:77–83. [DOI] [PubMed] [Google Scholar]
- Bettinger, B.T., D.M. Gilbert, and D.C. Amberg. 2004. Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 5:410–415. [DOI] [PubMed] [Google Scholar]
- Boyer, L.A., and C.L. Peterson. 2000. Actin-related proteins (Arps): conformational switches for chromatin-remodeling machines? Bioessays. 22:666–672. [DOI] [PubMed] [Google Scholar]
- Cramer, P., D.A. Bushnell, and R.D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 292:1863–1876. [DOI] [PubMed] [Google Scholar]
- de Lanerolle, P., T. Johnson, and W.A. Hofmann. 2005. Actin and myosin I in the nucleus: what next? Nat. Struct. Mol. Biol. 12:742–746. [DOI] [PubMed] [Google Scholar]
- DeMali, K.A., K. Wennerberg, and K. Burridge. 2003. Integrin signaling to the actin cytoskeleton. Curr. Opin. Cell Biol. 15:572–582. [DOI] [PubMed] [Google Scholar]
- Dvir, A. 2002. Promoter escape by RNA polymerase II. Biochim. Biophys. Acta. 1577:208–223. [DOI] [PubMed] [Google Scholar]
- Egly, J.M., N.G. Miyamoto, V. Moncollin, and P. Chambon. 1984. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 3:2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomproix, N., and P. Percipalle. 2004. An actin-myosin complex on actively transcribing genes. Exp. Cell Res. 294:140–148. [DOI] [PubMed] [Google Scholar]
- Gelles, J., and R. Landick. 1998. RNA polymerase as a molecular motor. Cell. 93:13–16. [DOI] [PubMed] [Google Scholar]
- Hall, A. 2005. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33:891–895. [DOI] [PubMed] [Google Scholar]
- Hofmann, W.A., L. Stojiljkovic, B. Fuchsova, G.M. Vargas, E. Mavrommatis, V. Philimonenko, K. Kysela, J.A. Goodrich, J.L. Lessard, T.J. Hope, et al. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 6:1094–1101. [DOI] [PubMed] [Google Scholar]
- Hu, P., S. Wu, and N. Hernandez. 2004. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 18:3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, A.P., Y. Aissouni, C. Palmberg, P. Percipalle, E. Nordling, B. Daneholt, H. Jornvall, and T. Bergman. 2001. Recovery of gel-separated proteins for in-solution digestion and mass spectrometry. Anal. Chem. 73:5370–5377. [DOI] [PubMed] [Google Scholar]
- Kiesler, E., M.E. Hase, D. Brodin, and N. Visa. 2005. Hrp59, an hnRNP M protein in Chironomus and Drosophila, binds to exonic splicing enhancers and is required for expression of a subset of mRNAs. J. Cell Biol. 168:1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva, E., S.P. Drummond, M.W. Goldberg, S.A. Rutherford, T.D. Allen, and K.L. Wilson. 2004. Actin- and protein-4.1-containing filaments link nuclear pore complexes to subnuclear organelles in Xenopus oocyte nuclei. J. Cell Sci. 117:2481–2490. [DOI] [PubMed] [Google Scholar]
- Kukalev, A., Y. Nord, C. Palmberg, T. Bergman, and P. Percipalle. 2005. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 12:238–244. [DOI] [PubMed] [Google Scholar]
- Kysela, K., A.A. Philimonenko, V.V. Philimonenko, J. Janacek, M. Kahle, and P. Hozak. 2005. Nuclear distribution of actin and myosin I depends on transcriptional activity of the cell. Histochem. Cell Biol. 124:347–358. [DOI] [PubMed] [Google Scholar]
- Martens, J.H.A., M. Verlaan, E. Kalkhoven, J.C. Dorsman, and A. Zantema. 2002. Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol. Cell. Biol. 22:2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, D., G. Carrero, C. Andrin, G. de Vries, and M.J. Hendzel. 2006. Nucleoplasmic β-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J. Cell Biol. 172:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave, I.A., S.L. Reck-Peterson, and G.R. Crabtree. 2002. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71:755–781. [DOI] [PubMed] [Google Scholar]
- Page, R., U. Lindberg, and C.E. Schutt. 1998. Domain motions in actin. J. Mol. Biol. 280:463–474. [DOI] [PubMed] [Google Scholar]
- Pederson, T., and U. Aebi. 2002. Actin in the nucleus: what form and what for? J. Struct. Biol. 140:3–9. [DOI] [PubMed] [Google Scholar]
- Pederson, T., and U. Aebi. 2005. Nuclear actin extends, with no contraction in sight. Mol. Biol. Cell. 16:5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle, P., J. Zhao, B. Pope, A. Weeds, U. Lindberg, and B. Daneholt. 2001. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J. Cell Biol. 153:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle, P., A. Jonsson, D. Nashchekin, C. Karlsson, T. Bergman, A. Guialis, and B. Daneholt. 2002. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res. 30:1725–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle, P., N. Fomproix, K. Kylberg, F. Miralles, B. Bjorkroth, B. Daneholt, and N. Visa. 2003. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA. 100:6475–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestic-Dragovich, L., L. Stojiljkovic, A.A. Philimonenko, G. Nowak, Y. Ke, R.E. Settlage, J. Shabanowitz, D.F. Hunt, P. Hozak, and P. de Lanerolle. 2000. A myosin I isoform in the nucleus. Science. 290:337–341. [DOI] [PubMed] [Google Scholar]
- Philimonenko, V.V., J. Zhao, S. Iben, H. Dingova, K. Kysela, M. Kahle, H. Zentgraf, W.A. Hofmann, P. de Lanerolle, P. Hozak, and I. Grummt. 2004. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 6:1165–1172. [DOI] [PubMed] [Google Scholar]
- Rando, O.J., K. Zhao, and G.R. Crabtree. 2000. Searching for a function for nuclear actin. Trends Cell Biol. 10:92–97. [DOI] [PubMed] [Google Scholar]
- Scheer, U., H. Hinssen, W.W. Franke, and B.M. Jockusch. 1984. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 39:111–122. [DOI] [PubMed] [Google Scholar]
- Schoenenberger, C.A., S. Buchmeier, M. Boerries, R. Sutterlin, U. Aebi, and B.M. Jockusch. 2005. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J. Struct. Biol. 152:157–168. [DOI] [PubMed] [Google Scholar]
- Sjölinder, M., P. Björk, E. Söderberg, N. Sabri, A.K. Östlund Farrants, and N. Visa. 2005. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev. 19:1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., W. Wang, O.J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G.R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 95:625–636. [DOI] [PubMed] [Google Scholar]