Abstract
Protein degradation, chromatin remodeling, and membrane trafficking are critically regulated by ubiquitylation. The presence of several coexisting ubiquitin-dependent processes, each of crucial importance to the cell, is remarkable. This brings up questions on how the usage of this versatile regulator is negotiated between the different cellular processes. During proteotoxic stress, the accumulation of ubiquitylated substrates coincides with the depletion of ubiquitylated histone H2A and chromatin remodeling. We show that this redistribution of ubiquitin during proteotoxic stress is a direct consequence of competition for the limited pool of free ubiquitin. Thus, the ubiquitin cycle couples various ubiquitin-dependent processes because of a rate-limiting pool of free ubiquitin. We propose that this ubiquitin equilibrium may allow cells to sense proteotoxic stress in a genome-wide fashion.
Introduction
The archetypical protein modifier ubiquitin is a ubiquitously expressed, highly conserved polypeptide best known as a marker for intracellular protein turnover (Varshavsky, 2005). Proteasomal degradation of proteins is generally preceded by covalent tagging of proteins with a ubiquitin polymer (Hershko and Ciechanover, 1998). Ubiquitin tagging is the result of an enzymatic cascade executed by a ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin-ligating enzymes (E3; Pickart, 2001). The E1, E2, and some E3 enzymes form a thiolester linkage with ubiquitin, which is eventually conjugated by an isopeptide bond either to an internal lysine residue or to the free NH2 terminus of a target protein.
Ubiquitylation plays a critical role in many other cellular events as well (Aguilar and Wendland, 2003). Histones were the first ubiquitin-modified proteins to be identified and are the predominant ubiquitin targets in the nuclei of metazoans (Hunt and Dayhoff, 1977). Ubiquitylated histone H2A (uH2A) is required for gene silencing (de Napoles et al., 2004; Wang et al., 2004; Baarends et al., 2005). The internalization of receptors and the delivery of proteins to the multivesicular bodies are also dependent on ubiquitylation (Di Fiore et al., 2003).
Although the roles of ubiquitin in these processes have been studied in detail, the dynamic exchange of ubiquitin between these different systems is less well understood. We followed the dynamics of fluorescently tagged ubiquitin in living cells and showed that histones and other ubiquitin substrates compete for a limited pool of free ubiquitin. This links ubiquitin-dependent processes, coupling protein degradation to chromatin remodeling, and adds a dynamic dimension to ubiquitin as a general regulator of the cellular proteome.
Results and discussion
We generated a construct encoding wild-type ubiquitin with an NH2-terminal GFP tag. It has been recently shown that GFP–ubiquitin (GFP-Ub) fusions are functionally conjugated to substrates and show similar localization as endogenous ubiquitin (Qian et al., 2002). A similar fusion was made with a conjugation-deficient mutant ubiquitin lacking all internal lysine residues and the COOH-terminal glycine residue (GFP-UbK0,G76V). Western blot analysis of the total lysates of human melanoma Mel JuSo cells stably expressing these fusions confirmed that GFP-Ub was present both as free monomers (∼33 kD) and in large ubiquitin conjugates, whereas GFP-UbK0,G76V was exclusively found as free monomers (Fig. 1 A, left). Importantly, comparing the signals that were obtained when both the parental and stable Mel JuSo cell lysates were probed with the antiubiquitin antibody showed that GFP-Ub and GFP-UbK0,G76V were expressed in minute amounts compared with endogenous ubiquitin (Fig. 1 A, right). Under nonreducing conditions, the levels of free GFP-Ub and ubiquitin were lower, suggesting that a major fraction of the monomeric GFP-Ub and ubiquitin is not free, but covalently linked by reducible thiolester linkage to ubiquitylation enzymes (Fig.1 A). We consistently found that fewer ubiquitin conjugates were recovered under nonreducing conditions, which may be caused by poorer solubility of the conjugates in the absence of reducing agents.
Figure 1.
Generation and characterization of cell lines for in vivo monitoring of ubiquitin. (A) Western blot analysis of cell lysates of parental Mel JuSo cells and Mel JuSo cells stably expressing GFP-Ub or GFP-UbK0,G76V. The samples were separated under reducing and nonreducing conditions and probed with an anti-GFP antibody (left) and an antiubiquitin antibody (right). The blots were reprobed with an anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody to check for protein loading. (B) Micrographs of living GFP-Ub and GFP-UbK0,G76V cells. (C) Fluorescence micrographs of live GFP-Ub cells stained with LysoTracker red. GFP fluorescence (left), LysoTracker fluorescence (middle), and merged images (right) are shown. (bottom) Magnification of the boxed regions. (D) Mel JuSo cells expressing GFP-Ub were stained with the ubiquitin-specific antibody FK2. Native GFP fluorescence, antiubiquitin staining, a DAPI nuclear staining, and the merge of the three images are shown. Bars, 10 μm.
Microscopic analysis of living cells showed that GFP-Ub was present in both nucleus and cytosol. Although GFP-UbK0,G76V was equally distributed throughout the cytosolic and nuclear compartments, GFP-Ub levels were highest in the nucleus, where it displayed a punctuate staining with irregular granular dots, and was lower in the nucleoli (Fig. 1 B). In the cytosol, GFP-Ub was distributed in a diffuse pattern and associated with a large number of mobile punctuate structures (Fig. 1 C, top), of which many appear to be lysosomes (Fig. 1 C, bottom). The staining that was obtained with a ubiquitin-specific antibody matched the GFP fluorescence in GFP-Ub–expressing Mel JuSo cells (Fig. 1 D). Notably, because GFP-Ub forms only a small fraction of the total ubiquitin pool in these cells, GFP-Ub apparently distributes like endogenous ubiquitin.
The ubiquitin–proteasome system was functional in the presence of the GFP–Ub fusions because the cell cycle distribution pattern and the cell surface expression of stable major histocompatibility class I molecules (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200510071/DC1), two events that strongly depend on ubiquitylation machinery that is intact, were not affected.
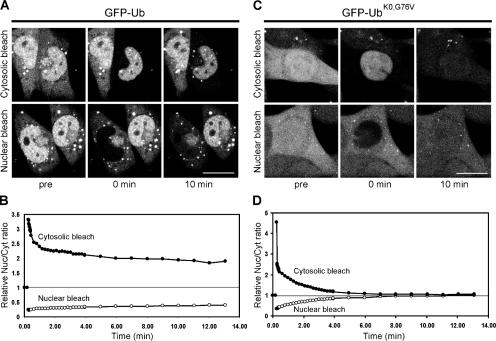
Our biochemical analysis (Fig. 1 A) and that of others (Carlson et al., 1987; Mimnaugh et al., 1997) suggested that cells contain only a limited pool of free ubiquitin. To test this in living cells, we took advantage of the fact that the molecular mass of free monomeric GFP-Ub is 33 kD, which allows passive diffusion through the nuclear pore (Talcott and Moore, 1999), unless it is incorporated into larger complexes. We photobleached GFP-Ub and GFP-UbK0,G76V in the cytosol or nucleus and quantified the redistribution of fluorescence from the nonbleached compartment in a fluorescence loss in photobleaching protocol, basically using the nuclear pore as a molecular sieve to distinguish free from conjugated GFP-Ub molecules (Koster et al., 2005). GFP-Ub displayed biphasic redistribution between the two compartments, with a fast component in the first minute followed by a major slow component (Fig. 2, A and B). The presence of a small fraction that is rapidly exchanged during the first minute is in agreement with a small amount of free monomeric GFP-Ub, as detected biochemically (Fig. 1 A). The slow exchange persisted with similar kinetics throughout the recording, suggesting the continuous generation of freely diffusing GFP-Ub. Rapid redistribution was observed with the GFP-UbK0,G76V, with a complete exchange of fluorescence within 6 min confirming that this monomeric form efficiently diffuses through the nuclear pore (Fig. 2, C and D).
Figure 2.
Limited exchange of ubiquitin between nuclear and cytosolic compartments. (A) GFP-Ub cells were photobleached in either the complete cytosol (top) or the complete nucleus (bottom) and recovery was measured in both compartments. (B) Quantification of the photobleaching experiments with GFP-Ub cells. The ratios of nuclear fluorescence to cytosolic fluorescence are plotted for cytosolic bleaching (shaded circles) and nuclear bleaching (open circles). (C) GFP-UbK0,G76V cells were photobleached in either the complete cytosol (top) or the complete nucleus (bottom) and recovery was measured in both compartments. (D) Quantification of the photobleaching experiments with GFP-UbK0,G76V cells. The relative ratios of nuclear fluorescence to cytosolic fluorescence are plotted for cytosolic bleaching (shaded circles) and nuclear bleaching (open circles). Bars, 20 μm.
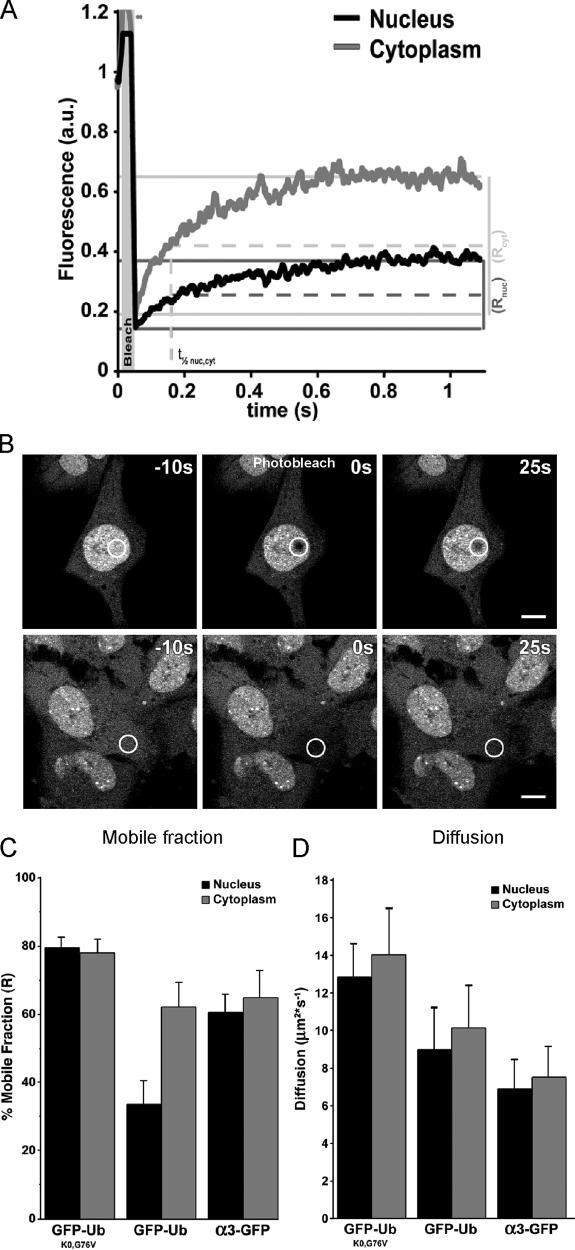
The slow exchange of GFP-Ub between the nuclear and cytosolic compartments suggests that the vast majority of ubiquitin is incorporated into large complexes that cannot pass the nuclear pore. We performed FRAP analysis, which allows determination of protein diffusion and mobility rates (Reits and Neefjes, 2001). The Brownian motion of particles is related to their size, and large polyubiquitin complexes are thus expected to diffuse considerably slower than free ubiquitin. For comparison, we included a Mel JuSo cell line expressing a GFP-tagged α3 subunit of the proteasome, which is a large, freely diffusible complex (Reits et al., 1997). Coimmunoprecipitation and sucrose gradient experiments confirmed that the α3 subunit is incorporated into the proteasome particle (unpublished data). FRAP analysis revealed both the diffusion rate and the fraction of mobile proteins. A large portion was mobile in the cytosol, unlike GFP-Ub in the nucleus, which is where the majority of GFP-Ub was immobile (Fig. 3, A and B). Quantitative analysis of the FRAP data revealed a much larger fraction of immobile nuclear GFP-Ub, as compared with the cytosolic GFP-Ub (Fig. 3 C). An immobile GFP-Ub fraction in the cytosol is likely to be a consequence in part of the role of ubiquitin in membrane trafficking (Di Fiore et al., 2003). Moreover, ubiquitylated proteins can bind to cytoskeletal-associated proteins (Kawaguchi et al., 2003) and form cytosolic clusters (Bjorkoy et al., 2005). Some 70% of GFP-Ub is immobile in the nucleus, which supports the notion that a major fraction of GFP-Ub is conjugated to histones (see Fig. 4).
Figure 3.
Dynamics of ubiquitin in nucleus and cytosol. (A) FRAP curves for GFP-Ub in the nucleus (black line) and cytoplasm (gray line). Mobile fractions (R) are indicated for both nuclear (dark gray) and cytoplasmic bleached area (light gray). The interpolations for the t 1/2 are indicated with dashed lines in the same respective gray values. (B) Confocal images of a FRAP experiment in the nucleus (top) or in the cytoplasm (bottom) before, immediately after, and 25 s after a 2-s photobleaching. Bars, 10 μm. (C) Mobile fractions (R) of GFP-UbK0,G76V, GFP-Ub, and proteasome α3-GFP in stably transfected Mel JuSo cells. Diffusion was measured in both the nucleus (black bars) and the cytoplasm (gray bars). Error bars are SD (n > 10). (D) Diffusion rates of GFP-UbK0,G76V, GFP-Ub, and α3-GFP in stably transfected Mel JuSo cells. Diffusion was measured in both the nucleus (black bars) and the cytoplasm (gray bars). Error bars are SD (n > 10).
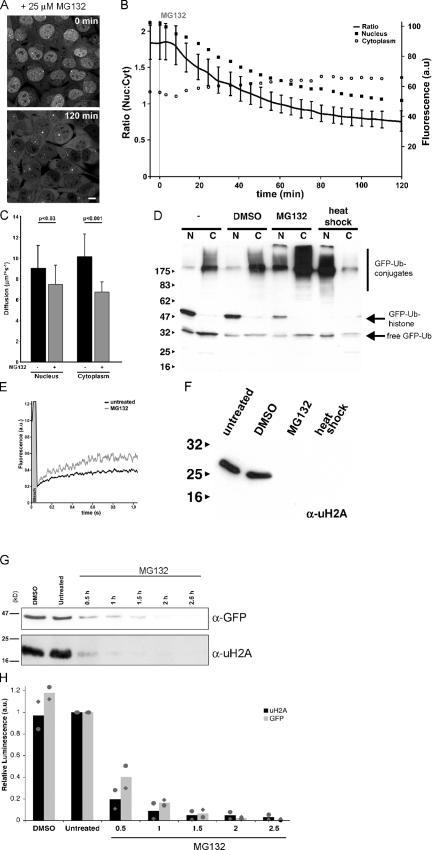
Figure 4.
Accumulation of polyubiquitylated proteins coincides with depletion of uH2A and chromatin remodeling. (A) Fluorescence images of living GFP-Ub cells before and after 2 h incubation with 25 μM MG132. Bar, 10 μm. (B) Quantification of GFP-Ub levels in the cytoplasm (open circles) and nucleus (closed squares) during proteasome inhibition with 25 μM MG132. Black line is the ratio of the nucleus to the cytoplasm as plotted on the left y axis. The relative fluorescence of the nucleus and cytoplasm is plotted on the right y axis. (C) Diffusion rates of GFP-Ub in nucleus and cytoplasm without treatment (black bars) and after 2 h of MG132 treatment (gray bars). P values are indicated (unpaired t tests). Error bars represent the mean and SD of 15 independent cells in one representative experiment. (D) Western blot analysis of nuclear (N) and cytosolic (C) fractions of GFP-Ub cells. Cells were left untreated or treated for 2 h with DMSO, MG132, or a heat shock, and nuclear (N) and cytosolic (C) fractions were isolated. Nuclear fractions contained, on average, two times less protein than the cytosolic fractions. For analysis, 15- and 30-μg proteins were loaded for the nuclear and cytosolic fractions, respectively. Membranes were probed with an anti-GFP antibody. Molecular mass markers are indicated. (E) FRAP curves for GFP-Ub in the nucleus of untreated cells (black line) and after 2 h incubation with MG132 (gray line). (F) Western blot analysis of uH2A in GFP-Ub cells left untreated or exposed for 2 h to DMSO, MG132, or a heat shock. The membranes were probed with an anti-uH2A antibody. Molecular mass markers are indicated. (G) Lysates of Mel JuSo cells expressing GFP-Ub that were treated for various time periods with MG132 were probed with an anti-GFP antibody (top) and uH2A antibody (bottom). (H) Quantification of two independent experiments as shown in G. The values of experiment 1 (circles), experiment 2 (diamonds), and the mean of the two experiments (bars) shown. The values were standardized to the intensities of the corresponding band in untreated cells. Deubiquitylation of GFP-Ub–histone and uH2A followed similar kinetics.
The monomeric GFP-UbK0,G76V diffused rapidly through the cytosol and nucleus, whereas the GFP-tagged proteasome moved relatively slow in both compartments, in line with their size differences. Consistent with the notion that ubiquitin is incorporated in large ubiquitin chains (Hershko and Ciechanover, 1998), the GFP-Ub pool had a surprisingly slow diffusion rate in the nucleus and cytosol, especially when compared with the proteasome (Fig. 3 D).
Biochemical analysis has revealed that proteasome inhibitor treatment and heat shock can deplete histones from ubiquitin (Carlson et al., 1987; Mimnaugh et al., 1997). To reveal the dynamics of this process, we monitored GFP-Ub in living cells after the administration of the proteasome inhibitor MG132. A rapid accumulation of GFP-Ub in the cytosol and the formation of aggresomes in the perinuclear region were observed within 2 h, which was accompanied by a profound loss of nuclear GFP-Ub (Fig. 4 A). Staining of fixed cells with the ubiquitin-specific FK2 antibody revealed a similar redistribution of endogenous ubiquitin (Qian et al., 2002; unpublished data). During the 2-h inhibitor treatment, we observed a steady and gradual decline in nuclear GFP-Ub, coinciding with an increase in cytosolic fluorescence (Fig. 4 B). FRAP analysis demonstrated that the mobile pool of GFP-Ub in the nuclear and cytosolic compartment was further decelerated by the inhibitor treatment (Fig. 4 C), which correlated with an accumulation of ubiquitin conjugates, as well as with a shift of the conjugates to higher molecular masses (Fig. 4 D). Both an increase in the amount of polyubiquitylated proteins, as well as an increase in the size of the polyubiquitin changes, is likely responsible for the reduced velocity of ubiquitin in MG132-treated cells. Notably, in the presence of MG132, the diffusion was reduced to velocities that were in the same range as proteasomes, emphasizing the considerable size of these polyubiquitin complexes or direct association with proteasomes (compare Fig. 3 B and Fig. 4 C). The putative GFP-Ub–modified histones were only found in the nucleus and strongly declined during inhibitor treatment (Fig. 4 D). A similar reduction in the GFP-Ub–histone band was observed during heat shock, which is another form of proteotoxic stress, although under this condition polyubiquitylated material primarily accumulated in the nucleus (Fig. 4 D). A gradual redistribution of endogenous ubiquitin from the nuclear to the cytosol compartment was also evident when lysates of cells harvested at various times after inhibitor administration were probed with a ubiquitin-specific antibody (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200510071/DC1). Notably, proteasome inhibitor treatment reduced the nuclear immobile pool of GFP-Ub, which is in line with a reduction in histone-conjugated ubiquitin (Fig. 4 E). Western blot analysis confirmed a decrease in endogenous uH2A levels under these stress conditions that was analogous to GFP-Ub–histone (Fig. 4 F). To further test whether GFP-Ub correctly reflected the behavior of endogenous ubiquitin in the process of MG132-driven histone deubiquitylation, cells were incubated with MG132 and histones were analyzed at various periods after proteasome inhibition. Both GFP-Ub–histone and uH2A were quantified and followed similar kinetics of deubiquitylation (Fig. 4, G and H). Half of the histones had released ubiquitin or GFP-Ub ∼30 min after proteasome inhibition. Chromatin of proteasome inhibitor–treated and heat-shocked cells was less sensitive to staphylococcal nuclease (Fig. S3), suggesting a general condensation of nucleosomes that is similar to what has been observed previously for cells subjected to heat shock (Levinger and Varshavsky, 1982).
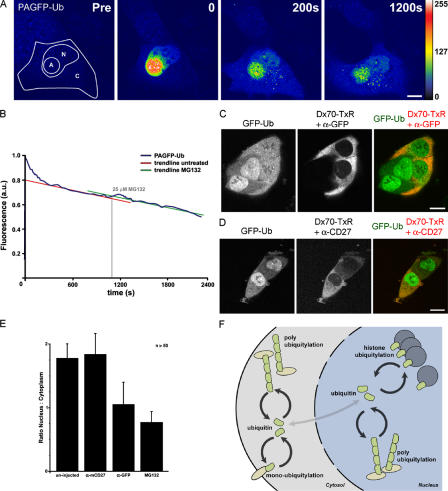
To gain insight into the mechanism responsible for depletion of uH2A, we followed the redistribution of ubiquitin during proteotoxic stress in living cells. The GFP in the fusion constructs was replaced by a photoactivatable GFP (PAGFP; Patterson and Lippincott-Schwartz, 2002), and PAGFP-Ub was photoactivated in a confined region in the nucleus (Fig. 5 A and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200510071/DC1). Although most of the fluorescence was maintained in the photoactivated region, a small fraction of the photoactivated PAGFP-Ub immediately diffused to other regions in the nucleus, probably because of rapid redistribution of a small pool of free PAGFP-Ub (Fig. 2, A and B). Subsequently, fluorescence slowly appeared in the cytosolic compartment coinciding with a gradual decrease in nuclear fluorescence. The fluorescent PAGFP-Ub distributed homogenously in the cytosol and on intracellular punctuate structures. In line with the notion that the vast majority of nuclear PAGFP-Ub was conjugated to histones, PAGFP-Ub only slowly disappeared from the photoactivated region in the nucleus. We monitored the disappearance of the immobile PAGFP-Ub as a measure for histone deubiquitylation. Administration of proteasome inhibitor did not affect the rate of disappearance of the immobile nuclear PAGFP-Ub from the photoactivated region (Fig. 5 B), which suggests that the rate of histone deubiquitylation is not altered by proteasome inhibition.
Figure 5.
Competition for free ubiquitin causes depletion of uH2A during proteotoxic stress. (A) Confocal images of PAGFP-Ub in Mel JuSo cells before photoactivation (Pre) and three time points after photoactivation (0, 200, and 1,200 s). In the image before activation (Pre), the contours of the cell (C), the nucleus (N), and the region to be activated (A) are indicated. The look-up table is provided on the right (see also Video 1). (B) Quantification of fluorescence in an activated nuclear region visualizes the decay of PAGFP-Ub before and after the addition of MG132. PAGFP fluorescence is depicted with the blue line, whereas trend lines derived from similar experiments with untreated or MG132-treated cells are shown in red and green, respectively. (C) Confocal images of GFP-Ub cells injected with dextran–Texas red and anti-GFP antibody or (D) injected with dextran–Texas red and anti-mCD27 antibody. (E) Quantifications of the relative fluorescence ratio between the nucleus and the cytoplasm in control, anti-mCD27–injected, anti-GFP–injected, or MG132-treated cells. Error bars are standard deviations (n > 50). (F) Schematic representation of the dynamic ubiquitin equilibrium in the cell. Video 1 is available at http://www.jcb.org/cgi/content/full/jcb.200510071/DC1. Bars: (A) 10 μm; (C and D) 20 μm.
Alternatively, the redistribution of ubiquitin may be the result of competition of two classes of ubiquitin substrates, i.e., proteasome substrates and histones, for the rate-limiting pool of free ubiquitin. If the loss of histone-conjugated ubiquitin in the nucleus is the result of limiting free ubiquitin levels, experimental introduction of another ubiquitin competitor should have a similar effect. Indeed, microinjection of a GFP-specific antibody in the cytosol of GFP-Ub–expressing cells caused the accumulation of GFP-Ub in the cytosol and the depletion of nuclear GFP-Ub, which is very similar to proteotoxic stress (Fig. 5, C and E). An irrelevant antibody did not affect the distribution of GFP-Ub (Fig. 5, D and E). These data show that changes in the ubiquitin equilibrium can dramatically affect various ubiquitin-dependent processes.
Our data reveal a new dimension of ubiquitin-dependent regulation as the result of a delicate ubiquitin equilibrium (Fig. 5 F). This ubiquitin equilibrium may be a reflection of the constraints of the heavily used ubiquitylation system by various ubiquitin-dependent processes. Alternatively, changes in the cellular proteome as a consequence of the depletion of ubiquitylated histones may aid the cellular stress response. It has been shown that the decrease in the levels of ubiquitylated histones during proteotoxic stress causes major changes in gene expression (Carlson et al., 1987; Mimnaugh et al., 1997). In fact, the depletion of ubiquitylated histones is a rapid response, and the first changes can already be observed within 5 min. Cellular stress is apparently rapidly translated into chromatin alterations, which are likely to affect gene expression. Cross-talk between these ubiquitin-dependent processes by means of limiting free ubiquitin levels may be of functional significance, as it may integrate diverse mechanisms in the combined effort to adapt the cellular proteome to the altering intracellular environment.
Materials and methods
Cell culture and constructs
Wild-type Ub and the UbK0,G76V mutant were cloned into EGFP-C1 vector (CLONTECH Laboratories, Inc.) and PAGFP-C1 vector (gift from J. Lippincott-Schwarz, National Institutes of Health, Bethesda, MD) and transfected into the human melanoma cell line Mel JuSo. Stable cell lines were generated under the selection of 1 mg/ml neomycin containing Iscove's DME supplemented with penicillin/streptomycin and 8% FCS (Invitrogen). For live cell imaging, cells were either cultured on 24-mm glass coverslips or cultivated in 0.17-mm Delta T dishes (Bioptechs). Before microscopic analysis, the culture medium was covered with a thin layer of mineral oil (Sigma-Aldrich) to prevent evaporation of the medium during recording. Lysosomes were stained by incubating cells with 50 nM LysoTracker red (Invitrogen). The proteasome inhibitor MG132 (Sigma-Aldrich) was dissolved in DMSO and used at a 25-μM concentration, unless otherwise stated. Heat shock was induced by incubating the cells for 3 h at 42°C.
Immunostainings
Cells were cultured on 15-mm glass coverslips, fixed with 3.7% formaldehyde for 10 min at room temperature, permeabilized with 0.5% Triton X-100 for 2 min, and immunostained in phosphate-buffered saline with 0.5% bovine serum albumin. FK2 antibody (Affinity BioReagents, Inc.) was used at a ratio of 1:1,000. 2 ng/ml DAPI (Sigma-Aldrich) was added during secondary antibody incubation with goat anti–mouse-TxR (Invitrogen).
Western blot analysis
Parental GFP-Ub, stable GFP-Ub, and GFP-UbK0,G76V Mel JuSo cells were washed with phosphate-buffered saline and trypsinized. Cells were lysed in SDS-PAGE sample buffer. Proteins were separated by SDS-PAGE, transferred onto nitrocellulose or PVDF membranes, and probed with two different rabbit polyclonal antibodies against GFP (Invitrogen; van Ham et al., 1997) or a rabbit polyclonal antibody against ubiquitin (DakoCytomation and Sigma-Aldrich, respectively). The filters were reprobed with a mouse monoclonal antibody against glyceraldehyde-3-phosphate dehydrogenase (Fitzgerald Industries, Intl.) as a control for equal protein loading. After incubation with peroxidase-conjugated secondary antibodies, the blots were developed by enhanced chemiluminescence (GE Healthcare).
For separation of nuclei and cytosol, cells were scraped in a buffer containing 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 50 mM Hepes, pH 7.0, 1 mM EGTA, and 0.2% Triton X-100 supplemented with protease inhibitors and 50 mM N-ethylmaleimide. Cells were lysed for 10 min, and nuclei were pelleted by centrifugation for 5 min at 1,000 g. The supernatant is the cytosolic fraction; nuclei were resuspended in a buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% SDS supplemented with 50 mM N-ethylmaleimide and sonicated on ice to disrupt DNA.
Live cell imaging
For fluorescence loss in photobleaching experiments, Mel JuSo cells were cultured in 0.17-mm Delta T dishes (Bioptechs). Confocal laser scanning microscopy was performed with an LSM 510 META with a Plan-Apochromat 63× oil objective, NA 1.4 (both Carl Zeiss MicroImaging, Inc.), equipped with a cell culture stage (Bioptechs) at 35°C. After photobleaching of the GFP fluorescence by exposure of selected regions to 488-nm laser with 100% intensity for 30 iterations, images were obtained every 10 s during a time frame of 4 min, followed by 10 images during a time frame of 10 min. Images were processed using the LSM software. Fluorescence intensities were measured using ImageJ software (National Institutes of Health). The relative fluorescence ratio between the nucleus and cytoplasm was averaged from three recordings. For line-scan FRAP experiments, we used a confocal system (TCS SP2; Leica) equipped with an external bleaching laser and a heating ring to keep the cells at 37°C. PAGFP-Ub was transiently expressed in Mel JuSo cells. In the photoactivation step, PAGFP was activated by applying a single pulse to a small region in the cell with 405-nm laser light at full intensity. For photoactivation experiments, we used a TCS SP2 AOBS system equipped with HCX PL APO and HCX PL APO lbd.bl 63× objective lenses, both with an NA of 1.4 (all Leica). Quantification was done with physiology software version 2.61 (Leica). FRAP data was analyzed as previously described (Reits and Neefjes, 2001).
Antibody injection
For antibody injection, cells were seeded on 15-mm glass coverslips. Cells were microinjected with a mixture containing 1 mg/ml lysine-fixable 70-kD Dextran–Texas red (Invitrogen) and 1 mg/ml of purified polyclonal rabbit anti-GFP antibody (van Ham et al., 1997) or purified polyclonal rabbit anti-mCD27 (gift from J. Borst, The Netherlands Cancer Institute, Amsterdam, Netherlands). Microinjections were done on an inverse epifluorescence microscope (Axiovert 200; Carl Zeiss MicroImaging, Inc.) equipped with a manipulator 5171/transjector 5246 system (Eppendorf) and a 37°C heated ring. After microinjection, cells were cultured for another 2 h and fixed with 3.7% formaldehyde for 10 min at room temperature.
Online supplemental material
Fig. S1 shows the analysis of functionality for the ubiquitin–proteasome system, Fig. S2 shows changes in ubiquitin distribution during proteasome inhibitor treatment, Fig. S3 shows changes in nucleosome condensation during proteotoxic stress, and Video 1 shows the distribution of PAGFP-Ub after photoactivation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200510071/DC1.
Supplementary Material
Acknowledgments
We thank Lauran Oomen and Lennert Janssen for technical assistance, Jennifer Lippincott-Schwartz for the PAGFP plasmid, Kees Jalink for support with line-scan FRAP, Jannie Borst for the CD27 antibody, Fred van Leeuwen for providing help with the nuclease assay, Coen Kuijl for his help with image analysis software, and Steven Bergink, Deborah Hoogstraten, Wim Vermeulen, and the members of the Dantuma and Neefjes laboratories for their helpful suggestions.
This work was supported by the Swedish Research Council, the Swedish Cancer Society, the Netherlands Cancer Society, the Wallenberg foundation, and the Karolinska Institutet. F.A. Salomons is supported by the Nordic Center of Excellence in Neurodegeneration and the Marie Curie Research Training Network (MRTN-CT-2004-512585). The Netherlands Organization for Scientific Research supported a sabbatical stay for N.P. Dantuma in the laboratory of J. Neefjes. N.P. Dantuma is supported by the Swedish Research Council. The authors declare that there is no conflict of interest.
N.P. Dantuma and T.A.M. Groothius contributed equally to this paper.
Abbreviations used in this paper: GFP-Ub, GFP-ubiquitin; PAGFP, photoactivatable GFP; uH2A, ubiquitylated histone H2A.
References
- Aguilar, R.C., and B. Wendland. 2003. Ubiquitin: not just for proteasomes anymore. Curr. Opin. Cell Biol. 15:184–190. [DOI] [PubMed] [Google Scholar]
- Baarends, W.M., E. Wassenaar, R. van der Laan, J. Hoogerbrugge, E. Sleddens-Linkels, J.H. Hoeijmakers, P. de Boer, and J.A. Grootegoed. 2005. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell. Biol. 25:1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy, G., T. Lamark, A. Brech, H. Outzen, M. Perander, A. Overvatn, H. Stenmark, and T. Johansen. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, N., S. Rogers, and M. Rechsteiner. 1987. Microinjection of ubiquitin: changes in protein degradation in HeLa cells subjected to heat-shock. J. Cell Biol. 104:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles, M., J.E. Mermoud, R. Wakao, Y.A. Tang, M. Endoh, R. Appanah, T.B. Nesterova, J. Silva, A.P. Otte, M. Vidal, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 7:663–676. [DOI] [PubMed] [Google Scholar]
- Di Fiore, P.P., S. Polo, and K. Hofmann. 2003. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell Biol. 4:491–497. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479. [DOI] [PubMed] [Google Scholar]
- Hunt, L.T., and M.O. Dayhoff. 1977. Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem. Biophys. Res. Commun. 74:650–655. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, Y., J.J. Kovacs, A. McLaurin, J.M. Vance, A. Ito, and T.P. Yao. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 115:727–738. [DOI] [PubMed] [Google Scholar]
- Koster, M., T. Frahm, and H. Hauser. 2005. Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies. Curr. Opin. Biotechnol. 16:28–34. [DOI] [PubMed] [Google Scholar]
- Levinger, L., and A. Varshavsky. 1982. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 28:375–385. [DOI] [PubMed] [Google Scholar]
- Mimnaugh, E.G., H.Y. Chen, J.R. Davie, J.E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry. 36:14418–14429. [DOI] [PubMed] [Google Scholar]
- Patterson, G.H., and J. Lippincott-Schwartz. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533. [DOI] [PubMed] [Google Scholar]
- Qian, S.B., D.E. Ott, U. Schubert, J.R. Bennink, and J.W. Yewdell. 2002. Fusion proteins with COOH-terminal ubiquitin are stable and maintain dual functionality in vivo. J. Biol. Chem. 277:38818–38826. [DOI] [PubMed] [Google Scholar]
- Reits, E.A., and J.J. Neefjes. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3:E145–E147. [DOI] [PubMed] [Google Scholar]
- Reits, E.A., A.M. Benham, B. Plougastel, J. Neefjes, and J. Trowsdale. 1997. Dynamics of proteasome distribution in living cells. EMBO J. 16:6087–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talcott, B., and M.S. Moore. 1999. Getting across the nuclear pore complex. Trends Cell Biol. 9:312–318. [DOI] [PubMed] [Google Scholar]
- van Ham, S.M., E.P. Tjin, B.F. Lillemeier, U. Gruneberg, K.E. van Meijgaarden, L. Pastoors, D. Verwoerd, A. Tulp, B. Canas, D. Rahman, et al. 1997. HLA-DO is a negative modulator of HLA-DM-mediated MHC class II peptide loading. Curr. Biol. 7:950–957. [DOI] [PubMed] [Google Scholar]
- Varshavsky, A. 2005. Regulated protein degradation. Trends Biochem. Sci. 30:283–286. [DOI] [PubMed] [Google Scholar]
- Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst, R.S. Jones, and Y. Zhang. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 431:873–878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.