Abstract
Sterols are moved between cellular membranes by nonvesicular pathways whose functions are poorly understood. In yeast, one such pathway transfers sterols from the plasma membrane (PM) to the endoplasmic reticulum (ER). We show that this transport requires oxysterol-binding protein (OSBP)–related proteins (ORPs), which are a large family of conserved lipid-binding proteins. We demonstrate that a representative member of this family, Osh4p/Kes1p, specifically facilitates the nonvesicular transfer of cholesterol and ergosterol between membranes in vitro. In addition, Osh4p transfers sterols more rapidly between membranes containing phosphoinositides (PIPs), suggesting that PIPs regulate sterol transport by ORPs. We confirmed this by showing that PM to ER sterol transport slows dramatically in mutants with conditional defects in PIP biosynthesis. Our findings argue that ORPs move sterols among cellular compartments and that sterol transport and intracellular distribution are regulated by PIPs.
Introduction
Though the proper distribution of lipids among organelles is critical for numerous cellular functions, exactly how lipids are sorted and moved among intracellular membranes remains poorly understood. Some lipid sorting likely occurs during vesicular trafficking (Sprong et al., 2001; Holthuis and Levine, 2005). For example, sphingomyelin and cholesterol are partially excluded from COPI-coated vesicles, a process which may contribute to the enrichment of these lipids in the late Golgi network and the plasma membrane (PM; Brugger et al., 2000). In addition, many classes of lipids are transferred between organelles by nonvesicular pathways. In most cases, the molecular mechanisms and regulation of these pathways have not been characterized.
It has been known for some time that sterols, such as cholesterol in mammals and ergosterol in yeast, can move between cellular compartments via nonvesicular pathways whose functions are poorly understood. Treating mammalian cells with brefeldin A, which inhibits protein secretion by disassembling the Golgi complex (Misumi et al., 1986; Doms et al., 1989; Lippincott-Schwartz et al., 1989), only slightly slows the delivery of newly synthesized cholesterol from the ER to the PM (Kaplan and Simoni, 1985; Urbani and Simoni, 1990; Heino et al., 2000). Similar results were obtained in yeast using sec mutants with conditional defects in the proteins required for vesicular trafficking (Baumann et al., 2005; Schnabl et al., 2005). We have found that the movement of exogenous sterols from the PM to the ER sterol in yeast is also not blocked in sec mutants (Li and Prinz, 2004). Nonvesicular transport pathways move sterols between other cellular compartments as well (Liscum and Munn, 1999; Maxfield and Wustner, 2002; Prinz, 2002; Soccio and Breslow, 2004; Holthuis and Levine, 2005). The proteins required for these nonvesicular transport pathways have not been identified. In mammals, nonvesicular cholesterol transport could be facilitated by lipid transfer proteins known to bind sterols, including some StART domain–containing proteins or SCP2 homologues (Holthuis and Levine, 2005). However, the yeast Saccharomyces cerevisiae lacks homologues of these proteins. We wondered if another class of lipid-binding proteins, the oxysterol-binding protein (OSBP)–related proteins (ORPs), might facilitate nonvesicular sterol transport in yeast.
OSBP, which is the founding member of the ORP family, was identified as a cytosolic receptor for oxysterols (Kandutsch and Shown, 1981). In mammals, these compounds down-regulate cholesterol biosynthesis and uptake and have been implicated in several other cellular processes (Schroepfer, 2000). Oxysterols may also modulate sterol biosynthesis in S. cerevisiae (Gardner et al., 2001). The cloning of OBSP (Dawson et al., 1989) was followed by the identification of a large family of ORPs, including 16 in humans and 7 in S. cerevisiae (Olkkonen and Levine, 2004). All of these proteins contain an OSBP-related domain, which binds oxysterols and other lipids. Most also contain one or more other domains, including pleckstrin homologue (PH) domains (which bind phosphoinositides [PIPs]), the ER-targeting motif FFAT, ankyrin repeats, and transmembrane domains; these domains serve to localize these proteins to various cellular compartments (Levine and Munro, 1998, 2001; Loewen et al., 2003; Olkkonen and Levine, 2004). In addition, the localization of ORPs is not static; OBSP alters its intracellular location in response to oxysterols (Ridgway et al., 1992).
Some ORPs may function as lipid sensors that integrate lipid metabolism with other cellular processes. It has recently been shown that OSBP acts as a sensor that regulates two phosphatases in a cholesterol-dependent manner (Wang et al., 2005a). In addition, two yeast ORPs (Osh6p and Osh7p) interact with Vps4p, which is a member of the AAA ATPase family, and could regulate its function in response to cellular sterol levels (Wang et al., 2005b). It has also been suggested that ORPs are lipid transfer proteins (Beh et al., 2001; Levine and Munro, 2001; Olkkonen, 2004). We have recently solved the structure of the yeast ORP Osh4p/Kes1p and found that it looks remarkably like other lipid transfer proteins; it binds a single sterol molecule in a hydrophobic binding pocket covered by a “lid” domain (Im et al., 2005). A role for some yeast ORPs (called Osh proteins) in sterol transport is also supported by studies on mutants missing one or more of these proteins; intracellular sterol distribution is severely altered in mutants missing all of these proteins (Beh et al., 2001; Beh and Rine, 2004). We investigated the role of these proteins in the nonvesicular transport of exogenous sterols from the PM to the ER in yeast.
Results
Osh proteins are needed for rapid PM to ER sterol transport
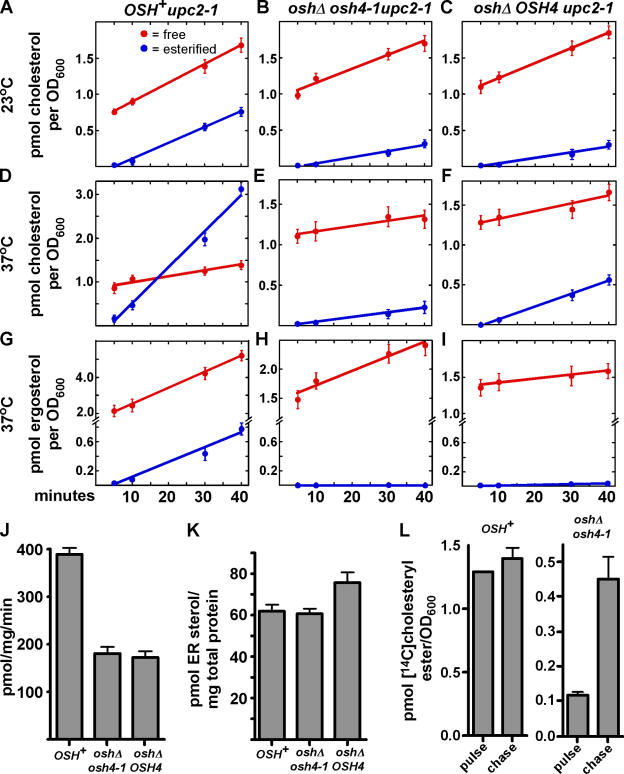
We measured the rate of PM to ER sterol transport in mutants lacking Osh proteins. These studies exploited the ER localization of the enzymes (known as acyl-coenzyme A:cholesterol acyltransferases [ACATs]) that convert free sterols to steryl esters (Zweytick et al., 2000); the esterification of exogenous radiolabeled sterol indicates that the sterol has been moved to the ER. Because S. cerevisiae does not take up exogenous sterol during aerobic growth, we use strains that have an altered allele of a transcription factor (upc2-1) that permits aerobic sterol uptake (Crowley et al., 1998). These strains take up and use several sterols, including cholesterol and ergosterol. Both of these sterols support the growth of yeast mutants that cannot make sterols. Because the Osh proteins likely have overlapping functions (Beh et al., 2001), we first measured PM to ER sterol transport in a mutant lacking all of these proteins. Yeast requires any one of the seven Osh proteins for viability (Beh et al., 2001). Therefore, we used a strain (oshΔ osh4-1) that has a temperature-sensitive allele of one of the OSH genes (osh4-1) and deletions of the other six OSH genes (Beh and Rine, 2004). At permissive temperatures, this strain esterifies exogenous cholesterol at less than half the rate of a strain that has all the Osh proteins (OSH +; Fig. 1, A and B). At a nonpermissive temperature the oshΔ osh4-1 strain esterifies exogenous cholesterol more than seven times more slowly than the OSH + strain, despite the fact that the two strains take up similar amounts of free cholesterol (Fig. 1, D and E). We also examined the esterification rate of exogenous ergosterol, which we previously showed is transported to the ER much more slowly than exogenous cholesterol (Li and Prinz, 2004). The esterification of exogenous 3H-ergosterol dramatically slows in oshΔ osh4-1 cells; at 37°C, 3H-ergosteryl esters were not detectable (Fig. 1, G and H). To facilitate comparisons of the strains in different conditions, we have calculated their relative rates of esterification of exogenous sterols (Table I). It should be noted that we have previously shown that the fraction of exogenous sterol that becomes esterified over time is not affected by the total amount taken up; therefore, comparisons between strains that take up slightly different amounts of sterol are valid (Li and Prinz, 2004). The small amount of exogenous cholesterol that becomes esterified in oshΔ osh4-1 at elevated temperature suggests that other pathways may also move cholesterol to the ER inefficiently. Interestingly, we also noted that OSH + cells take up and esterify cholesterol more slowly at low temperatures (compare Fig. 1 A to Fig. 1 D and Fig. 2 A), perhaps because steryl ester synthease activity decreases or membrane fluidity changes. Collectively, these findings show that efficient esterification of exogenous sterols requires Osh4p and likely other Osh proteins, particularly at elevated temperatures.
Figure 1.
Osh4p and other Osh proteins are required for efficient nonvesicular PM to ER sterol transport. (A–I) Cells were grown at either 23°C (A–C) or shifted to 37°C for 1 h (D–I) and 14C-cholesterol or 14C-ergosterol was added to medium. To ensure that the strains took up similar amounts of 14C-cholesterol, the OSH + strain (A and D) was given 1.0 μM 14C-cholesterol and the oshΔ strains (B,C,E,F) were given 4.0 μM 14C-cholesterol. All three strains were given14C-ergosterol at 2.5 μM. At the indicated times, cells were harvested, lipids were extracted, and the amount of free and esterified radiolabeled sterol in the cells was quantitated. (J) ACAT activity of the OSH + and oshΔ strains. Cells were grown at 25°C and shifted to 37°C for 1 h, and the ACAT activity of protein extracts from each strain was determined. (K) To estimate the amount of endogenous sterol in the ER, lysates from the OSH + and oshΔ cells were reacted to completion with 14C-oleoyl CoA. The amount of sterol in the ER was calculated from the amount of 14C-steryl ester formed. (L) To determine if 14C-cholesterol esters are hydrolyzed after they are formed, oshΔ osh4-1 and OSH + cells were grown at 23°C, shifted to 37°C for 30 min, and 2.0 μM 14C-cholesterol was added to the medium. After 30 min, the 250-μM unlabeled cholesterol was added to the medium and cells grown for an additional 40 min. The amount of 14C-cholesteryl esters per OD600 of cells before (pulse) and after addition of unlabeled cholesterol (chase) was determined. In all graphs, values are the mean of two determinations. Error bars indicate the SEM. n = 2.
Table I.
Relative esterification rate of strains used in this study
| Strain | Exogenous sterol |
Growth temperature |
Relative esterification ratea |
|---|---|---|---|
| °C | % | ||
| upc2-1 | cholesterol | 23 | 0.94 ± 0.23 |
| upc2-1 oshΔ osh4-1 | cholesterol | 23 | 0.37 ± 0.042 |
| upc2-1 oshΔ OSH4 | cholesterol | 23 | 0.37 ± 0.072 |
| upc2-1 | cholesterol | 37 | 1.9 ± 0.27 |
| upc2-1 oshΔ osh4-1 | cholesterol | 37 | 0.27 ± 0.064 |
| upc2-1 oshΔ OSH4 | cholesterol | 37 | 0.44 ± 0.10 |
| upc2-1 | ergosterol | 37 | 0.39 ± 0.067 |
| upc2-1 oshΔ osh4-1 | ergosterol | 37 | N.D. |
| upc2-1 oshΔ OSH4 | ergosterol | 37 | 0.042 ± 0.003 |
| upc2-1 | cholesterol | 37 | 2.1 ± 0.24 |
| upc2-1 pik1-83 | cholesterol | 37 | 0.64 ± 0.057 |
| upc2-1 stt4-4 | cholesterol | 37 | 0.63 ± 0.090 |
| upc2-1 mss4-2ts | cholesterol | 37 | 0.23 ± 0.017 |
| upc2-1 | cholesterol | 30 | 2.3 ± 0.25 |
| upc2-1 osh1Δ | cholesterol | 30 | 2.5 ± 0.046 |
| upc2-1 osh2Δ | cholesterol | 30 | 2.6 ± 0.098 |
| upc2-1 osh3Δ | cholesterol | 30 | 1.5 ± 0.11 |
| upc2-1 osh4Δ | cholesterol | 30 | 2.4 ± 0.19 |
| upc2-1 osh5Δ | cholesterol | 30 | 1.8 ± 0.25 |
| upc2-1 osh6Δ | cholesterol | 30 | 2.1 ± 0.091 |
| upc2-1 osh7Δ | cholesterol | 30 | 2.2 ± 0.10 |
| upc2-1 osh3Δ osh5Δ | cholesterol | 30 | 1.2 ± 0.12 |
N.D., no ergosteryl ester detected.
Esterification rate, percentage of total sterol taken up that is esterified per minute.
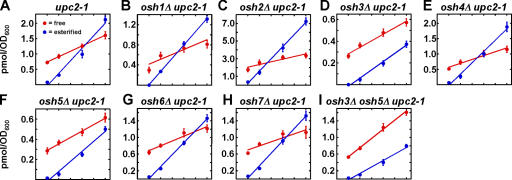
Figure 2.
Uptake and esterification of cholesterol by osh mutants. (A–I) Cells with the indicated genotypes were grown at 30°C and 2.0 μM 14C-cholesterol was added to the medium. At the indicated times, cells were harvested, the lipids were extracted, and the amount of free and esterified radiolabeled sterol in the cells was quantitated. In all graphs, values are the mean of two determinations. Error bars indicate the SEM. n = 2.
To estimate the contribution of Osh proteins other than Osh4p, we performed the same experiment with a strain (oshΔ OSH4) that is the same as oshΔ osh4-1, except that it has a wild-type OSH4 allele. This strain does not have a conditional growth defect. At low temperatures, oshΔ OSH4 esterifies exogenous cholesterol at a rate similar to oshΔ osh4-1 (Fig. 1 C). At 37°C, esterification is slightly faster than the oshΔ osh4-1 strain, but still significantly slower than a strain that has all the Osh proteins (Fig. 1 F). The oshΔ OSH4 cells also esterify exogenous ergosterol significantly more slowly than OSH + cells. (Fig. 1 I). Therefore, Osh proteins other than Osh4p are required for most of the esterification of exogenous sterol, though Osh4p makes some contribution.
The slow esterification of exogenous sterols in the oshΔ strains suggests that Osh4p and other Osh proteins are required for the efficient transfer of exogenous cholesterol from the PM to the ER. We performed several control experiments to rule out other explanations of the slow esterification. First, we found that this difference is not explained by low ACAT activity in the oshΔ cells (Fig. 1 J). Though these strains have about half the ACAT activity of the OSH + cells, this difference is not enough to explain the dramatically slower esterification of exogenous cholesterol by oshΔ cells. This difference might also be explained if there were substantially more endogenous sterol in the ER of oshΔ strains than in OSH + cells, which is a possibility because intracellular sterol distribution is altered in oshΔ cells (Beh and Rine, 2004). To rule this out, we measured the amount of sterol in the ER of the OSH + and oshΔ strains using the method of Lange and Steck, which makes use of the fact that ACAT only has access to sterols in the ER (Lange and Steck, 1997). Whole-cell lysates are reacted to completion with an excess of 14C-oleoyl-CoA; the amount of 14C-steryl oleate formed indicates the quantity of sterol in the ER. Using this method, we found no significant difference in the ER–sterol levels of oshΔ and OSH + cells (Fig. 1 K). Thus, the slow esterification of exogenous cholesterol by oshΔ cells is not caused by elevated amounts of endogenous sterol in the ER. Finally, our findings would be explained if cholesteryl ester was more rapidly hydrolyzed in the oshΔ mutants than in the OSH + cells. To address this possibility, we performed pulse-chase experiments. The oshΔ osh4-1 and OSH + strains were grown at a nonpermissive temperature for 30 min with radiolabeled cholesterol and then chased with a large excess of unlabeled cholesterol for 40 min. If steryl esters were rapidly hydrolyzed in the oshΔ osh4-1 strain then the amount of radiolabeled cholesteryl ester should have decreased after the chase. However, this was not the case (Fig. 1 L), indicating that cholesteryl ester is not rapidly hydrolyzed in the oshΔ osh4-1 cells.
Collectively, these findings strongly argue that oshΔ cells esterify exogenous sterols substantially slower than OSH + cells because they have impaired sterol transport to the ER. Because numerous mutations that severely inhibit vesicular transport do not block the movement of exogenous cholesterol to the ER (Li and Prinz, 2004), we conclude that the Osh proteins perform overlapping functions necessary for nonvesicular transport of exogenous cholesterol to the ER.
Role of individual Osh proteins in PM to ER sterol transfer
Our findings suggest that, in addition to Osh4p, other Osh proteins are required for the efficient transfer of exogenous cholesterol from the PM to the ER. To estimate the role of these proteins, we determined the rate at which exogenous cholesterol is esterified in mutants missing each of the Osh proteins individually (Fig. 2). The relative rate of esterification slowed somewhat in cells missing either Osh3p or Osh5p (Fig. 2, D and F), but was not significantly altered in mutants lacking the other Osh proteins. These defects were additive, i.e., a mutant lacking both Osh3p and Osh5p esterified cholesterol more slowly than cells missing just one of these proteins (Fig. 2 I). These findings suggest that Osh3p and Osh5p, in addition to Osh4p, may move sterols from the PM to the ER. However, because cells missing both Osh3p and Osh5p still transfer exogenous cholesterol from the PM to the ER more rapidly than oshΔ OSH4 cells (compare Fig. 1 F and Fig. 2 I ), other Osh proteins may also facilitate PM to ER sterol transfer.
Osh4p transports sterols in vitro
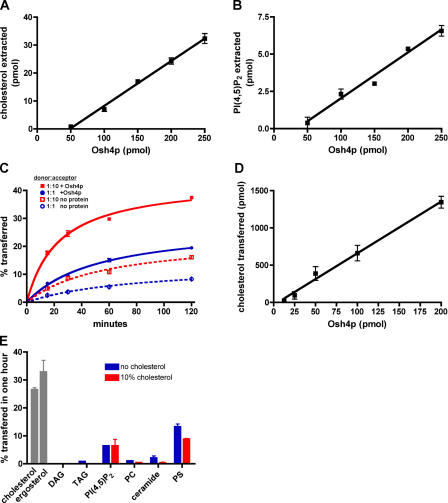
Because the Osh proteins are needed for PM to ER sterol transport in vivo, we wanted to determine if one of the Osh proteins could extract sterols from donor membranes and transfer them to acceptor membranes in vitro. Osh4p was chosen because it is one of the best characterized of the Osh proteins and because it binds to phosphatidylinositol(4,5) bisphosphate (PI(4,5)P2; Li et al., 2002), which is a lipid enriched in the PM. Consistent with a role for Osh4p in sterol transport, purified Osh4p-extracted cholesterol from liposomes (Fig. 3 A). We also examined its ability to extract other lipids and found it could also extract PI(4,5)P2 (1-arachidonyl-2-steroyl-PI(4,5)P2), though approximately five times less efficiently than cholesterol (Fig. 3 B). Osh4p could not extract any of the other lipids we tested, including the following: dipalmitoyl phosphatiylcholine (PC), phosphatidylserine (PS), a ceramide (oleoyl phytosphingosine), dioleoyl glycerol (DAG), and triolein (TAG; unpublished data). Thus, Osh4p extracts cholesterol, but not most other lipids, from membranes, suggesting Osh4p directly moves sterols.
Figure 3.
Extraction and transfer of sterol from liposomes by purified Osh4p. (A) 50 μL of 1 mM liposomes containing DOPC and 14C-cholesterol (99:1 mol%) in TS were mixed with the indicated amount of Osh4p at 30°C for 1 h. The amount of 14C-cholesterol extracted was determined as described in Materials and methods. (B) Liposomes containing DOPC and 3H-PI(4,5)P2 (99:1 mol%) in TS were incubated with the indicated amount of Osh4p for 1 h at 30°C. The amount of 3H-PI(4,5)P2 extracted was determined. (C) Donor vesicles (DOPC/egg PE/lactosyl-PE/14C-cholesterol/PI(4,5)P2; 59.5:20:10:10:0.5 mol%) were mixed with acceptor vesicles (DOPC/PE; 80:20 mol%) at a molar ratio of 1:10 or 1:1, mixed with 40 picomols (pmol) of Osh4p, and incubated at 30°C. At various times, samples were place on ice and the amount of 14C-cholesterol transferred to the donor was determined. The amount of 14C-cholesterol transferred in the absence of protein is also shown. (D) Donor vesicles were prepared as described in C and mixed with acceptor membranes at a molar ratio of 1:10. Different amounts of Osh4p were added and the reactions were incubated at 30°C for 15 min. The amount of radiolabeled sterol transferred to acceptor membranes minus the amount transferred in the absence of protein was determined. (E) Donor membranes were prepared as in C with either 10% 14C-cholesterol or 10% 3H-ergosterol (gray bars) or with either 1.0% 14C-DAG, 0.01% 3H-TAG, 0.5% 3H-PI(4,5)P2, 0.008% 3H-PC, 1.0% 14C-ceramide, or 10% 3H-PS and either no (blue bars) or 10% (red bars) cholesterol. These vesicles were mixed with acceptor vesicles at molar ratio of 1:10 and incubated with 40 pmol Osh4p at 30°C for 1 h. The amount of radiolabeled lipid transferred to acceptor membranes was calculated by subtracting the amount of transfer in control reactions incubated at 4°C. In all graphs, the values are the mean of three determinations. Error bars indicate the SEM. n = 3.
We next determined the ability of Osh4p to transfer sterols between membranes. Purified Osh4p moved cholesterol between liposomes in vitro in a time- (Fig. 3 C) and concentration-dependent (Fig. 3 D) manner. It also transported ergosterol, which is the primary sterol in yeast (Fig. 3 E). Thus, Osh4p can function as a sterol transport protein for both cholesterol and ergosterol. Osh4p transferred most other lipids poorly, though it could mediate some transfer of PI(4,5)P2 and PS (Fig. 3 E). Consistent with a role for Osh4p in sterol transport, it moved even less of most other lipids from liposomes containing 10% cholesterol than from those that lacked cholesterol (Fig. 3 E). In contrast, as discussed in the section Sterol transport by Osh4p is regulated by PIPs, liposomes including 10% PS in donor membranes stimulated cholesterol transfer by Osh4p (Fig. 5 C). We conclude that Osh4p likely preferentially transports sterols between membranes, but can also move PI(4,5)P2 and PS.
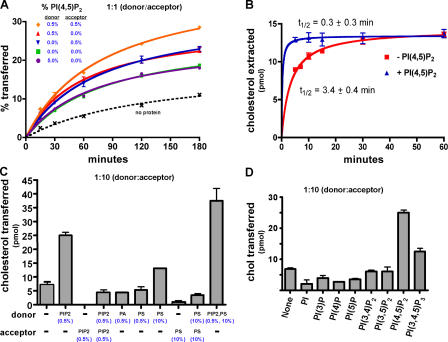
Figure 5.
Stimulation of sterol transport and binding by PIPs. (A) Donor and acceptor vesicles were prepared as in Fig. 3 C, except that they contained PI(4,5)P2 (as indicated). They were mixed at a molar ratio of 1:1, together with 40 pmol of Osh4p. The amount of 14C-cholesterol transferred to acceptor membranes was determined as described. Because the amount of 14C-cholesterol transferred in the absence of Osh4p was similar in all cases only one is shown. (B) The amount of 14C-cholesterol extracted from liposomes either with or without 0.5% PI(4,5)P2 by 200 pmol Osh4p was determined as in Fig. 3 A. The half-time (t1/2) and standard error of cholesterol extraction was calculated by nonlinear regression analysis. (C) Donor membranes were prepared containing DOPC/egg PE/lactosyl-PE/ 14C-cholesterol (60:20:10:10 mol%) with either no addition (−) or the indicated amount of either PI(4,5)P2, egg phosphatidic acid, or brain PS. Acceptor membranes were prepared as in Fig. 3 C or with 0.5% PI(4,5)P2 or 10% brain PS. Donor and acceptor membranes were mixed at molar ratio of 1:10 and incubated at 30°C for 1 h together with 40 pmol of Osh4p. The amount of transfer in control reactions lacking protein was subtracted to give the total amount of cholesterol transferred. (D) Donor membranes were prepared as in Fig. 3 C with either no addition (−) or 0.5% of either PI, PI(3)P, PI(4)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, or PI(3,4,5)P3. Cholesterol transfer to acceptor membranes was determined as in A. In all graphs, values are the mean of three determinations. Error bars indicate the SEM. n = 3.
Lipid transport by Osh4p mutants
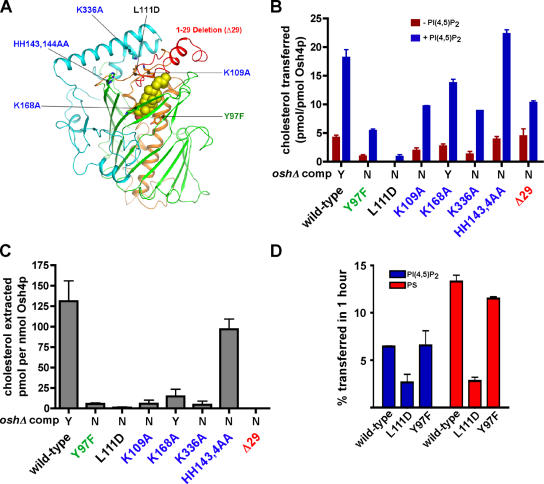
If sterol transfer is one of the functions of Osh4p, then mutations in Osh4p that render it nonfunctional by complementation analysis might disrupt the ability of the protein to transport sterols. We studied proteins with mutations in the following three regions of Osh4p: the binding tunnel (Y97F and L111D), the deletion of the conserved 29–amino acid lid domain (Δ29) that covers the binding tunnel, and the conserved charged residues near the mouth of the cholesterol-binding tunnel might be needed for interaction of Osh4p with bilayers (K109A, K168A, K336A, HH143, and 144AA; Fig. 4 A). With the exception of K168A, all of these mutations render Osh4p nonfunctional by complementation analysis (Im et al., 2005). All but one of the mutations decreases the ability of Osh4p to transport cholesterol (Fig. 4 B) and substantially reduce its ability to extract cholesterol (Fig. 4 C). Therefore, we conclude that lipid transfer is likely one of the physiological functions of Osh4p.
Figure 4.
Effect of mutations in Osh4p on cholesterol transport and extraction. (A) Location of the mutations shown in a stick model of Osh4p. The green residue is involved in ligand binding, the black residue is in the hydrophobic core, the blue residues are basic conserved residues, and the deletion of the NH2-terminal lid domain is red. A space-filling model of the cholesterol ligand is shown. (B) Cholesterol transport by wild-type and mutant Osh4p was measured. Donor and acceptor vesicles (1:10) were prepared as in Fig. 3 C, except that donor liposomes were prepared either with (blue bars) or without (red bars) 0.5% PI(4,5)P2. Vesicles were incubated at 30°C for 1 h, together with 40 pmol of Osh4p. The amount of transfer in control reactions lacking protein was subtracted to give the total amount of cholesterol transferred. (C) The amount of 14C-cholesterol extracted from liposomes (DOPC/14C-cholesterol, 99:1 mol%) by 200 pmol of wild-type or mutant Osh4p was determined as in Fig. 3 A. (D) Donor vesicles containing DOPC/egg PE/lactosyl-PE/3H-PI (60:20:10:1 mol%) or DOPC/egg PE/lactosyl-PE/3H-PS (60:20:10:10 mol%) were mixed with acceptor vesicles at a molar ratio of 1:10 and 40 pmol of wild-type or mutant Osh4p. Reactions were incubated for 1 h at 30°C. The amount of radiolabeled lipid transferred to acceptor vesicles minus the amount transferred in control reactions at 4°C was calculated. In B and C, the ability of the indicated mutants to complement oshΔ osh4-1 cells is shown (Y, growth; N, no growth; Im et al., 2005). In all graphs, values are the mean of three determinations. Error bars indicate the SEM. n = 3.
We found that PI(4,5)P2 stimulates sterol transfer by Osh4p. Because cholesterol transport was still stimulated by this PIP in all of the Osh4p mutants (Fig. 4 B, compare red and blue bars), none of the residues we altered is required for the PIP stimulation of sterol transport by Osh4p.
Because Osh4p can also transfer some PI(4,5)P2 and PS between membranes, we wondered if Osh4p mutants that were unable to transfer cholesterol efficiently also transferred these lipids poorly. PI(4,5)P2 and PS transport were not affected by Y97F, but were reduced by L111D (Fig. 4 D). Because Y97 interacts with the hydroxyl group of sterols (Im et al., 2005), it is perhaps not surprising that altering this residue does not affect the ability of Osh4p to move other lipids. We speculate that the acyl moieties of PI(4,5)P2 and PS bind in place of sterol in the hydrophobic tunnel and that the charged headgroups of these lipids are accommodated by an open conformation of the lid (Im et al., 2005).
Sterol transport by Osh4p is regulated by PIPs
Because Osh4p has been shown to bind PIPs (Li et al., 2002), we wondered if they might regulate sterol transfer by Osh4p. It has previously been shown that the PIP-binding domain of a ceramide transfer protein is needed for it to function in vivo (Hanada et al., 2003). To determine if PIPs affect sterol transport by Osh4p in vitro, donor and acceptor liposomes were prepared either with or without 0.5 mol% PI(4,5)P2 because Osh4p binds this PIP (Li et al., 2002). We found that PI(4,5)P2 stimulates cholesterol transport by Osh4p when it is on either donor or acceptor vesicles (Fig. 5 A). These effects were additive because transfer was fastest when both donor and acceptor vesicles contained PI(4,5)P2. To further characterize the affect of PI(4,5)P2 on Osh4p, we measured the rate of cholesterol extraction from liposomes by Osh4p and found it was stimulated ∼10-fold when the liposomes contained 0.5 mol% PI(4,5)P2 (Fig. 5 B).
We determined the ability of other lipids to stimulate sterol transfer by Osh4p (Fig. 5 C). No enhancement of transport was found when the donor membrane contained 0.5 mol% of other acidic phospholipids such as phosphatidic acid or PS, suggesting that the enhancement of transport with PI(4,5)P2 is specific for this PIP. A smaller enhancement of transport was also found when donor membranes contained 10 mol% PS. The stimulatory affects of 10 mol% PS and 0.5 mol% PI(4,5)P2 were additive. In addition, we found that only PI(4,5)P2, and, to a lesser extent PI(3,4,5)P3, were able to increase the rate of cholesterol transport by Osh4p (Fig. 5 D). It should be noted, however, that S. cerevisiae lacks detectable levels of PI(3,4,5)P3. Therefore, we conclude that PI(4,5)P2 is the primary PIP species able to stimulate sterol transport by Osh4p in cells. Sterol transfer is also slightly stimulated by PS. Because PI(4,5)P2 is enriched in the PM and is also found in other compartments including Golgi membranes, it likely stimulates sterol transfer at these compartments.
PI(4,5)P2 may stimulate sterol transfer by Osh4p by increasing the affinity of Osh4p for PI(4,5)P2-containing membranes, probably by binding to PI(4,5)P2 headgroups on the bilayer surface. Although low levels of PI(4,5)P2 stimulate transport, we wondered if high levels of PI(4,5)P2 might inhibit transfer by effectively trapping Osh4p on membranes. Consistent with this, we found that, with a 1:1 ratio of donor to acceptor vesicles, increasing the amount of PI(4,5)P2 in donor vesicles by 10-fold slowed transport (Fig. 5 A; 5.0% PIP2 in donor). In addition, even low levels of PI(4,5)P2 in acceptor vesicles can inhibit transfer when the ratio of donor to acceptor vesicles is 1:10 (Fig. 5 C, column 3). Collectively, these results suggest that PI(4,5)P2 modulates sterol transfer by Osh4p, likely by affecting its affinity for membranes.
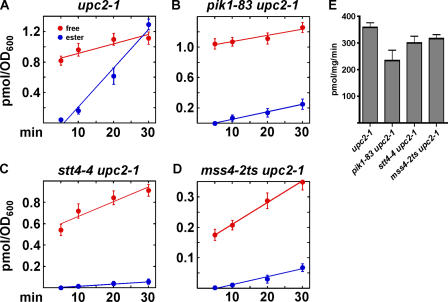
PM to ER sterol transfer slows in PIP biosynthesis mutants
Our findings suggest that PIPs regulate sterol transport by Osh4p and possibly other Osh proteins, some of which also bind PIPs (Levine and Munro, 2001; Roy and Levine, 2004; Wang et al., 2005b). To confirm the requirement of PIPs for efficient PM to ER sterol transport in cells, we used mutants with conditional defects in PI(4)P and PI(4,5)P2 synthesis. Stt4p and Pik1p are essential PI-4 kinases in yeast. Mutants with conditional defects in these proteins have reduced levels of PI(4)P and PI(4,5)P2 at elevated temperature (Audhya et al., 2000). These mutants transport exogenous cholesterol to the ER significantly more slowly than an isogenic STT4 + PIK1 + strain (Fig. 6, A–C). The differences in the total amount of sterol taken up by the strains does not affect the analysis because we have previously shown that the fraction of exogenous sterol that becomes esterified over time is not affected by the total amount taken up (Li and Prinz, 2004). To look more directly at the requirement of PI(4,5)P2, we used a mss4-2ts strain, which has a conditional defect in PI(4)P 5 kinase. At elevated temperature, mss4 mutants have reduced levels of PI(4,5)P2, but normal amounts of other PIPs (Desrivieres et al., 1998; Audhya and Emr, 2003). We found that cells depleted of PI(4,5)P2 had defects in sterol transfer to the ER (Fig. 6 D), suggesting that this PIP is particularly important for rapid sterol transport. We ruled out that these differences are caused by differences in ACAT levels in these strains (Fig. 6 E). Therefore, normal levels of PI(4)P and PI(4,5)P2 are required for efficient transfer of exogenous cholesterol to the ER. PIP depletion likely disrupts nonvesicular PM to ER sterol transport because mutations that dramatically slow vesicular trafficking do not affect the movement of exogenous sterols to the ER (Li and Prinz, 2004). In addition, because yeast lacks putative nonvesicular sterol transporters other than the Osh proteins (i.e., it does not have StART or SCP2 homologues), PIP depletion may affect sterol transport by Osh4p and other PIP-responsive Osh proteins.
Figure 6.
PM to ER sterol transport slows in PIP mutants. (A–D) Cells with conditional defects in PIP biosynthesis and an isogenic wild-type strain were grown in yeast extract/peptone/dextrose medium at 25°C and then shifted to 37°C for 1 h. 3.0 μM 14C-cholesterol was added to the medium and the amount of free and esterified 14C-cholesterol determined. (E) Cells were grown at 25°C, shifted to 37°C for 1 h, and the ACAT activity of microsomal extracts of each strain determined. Values are the mean of two determinations. Error bars indicate the SEM. n = 2.
Osh proteins are not needed for nonvesicular PS transfer to PS decarboxylase (Psd)
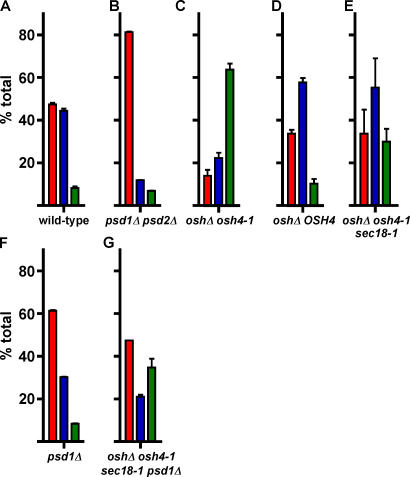
Because Osh4p can transfer PS between liposomes in vitro, we wondered if some of the Osh proteins might transport PS in cells. PS is synthesized in the ER from cytidine diphosphate-diacylglycerol and serine and can subsequently be converted to PE by Psd (Daum et al., 1998). Yeast has two Psds, one in the Golgi complex (Psd2p) and the other in mitochondria (Psd1p; Kuchler et al., 1986; Trotter and Voelker, 1995). Because these enzymes are not in the ER, the conversion of PS to PE requires the transfer of PS to the Golgi complex or mitochondria. Nonvesicular transport pathways mediate this transfer (Voelker, 2005). The PE generated by the Psds can be returned to the ER and converted to PC. Though some of the components of this pathway have been identified (Voelker, 2005), the lipid transfer proteins that move PS have not been determined. If the Osh proteins were required for nonvesicular PS transfer in vivo, then the conversion of newly synthesized PS to PE might be expected to slow in cells lacking Osh proteins.
When wild-type cells were labeled with 3H-serine for 30 min, ∼50% of the PS synthesized was converted into PE and PC (Fig. 7 A). To confirm that the radiolabeled PE and PC were generated from 3H-PS, we found that a strain missing the two Psds (psd1Δ psd2Δ) makes small amounts of radiolabeled PE or PC (Fig. 7 B). The conversion of PS to PE and PC was not blocked in the oshΔ strains (Fig. 7, C and D). It is not clear why these strains convert a greater proportion of newly synthesized PS to PE and PC than a wild-type strain, but lipid metabolism may be altered in these strains. The finding that the conversion of PS to PE is not blocked in oshΔ strains suggests that Osh proteins may not be needed for the nonvesicular transfer of PS to the Psds. However, because Psd2p is located in the Golgi complex, newly synthesized PS might still reach Psd2p by vesicular transport. To address this possibility, we introduced a temperature-sensitive sec18-1 allele into the oshΔ osh4-1 strain. Sec18p is the yeast homologue of N-ethylmaleimide-sensitive factor and is essential for most vesicular trafficking (Wilson et al., 1989; Graham and Emr, 1991). The oshΔ osh4-1/ sec18-1 cells were still able to convert newly synthesized PS to PE and PC after half an hour at a nonpermissive temperature (Fig. 7 E), which are conditions that result in a severe block in vesicular transport (Graham and Emr, 1991). It is also possible that Osh proteins might be needed to move PS to only one of the Psds. Because several Osh proteins have been localized to the Golgi complex, but none to the mitochondria, we thought it more likely that they would transfer PS to Psd2p than Psd1p. To investigate this possibility, we determined the rate of conversion of PS to PE and PC in cells missing Psd1p. Cells lacking Psd1p still convert PS to PE and PC, though slightly less efficiently than a wild-type strain (Fig. 7 F). To determine if the Osh proteins are needed to transport PS to Psd2p in conditions that block vesicular transport, we constructed an oshΔ osh4-1/sec18-1 psd1Δ strain. At nonpermissive temperatures, this strain is still able to convert PS to PE and PC (Fig. 7 G). Collectively, these results suggest that the Osh proteins are not required for the nonvesicular transfer of PS in the ER to Psds in the Golgi complex or mitochondria.
Figure 7.
Osh proteins are not required for PS transport to mitochondria and the Golgi complex. (A–G) Strains were labeled with 3H-serine at nonpermissive temperature as described in Materials and methods. Lipids were extracted and the amounts of radiolabeled PS, PE, and PC were determined. The percent of the total of these three lipids was calculated. Values are the mean of at least three determinations. Error bars indicate the SEM. n = 3.
Discussion
The transport of exogenous sterols from the PM to the ER in yeast is not blocked in numerous mutants that have severe defects in vesicular trafficking (Li and Prinz, 2004). We demonstrate that this transport slows significantly in cells lacking Osh proteins. Consistent with a direct role for these proteins in sterol transport, we find that a representative member of this family, Osh4p, transfers sterols between liposomes in vitro. This is the first demonstration that ORPs are lipid transfer proteins. We propose that nonvesicular transport of endogenous sterols is one of the functions of the ORPs in yeast, and probably in higher eukaryotes as well.
It seems likely that some Osh proteins have other functions. They may transport other classes of lipids in addition to sterols. We found that Osh4p also transfers PI(4,5)P2 and PS between liposomes, though less efficiently than cholesterol and ergosterol. However, Osh proteins may not transport significant amounts of PS in vivo because they are not required for nonvesicular PS transport to mitochondria or the Golgi complex. It is also likely that some of the Osh proteins function as lipid sensors that regulate other proteins, a function that has recently been demonstrated for OSBP in mammals (Wang et al., 2005a).
PM to ER sterol transport is likely mediated by several Osh proteins because it is unaffected or only moderately slower in cells missing any one of them. Our findings suggest that, in addition to Osh4p, Osh3p and Osh5p may also transfer sterols from the PM to ER in vivo. A role for Osh3p and Osh5p in PM to ER sterol transport is consistent with their largely cytoplasmic localization and the finding that the Osh3p PH domain (without the OSBP-related domain) localizes to the PM (Levine and Munro, 2001; Habeler et al., 2002). Because cells missing Osh3p and Osh5p still move exogenous sterols to the ER more rapidly than oshΔ cells, other Osh proteins, including Osh4p, probably also transfer PM sterols to the ER in vivo.
We have previously shown that efficient PM to ER transfer requires either of two ATP-binding cassette transporters in the PM (Li and Prinz, 2004). These transporters are also needed for efficient uptake of exogenous sterols by yeast (Wilcox et al., 2002). How the ATP-binding cassette transporters affect the rate of PM to ER sterol transport is not known. They might directly facilitate the movement of sterols out of the PM, perhaps to Osh proteins. Alternatively, they might affect the composition or distribution of lipids in the PM in such a way that Osh proteins can more easily extract sterols and move them to internal compartments.
Why yeast has such a large number of Osh proteins remains an open question. Several Osh proteins localize to various compartments throughout the cell (Levine and Munro, 2001; Li et al., 2002; Olkkonen and Levine, 2004; Wang et al., 2005c). Therefore, some Osh proteins may transport sterols or other lipids primarily to or from specific organelles. Genetic analysis has revealed that all seven Osh proteins have a single overlapping essential function and suggests that loss of the Osh proteins causes a defect in the recycling of ergosterol back to the PM (Beh et al., 2001; Beh and Rine, 2004). Thus, while individual Osh proteins may move lipids primarily to or from particular organelles, they may all have the ability to transfer sterols from endosomes back to the PM. Alternatively, the Osh proteins could have a shared essential function as lipid sensors.
Osh4p is partially localized to the Golgi complex and has been genetically implicated in Golgi function (Fang et al., 1996; Li et al., 2002). Sec14p is an essential PI-PC transfer protein that is required for proper Golgi function. However, cells lacking Osh4p do not require Sec14p for viability. It has been proposed that Sec14p acts as a lipid-sensor that regulates a functionally important pool of DAG in the Golgi complex by modulating enzymes that consume or generate DAG (Kearns et al., 1998; Routt and Bankaitis, 2004). What role Osh4p plays in this process is not clear. Like Sec14p, it could act as a lipid sensor at the Golgi complex. Alternatively, Osh4p could affect the lipid composition and function of the Golgi by moving sterols, PIPs, or some other lipid to or from this organelle.
We have also found that PIPs stimulate sterol transfer by Osh4p. PIP stimulation is probably a wide-spread property of ORPs. Indeed, we find that PIP-depleted cells have defects in PM to ER sterol transport. Some large mammalian and yeast ORPs interact with PIPs via PH domains (Levine and Munro, 2001; Holthuis and Levine, 2005). The fact that Osh4p and some other Osh proteins lack these domains (Im et al., 2005) but bind PIPs (Li et al., 2002; Wang et al., 2005b) suggests that PIPs may interact with ORPs via multiple mechanisms. Osh4p likely binds to PIP headgroups using some part of its external surface. Because altering the conserved charged residues near the mouth of the sterol-binding tunnel did not affect the ability of PI(4,5)P2 to stimulate transport, these residues are probably not required for PIP binding. Bankaitis and coworkers showed that an Osh4p with mutations in three basic residues fails to bind PI(4,5)P2 (Li et al., 2002). When we altered these residues, the resulting protein was largely insoluble (unpublished data) and we were not able to obtain enough protein to assess sterol transport by this mutant.
Our findings suggest that PIPs stimulate sterol transport by Osh4p, and likely other ORPs, by increasing the affinity of Osh4p for PIP-containing membranes. Because high levels of PIPs can also inhibit sterol transfer by Osh4p, changes in PIP levels, perhaps in response to cell-signaling events, could regulate sterol transfer by ORPs. In addition, because different PIP species are enriched in various cellular compartments, PIP stimulation of ORPs likely serves to regulate the movement and distribution of sterols (and possibly other lipids) among cellular compartments by ORPs. The identification of ORPs as PIP-regulated nonvesicular lipid transfer proteins in yeast provides a good model system to help us begin to understand the role of these proteins in maintaining intracellular lipid distribution.
Materials and methods
Strains and materials
The strains used in this study are listed in Table II. To allow these strains to take up exogenous cholesterol during aerobic growth, their UPC2 alleles were replaced with upc2-1 by homologous recombination, as follows. A 757-bp fragment of the upc2-1 allele (from nucleotide 1985 to the end of the gene) was cloned into YIPlac211 (Gietz and Sugino, 1988). The resulting plasmid was used as a template for a PCR reaction with primers UPC2-A2 and UPC-C (Table III). The product of this reaction was used to transform cells, and the transformants were selected on medium that lacked uracil. The presence of the upc2-1 allele was confirmed, as previously described (Wilcox et al., 2002). To introduce sec18-1 into cells, a 901-bp fragment of sec18-1 containing the mutation in this allele was cloned into YIPlac211. The resulting plasmid was used as template for a PCR reaction with the primers SEC18-C and SEC18-B1 (Table III). The product of this reaction was used to transform cells, and the transformants were selected on medium that lacked uracil. To delete PSD1 in cells, a PCR reaction using primers PSD1∷URA-f and PSD1∷URA-r (Table III) using the plasmid pRS416 (Sikorski and Hieter, 1989) as a template. The resulting product was used to transform cells, and the transformants were selected on medium that lacked uracil. Replacement of PSD1 with psd1Δ ∷URA3 was confirmed by PCR. Before replacing PSD1 with psd1Δ ∷URA3 in WPY817, this strain was plated on medium containing 5-fluoroortic acid (Sikorski and Boeke, 1991) to select cells unable to grow without uracil.
Table II.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SEY6210 | MATα ura3-52, his3-Δ 200, leu2-3, -112 trp1-Δ 901, suc2-Δ 9, lys2-801 | C. Beha |
| CBY924 | SEY6210 osh1Δ ∷kan-MX4 osh2Δ ∷kan-MX4 osh3Δ ∷LYS2 osh4Δ ∷HIS3 osh5Δ ∷LEU2 osh6Δ ∷LEU2 osh7Δ ∷HIS / OSH4, CEN-TRP | C. Beh |
| CBY926 | SEY6210 osh1Δ ∷kan-MX4 osh2 Δ∷kan-MX4 osh3Δ ∷LYS2 osh4Δ ∷HIS3 osh5Δ ∷LEU2 osh6Δ ∷LEU2 osh7Δ ∷HIS3 / osh4-1, CEN-TRP | C. Beh |
| SEY6218 | MATα ura3-52, his3-Δ 200, leu2-3, -112 trp1-Δ 901, suc2-Δ 9, lys2-801 | S. Emrb |
| AAY102 | SEY6218 stt4∷HIS3 / pRS415 CEN-LEU2-stt4-4 | S. Emr |
| AAY104 | SEY6218 pik1∷HIS3 / pRS314 CEN-TRP1-pik1-83 | S. Emr |
| SD102 | MATa leu2 ura3 rme1 trp1 his3Δ GAL+ TOF3 mss4∷HIS3MX6 / YCplac111∷mss4-2ts | C. Jacksonc |
| NDY202 | SEY6210 upc2-1:URA3 | This study |
| NDY222 | CBY 924 upc2-1:URA3 | This study |
| NDY75 | CBY926 upc2-1:URA3 | This study |
| NDY234 | SEY6218 upc2-1:URA3 | This study |
| NDY231 | AAY102 upc2-1:URA3 | This study |
| NDY233 | AAY104 upc2-1:URA3 | This study |
| NDY236 | SD102 upc2-1:URA3 | This study |
| WPY817 | CBY926 sec18-1:URA3 | This study |
| WPY833 | MATa his3Δ leu2Δ met15Δ ura3Δ upc2-1:URA3 | This study |
| YFY248 | WPY833 osh1Δ ∷kan-MX4 | This study |
| YFY249 | WPY833 osh2Δ ∷kan-MX4 | This study |
| YFY250 | WPY833 osh3Δ ∷kan-MX4 | This study |
| YFY251 | WPY833 osh4Δ ∷kan-MX4 | This study |
| YFY252 | WPY833 osh5Δ ∷kan-MX4 | This study |
| YFY253 | WPY833 osh6Δ ∷kan-MX4 | This study |
| YFY254 | WPY833 osh7Δ ∷kan-MX4 | This study |
| YFY435 | WPY833 osh3Δ ∷kan-MX4 osh5Δ ∷HIS3-MX6 | This study |
| NDY275 | SEY6210 psd1Δ ∷URA3 | This study |
| WPY832 | WPY817 psd1Δ ∷URA3 | This study |
Simon Fraser University, Vancouver, British Columbia, Canada.
University of California, San Diego, La Jolla, CA.
National Institues of Health, Bethesda, MD.
Table III.
Primers used in this study
| Primer | Sequence |
|---|---|
| UPC2-A2 | gaggactgaaactggactggagc |
| UPC2-C | ctgcagttctttttgtatttgcattacacataaaaaaaaaccacctgacgtctaagaaacc |
| SEC18-B1 | ctatcccagttgcaacagcgg |
| SEC18-C | taatgagcgcgctaaatagttgaatattttatctttaattcacctgacgtctaagaaacc |
| PSD1∷URA-f | ggtcgttattttttgaagaagaaggaaaagcaaagccagcgattcggtaatctccgaaca |
| PSD1∷URA-r | tatacagcaaaataatgctaactttacatatgattgctt ttagttttgctggccgcatc |
Lipids were purchased from Avanti Polar Lipids, Inc. Radiolabeled lipids were purchased from American Radiolabeled Chemicals, Inc., except for 3H-ergosterol, which was isolated as previously described (Li and Prinz, 2004), and 3H-PS, which was made as follows: yeast cells were labeled with 3H-acetic acid, and phospholipids were extracted and separated as previously described (Wang et al., 2003) using an Agilent 1100 series HPLC. The peak containing 3H-PS (detected at 203 nm) was identified by comparison to known standards and collected. The species of purchased radiolabeled lipids used were as follows: 1-arachidonyl-2-steroyl-PI(4,5)P2; PC; oleoyl phytosphingosine; dioleoyl glycerol; and triolein. Purified Osh4p was obtained as previously described (Im et al., 2005).
Cholesterol uptake and esterification
The uptake and esterification of 14C-cholesterol was determined as previously described (Li and Prinz, 2004). In brief, 14C-cholesterol in Tween80/ethanol (1:1) was added at the indicated concentration to the medium of growing cells so that the final Tween80 concentration was 0.5%. For pulse-chase experiments, cells were grown and 23°C, shifted to 37°C for 30 min, and 2.0 μM 14C-cholesterol added to the medium. After 30 min, 250 μM of unlabeled cholesterol was added to the medium and cells were grown for an additional 40 min. Samples were removed at the indicated times and lipids were extracted and analyzed as previously described (Li and Prinz, 2004).
ACAT activity measurements
ACAT activity of the strains was measured as previously described (Li and Prinz, 2004).
Determination of ER sterol concentration
The relative amount of sterol in the ER was determined basically as previously described (Lange and Steck, 1997). Cells from rapidly growing cultures were pelleted, washed once with ice-cold H2O, resuspended in 20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DTT, and lysed by homogenization with glass beads in a Bead-Beater 8 (BioSpec Products, Inc.). The lysate was centrifuged for 5 min at 500 g to remove debris and unlysed cells. For the assay, 200 μg lysate was mixed with 25 μM 14C-oleoyl-CoA in a total volume of 100 μL of 20 mM Tris, pH 7.5, and 100 mM NaCl. The reaction was incubated at 37°C for 10 min and stopped by the addition of CHCl3 and MeOH. Lipids were extracted and quantitated as previously described (Li and Prinz, 2004).
Cholesterol extraction assay
Liposomes were prepared by mixing dioleoyl phosphatidyl choline (DOPC) and 14C-cholesterol at a molar ratio of 99:1. The lipids were dried under a stream of nitrogen and resuspended in 20 mM Tris-HCl, pH 7.4, and 100 mM NaCl (TS) at a final concentration of 1 mM. The membranes were allowed to swell at room temperature for at least 1 h and vigorously vortexed, and then liposomes with a mean diameter of 1 μm were made using a mini-extruder from Avanti Polar Lipids, Inc. For the extraction assay, purified Osh4p was incubated with 50 μL of membranes at 30°C for the indicated times, after which the membranes were kept at 4°C and pelleted by centrifugation in a rotor (model TLA100; Beckman Coulter) at 70,000 rpm (191,000 g) for 1 h. The amount of 14C-cholesterol in the supernatant was determined by scintillation counting and was taken to be cholesterol that had been extracted from the liposomes. The amount of radiolabel in the supernatant in control reactions without Osh4p (usually ∼2% of the total) was subtracted from the amount measured with Osh4p.
Lipid transport assay
Lipid transport was assayed as previously described (Hanada et al., 2003), with the following modifications. Unless otherwise indicated, the lipid composition of the donor membranes was DOPC/egg PE/lactosyl-PE/cholesterol/PI(4,5)P2 (59.5:20:10:10:0.5). Acceptor liposomes were prepared by mixing DOPC and egg PE at a ratio of 80:20, unless otherwise indicated. The lipids were dried under a stream of nitrogen, resuspended in 20 mM Hepes, pH 7.4, 100 mM NaCl, and 1 mM EDTA (HES) at either 1 or 10 mM (acceptor vesicles) and allowed to swell at room temperature for at least 1 h; they were then vigorously vortexed. Acceptor vesicles to be used for 1:10 donor to acceptor lipid transport assays were prepared by sonication with a Branson tip sonicator. Before use, the liposome solution was cleared by centrifugation at 15,000g at 4°C for 10 min. All donor vesicles and acceptor vesicles used in 1:1 donor to acceptor transport assays were prepared by extrusion using a filter with 0.2 μm pore size. All vesicles were used within 24 h of preparation. For the transport assay, 50 μL of donor membranes and acceptor membranes were mixed together with Osh4p in a total volume of 110 μL. After incubation at 30°C, 30 μg of ricinus communis agglutinin (Vector Laboratories) was added, and the reactions were placed on ice for 15 min, with occasional mixing. The donor membranes were then pelleted by centrifugation at 15,000 g for 5 min at 4°C. The amount of radiolabel in the supernatant was determined by scintillation counting and taken to be lipid that had been transferred to acceptor membranes. The amount of radiolabel in the supernatant in control reactions without Osh4p or with Osh4p and incubated on ice was subtracted from the amount measured with Osh4p.
Phospholipid labeling with 3H-serine
30-ml cultures were grown in synthetic complete medium with 1 mM ethanolamine at 25°C. They were shifted to 37°C, grown for 25 min and then 1 mM S-adenosyl methionine, 2 mM methionine, and 10 μg/ml cerulenin (from a 5 mg/ml stock in DMSO) were added to the medium. Control experiments showed that these compounds were needed to prevent radiolabel from being incorporated into PE and PC in cells missing Psd1p and Psd2p. The cultures were grown for 5 min, 50 μCi 3H-serine (American Radiolabeled Chemicals, Inc.) was added, and the cultures were grown for 30 min more. They were placed at 4°C and washed once with ice-cold H2O. Lipids were extracted (Li and Prinz, 2004) and phospholipids were separated as previously described (Wang et al., 2003). The peaks containing PS, PE, and PC were collected and counted in a scintillation counter.
Acknowledgments
We thank C. Beh, C. Jackson, and S. Emr for providing strains, N. DeAngelis for excellent technical assistance, and J. Hanover and T. Rapoport for reading the manuscript.
This work was supported by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Y.J. Im's present address is Dept. of Life Science, Gwangju Institute of Science and Technology, Gwangju City, 500-712, South Korea.
Abbreviations used in this paper: ACAT, acyl-coenzyme A:cholesterol acyltransferase; DAG, dioleoyl glycerol; DOPC, dioleoyl phosphatidyl choline; ORP, OSBP-related protein; OSBP, oxysterol-binding protein; PC, dipalmitoyl phosphatiylcholine; PH, pleckstrin homologue; PI, phosphatidylinositol; PI(4,5)P2, phosphatidylinositol(4,5) bisphosphate; PIP, phosphoinositide; PM, plasma membrane; pmol, picomols; PS, phosphatidylserine; Psd, PS decarboxylase; TAG, triolein.
References
- Audhya, A., and S.D. Emr. 2003. Regulation of PI4,5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. EMBO. J. 22:4223–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya, A., M. Foti, and S.D. Emr. 2000. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 11:2673–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, N.A., D.P. Sullivan, H. Ohvo-Rekila, C. Simonot, A. Pottekat, Z. Klaassen, C.T. Beh, and A.K. Menon. 2005. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 44:5816–5826. [DOI] [PubMed] [Google Scholar]
- Beh, C.T., and J. Rine. 2004. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 117:2983–2996. [DOI] [PubMed] [Google Scholar]
- Beh, C.T., L. Cool, J. Phillips, and J. Rine. 2001. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 157:1117–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger, B., R. Sandhoff, S. Wegehingel, K. Gorgas, J. Malsam, J.B. Helms, W.D. Lehmann, W. Nickel, and F.T. Wieland. 2000. Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J. Cell Biol. 151:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley, J.H., F.W. Leak, K.V. Shianna, S. Tove, and L.W. Parks. 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180:4177–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, G., N.D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 14:1471–1510. [DOI] [PubMed] [Google Scholar]
- Dawson, P.A., N.D. Ridgway, C.A. Slaughter, M.S. Brown, and J.L. Goldstein. 1989. cDNA cloning and expression of oxysterol-binding protein, an oligomer with a potential leucine zipper. J. Biol. Chem. 264:16798–16803. [PubMed] [Google Scholar]
- Desrivieres, S., F.T. Cooke, P.J. Parker, and M.N. Hall. 1998. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 273:15787–15793. [DOI] [PubMed] [Google Scholar]
- Doms, R.W., G. Russ, and J.W. Yewdell. 1989. Brefeldin A redistributes resident and itinerant Golgi proteins to the endoplasmic reticulum. J. Cell Biol. 109:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, M., B.G. Kearns, A. Gedvilaite, S. Kagiwada, M. Kearns, M.K. Fung, and V.A. Bankaitis. 1996. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO. J. 15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- Gardner, R.G., H. Shan, S.P. Matsuda, and R.Y. Hampton. 2001. An oxysterol-derived positive signal for 3-hydroxy-3-methylglutaryl-CoA reductase degradation in yeast. J. Biol. Chem. 276:8681–8694. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. [DOI] [PubMed] [Google Scholar]
- Graham, T.R., and S.D. Emr. 1991. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J. Cell Biol. 114:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeler, G., K. Natter, G.G. Thallinger, M.E. Crawford, S.D. Kohlwein, and Z. Trajanoski. 2002. YPL.db: the yeast protein localization database. Nucleic Acids Res. 30:80–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada, K., K. Kumagai, S. Yasuda, Y. Miura, M. Kawano, M. Fukasawa, and M. Nishijima. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426:803–809. [DOI] [PubMed] [Google Scholar]
- Heino, S., S. Lusa, P. Somerharju, C. Ehnholm, V.M. Olkkonen, and E. Ikonen. 2000. Dissecting the role of the Golgi complex and lipid rafts in biosynthetic transport of cholesterol to the cell surface. Proc. Natl. Acad. Sci. USA. 97:8375–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis, J.C., and T.P. Levine. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6:209–220. [DOI] [PubMed] [Google Scholar]
- Im, Y.J., S. Raychaudhuri, W.A. Prinz, and J.H. Hurley. 2005. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 437:154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandutsch, A.A., and E.P. Shown. 1981. Assay of oxysterol-binding protein in a mouse fibroblast, cell-free system. Dissociation constant and other properties of the system. J. Biol. Chem. 256:13068–13073. [PubMed] [Google Scholar]
- Kaplan, M.R., and R.D. Simoni. 1985. Transport of cholesterol from the endoplasmic reticulum to the plasma membrane. J. Cell Biol. 101:446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, B.G., J.G.J. Alb, and V. Bankaitis. 1998. Phosphatidylinositol transfer proteins: the long and winding road to physiological function. Trends Cell Biol. 8:276–282. [DOI] [PubMed] [Google Scholar]
- Kuchler, K., G. Daum, and F. Paltauf. 1986. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 165:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, Y., and T.L. Steck. 1997. Quantitation of the pool of cholesterol associated with acyl-CoA:cholesterol acyltransferase in human fibroblasts. J. Biol. Chem. 272:13103–13108. [DOI] [PubMed] [Google Scholar]
- Levine, T.P., and S. Munro. 1998. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 8:729–739. [DOI] [PubMed] [Google Scholar]
- Levine, T.P., and S. Munro. 2001. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell. 12:1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., M.P. Rivas, M. Fang, J. Marchena, B. Mehrotra, A. Chaudhary, L. Feng, G.D. Prestwich, and V.A. Bankaitis. 2002. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and W.A. Prinz. 2004. ABC-transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the ER. J. Biol. Chem. 279:45226–45234. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., L.C. Yuan, J.S. Bonifacino, and R.D. Klausner. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 56:801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, L., and N.J. Munn. 1999. Intracellular cholesterol transport. Biochim. Biophys. Acta. 1438:19–37. [DOI] [PubMed] [Google Scholar]
- Loewen, C.J., A. Roy, and T.P. Levine. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO. J. 22:2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield, F.R., and D. Wustner. 2002. Intracellular cholesterol transport. J. Clin. Invest. 110:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi, Y., K. Miki, A. Takatsuki, G. Tamura, and Y. Ikehara. 1986. Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J. Biol. Chem. 261:11398–11403. [PubMed] [Google Scholar]
- Olkkonen, V.M. 2004. Oxysterol binding protein and its homologues: new regulatory factors involved in lipid metabolism. Curr. Opin. Lipidol. 15:321–327. [DOI] [PubMed] [Google Scholar]
- Olkkonen, V.M., and T.P. Levine. 2004. Oxysterol binding proteins: in more than one place at one time? Biochem. Cell Biol. 82:87–98. [DOI] [PubMed] [Google Scholar]
- Prinz, W. 2002. Cholesterol trafficking in the secretory and endocytic systems. Semin. Cell Dev. Biol. 13:197–203. [DOI] [PubMed] [Google Scholar]
- Ridgway, N.D., P.A. Dawson, Y.K. Ho, M.S. Brown, and J.L. Goldstein. 1992. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 116:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routt, S.M., and V.A. Bankaitis. 2004. Biological functions of phosphatidylinositol transfer proteins. Biochem. Cell Biol. 82:254–262. [DOI] [PubMed] [Google Scholar]
- Roy, A., and T.P. Levine. 2004. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279:44683–44689. [DOI] [PubMed] [Google Scholar]
- Schnabl, M., G. Daum, and H. Pichler. 2005. Multiple lipid transport pathways to the plasma membrane in yeast. Biochim. Biophys. Acta. 1687:130–140. [DOI] [PubMed] [Google Scholar]
- Schroepfer, G.J.J. 2000. Oxysterols: modulators of cholesterol metabolism and other processes. Physiol. Rev. 80:361–554. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and J.D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned genes to mutant yeast. Methods Enzymol. 194:302–318. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio, R.E., and J.L. Breslow. 2004. Intracellular cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 4:1150–1160. [DOI] [PubMed] [Google Scholar]
- Sprong, H., P. van der Sluijs, and G. van Meer. 2001. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2:504–513. [DOI] [PubMed] [Google Scholar]
- Trotter, P.J., and D.R. Voelker. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270:6062–6070. [DOI] [PubMed] [Google Scholar]
- Urbani, L., and R.D. Simoni. 1990. Cholesterol and vesicular stomatitis virus G protein take separate routes from the endoplasmic reticulum to the plasma membrane. J. Biol. Chem. 265:1919–1923. [PubMed] [Google Scholar]
- Voelker, D.R. 2005. Bridging gaps in phospholipid transport. Trends Biochem. Sci. 30:396–404. [DOI] [PubMed] [Google Scholar]
- Wang, F., Y. Zhao, and P. Wang. 2003. Separation and determination of phospholipids in biological samples by high-performance liquid chromatography. J. Chromatogr. Sci. 41:142–144. [DOI] [PubMed] [Google Scholar]
- Wang, P.Y., J. Weng, and R.G.W. Anderson. 2005. a. OSBP is a cholesterol-regulated scaffolding protein in control of ERK1/2 activation. Science. 307:1472–1476. [DOI] [PubMed] [Google Scholar]
- Wang, P., Y. Zhang, H. Li, H.K. Chieu, A.L. Munn, and H. Yang. 2005. b. AAA ATPases regulate membrane association of yeast oxysterol binding proteins and sterol metabolism. EMBO. J. 24:2989–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., W. Duan, A.L. Munn, and H. Yang. 2005. c. Molecular characterization of Osh6p, an oxysterol binding protein homolog in the yeast Saccharomyces cerevisiae. FEBS J. 272:4703–4715. [DOI] [PubMed] [Google Scholar]
- Wilcox, L.J., D.A. Balderes, B. Wharton, A.H. Tinkelenberg, G. Rao, and S.L. Sturley. 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277:32466–32467. [DOI] [PubMed] [Google Scholar]
- Wilson, D.W., C.A. Wilcox, G.C. Flynn, E. Chen, W.J. Kuang, W.J. Henzel, M.R. Block, A. Ullrich, and J.E. Rothman. 1989. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 339:355–359. [DOI] [PubMed] [Google Scholar]
- Zweytick, D., E. Leitner, S.D. Kohlwein, C. Yu, J. Rothblatt, and G. Daum. 2000. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267:1075–1082. [DOI] [PubMed] [Google Scholar]