Abstract
Transcriptional control plays a key role in regulating epidermal proliferation and differentiation. Although ample information has been obtained on how epidermal homeostasis is controlled in adult skin, less is known about the control of proliferation/differentiation of epidermal stem/progenitor cells in the developing embryo. Ovol1, encoding a zinc finger protein homologous to Drosophila melanogaster Ovo, is expressed in embryonic epidermal progenitor cells that are transiting from proliferation to terminal differentiation. In this study, we demonstrate a function for Ovol1 in interfollicular epidermal development. In its absence, developing epidermis fails to properly restrict the proliferative potential of progenitor cells, and cultured keratinocytes fail to efficiently undergo growth arrest in response to extrinsic growth-inhibitory signals. We present molecular evidence that c-myc expression is up-regulated in Ovol1-deficient suprabasal cells and that Ovol1 represses c-myc transcription by directly binding to its promoter. Collectively, our findings indicate that Ovol1 is required for proliferation exit of committed epidermal progenitor cells and identify c-myc as an Ovol1 target.
Introduction
Terminal differentiation within the interfollicular epidermis ultimately leads to the formation of a functional skin barrier that protects organisms from water loss, infections, and various insults. This barrier is continuously renewed throughout the animal's postnatal life span as a result of the presence of stem cells that are capable of self-renewal and of producing transiently amplifying progenitor cells, which subsequently exit the cell cycle and embark on a terminal differentiation pathway as they migrate toward the skin surface (Fuchs and Raghavan, 2002). This process is recapitulated during embryogenesis, when a single layer of multipotent surface ectodermal cells develops into a stratified epidermis. Keratins 5 (K5) and 14 (K14), which are expressed in the proliferating basal layer of the mature epidermis, are among the earliest epidermal markers that are activated in the single-layered ectoderm of the embryonic skin (Fuchs, 1993; Koster and Roop, 2004). As development proceeds, the presumptive suprabasal layers arise and express differentiation-specific keratins K1 and K10 in the intermediate spinous layers as well as loricrin, a major component of the future cornified envelope (Byrne et al., 1994), in the upper granular layers. Associated with a spatiotemporally ordered change of gene expression are morphological transformations and biochemical events culminating in the formation of a barrier at the outermost layer of the epidermis that is impermeable after embryonic day (E) 17 (Hardman et al., 1998).
Transcriptional regulation is key to a successful epidermal development/differentiation program (for review see Dai and Segre, 2004). Among transcription factors that control the balance between proliferation and differentiation of keratinocytes, the c-myc proto-oncoprotein and the inhibitor of differentiation (Id) family of proteins surfaced as positive regulators of a cycling and nondifferentiating progenitor state. However, little is known about how the expression of these factors is regulated in skin. Although existing studies have provided insights into how homeostasis is achieved in mature epidermis, few examine the genetic pathways and molecular mechanisms that govern the growth and differentiation of stem/progenitor cells of the developing epidermis (Gugasyan et al., 2004; Okuyama et al., 2004).
ovo is an evolutionally conserved family of genes encoding C2H2 zinc finger transcription factors in animals. Functional studies in Caenorhabditis elegans, Drosophila melanogaster, and mice suggest that this gene family plays important roles in the development of epithelial tissues and germ cells (Oliver et al., 1987; Mevel-Ninio et al., 1995; Dai et al., 1998; Johnson et al., 2001; Mackay et al., 2006). Genetic and biochemical studies suggest that at least two members of this gene family, Drosophila ovo and Ovol1, act downstream of the Wnt– β-catenin–lymphoid enhancer factor/T cell factor signaling pathway (Payre et al., 1999; Li et al., 2002). Recently, OVOL1 was identified as a downstream target of the TGF-β/BMP7–Smad4 signaling pathway, a growth-inhibitory pathway in keratinocytes (Kowanetz et al., 2004). Therefore, the ovo gene family members appear to be important integrators of upstream developmental signals and key regulators of epithelial development and differentiation.
Ovol1, the first mouse ovo that was functionally characterized, is expressed in multiple somatic epithelial tissues, including skin (hair follicles and interfollicular epidermis) and kidney, as well as in the male germinal epithelium (Dai et al., 1998). Ovol1-deficient mice showed ruffled hairs, cystic kidneys, and defective spermatogenesis (Dai et al., 1998). In this study, we describe a functional requirement for Ovol1 in epidermal development. Specifically, we show that Ovol1 is required to restrict the proliferation potential of embryonic epidermal progenitor cells in vivo and in vitro. We also present molecular evidence indicating that Ovol1 represses the expression of c-myc by direct binding to its promoter, providing a possible mechanism by which Ovol1 regulates the proliferation arrest of developing epidermal cells.
Results
The K1-positive layers containing progenitor cells are expanded in the developing Ovol1 −/− epidermis
The initial characterization of Ovol1-deficient mice was performed in a 129Sv (129) × C57BL/6 (B6) mixed (50:50) genetic background (Dai et al., 1998). Upon close examination, we noticed that the epidermis of these mutant animals was often slightly thicker than that of the wild type (unpublished data). Because a “pure” B6 strain background can sometimes enhance the phenotypic manifestation of a particular mutation (McGowan et al., 2002), we transferred the Ovol1 mutant allele into a B6 strain background and analyzed the skin morphology of offspring from Ovol1 +/− intercrosses. We note that previously described phenotypes, including ruffled hairs and cystic kidneys, persisted in this new background and that a subset of the Ovol1-deficient pups died perinatally, with the surviving ones exhibiting flaky skin (unpublished data).
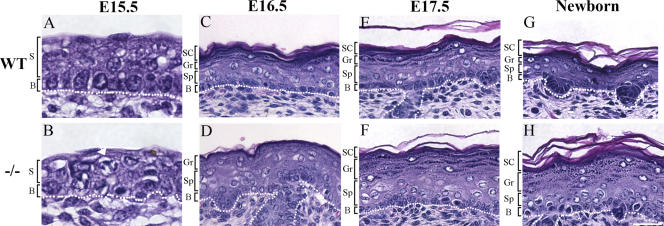
During normal epidermal development, presumptive suprabasal cells appear at ∼E15.5 and are morphologically distinct from the underlying presumptive basal cells (Fig. 1 A). Different from those in mature skin, these developing suprabasal cells express differentiation marker K1 but retain their proliferative potential for another 2–3 d (see below; Byrne et al., 1994; Okuyama et al., 2004) and are, therefore, embryonic epidermal progenitor cells. Although stratification occurred in Ovol1 −/− epidermis at E15.5, the morphological distinction between the presumptive suprabasal and basal layers was not apparent in many areas (Fig. 1 B), and more mitotic figures were seen than the wild type (Fig. 1 B, arrow; also see below). By E16.5, the Ovol1 −/− epidermis, where a morphological stratification had now become obvious, was considerably thicker than the controls, resembling acanthosis described in human patients (Fig. 1 D). This defect was not caused by a transient delay in development, as it was also observed at later stages (Fig. 1, E–H). Furthermore, there was impaired enucleation, flattening, and compaction of the developing granular cells in mutant epidermis (Fig. 1, D, F, and H), suggesting subtle, late differentiation defects. No histological defects were apparent in the Ovol1 +/− epidermis at all stages examined (unpublished data).
Figure 1.
Histological abnormalities of the developing Ovol1 mutant skin. White dotted line denotes the basement membrane. Arrow in B indicates a mutant mitotic cell in the presumptive suprabasal layer (S). B, basal layer; SC, stratum cornea; Gr, granular layer; Sp, spinous layer. Bar (A and B), 20 μm; (C–H) 30 μm.
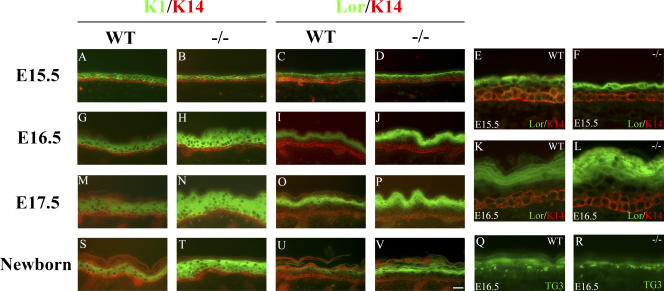
To investigate whether the thickening of Ovol1 −/− mutant epidermis was caused by an expansion of the presumptive basal or suprabasal layers, we stained the developing epidermis for K14, K1, and loricrin. As development proceeded from E15.5 to the newborn stage, mutant epidermis acquired the expected spatial arrangement of K14, K1, and loricrin- positive layers (Fig. 2). However, subtle abnormalities were seen such that at E15.5, the loricrin-positive layer was closer in space to the K14-positive basal layer than that in the wild type (Fig. 2, D and F; compare with C and E). More importantly, beginning at E16.5, the mutant K1-positive layers were significantly expanded compared with their wild-type counterparts (Fig. 2, H, N, and T). A slight expansion of the loricrin-positive layers was also observed (Fig. 2, J, L, P, and V), but this was unlikely caused by a general expansion of the presumptive granular layers, as no expansion was observed for cells that were positive for transglutaminase 3 (TG3), another marker for granular cells (Fig. 2, Q and R). No consistent expansion of the K14-positive layer was observed in the mutant. The mutant defects were not associated with any apparent change in the expression of K6 (unpublished data), which is generally regarded as a wound-healing keratin (Mazzalupo et al., 2003). Collectively, these data demonstrate that the K1-positive embryonic progenitor cell population was expanded in the absence of Ovol1, which underlies the thickening of the mutant interfollicular epidermis.
Figure 2.
Expansion of the K1-positive progenitor layers in the developing Ovol1−/− epidermis. Results of immunofluorescence staining of skin from wild-type (A, C, E, G, I, K, M, O, Q, S, and U) and Ovol1 −/− (B, D, F, H, J, L, N, P, R, T, and V) mice at E15.5 (A–F), E16.5 (G–L, Q, and R), E17.5 (M–P), and newborn (S–V) stages using a FITC-conjugated α-K1 (A, B, G, H, M, N, S, and T) or α-loricrin antibody (green; C–F, I–L, O, P, U, and V) and a rhodamine-conjugated α-K14 antibody (red) or a FITC-conjugated α-TG3 antibody (green; Q and R). Bar (A–D, G–J, M–P, and S–V), 30 μm; (E, F, K, L, Q, and R) 10 μm.
Increased proliferation in the developing Ovol1 −/− epidermis
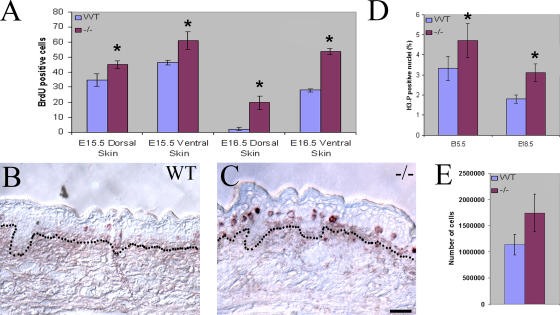
The continued expansion of K1-positive progenitor cells in developing Ovol1 −/− epidermis led us to wonder whether the developmental transition between proliferation and differentiation was affected. As measured by BrdU incorporation, epidermal proliferation in control embryos slows down as development proceeds (Fig. 3 A, compare E15.5 with E16.5; Okuyama et al., 2004). In its extreme, very little proliferation was observed on the dorsal side of the E16.5 wild-type epidermis (Fig. 3 B), which is consistent with the fact that the barrier first forms in this region (Hardman et al., 1998). In the mutant, an increase in the number of BrdU-positive cells was already evident at E15.5 (Fig. 3 A and not depicted). By E16.5, mutant epidermis still retained a considerable level of proliferation both dorsally and ventrally (Fig. 3 A), with the difference between wild type and mutant most prominent in the suprabasal compartment of the dorsal region (Fig. 3 C; compare with B). Using an antibody that detects phosphorylated histone H3 (H3-P), a marker for cells in mitosis (Gurley et al., 1978), we observed a higher average mitotic index in the mutant than the wild type, particularly at later developmental stages (Fig. 3 D and not depicted). In keeping with these in vivo findings, the initial plating of comparable numbers of primary keratinocytes isolated from newborn pups consistently yielded more attached and growing keratinocytes for the mutant when compared with their wild-type control littermates (Fig. 3 E).
Figure 3.
Increased number of cycling cells in the developing Ovol1 mutant epidermis. (A) Comparison of the number of BrdU-positive cells (average number per 1,000-μm distance) in dorsal and ventral epidermis of wild type and mutant at the indicated ages. (B and C) Representative stained sections from the dorsal region of wild-type (B) and mutant (C) skin at E16.5. Dotted lines denote the basement membrane. (D) Mitotic index of wild-type and Ovol1-deficient embryonic skin at the indicated ages. The mitotic index was calculated as the ratio of the number of cells in the developing interfollicular epidermis that stained positive for H3-P over the total number of cells as determined by DAPI staining. (E) More attached and growing keratinocytes were recovered from newborn Ovol1 mutant epidermis. Bars represent an average of three homozygous mutant pups or two wild-type controls, all from a single litter. Similar results were obtained from additional litters (not depicted). *, P < 0.03 difference from the wild-type counterparts. Error bars represent SEM. Bar (B and C), 30 μm.
Ovol1-deficient keratinocytes fail to efficiently exit proliferation upon induction with extrinsic growth-inhibitory signals
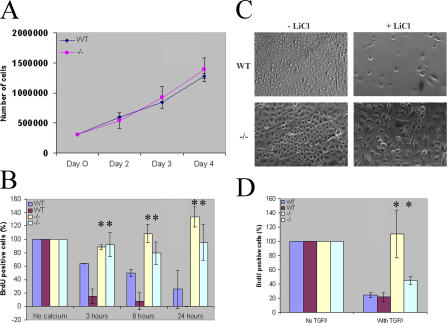
A priori, multiple possibilities may account for the observation of an increased number of proliferating keratinocytes in mutant epidermis. Among these are the precocious activation of a population of presumably slow-cycling embryonic epidermal stem cells, enhanced rate of proliferation of transit-amplifying progenitor cells, or impaired growth arrest of progenitor cells. Ovol1 expression was observed in the presumptive suprabasal layers but was barely detectable in the presumptive basal layer (Dai et al., 1998) where embryonic epidermal stem cells presumably reside, making the first possibility highly unlikely. FACS analysis identified a similar percentage of cells that are positive for α6 integrin, a marker for basal keratinocytes (Sonnenberg et al., 1991), in keratinocyte preparations from wild-type and Ovol1-deficient newborn epidermis (unpublished data). Furthermore, a growth analysis of actively proliferating keratinocytes of secondary passages in the absence of any growth-inhibitory treatment revealed no difference between the wild type and mutant (Fig. 4 A). Together, these results argue against an involvement of Ovol1 in embryonic epidermal stem cell activation and in regulating the rate of keratinocyte proliferation itself.
Figure 4.
Inefficient growth arrest of Ovol1-deficient keratinocytes. (A) Wild-type and mutant keratinocytes show a comparable proliferation rate. (B and D) BrdU-labeling index of keratinocytes cultured in the absence or presence of Ca2+ (B) or TGF-β (D). Error bars were calculated from three replicate samples of each condition. (C) Relative confluency of keratinocytes cultured in the absence or presence of LiCl. Results from WT and mutant offspring of a single litter are shown. Similar findings were obtained from multiple litters. *, P < 0.02 difference from two independently derived wild-type keratinocyte preparations.
Several treatments have been shown to induce the exit of proliferation and possibly terminal differentiation of cultured keratinocytes. These include high concentrations of Ca2+ (Hennings et al., 1980), LiCl, which is thought to mimic activated canonical Wnt signaling by inhibiting GSK3β (Olmeda et al., 2003), and TGF-β (Shipley et al., 1986). Therefore, we examined the ability of Ovol-deficient keratinocytes to exit proliferation in response to these reagents. First, wild-type and mutant keratinocytes were cultured in the presence of low (∼0.09 mM) or high (1.2 mM) Ca2+, and their BrdU-labeling index was determined. Although a time-dependent decrease in the number of BrdU-labeled cells was observed for the wild type upon Ca2+ treatment, the BrdU-labeling index remained high in Ovol1-deficient keratinocytes even 24 h after Ca2+ addition (Fig. 4 B). Second, LiCl, which induced efficient growth arrest of wild-type keratinocytes (as indicated by the <10% confluency of treated plates at the time when untreated replicate plates reached 100% confluency), failed to do so with the mutant cells (Fig. 4 C). Finally, a concentration of TGF-β that caused a significant reduction in the BrdU-labeling index in wild-type cells failed to cause Ovol1-deficient keratinocytes to stop cycling (Fig. 4 D). Collectively, our results indicate that the loss of Ovol1 renders the proliferating keratinocytes in culture less sensitive to extrinsic growth-inhibitory signals.
c-myc expression in the presumptive suprabasal layers of Ovol1-deficient epidermis was not properly down-regulated
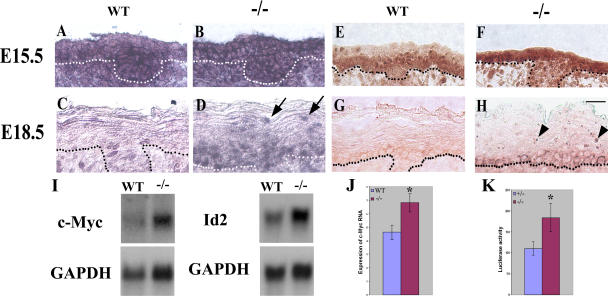
The inability of Ovol1-deficient epidermal progenitor cells to exit proliferation implies that Ovol1 normally functions in the developing epidermis to ensure the growth arrest of these cells. As it has been shown that a down-regulation of c-myc expression in suprabasal cells is important to maintain a postmitotic status (Pelengaris et al., 1999; Waikel et al., 1999), we hypothesized that Ovol1 may function by down-regulating c-myc expression. To test this hypothesis, we performed in situ hybridization experiments on developing epidermis using a c-myc cRNA probe. Although hybridization signals were observed in all layers of the interfollicular epidermis of both wild-type and Ovol1-deficient E15.5 embryos, a slight reduction in signal intensity was often apparent in the newly formed presumptive suprabasal layers of the wild type but not mutant (Fig. 5, A and B). As development proceeded to E18.5, the intensity of c-myc hybridization signals became weaker overall, with only a few scattered basal cells showing detectable expression (Fig. 5 C). In the mutant, however, many suprabasal cells showed clearly detectable hybridization signals, including those that are close to the skin surface (Fig. 5 D, arrows). The number of c-myc–expressing basal cells was also significantly higher. Additionally, our analysis of c-myc protein expression in embryonic skin revealed differences between the wild-type and mutant epidermis that are similar to those observed at the RNA level (Fig. 5, E–H). Of particular note is that although no staining was observed in the wild-type suprabasal cells at E18.5, those in the mutant epidermis retained strong nucleolar signals that are characteristic of the c-myc protein (Fig. 5 H, arrowheads; Arabi et al., 2005; Sanders and Gruppuso, 2005). To better quantify the difference in c-myc expression, we performed Northern blot analysis on RNA isolated from E16.5 embryonic skin and observed an ∼1.7-fold higher level of c-myc transcripts in the mutant than the wild type (Fig. 5 I). The expression of Id2, which was previously shown to be a target of c-myc transcriptional activation in skin and a target of Ovol1 transcriptional repression in the testis (Lasorella et al., 2000; Murphy et al., 2004; Li et al., 2005a), was also up-regulated by ∼1.5-fold in Ovol1-deficient skin (Fig. 5 I).
Figure 5.
c-myc expression is up-regulated in developing suprabasal layers of the Ovol1-deficient epidermis. Results of situ hybridization (A–D) and immunostaining (E–H) of wild-type (A, C, E, and G) and mutant (B, D, F, and H) embryonic skin using a c-myc cRNA probe and a mouse anti–c-myc antibody, respectively. Note that at E18.5, c-myc protein is cytoplasmic in the basal layer, and at least some c-myc transcripts appeared nuclear, which probably stems from the reported delicate control of subcellular localizations of c-myc gene products to tailor the needs of cell cycle control in these cells (Vriz et al., 1992; Bond and Wold, 1993). Dotted lines denote the basement membrane. Arrows and arrowheads indicate mutant suprabasal cells expressing c-myc mRNA and protein, respectively. (I) Results of Northern blot analysis of E16.5 wild-type and mutant skin. Each blot was stripped and reprobed with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. (J and K) Real-time PCR analysis of c-myc transcript levels (J) and reporter assays of c-myc promoter activity (K) in wild-type and mutant keratinocytes 24 h after Ca2+ induction. Average values obtained on cells from three mutant and three control animals are shown. Luciferase activities were normalized for transfection efficiency by using a β-actin promoter driving lacZ as an internal control. *, P < 0.03 difference from the wild type. Error bars represent SEM. Bar, 20 μm.
Although a failure to down-regulate c-myc expression in the mutant presumptive suprabasal cells could be a primary consequence of the loss of Ovol1 as it is expressed in these cells, the increase of c-myc expression in basal cells that normally do not express an appreciable amount of Ovol1 is curious and may be a secondary effect. To begin to distinguish between primary and secondary changes, we isolated primary keratinocytes from wild-type and mutant newborns and performed real-time PCR analysis to determine the level of endogenous c-myc transcripts in these isolated epidermal cells. No statistically significant difference was observed between wild-type and mutant keratinocytes that were basal-like when they were cultured under undifferentiating conditions (unpublished data). However, after these cells were treated with differentiation-inducing Ca2+, a significantly higher level of c-myc transcripts was detected in the mutant preparations (Fig. 5 J). To determine whether the increase in the c-myc transcript level was a result of increased transcription, we cloned a 2.3-kb human c-myc promoter fragment upstream of a luciferase reporter gene (see Fig. 7 A) and transfected the reporter construct into wild-type and mutant keratinocytes. Again, Ca2+-treated mutant keratinocytes allowed higher promoter activity than the wild type, whereas no difference was observed in untreated cells (Fig. 5 K and not depicted). Collectively, our results strongly suggest that Ovol1 is required to down-regulate c-myc transcription during epidermal development in the growth-restricted suprabasal cells.
Figure 7.
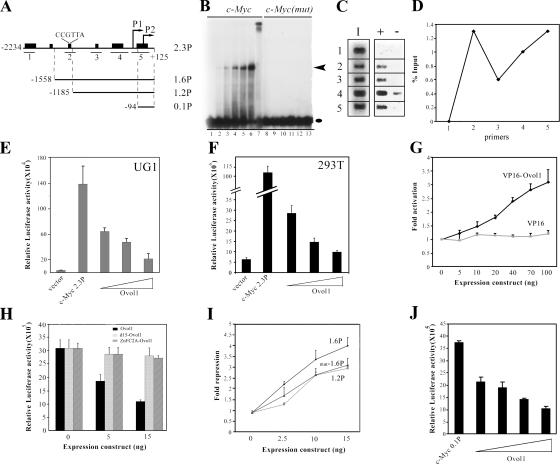
Ovol1 represses c-myc transcription by associating with two regions in its promoter. (A) Diagram of the human c-myc promoter in plasmid 2.3P (pGL3–c-myc) and in the deletion constructs 1.6P, 1.2P, and 0.1P. Transcription start sites are indicated as P1 and P2. Heavy bars indicate regions with significant similarity to the mouse c-myc gene. (B) EMSA and supershift assays showing binding of recombinant Ovol1 to a CCGTTA-containing sequence in the c-myc promoter. Black oval and arrowhead indicate the positions of Ovol1–DNA complexes and free probes, respectively. Lanes 1–6: c-myc oligonucleotide in the absence (lane 1) and presence of increasing concentrations of the recombinant Ovol1 protein (lanes 2–6). Lane 7: Ovol1 binding to c-myc oligonucleotide in the presence of an anti-Ovol1 antibody. Lanes 8–13: a mutant version of c-myc oligonucleotide in which the CCGTTA sequence is replaced by ATGCGC with (lanes 9–13) and without (lane 8) recombinant Ovol1. (C) Results of PCR amplifications using primer sets Cm-1 (1), Cm-2 (2), Cm-3 (3), Cm-4 (4), and Cm-5 (5; see A for locations of regions amplified by these primers). I, input; +, with anti-Ovol1 antibody; −, with IgG control. (D) Quantitative analysis of Ovol1 occupancy at various positions (see 1–5 in C) of the human c-myc promoter. (E and F) Dosage-dependent repression of c-myc promoter luciferase reporter expression by Ovol1 in UG1 (E) and 293T (F) cells. The triangles indicate increasing concentrations (0.01, 0.02, and 0.05 μg) of the Ovol1-expressing vector. (G) Dosage-dependent activation of reporter expression by VP16-Ovol1. (H) Repression of c-myc 2.3P depends on the presence of the first 15 amino acids of Ovol1 and the DNA-binding zinc finger domain. Expression levels of the mutant proteins were comparable with that of the wild type, as shown by Western blot analysis (not depicted). (I) Deletion (1.2P) and point mutations (mut-1.6P) of the CCGTTA Ovol1-binding site led to a significant reduction in repression by Ovol1. (J) Residual repression of a minimum c-myc promoter fragment (0.1P) by Ovol1. Each bar represents the average of triplicate samples in a single experiment, and the results are representative of several independent experiments. Error bars represent SEM. Luciferase activities were normalized for transfection efficiency by using a β-actin promoter driving lacZ as an internal control.
Biochemical evidence that Ovol1 represses c-myc transcription by direct binding to the c-myc promoter
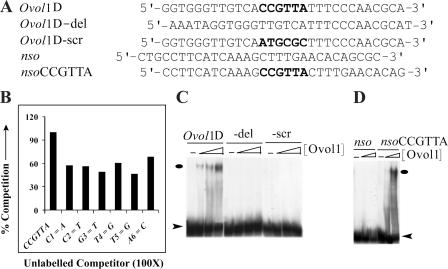
Does Ovol1 protein directly repress c-myc transcription, or are intermediate factors involved? To address this issue, we turned to study the nucleotide sequence determinants of Ovol1–DNA interaction. Previously, we showed that Ovol1 is able to bind to a Drosophila Ovo consensus sequence (Li et al., 2002). Since then, we have identified mouse genomic sequences to which Ovol1 binds in vitro and arrived at a putative consensus motif, CCGTTA (unpublished data). Although single nucleotide mutation in this motif (C1C2G3T4T5A6: C1→A, C2→T, G3→T, T4→G, T5→G, or A6→C) resulted in diminished Ovol1 binding (Fig. 6, A and B), deletion or scramble of this hexamer motif totally abolished binding (Fig. 6, A and C). On the other hand, the insertion of a CCGTTA sequence into an oligonucleotide to which Ovol1 does not bind (nso; Li et al., 2002) was able to confer binding (Fig. 6, A and D). Together, these results indicate that a CCGTTA sequence is necessary and sufficient to confer high-affinity Ovol1–DNA interaction in vitro.
Figure 6.
The CCGTTA hexamer is necessary and sufficient for Ovol1 recognition. (A) Sequence of oligonucleotides used in EMSA assays. (B) Competition assays were performed with Ovol1D (an oligonucleotide containing a high-affinity Ovol1-binding site) as the labeled probe and Ovol1D or oligonucleotides with indicated point mutations within the CCGTTA sequence as unlabeled competitors (present at a 100-fold excess). (C) EMSA assays using the Ovol1D oligonucleotide or its mutant derivatives showing that the CCGTTA hexamer sequence was necessary for Ovol1–DNA interaction. (D) EMSA assays using the nso oligonucleotide or its derivative showing that the CCGTTA hexamer sequence was sufficient for Ovol1–DNA interaction. Ovals and arrowheads indicate the positions of Ovol1–DNA complexes and free probes, respectively.
We next examined the sequences of gene-regulatory regions of both mouse and human c-myc genes and found that both contained a single CCGTTA motif, the position of which is also conserved (Fig. 7 A). In gel shift assays, recombinant Ovol1 bound to oligonucleotide c-myc, which contains human c-myc sequences including the CCGTTA motif (Fig. 7 B, lanes 2–6). The identity of the Ovol1–DNA complex was confirmed by the observation of a band supershift when anti-Ovol1 antibody was added to the binding reaction (Fig. 7 B, lane 7), and binding was abolished when the CCGTTA sequence was mutated (Fig. 7 B, lanes 9–13). Together, these results identify a bone fide Ovol1-binding site in the c-myc promoter.
We next used chromatin immunoprecipitation (ChIP) assays to determine whether Ovol1 was engaged at the endogenous human c-myc promoter (see Materials and methods). The anti-Ovol1 antibody-precipitated DNA was subject to PCR amplification with five pairs of primers spanning five long sequence blocks of the c-myc promoter that are conserved between mice and humans (Fig. 7 A). Ovol1 occupancy was detected with primer pairs Cm-2, Cm-3, Cm-4, and Cm-5 but not with Cm-1 (Fig. 7 C). No signal was observed when normal rabbit IgG was used for precipitation, showing the specificity of this assay. Quantitative analysis revealed two peaks of Ovol1 binding: one encompassed the CCGTTA sequence (primer set Cm-2), and the other was located in the proximal promoter region, which overlaps the sixth conserved block (primer set Cm-5; Fig. 7, A and D). These results indicate that Ovol1 physically associates with the endogenous c-myc promoter at the predicted distal site inside cells; however, they also reveal additional sites in the proximal promoter region that are occupied by Ovol1. We performed gel shift experiments on overlapping fragments encompassing this entire proximal region but observed no evidence of binding of recombinant Ovol1 to any fragment in this in vitro assay (unpublished data), raising the possibility that an alternative mechanism (e.g., assistance from auxiliary factors) exists in vivo to bring Ovol1 protein to this region (see Discussion).
Having established that Ovol1 binds to the c-myc promoter in vitro and in cells, we next used reporter assays to investigate whether Ovol1 directly represses c-myc transcription. The 2.3-kb human c-myc promoter fragment was able to direct active transcription in both UG1 mouse keratinocytes (Dai et al., 1998) and 293T cells (Fig. 7, A, E, and F). Cotransfection of an Ovol1 expression vector repressed reporter expression in both culture systems in a dosage-dependent manner (Fig. 7, E and F). To confirm that the observed repression is an Ovol1 protein–dependent event, we created the chimeric protein VP16-Ovol1 in which Ovol1 was fused to a strong, well-characterized transactivation domain from VP16 with the assumption that this activation domain might override the intrinsic transcriptional regulatory activity of Ovol1 and result in an activator. Indeed, VP16-Ovol1 activated the c-myc promoter in a dosage- dependent manner (Fig. 7 G). In contrast, the VP16 domain alone had no effect at all concentrations tested, indicating that the fusion protein was recruited to the c-myc promoter by its Ovol1 moiety. The specificity of the effect of Ovol1 was further demonstrated by the finding that a truncated Ovol1 protein (d15-Ovol1) lacking the first 15 amino acids at the NH2 terminus, which resembles the known repression domain SNAG (Nieto, 2002), failed to efficiently repress the c-myc promoter activity (Fig. 7 H).
To determine whether repression depends on the DNA binding ability of Ovol1, we generated a construct expressing a mutant form of the Ovol1 protein in which the cysteine amino acids in the first three zinc fingers were replaced by alanine (ZnFC2A). This mutant protein, which is no longer able to bind DNA (not depicted), failed to repress the c-myc promoter (Fig. 7 H). To further explore the dependence of repression on DNA binding, we generated mutant promoters in which upstream sequences were deleted or mutated. Although the deletion of sequences from −2.3 to −1.6 kb had no effect (not depicted), a partial release of repression was observed when a 373-bp sequence containing the CCGTTA site was removed (Fig. 7, A and I). Moreover, replacing the CCGTTA motif with a non-Ovol1 binding sequence, ATGCGC, led to a similar reduction in repression by Ovol1 (promoter construct mut-1.6P in Fig. 7 I), confirming that this site is indeed required for mediating Ovol1 repression. This said, considerable residual repression was still observed, implicating the contribution of other cis-elements. Analysis of additional deletion constructs mapped the minimum Ovol1 responsive region to within the smallest promoter fragment tested (the 0.1P construct; Fig. 7, A and J; and not depicted). The position of this region coincides with that of the proximal Ovol1-binding site identified by the aforementioned ChIP assays, implying that Ovol1 repressed this minimum promoter by binding to it. Collectively, our data suggest that Ovol1 represses c-myc transcription by binding to its promoter.
Aberrant loricrin expression in Ovol1-deficient skin
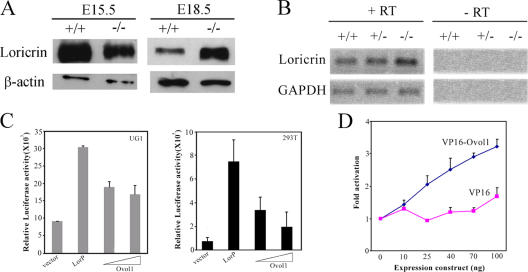
The apparent expansion of loricrin-positive layers in Ovol1-deficient skin in the absence of any concomitant expansion of the TG3-positive layers raises the possibility that loricrin expression is aberrantly activated in intermediate suprabasal cells when Ovol1 is ablated. We next used Western blot analysis to determine loricrin protein levels in wild-type and Ovol1 mutant skin from different developmental stages. Although mutant skin started out expressing a slightly lower level of loricrin protein (75% of that in wild type at E15.5 after normalization against actin levels), possibly because of a transient delay in the proliferation–differentiation switch, it produced a much higher level of the protein at later stages (approximately fourfold higher than the wild type at E18.5; Fig. 8 A). Using semiquantitative RT-PCR, we detected a twofold increase in loricrin RNA levels in the mutant skin taken from E16.5 embryos (Fig. 8 B), confirming that increased loricrin expression occurred at a transcriptional level.
Figure 8.
Up-regulated loricrin expression in Ovol1-deficient skin and repression of loricrin promoter activity by Ovol1 in reporter assays. (A) Western blot analysis of loricrin protein levels in wild-type and mutant skin. β-actin was used as a loading control. (B) RT-PCR analysis showing an up-regulation of loricrin transcripts in E16.5 Ovol1 −/− mutant skin. (C) Ovol1 represses loricrin promoter–luciferase reporter gene expression. (D) VP16-Ovol1 activated reporter expression in a dosage-dependent manner. Data treatment and presentation are as described in Fig. 6. Error bars represent SEM.
To better understand the effect of Ovol1 ablation on loricrin expression, we examined the effect of Ovol1 protein on loricrin promoter activity using reporter assays. We cloned a mouse loricrin promoter fragment (DiSepio et al., 1995) upstream of a luciferase reporter gene and found that this fragment directed active transcription in both UG1 and 293T cells (Fig. 8 C). This observation is consistent with previous findings that a core loricrin promoter fragment directs expression in not only granular cells but also basal and spinous cells of transgenic mice (DiSepio et al., 1995). Cotransfection of an Ovol1 expression vector repressed loricrin promoter activity in both cell systems (Fig. 8 C). However, at high Ovol1 concentrations, we often observed a release of repression, which was likely a result of “squelching” caused by Ovol1 binding to specific trans-acting factors that might be limiting in these cells. To confirm that the observed regulation is an Ovol1 protein–dependent event, we tested VP16-Ovol1, assuming that this chimeric activator would likely bypass the requirement for those limiting factors needed for repression. Indeed, VP16-Ovol1 activated loricrin promoter–luciferase reporter expression in a clearly dosage-dependent manner, whereas the VP16 activation domain alone had no significant effect (Fig. 8 D). A CCGTTA sequence was found in the loricrin promoter; however, this site is very close to the transcription start so that its mutation abolished promoter activity, preventing us from examining the DNA site dependence of Ovol1 repression (unpublished data). Nonetheless, our data demonstrate that Ovol1 is capable of repressing loricrin expression in a cell-autonomous manner.
Discussion
Ovol1 and the proliferation potential of embryonic epidermal progenitor cells
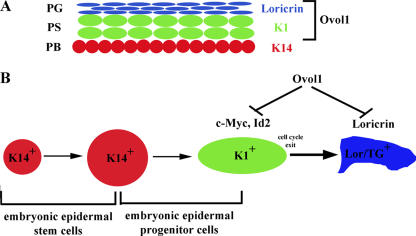
Our studies identified a novel function of Ovol1 in epidermal development. The expansion of K1-positive layers and increase in the number of actively proliferating cells in the developing epidermis of Ovol1-deficient mice are consistent with a hyperproliferative defect. A priori, hyperproliferation may occur as a primary cell-autonomous consequence of Ovol1 ablation in the proliferative compartment or as a secondary consequence of abnormal terminal differentiation and/or barrier defects, as frequently observed in the study of adult epidermis. Several lines of findings support the former possibility. First, increased proliferation was already evident at E15.5, which is before late-terminal differentiation events occur and a barrier forms. Although a transient delay in barrier acquisition was observed in Ovol1-deficient embryos, all were fully impermeable to dye penetration at the age of E18.5–newborn (unpublished data), suggesting that barrier development is largely normal (Hardman et al., 1998). Late-differentiation defects and delayed barrier formation were also observed in loricrin knockout mice, yet loricrin-deficient epidermis is not acanthotic (Koch et al., 2000), indicating that these defects are not sufficient to trigger a compensatory hyperproliferative response in the embryo. Second, the expression of K6, which is typically up-regulated in repair-associated hyperproliferation (Mazzalupo et al., 2003), was not affected in the developing Ovol1 mutant epidermis. Finally, the most direct evidence came from our observation that keratinocytes isolated from mutant animals could not be efficiently induced to exit the cell cycle in response to extrinsic growth-inhibitory signals such as Ca2+, LiCl, and TGF-β. As these cells were cultured independently of feeders, this analysis allowed a direct assessment of the intrinsic proliferative capacity of the mutant epidermal cells. Based on these results and the onset of Ovol1 expression in early presumptive suprabasal cells (Fig. 9 A), we propose that Ovol1 is required for the growth arrest of embryonic progenitor cells during epidermal development (Fig. 9 B). It is worth mentioning that similar to the epidermis, Ovol1-deficient male germ cells were sluggish in exiting mitosis (Li et al., 2005a). Therefore, Ovol1 might play a general role in down-regulating the proliferation of developmental progenitor cells in tissues that require its function.
Figure 9.
Working model of the role of Ovol1 in epidermal development. The location of Ovol1 expression in developing epidermis with respect to differentiation markers is indicated in A. PB, presumptive basal layer; PS, presumptive spinous layer; PG, presumptive granular layer. The model of the role of Ovol1 is shown in B.
While this study was in revision, Lechler and Fuchs (2005) published that epidermal stratification during mid–late gestation, where p63 is critically involved (Mills et al., 1999; Koster et al., 2004), entails asymmetric divisions of the ectodermal cells, yielding a basal cell and a suprabasal cell with a short-lived proliferation potential. Ovol1 expression is activated immediately upon stratification in these transit-amplifying suprabasal layers (Dai et al., 1998). Although stratification occurred in the absence of Ovol1, the newly formed mutant suprabasal cells appeared different in size and shape from their wild-type counterparts despite their ability to express K1 (Figs. 1 and 2). Is it possible that Ovol1 is an intrinsic molecular “clock” built into the suprabasal daughter cell that somehow makes it different from its long-lived parent and its postmitotic successors in terms of proliferation potential, so that in its absence, this daughter cell is improperly programmed upon stratification and, therefore, carries out just a few more rounds of proliferation than it should? What signals lie upstream of Ovol1? Existing evidence tentatively places Ovol1 downstream of Wnt as well as TGF-β1/BMP signaling. The suprabasal hyperproliferative phenotype of Ovol1-deficient epidermis is similar to that observed in Ikkα-deficient mice (Hu et al., 1999; Takeda et al., 1999) and in mice with the repeated epilation (Er) mutation, which is a mutation in 14-3-3σ (Li et al., 2005b). However, loricrin expression is completely blocked in the absence of Ikkα or in Er mice but only slightly delayed in the absence of Ovol1, suggesting that Ovol1 lies downstream of Ikkα and 14-3-3σ in the epidermal differentiation process. It is tempting to speculate that Ovol1 might be a key integrator of upstream developmental signals/molecular triggers like Wnt, TGF-β/BMP, and Ikkα in negative growth regulation during embryonic development. As little is known about the signaling and transcriptional network regulating the proliferation to differentiation transition during epidermal development, our elucidation of an in vivo role for Ovol1, a target of well-known signaling pathways, offers interesting new angles to understand these cellular and developmental processes.
The molecular mechanism of Ovol1's function in growth arrest
How does Ovol1 down-regulate the proliferation of embryonic epidermal progenitor cells? We probed this important question by characterizing the DNA binding specificity of the protein and looking for possible downstream targets. These studies led us to the discovery of c-myc as a direct Ovol1 target. Repression of the c-myc promoter by Ovol1 in reporter assays as well as results of ChIP assays detecting a physical association of Ovol1 to the endogenous c-myc promoter in its chromatin context indicate that Ovol1 can directly repress c-myc expression. This provides at least one possible molecular mechanism by which Ovol1 down-regulates proliferation (Fig. 9 B). The up-regulation of c-myc expression in Ovol1-deficient suprabasal cells as well as the phenotypic parallel, namely abnormal suprabasal proliferation, between Ovol1-deficient mice and transgenic mice that overexpress c-myc under the suprabasal-expressing involucrin or loricrin promoter provide in vivo validation for this model (Pelengaris et al., 1999; Waikel et al., 1999; Flores et al., 2004).
The underlying mechanism by which Ovol1 is recruited to the c-myc promoter appears complex, as we found two regions in the promoter that mediate Ovol1 repression and are bound by Ovol1 inside cells, yet only one region contains a detectable in vitro binding site. Alternative mechanisms, such as the use of DNA-binding partners to enhance binding affinity/specificity or via protein–protein interactions (Massague, 2000), likely exist as additional means to recruit Ovol1 to its target promoters in vivo. Future work is necessary to systematically explore these “hidden” cis-elements to fully understand the biochemical mechanism of Ovol1 repression.
It is unlikely that c-myc serves as the only Ovol1 target to mediate its negative effect on proliferation. Our previous study on the role of Ovol1 in male germ cell differentiation identified Id2 as a direct target of the Ovol1 protein (Li et al., 2005a). The observation of increased Id2 expression in Ovol1-deficient skin suggests that Id2 is also repressed by Ovol1 during epidermal development, probably both directly by Ovol1 binding to its promoter and indirectly because of increased c-myc gene products, as it has been shown that c-myc induces Id2 expression in epidermis (Murphy et al., 2004). As both c-myc and Id2 have been implicated in tumorigenesis, these findings also raise the possibility that Ovol1 might play a negative role in malignant growth. Although the biological function of ovo genes has been studied in various organisms, our study reports the first identification of candidate molecular targets of ovo, namely c-myc and Id2, two key positive regulators of proliferation and negative regulators of differentiation, that bear relevance to the cellular process that they regulate.
Ovol1 and terminal differentiation in the developing epidermis
Is Ovol1 also required for terminal differentiation itself in the developing epidermis? The expression of differentiation markers such as K1, loricrin, and TG3 is detected in the Ovol1 mutant, indicating that the epidermis is able to execute a largely normal terminal differentiation process in the absence of a functional Ovol1 gene. This said, morphological defects in the granular layers as well as a premature activation of loricrin expression in the intermediate layers were observed. These abnormalities are subtle and are apparently not translated into severe functional impairment of the skin, as mutant embryos acquire a functional barrier with only a transient delay of ∼1 d or so (unpublished data).
The premature activation of loricrin expression in the absence of Ovol1 together with the observation that Ovol1 represses the activity of the loricrin promoter in reporter assays led us to propose that Ovol1 might normally act to transiently repress loricrin expression in late presumptive spinous layers (Fig. 9). As the developing epidermis switches from a growing to a differentiating mode during late embryogenesis, a proposed involvement of Ovol1 in preventing premature terminal differentiation events at the critical cross-road might be important to ensure an orderly progression of the terminal differentiation-associated gene expression program. Alternatively, the up-regulated loricrin expression in Ovol1-deficient epidermis from E16.5 onward might be a secondary consequence of the mutation. Clearly, future studies are necessary to distinguish between these possibilities.
Materials and methods
Mouse breeding
Ovol1 +/− mice in a 129 × B6 mixed genetic background were backcrossed with B6 mice for 8–10 sequential generations, which, under no selection, is expected to generate a genetic background that is ∼99.9% B6. Ovol1 +/− mice with an enriched B6 genetic background were then intercrossed to produce homozygous mutant progeny for study.
Histology and immunofluorescence
Embryos or backskin samples were fixed in Bouin's fixative for 12–24 h at room temperature or in 4% PFA overnight at 4°C. 5 μm of paraffin or 5 μm of frozen sections were prepared and stained with hematoxylin and eosin or the appropriate polyclonal antibodies as described previously (Dai et al., 1998): rabbit K1 (1:500; Covance), rabbit K14 (1:1,000; Covance), guinea pig K14 (1:50; a gift from D. Roop, Baylor College of Medicine, Houston, TX; Waikel et al., 2001), rabbit loricrin (1:50; Mehrel et al., 1990), rabbit TG3 (1:100; a gift from L. Milstone, Yale University School of Medicine, New Haven, CT), rabbit K6 (1:200; a gift from P. Coulombe, Johns Hopkins University School of Medicine, Baltimore, MD; Mazzalupo et al., 2003), mouse c-myc (1:500; Abcam), and rabbit phosphorylated histone-H3 (1:1,000; Upstate Biotechnology). Images were acquired with a microscope (Eclipse E600; Nikon).
BrdU labeling
Pregnant females were injected intraperitoneally with BrdU (Sigma-Aldrich) at a dosage of 50 μg/g of body weight, killed 2 h after injection, and embryos were dissected and fixed in 4% PFA. Embryos were then washed in PBS and frozen in optimal cutting temperature (Tissue-Tek). Frozen sections were treated with 50% formamide in 2× SSC at 65°C for 2 h followed by two brief 5-min rinses in 2× SSC and were incubated in 2N HCl at 37°C for 30 min. Samples were neutralized by incubation in 0.1M boric acid, pH 8.5, for 10 min, rinsed briefly in PBS, and endogenous peroxidase was quenched by incubating in freshly prepared 3% H2O2 for 15 min. After three 5-min washes in PBS, samples were subjected to immunohistochemical analysis using a mouse monoclonal anti-BrdU antibody (Roche) according to the manufacturer's instructions.
Culture and analysis of primary keratinocytes
Keratinocytes were isolated from newborn backskin of mutant and wild-type littermates using an established protocol (Caldelari et al., 2000). About 4–6 × 106 cells were recovered from each mouse and were plated at comparable cell densities (passage 0). The total number of attached cells obtained from each mouse was counted after 5 d of the initial plating and subsequently normalized against the total number of cells plated. For growth curve analysis, 3 × 105 cells of passage 1 were plated in replicate wells of six-well plates, and the total number of cells at 2, 3, and 4 d after plating were counted.
For determination of the BrdU-labeling index in culture, wild-type and mutant keratinocytes were seeded in chamber slides precoated with collagen (36.9 μg/ml in PBS) and fibronectin (5 μg/ml in PBS) and were allowed to grow overnight. CaCl2 (final concentration of 1.2 mM), LiCl (final concentration of 20 mM), or TGF-β (final concentration of 1 ng/ml; Research Diagnostic) was added to the culture, and samples were fixed at various time points after the addition as indicated in the figures. BrdU was added 1 h before fixation at a final concentration of 10 μM, and fixation was in 100% methanol at −20°C for 10 min followed by three 5-min washes with 1× PBS. Keratinocytes were subsequently treated with 1N HCl for 30 min at 37°C followed by brief washes in PBS and were subject to immunohistochemical analysis as described in the previous section.
EMSA
Electrophoretic mobility shift assays (EMSAs) were performed using different amounts of partially purified recombinant His6-Ovol1 (final concentrations of Ovol1 were in the range of 47–600 nM) and ∼20 fmol (∼3 × 104 cpm) of gel-purified, 5′ 32P end-labeled double-stranded oligonucleotides. Typically, binding reactions were performed in a 20-μl volume containing 20 mM Hepes, pH 7.9, 75 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 1 mM EDTA, 12% glycerol, and 1 μg of poly(dI-dC) for 30 min at room temperature. In competition experiments, a 100-fold molar excess of unlabeled competitor was used. The protein–DNA complexes were resolved on 6% nondenaturing polyacrylamide gels and visualized by autoradiography.
Reporter assays
UG1 keratinocytes were cultured and transfected as previously described (Li et al., 2002). 293T cells were seeded in 24-well plates and transfected at 12–15% confluence with calcium phosphate as described previously (Pear et al., 1993). A typical transfection mixture contained a total of 0.5 μg of plasmids, including 0.05 μg of a promoter construct (pGL3–c-myc, in which a 2.3-kb human c-myc promoter fragment [a gift from G. Radziwill, University of Zurich, Zurich, Switzerland] drives the luciferase reporter [Hay et al., 1987], or pGL3-loricrin, in which a 1.3-kb mouse loricrin promoter fragment encompassing both the upstream and downstream transcription start sites drives the luciferase reporter [DiSepio et al., 1995]), with varying amounts of pCB6-Ovol1, an Ovol1 expression vector (Dai et al., 1998), and 0.04 μg of a β-actin–β- galactosidase construct or a total of 0.5 μg of plasmids, including 10 ng of a promoter construct with varying amounts of the VP16-Ovol1–expressing vector and 0.04 μg of a β-actin promoter–β-galactosidase construct. pCB-6 (+) (empty vector containing the cytomegalovirus promoter) was used as stuffer DNA. Luciferase activity was measured in whole cell extracts using the Luciferase Assay System (Promega), and β-galactosidase activity was measured as previously described (Eustice et al., 1991). For transfecting primary keratinocytes, cells isolated from each newborn were plated in two 35-mm plates, one of which was treated with CaCl2 (final concentration of 1.2 mM) 24 h after plating. 3 h after calcium addition, each plate was transfected with 700 ng pGL3–c-myc and 350 ng β-actin–β-galactosidase using the helium-driven gene gun system (Biolistic PDS-1000; Bio-Rad Laboratories). Cells were collected 24 h later, luciferase activity was measured using the Luciferase Assay System (Promega), and β-galactosidase activity was measured using the Galacto-Light system (Tropix).
ChIP assays
293T cells (a human kidney epithelial cell line) were seeded in 10-cm plates, and each plate was transfected with 6 μg pCB6-Ovol1. PCR amplification of the chromatin immunoprecipitates, prepared using the ChIP Assay Kit (Upstate Biotechnology) and anti-Ovol1 antibody (Dai et al., 1998) according to the manufacturer's instructions, was performed using the following primers containing sequences of the human c-myc promoter: 1F, 5′-AAGGAACCGCCTGTCCTTCC-3′; 1R, 5′-GCAACCAATCGCTATGCTGGA-3′; 2F, 5′-GGGAAAGAGGACCTGGAAAGG-3′; 2R, 5′-AGAGACAAATCCCCTTTGCGC-3′; 3F, 5′-ATCCAATCCAGATAGCTGTGC-3′; 3R, 5′-AAGAAGGGTATTAATGGGCGC-3′; 4F, 5′-ATCCTCTCTCGCTAATCTCCG-3′; 4R, 5′-TATTCGCTCCGGATCTCCCTT-3′; 5F, 5′-CCGCCTGCGATGATTTATACT-3′; and 5R, 5′-TTCTTTTCCCCCACGCCCT-3′. The following PCR program was used: 94°C for 1 min followed by 30 cycles of 94°C for 45 s, 60°C for 45 s, and 72°C for 1 min followed by a final extension at 72°C for 7 min. The percent input value was calculated for each specific primer set as follows: (PCR band intensity from anti-Ovol1 immunoprecipitate – PCR band intensity in IgG sample)/PCR band intensity from the input sample before immunoprecipitation.
Northern blot analysis, in situ hybridizations, and RT-PCR
Total RNA was extracted from embryonic skin, and Northern analysis was performed as described previously (Dai et al., 1998) using the following cDNA probes: a 304-bp fragment containing sequences corresponding to 222–535 (5′-UTR) of c-myc mRNA (GenBank/EMBL/DDBJ accession no. X01023) and a 407-bp fragment containing sequences corresponding to 1,062–1,468 of Id2 mRNA (GenBank/EMBL/DDBJ accession no. AF077860; a gift from S. Sinha, State University of New York at Buffalo, Buffalo, NY). In situ hybridizations were performed as described previously (Dai et al., 1998) using digoxigenin-labeled antisense and sense cRNA probes synthesized from a c-myc EST clone (Invitrogen). For RT-PCR, 5 μg RNA was reverse transcribed into cDNA using Superscript II RNase H reverse transcriptase (Invitrogen). PCR reactions were performed using the following primer set for loricrin: 5′-GTTCCTATGGAGGTGGTTCCAGCTG-3′ and 5′-TCCGTAGCTCTGGCACTGATACTGT-3′. For real-time PCR analysis, total RNA was extracted from primary keratinocytes cultured under low Ca2+ conditions or treated for 24 h with CaCl2 (final concentration of 1.2 mM) and reversed transcribed into cDNA. PCR reactions were set up using the iQ SYBR Green Supermix (Bio-Rad Laboratories) and gene-specific primer pairs for c-myc (F: CTCGCTCTCCATCCTATG; R: CAAGTAACTCGGTCATCATC) and glyceraldehyde-3-phosphate dehydrogenase (F: CCTGCCAAGTATGATGAC; R: GGAGTTGCTGTTGAAGTC). Reactions were completed on a real-time PCR machine (iCycler; Bio-Rad Laboratories) according to the manufacturer's recommendations.
Acknowledgments
We are grateful to Elaine Fuchs, Dennis Roop, Leonard Milstone, Pierre Coulombe, Satrajit Sinha, and Gerald Radziwill for providing reagents and to Julie Segre, Rob Steele, and Bogi Andersen for critical reading of this manuscript. We thank Magid Fallahi for technical assistance with mouse breeding.
This work was supported by National Institutes of Health (NIH) grants R01-AR47320 and K02-AR51482 to X. Dai. M. Nair was partially supported by an institutional predoctoral NIH Training Program in Developmental Mechanisms Underlying Congenital Defects.
M. Nair and A. Teng contributed equally to this paper.
B. Li's present address is School of Life Sciences, Xiamen University, Xiamen, Fujian 361005, China.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; Id, inhibitor of differentiation; TG3, transglutaminase 3.
References
- Arabi, A., S. Wu, K. Ridderstrale, H. Bierhoff, C. Shiue, K. Fatyol, S. Fahlen, P. Hydbring, O. Soderberg, I. Grummt, et al. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7:303–310. [DOI] [PubMed] [Google Scholar]
- Bond, V.C., and B. Wold. 1993. Nucleolar localization of myc transcripts. Mol. Cell. Biol. 13:3221–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, C., M. Tainsky, and E. Fuchs. 1994. Programming gene expression in developing epidermis. Development. 120:2369–2383. [DOI] [PubMed] [Google Scholar]
- Caldelari, R., M.M. Suter, D. Baumann, A. De Bruin, and E. Muller. 2000. Long-term culture of murine epidermal keratinocytes. J. Invest. Dermatol. 114:1064–1065. [DOI] [PubMed] [Google Scholar]
- Dai, X., and J.A. Segre. 2004. Transcriptional control of epidermal specification and differentiation. Curr. Opin. Genet. Dev. 14:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X., C. Schonbaum, L. Degenstein, W. Bai, A. Mahowald, and E. Fuchs. 1998. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 12:3452–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSepio, D., A. Jones, M.A. Longley, D. Bundman, J.A. Rothnagel, and D.R. Roop. 1995. The proximal promoter of the mouse loricrin gene contains a functional AP-1 element and directs keratinocyte-specific but not differentiation-specific expression. J. Biol. Chem. 270:10792–10799. [DOI] [PubMed] [Google Scholar]
- Eustice, D.C., P.A. Feldman, A.M. Colberg-Poley, R.M. Buckery, and R.H. Neubauer. 1991. A sensitive method for the detection of beta-galactosidase in transfected mammalian cells. Biotechniques. 11:739–740, 742–743. [PubMed] [Google Scholar]
- Flores, I., D.J. Murphy, L.B. Swigart, U. Knies, and G.I. Evan. 2004. Defining the temporal requirements for Myc in the progression and maintenance of skin neoplasia. Oncogene. 23:5923–5930. [DOI] [PubMed] [Google Scholar]
- Fuchs, E. 1993. Epidermal differentiation and keratin gene expression. J. Cell Sci. Suppl. 17:197–208. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and S. Raghavan. 2002. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 3:199–209. [DOI] [PubMed] [Google Scholar]
- Gugasyan, R., A. Voss, G. Varigos, T. Thomas, R.J. Grumont, P. Kaur, G. Grigoriadis, and S. Gerondakis. 2004. The transcription factors c-rel and RelA control epidermal development and homeostasis in embryonic and adult skin via distinct mechanisms. Mol. Cell. Biol. 24:5733–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley, L.R., J.A. D'Anna, S.S. Barham, L.L. Deaven, and R.A. Tobey. 1978. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 84:1–15. [DOI] [PubMed] [Google Scholar]
- Hardman, M.J., P. Sisi, D.N. Banbury, and C. Byrne. 1998. Patterned acquisition of skin barrier function during development. Development. 125:1541–1552. [DOI] [PubMed] [Google Scholar]
- Hay, N., J.M. Bishop, and D. Levens. 1987. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1:659–671. [DOI] [PubMed] [Google Scholar]
- Hennings, H., D. Michael, C. Cheng, P. Steinert, K. Holbrook, and S.H. Yuspa. 1980. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 19:245–254. [DOI] [PubMed] [Google Scholar]
- Hu, Y., V. Baud, M. Delhase, P. Zhang, T. Deerinck, M. Ellisman, R. Johnson, and M. Karin. 1999. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 284:316–320. [DOI] [PubMed] [Google Scholar]
- Johnson, A.D., D. Fitzsimmons, J. Hagman, and H.M. Chamberlin. 2001. EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development. 128:2857–2865. [DOI] [PubMed] [Google Scholar]
- Koch, P.J., P.A. de Viragh, E. Scharer, D. Bundman, M.A. Longley, J. Bickenbach, Y. Kawachi, Y. Suga, Z. Zhou, M. Huber, et al. 2000. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J. Cell Biol. 151:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster, M.I., and D.R. Roop. 2004. The role of p63 in development and differentiation of the epidermis. J. Dermatol. Sci. 34:3–9. [DOI] [PubMed] [Google Scholar]
- Koster, M.I., S. Kim, A.A. Mills, F.J. DeMayo, and D.R. Roop. 2004. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 18:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz, M., U. Valcourt, R. Bergstrom, C.H. Heldin, and A. Moustakas. 2004. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol. Cell. Biol. 24:4241–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella, A., M. Noseda, M. Beyna, Y. Yokota, and A. Iavarone. 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 407:592–598. [DOI] [PubMed] [Google Scholar]
- Lechler, T., and E. Fuchs. 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 437:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., D.R. Mackay, Q. Dai, T.W.H. Li, M. Nair, M. Fallahi, C. Schonbaum, J. Fantes, A. Mahowald, M.L. Waterman, et al. 2002. The LEF1/β-catenin complex activates movo1, a mouse homolog of Drosophila ovo gene required for epidermal appendage differentiation. Proc. Natl. Acad. Sci. USA. 99:6064–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., M. Nair, D.R. Mackay, V. Bilanchone, M. Hu, M. Fallahi, H. Song, Q. Dai, P.E. Cohen, and X. Dai. 2005. a. Ovol1 regulates meiotic pachytene progression during spermatogenesis by repressing Id2 expression. Development. 132:1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Q. Lu, G. Estepa, and I.M. Verma. 2005. b. Identification of 14-3-3sigma mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proc. Natl. Acad. Sci. USA. 102:15977–15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, D.R., M. Hu, B. Li, C. Rheaume, and X. Dai. 2006. The mouse Ovol2 gene is required for cranial neural tube development. Dev. Biol. 291:38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague, J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1:169–178. [DOI] [PubMed] [Google Scholar]
- Mazzalupo, S., P. Wong, P. Martin, and P.A. Coulombe. 2003. Role for keratins 6 and 17 during wound closure in embryonic mouse skin. Dev. Dyn. 226:356–365. [DOI] [PubMed] [Google Scholar]
- McGowan, K.M., X. Tong, E. Colucci-Guyon, F. Langa, C. Babinet, and P.A. Coulombe. 2002. Keratin 17 null mice exhibit age- and strain-dependent alopecia. Genes Dev. 16:1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrel, T., D. Hohl, J.A. Rothnagel, M.A. Longley, D. Bundman, C. Cheng, U. Lichti, M.E. Bisher, A.C. Steven, P.M. Steinert, et al. 1990. Identification of a major keratinocyte cell envelope protein, loricrin. Cell. 61:1103–1112. [DOI] [PubMed] [Google Scholar]
- Mevel-Ninio, M., R. Terracol, C. Salles, A. Vincent, and F. Payre. 1995. ovo, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech. Dev. 49:83–95. [DOI] [PubMed] [Google Scholar]
- Mills, A.A., B. Zheng, X.J. Wang, H. Vogel, D.R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 398:708–713. [DOI] [PubMed] [Google Scholar]
- Murphy, D.J., L.B. Swigart, M.A. Israel, and G.I. Evan. 2004. Id2 is dispensable for Myc-induced epidermal neoplasia. Mol. Cell. Biol. 24:2083–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto, M.A. 2002. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 3:155–166. [DOI] [PubMed] [Google Scholar]
- Okuyama, R., B.C. Nguyen, C. Talora, E. Ogawa, A. Tommasi di Vignano, M. Lioumi, G. Chiorino, H. Tagami, M. Woo, and G.P. Dotto. 2004. High commitment of embryonic keratinocytes to terminal differentiation through a Notch1-caspase 3 regulatory mechanism. Dev. Cell. 6:551–562. [DOI] [PubMed] [Google Scholar]
- Oliver, B., N. Perrimon, and A.P. Mahowald. 1987. The ovo locus is required for sex-specific germ line maintenance in Drosophila. Genes Dev. 1:913–923. [DOI] [PubMed] [Google Scholar]
- Olmeda, D., S. Castel, S. Vilaro, and A. Cano. 2003. Beta-catenin regulation during the cell cycle: implications in G2/M and apoptosis. Mol. Biol. Cell. 14:2844–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payre, F., A. Vincent, and S. Carreno. 1999. ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature. 400:271–275. [DOI] [PubMed] [Google Scholar]
- Pear, W.S., G.P. Nolan, M.L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 90:8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris, S., T. Littlewood, M. Khan, G. Elia, and G. Evan. 1999. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell. 3:565–577. [DOI] [PubMed] [Google Scholar]
- Sanders, J.A., and P.A. Gruppuso. 2005. Nucleolar localization of hepatic c-Myc: a potential mechanism for c-Myc regulation. Biochim. Biophys. Acta. 1743:141–150. [DOI] [PubMed] [Google Scholar]
- Shipley, G.D., M.R. Pittelkow, J.J. Wille Jr., R.E. Scott, and H.L. Moses. 1986. Reversible inhibition of normal human prokeratinocyte proliferation by type beta transforming growth factor-growth inhibitor in serum-free medium. Cancer Res. 46:2068–2071. [PubMed] [Google Scholar]
- Sonnenberg, A., J. Calafat, H. Janssen, H. Daams, L.M. van der Raaij-Helmer, R. Falcioni, S.J. Kennel, J.D. Aplin, J. Baker, M. Loizidou, et al. 1991. Integrin α 6β4 complex is located in hemidesmosomes, suggesting a major role in epidermal cell-basement membrane adhesion. J. Cell Biol. 113:907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, K., O. Takeuchi, T. Tsujimura, S. Itami, O. Adachi, T. Kawai, H. Sanjo, K. Yoshikawa, N. Terada, and S. Akira. 1999. Limb and skin abnormalities in mice lacking IKKalpha. Science. 284:313–316. [DOI] [PubMed] [Google Scholar]
- Vriz, S., J.M. Lemaitre, M. Leibovici, N. Thierry, and M. Mechali. 1992. Comparative analysis of the intracellular localization of c-Myc, c-Fos, and replicative proteins during cell cycle progression. Mol. Cell. Biol. 12:3548–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel, R.L., X.J. Wang, and D.R. Roop. 1999. Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene. 18:4870–4878. [DOI] [PubMed] [Google Scholar]
- Waikel, R.L., Y. Kawachi, P.A. Waikel, X.J. Wang, and D.R. Roop. 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat. Genet. 28:165–168. [DOI] [PubMed] [Google Scholar]