Abstract
Eukaryotic pre-ribosomes go through cytoplasmic maturation steps before entering translation. The nucleocytoplasmic proteins participating in these late stages of maturation are reimported to the nucleus. In this study, we describe a functional network focused on Rei1/Ybr267w, a strictly cytoplasmic pre-60S factor indirectly involved in nuclear 27S pre-ribosomal RNA processing. In the absence of Rei1, the nuclear import of at least three other pre-60S factors is impaired. The accumulation in the cytoplasm of a small complex formed by the association of Arx1 with a novel factor, Alb1/Yjl122w, inhibits the release of the putative antiassociation factor Tif6 from the premature large ribosomal subunits and its recycling to the nucleus. We propose a model in which Rei1 is a key factor for the coordinated dissociation and recycling of the last pre-60S factors before newly synthesized large ribosomal subunits enter translation.
Introduction
Ribosome biogenesis is a very conserved process in the eukaryotic kingdom. In Saccharomyces cerevisiae, the pathway begins with transcription of the 35S and 5S ribosomal RNA (rRNA) precursors by RNA polymerases I and III, respectively. The association of ribosomal proteins and pre-ribosomal factors with nascent pre-rRNAs gives birth to a 90S pre-ribosomal complex that undergoes various steps of maturation. The 90S complex separates into a pre-60S complex, which will generate the large ribosomal subunit containing mature 25S, 5.8S, and 5S rRNAs, and a pre-40S complex, which will generate the small ribosomal subunit containing 18S rRNA. The maturation of both particles follows two distinct pathways, first in the nucleolus and then in the nucleoplasm, and finally in the cytoplasm after Crm1-dependent export through the nuclear pores (Hurt et al., 1999; Moy and Silver, 1999; for review see Johnson et al., 2002). Several factors are necessary for correct modification, cleavage, and processing of pre-rRNAs, positioning of ribosomal proteins, and export of the pre-60S and pre-40S particles (for review see Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Tschochner and Hurt, 2003).
For the large ribosomal subunit, apart from the 46 ribosomal proteins that are found in the mature particle, ∼100 pre-ribosomal factors have been identified to date. Except for a few factors with known enzymatic activity, the function of most of the pre-ribosomal factors biochemically associated with pre-60S complexes remains to be defined.
Throughout the pathway of biogenesis, the composition of the pre-60S particles changes. At the beginning of the pathway, they contain many pre-60S factors; these proteins gradually dissociate from the complexes in the nucleolus, the nucleoplasm, or the cytoplasm. Even if other pre-60S factors load later on the complexes, the average number of pre-60S factors is significantly lower at the end of biogenesis than at the beginning (Nissan et al., 2002). The whole process is highly dynamic; as they dissociate from the precursors of the large ribosomal subunits, pre-60S factors can be recycled and participate in new rounds of biogenesis.
In the late cytoplasmic events of 60S biogenesis, only a few pre-ribosomal factors are present on the pre-60S particles (for review see Fromont-Racine et al., 2003; Dez and Tollervey, 2004). Some of these factors are shuttling proteins such as Tif6 (Senger et al., 2001), Rlp24 (Saveanu et al., 2003), Nmd3 (Ho et al., 2000a; Gadal et al., 2001), and Arx1 (Nissan et al., 2002), which are loaded on the pre-60S complexes in the nucleus and undergo export toward the cytoplasm together with the particle. Other factors seem exclusively cytoplasmic, such as the GTPases Lsg1 (Kallstrom et al., 2003) and Efl1 (Senger et al., 2001).
Existing data reveal that recycling shuttling factors and triggering the end of biogenesis are tightly intertwined events. Indeed, since the beginning of the 1980s, the pre-60S factor Tif6 is thought to act as an antiassociation factor between the small and large subunits (Valenzuela et al., 1982); the GTPase Efl1 triggers the recycling of Tif6 (Senger et al., 2001). Recycling of the Nmd3 shuttling protein also requires the GTPase Lsg1 and the late-associating ribosomal protein Rpl10 (Gadal et al., 2001; Hedges et al., 2005).
The shuttling of pre-60S factors between the cytoplasm and the nucleus involves the general import machinery. Recycling of Nmd3 is mediated by the β-karyopherin Kap123 (Sydorskyy et al., 2003), which also accounts for the import of various ribosomal proteins to the nucleus through the nuclear pore complexes (Rout et al., 1997). At least three β-karyopherins participate in the import of ribosomal proteins and pre-ribosomal factors— Kap123, Kap121/Pse1, and Kap104 (Leslie et al., 2004)—with a significant overlap as far as their specificity for cargoes is concerned.
In this study, we investigate late events in the maturation of the large ribosomal subunit in which a newly described, exclusively cytoplasmic pre-60S factor, Rei1/Ybr267w, plays a central role. Our results indicate that coordinated dissociation and return to the nucleus of shuttling pre-60S factors are essential for correct maturation of the large ribosomal subunits and participate in their activation for subsequent entry into translation.
Results
Rei1 is physically linked to Rpl24
Most ribosomal proteins of the large subunit are loaded on 60S pre-ribosomal particles in the early nuclear steps of 60S biogenesis. However, a few proteins, such as Rpl10 and Rpl24, appear to associate later to the pre-60S particles (Kruiswijk et al., 1978). Rpl24 is a late-associating ribosomal protein that shares significant homology with the essential pre-ribosomal factor Rlp24 (Saveanu et al., 2003). Rlp24 is present on pre-60S particles from the nucleolus to the cytoplasm, where it is believed to dissociate when Rpl24 associates with the particle.
To identify potential partners required for the loading of Rpl24 to the particles, a two-hybrid screen using Rpl24b as bait was performed. The most frequent prey selected in the screen was REI1/YBR267W, which is found as seven distinct inserts. All of the inserts containing the REI1 ORF shared a minimal interacting domain from amino acids 270 to 342 (Fig. 1 A).
Figure 1.
Rei1 is physically associated with nontranslating, Rpl24-containing complexes. (A) Rei1 is a two-hybrid prey of Rpl24b. YBR267W (REI1) was found as prey ORF in a genomic two-hybrid screen performed with RPL24B as bait. We represented the 393–amino acid–long protein and pointed out its putative motifs: three conserved C2H2 Zn fingers (dark gray) and the PFAM E-MAP-115 motif (stripes). Underneath, displayed as black rectangles, are the seven distinct DNA portions of REI1 that were found as preys. The minimal interacting domain is indicated in light gray. (B) Rei1-myc copurifies specifically with Rpl24b-TAP. TAPs were performed with strains expressing Rei1-13Myc together with Ssf1-TAP, Rlp24-TAP, Rpl24-TAP, or in a nontagged strain. The components of the purified complexes were separated by SDS-PAGE as shown on the Coomassie staining of the TEV eluate. Rei1-13Myc and Nog1 were detected in the TEV eluates by immunoblotting. As a loading control, Rei1-13Myc was detected in the input fractions (top). (C) Rei1 is associated with 60S particles. Whole cell extracts were prepared from a strain expressing Rpl24b-TAP and separated on a sucrose gradient by ultracentrifugation. Absorbance at 254 nm was measured, and 0.5-ml fractions of the gradient were collected. Peaks corresponding to the 40S, 60S, and 80S particles or to the polysomes are indicated. The proteins of each fraction were TCA precipitated and analyzed by Western blotting with antibodies directed against Rei1 or with PAP for the detection of the TAP fusion protein.
Rei1 is a 393–amino acid–long protein that contains three conserved C2H2 Zn finger motifs extending from amino acids 7 to 31, 162 to 187, and 215 to 239. The domain extending from amino acids 57 to 202 displays similarity with the PFAM E-MAP-115 domain characteristic of ensconsin, a microtubule-associated protein expressed in higher eukaryotes (Masson and Kreis, 1993). The minimal two-hybrid interaction domain between Rei1 and Rpl24b is located in the COOH-terminal part of the protein apart from all of these motifs. This region is quite conserved in Rei1 homologues among eukaryotes, mainly in the portion extending from amino acids 300 to 330 (unpublished data).
Rei1 is associated with Rpl24-containing 60S complexes but is absent from the polysomes
The two-hybrid link between Rei1 and Rpl24 suggested that Rei1 might participate in molecular events involving the large ribosomal subunit together with the late-associating ribosomal protein Rpl24. This hypothesis was supported by previous tandem affinity purifications (TAPs), which had revealed the presence of Rei1 in several pre-60S complexes (Gavin et al., 2002; Nissan et al., 2002). The clustering of these TAP data (for review see Fromont-Racine et al., 2003) had already shed light on Rei1 as a putative late pre-60S factor. Therefore, we further investigated the possible role of Rei1 in the cytoplasmic events of 60S biogenesis.
In addition to its cytoplasmic localization (Iwase and Toh-e, 2004), the interaction of Rei1 with Rpl24 prompted us to determine whether this factor was associated with ribosomal or pre-ribosomal complexes in the cytoplasm. We performed TAP experiments on strains producing a Rei1 protein fused to a 13Myc epitope and either Rlp24 or Rpl24b fused to the TAP tag. Rei1-13Myc was enriched in the Rpl24b-TAP–associated complexes compared with the Rlp24-TAP–associated complexes or to the strictly nuclear Ssf1-TAP–associated complexes used as negative controls (Fig. 1 B). In contrast, the presence of Nog1 was observed, as expected, in both Ssf1-TAP– and Rlp24-TAP–associated pre-ribosomal complexes but not in the Rpl24b-TAP–associated complex. These data suggest that Rei1 is specifically associated with Rpl24-containing cytoplasmic complexes.
To analyze a potential association of Rei1 with mature translating ribosomes, total cellular extracts from cells producing Rpl24b-TAP were fractionated on a sucrose gradient. As expected, Rpl24b-TAP was found in fractions corresponding to the 60S, 80S, and polysomes. We observed that Rei1 was absent from the 80S and polysomal fractions but present in the 60S fraction (Fig. 1 C), indicating that in contrast to Rpl24, Rei1 is not associated with mature ribosomes during translation. Exclusive association with 60S particles supported the hypothesis that Rei1 is a late pre-60S factor that transiently binds to pre-ribosomal complexes in the cytoplasm after the release of Rlp24 from the particles.
Rei1 is a late cytoplasmic 60S pre-ribosomal factor
The presence of Rei1 in the 60S fractions of a sucrose gradient and not in polysome fractions suggested that it might participate in the late events of the large subunit biogenesis or be involved in the very early steps of translation initiation.
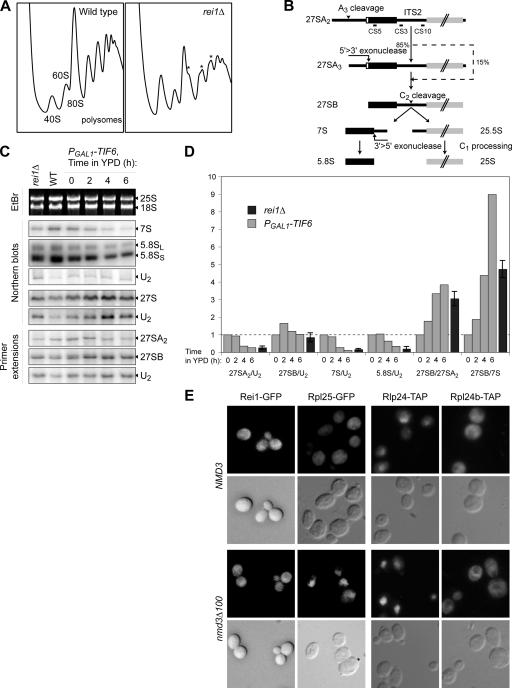
To determine whether Rei1 could be involved in ribosome biogenesis or in translation, the rei1Δ strain and the corresponding wild-type strain were used for sucrose gradient analysis. The rei1Δ strain displayed a slow growth phenotype at 30°C and was cold sensitive as previously observed in another genetic background (Iwase and Toh-e, 2004). In the rei1Δ strain, the polysome profile was clearly affected. At 30°C, the amount of 60S subunit decreased significantly (unpublished data). At 23°C, not only a decrease of the 60S subunit amount was observed, but also a drop in the average level of polysomes and the appearance of half-mers, which can be explained by abortive 48S preinitiation complexes being formed but remaining complexes being blocked on mRNA because of a lack of mature large ribosomal subunits (Fig. 2 A).
Figure 2.
Absence of Rei1 affects the 60S maturation. (A) The rei1Δ strain displays an altered polysome profile. The disrupted strain and wild-type strain were grown for 16 h at 23°C in yeast extract–peptone–d-glucose (YPD). Whole cell extracts were separated on sucrose gradients as described in Fig. 1 C. In the rei1Δ strain (right), the position of half-mers is highlighted by asterisks. (B) Schematic representation of the maturation of the large subunit rRNA from 27SA2 to 5.8S and 25S. The positions of the oligonucleotide probes used in this study are indicated. (C) Effects of rei1Δ on pre-rRNA processing are similar to those of a TIF6 repression. Total RNAs were extracted from a rei1Δ or wild-type strain grown for 16 h at 23°C in YPD or from a PGAL1-TIF6 strain shifted to YPD from 0 to 6 h. 25S and 18S rRNAs were detected by ethidium bromide staining. 27SA2 and 27SB pre-rRNAs were detected by primer extension with probe CS10 (see Fig. 2 B). 7S pre-rRNA, 5.8S rRNA, and total 27S species were detected by Northern blotting with the CS3, CS5, and CS10 probes, respectively. (D) The absence of Rei1 or Tif6 correlates with a defect in the C1 + C2 rRNA processing step. Ratios were calculated either between two mature rRNA or pre-rRNA detected in the same samples on the same gel or after normalization with a control RNA (U2 small nuclear RNA) detected with oligonucleotide MFR457. RNA levels in the rei1Δ strain were standardized to the wild-type levels, indicated as a dotted line; for the TIF6 repression, the levels were indicated relative to those at time = 0. For rei1Δ, the calculation was performed on four distinct experiments; error bars represent SD. (E) Rei1 is not a shuttling factor. Strains producing Rei1-GFP, Rpl25-GFP, Rlp24-TAP, or Rpl24b-TAP were transformed with plasmids pAJ544 and pAJ368 overexpressing wild-type NMD3 or the dominant-negative nmd3Δ100 allele, respectively. The TAP fusion proteins were revealed by immunocytochemistry, and all samples were observed by epifluorescence microscopy.
The clearly affected polysome profiles in the rei1Δ strain correlated with a defect in rRNA maturation (Fig. 2, C and D). Ethidium bromide staining of agarose gels, Northern blots, and primer extensions were performed to determine the relative levels of various rRNA intermediates and mature species. The rRNA defect was already significant at 30°C (unpublished data) but increased at 23°C. At this temperature, we observed a drop in the levels of total 27S species, 27SA2 and 7S nuclear pre-rRNAs, as well as mature 5.8S rRNA normalized relatively to U2 small nuclear RNA levels. The 27SB/27SA2 pre-rRNA ratio was increased by 3.1 ± 0.4, and the 27SB/7S ratio was increased by 4.7 ± 0.5. Altogether, these data are typical of a defect in the ITS2 processing step (Fig. 2 B). They are in correlation with the drastic decrease in the 60S peak on sucrose gradients, which results in a stoichiometric imbalance between the small/large ribosomal subunit (Fig. 2 A).
Curiously, although Rei1 appears to be a cytoplasmic protein, its absence displayed rRNA maturation defects at the nuclear level. To investigate whether Rei1 might be a shuttling factor, we monitored Rei1-GFP localization in a strain defective for the export of pre-60S particles (Fig. 2 E). We used a strain in which a version of the nmd3Δ100 dominant-negative mutant was overexpressed compared with the same strain overexpressing a wild-type NMD3 version (Ho et al., 2000b). As a control, we monitored the localization of the ribosomal protein Rpl25-GFP. As expected, Rpl25-GFP, which is normally mainly cytoplasmic, was accumulated in the nucleus when nmd3Δ100 was overexpressed. In contrast, no accumulation was seen for Rei1-GFP, which appears to be an exclusively cytoplasmic pre-60S factor.
We additionally tested the shuttling ability of pre-ribosomal factor Rlp24-TAP and of ribosomal protein Rpl24b-TAP (Fig. 2 E). In the nmd3Δ100 dominant-negative mutant, Rlp24-TAP accumulated in the yeast nuclei compared with wild-type conditions. Meanwhile, Rpl24b-TAP remained exclusively cytoplasmic. This confirmed previous data (Saveanu et al., 2003) in favor of Rlp24 being a shuttling nucleocytoplasmic pre-60S factor, whereas Rpl24 appears to be a ribosomal protein that associates with pre-60S particles in the very last cytoplasmic steps of ribosome biogenesis.
As a late pre-60S factor, Rei1 was a candidate for participating in the export of pre-60S particles from the nucleus to the cytoplasm. Therefore, we monitored the localization of an Rpl25-GFP reporter construct in a rei1Δ strain. Because no export defect was observed (unpublished data), we assume that Rei1 is not required for the export of precursors of the large ribosomal subunit.
Altogether, these results are consistent with Rei1 being involved in the large ribosomal subunit biogenesis. Though it is a cytoplasmic nonshuttling protein, its absence results in defects in nuclear steps of the 60S maturation, suggesting that Rei1 could be required for the recycling of shuttling pre-60S factors from the cytoplasm to the nucleus.
The absence of Rei1 affects nuclear import of the shuttling factors Arx1 and Tif6
Considering the previous hypothesis, we investigated the possible role for Rei1 in the recycling of shuttling pre-60S factors such as Tif6 (Senger et al., 2001), Rlp24 (Saveanu et al., 2003), and Arx1 (Nissan et al., 2002).
We looked at the localization at 23°C of the Arx1-GFP, Tif6-GFP, or Rlp24-TAP fusion proteins in a rei1Δ strain compared with a wild-type strain (Fig. 3 A). In the wild-type strains, Arx1-GFP and Tif6-GFP were mainly observed in the nucleus, and a small fraction of the proteins was found in the cytoplasm. In the absence of Rei1, Arx1-GFP or Tif6-GFP were mainly cytoplasmic. This confirmed our hypothesis that nuclear import of some shuttling factors could be impaired in the absence of Rei1. In contrast, no recycling defect was observed for Rlp24-TAP, which even appeared somewhat concentrated in the nucleolus of the rei1Δ strain compared with the wild-type strain, where it appeared more equally distributed between the nucleolus, nucleoplasm, and cytoplasm.
Figure 3.
Rei1 is functionally linked to the shuttling factors Arx1 and Tif6. (A) In the absence of Rei1, Arx1 and Tif6 are redistributed to the cytoplasm. The localization of Arx1-GFP, Tif6-GFP, or Rlp24-TAP was observed after the growth of rei1Δ or wild-type strains for 8 h in minimal medium at 23°C. (B) Rei1 is required for the binding of Arx1 to pre-60S complexes. rei1Δ or wild-type strains expressing ARX1-GFP or TIF6-GFP were grown for 8 h at 23°C, and whole cell extracts were prepared. The extracts were separated on sucrose gradients and analyzed as described in Fig. 1 C. The immunoblots, which were performed using antibodies against GFP, are displayed below the corresponding profile.
These data offer a simple explanation for the nuclear rRNA maturation defect observed in the rei1Δ strain. Indeed, the absence of Rei1 affects the processing of ITS2 in a manner similar to the repression of Tif6 (Basu et al., 2001). To better illustrate this, a strain in which TIF6 was placed under the control of a PGAL1 promoter was shifted to glucose from 0 to 6 h, and pre-rRNA species were analyzed and quantified along these kinetics (Fig. 2, C and D). The rRNA intermediates ratio of the rei1Δ strain to the wild type were comparable with the effects of a TIF6 repression for 4–6 h.
We also tested the phenotype of an ARX1 deletion, but no obvious rRNA maturation impairment was detected (unpublished data). Therefore, the 60S maturation defects observed in the rei1Δ strain are likely caused by a decrease of nuclear Tif6. We tested whether the overexpression of TIF6 could compensate this phenotype, but TIF6 overexpression appeared to be toxic in wild-type as well as in rei1Δ strains (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200510080/DC1).
As Arx1-GFP and Tif6-GFP accumulated in the cytoplasm in the absence of Rei1, we wondered whether they were still associated with pre-60S complexes. To address this question, we separated extracts from a rei1Δ or wild-type strain expressing chromosomal ARX1-GFP or TIF6-GFP on sucrose gradients (Fig. 3 B). In the wild-type strain, we could detect Arx1-GFP in the 60S fractions of the gradient. In the absence of Rei1, Arx1 appeared to sediment closer to the top of the gradient, which suggested that in such conditions, Arx1-GFP was present in the cytoplasm as a small complex or as an oligomeric protein. In contrast, in the absence of Rei1, the Tif6-GFP fusion protein was still associated with particles of the size of 60S. Although the absence of Rei1 leads to a cytoplasmic retention of both shuttling pre-60S factors Tif6 and Arx1, Tif6 is still stably associated with 60S particles, whereas Arx1 accumulates as a small complex in the cytoplasm.
Yjl122w/Alb1 is a novel 60S-associated factor that is tightly linked with Arx1
The presence of Arx1 in a small cytoplasmic complex in the absence of Rei1 suggested the possibility that Arx1 could associate and act together with other proteins. We performed a two-hybrid screen using Arx1 as bait to identify the physical partners of Arx1. The most frequent prey we selected was YJL122W, which is hereafter referred to as ALB1 (ARX1 little brother 1). This prey was found as seven distinct inserts; all of the inserts shared a minimal interacting domain from amino acids 148 to 175 (Fig. 4 A). The deletion of ALB1 had little effect on growth and displayed no obvious rRNA maturation impairment. In the arx1Δ, alb1Δ double mutant, the growth phenotype as well as the polysome profile resembled that of the arx1Δ single mutant (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.2005010080/DC1).
Figure 4.
Yjl122w/Alb1 is a physical partner of Arx1. (A) Alb1 is a two-hybrid prey of Arx1. YJL122W (ALB1) was found as prey ORF in a genomic two-hybrid screen performed with ARX1 as bait. The results are presented as in Fig. 1 A. (B) Rei1, Arx1, and Alb1 are associated with the same complexes. Rei1-, Arx1-, and Alb1-TAP–associated complexes were TAP purified. The Coomassie staining is displayed on the left. The presence of Arx1, Rei1, or Rlp24 in the TEV eluates was assessed by immunoblotting with specific antibodies against each of these proteins, as shown on the right, with the corresponding Ponceau red staining. (C) Alb1-TAP is associated with 60S particles. Whole cell extracts were prepared from a strain expressing Alb1-TAP, separated on a sucrose gradient, and analyzed as described in Fig. 1 C. Alb1-TAP was detected by Western blotting with PAP. (D) Arx1 and Alb1 interact directly in vitro. GST, GST-Rei1, GST-Alb1, and His6-Arx1 fusion proteins were produced in Escherichia coli BL21. Crude extracts containing the GST-tagged baits were mixed with equal amounts of crude extracts containing the His6-Arx1 putative prey and were purified on glutathione–Sepharose beads. The input samples as well as the eluted proteins were separated on a 10% polyacrylamide-SDS gel. The bait proteins were detected in the eluted fractions by Western blotting with antibodies against GST (top). His6-Arx1 was detected with antibodies against Arx1 in the fractions where it is coeluted (middle). As a charge control, His6-Arx1 was also detected in the input (bottom).
To confirm the physical interaction between Arx1 and Alb1, we purified the complexes associated with Alb1 (Fig. 4 B). Arx1- and Alb1-TAP–associated complexes look very similar on the Coomassie staining compared with Rei1-TAP. We could detect Arx1 and Rei1 in the three complexes. These results corroborate previous affinity precipitations (Ho et al., 2002; Krogan et al., 2004) where Alb1 had been identified in both Rei1- and Arx1-associated complexes. They not only confirm that Arx1 and Alb1 are physically associated under physiological conditions but also suggest that Alb1 could be a pre-60S–associated factor. Note that Rlp24 was present in Arx1- and Alb1-TAP–associated complexes but was absent from Rei1-TAP complexes, which confirms that Rei1 loads onto the particles after the release of Rlp24. In addition, we observed that Alb1-TAP sedimented around the 60S peak but was absent from the polysomal fractions on a sucrose gradient (Fig. 4 C); therefore, it is likely to associate with precursors of the large ribosomal subunit.
To determine whether any direct interaction could be detected between these factors, we performed in vitro GST pull-down experiments with GST-tagged baits (GST-Alb1, GST-Rei1, or GST) and the His6-Arx1 fusion protein as prey (Fig. 4 D). His6-Arx1 copurified with GST-Alb1 but not with GST or GST-Rei1 used as negative controls. Thus, we conclude that Arx1 and Alb1 are able to interact directly even in the absence of other yeast components.
The absence of Rei1 affects nuclear import of Alb1 and its binding to pre-60S particles
Because the nuclear import of Arx1 was affected in the absence of Rei1, we wondered whether Alb1 participated with Arx1 in the small cytoplasmic complex detected in these conditions. Therefore, we assessed the effects of the absence of Rei1 on Alb1 localization and association with 60S particles.
First, we tested whether Alb1 was an additional shuttling factor by monitoring the localization of an Alb1-GFP reporter construct in a wild-type or rei1Δ strain at 23°C (Fig. 5 A). In the wild-type strain, Alb1-GFP localization was similar to that of Arx1-GFP, with a strong nuclear–nucleolar signal and a weak cytoplasmic signal. In the rei1Δ mutant, Alb1 was relocalized to the cytoplasm, as was the case for Arx1.
Figure 5.
Alb1 is an additional shuttling pre-60S factor and target of Rei1. (A) In the absence of Rei1, Alb1 is blocked in the cytoplasm. The localization of Alb1-GFP was observed in rei1Δ or wild-type strains and grown for 8 h in minimal medium at 23°C. (B) Rei1 is required for the anchoring of Alb1 to pre-60S complexes. rei1Δ or wild-type strains expressing ALB1-GFP were grown for 16 h at 23°C, and whole cell extracts were prepared. The extracts were separated on sucrose gradients and analyzed as described in Fig. 1 C. The immunoblots, performed using antibodies against GFP or Arx1, are displayed below the corresponding profile. (C) Arx1 and Alb1 form a small nonribosomal complex in the absence of Rei1. Arx1- and Alb1-TAP–associated complexes were purified in wild-type or rei1Δ strains grown at 23°C. The Coomassie staining is shown on the left; the right panel displays the results of immunoblots using antibodies against Arx1, Nog1, Rei1, and Tif6 against the TEV eluates. Protein amounts were adjusted relative to Arx1. Positions of the tagged proteins are indicated by asterisks. (D) Arx1 and Kap121 cosediment with Alb1-TAP in rei1Δ strains. The TEV eluate from a purification of Alb1-TAP in the absence of Rei1 was further separated on sucrose gradients either directly (left) or after preincubation with calmodulin affinity resin (right; Alb1 complex subtracted). The components of the fractions were analyzed by immunoblotting using Arx1 and Kap121 antibodies after membrane staining by Ponceau red.
Second, in a sucrose gradient analysis (Fig. 5 B), Alb1-GFP could be detected in the 60S fractions in the wild-type strain together with Arx1, whereas in the rei1Δ strain, it was found mainly in the same fractions as Arx1 at the top of the gradient. Thus, the association of Alb1 to nuclear pre-60S particles was impaired, as it was for Arx1, in a rei1Δ strain.
We tried to further characterize the small complex observed in the absence of Rei1 by purifying the Arx1- and Alb1-TAP–associated complexes in these conditions (Fig. 5 C). The Coomassie staining showed a weak decrease of the levels of most proteins in the rei1Δ strain compared with the wild-type profiles, except for Arx1. In the absence of Rei1, immunoblots performed on the purified fractions revealed a loss of the pre-ribosomal factors Nog1 and Tif6. These results suggest that Arx1 and Alb1 can form cytoplasmic subcomplexes in the absence of Rei1 independently from pre-ribosomal particles.
To confirm these results, purified complexes associated with Alb1-TAP were separated on a sucrose gradient in the absence of Rei1 (Fig. 5 D). These complexes sedimented in the first five fractions of the gradient and contained Arx1 and Kap121, as discussed in the following paragraphs. In contrast to the other Ponceau-stained bands, these two proteins could be depleted by preincubating the TEV eluate with calmodulin affinity resin, which retains Alb1-CBP, indicating that they are strongly associated with this bait.
Reimport of shuttling factors into the nucleus in the absence of Rei1 restores growth
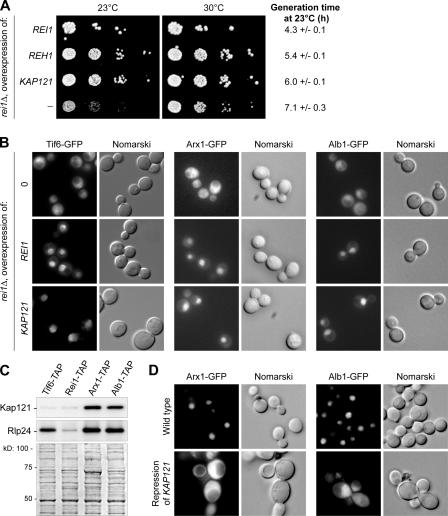
To gain more insights into the molecular events involved in the observed phenotypes in the absence of Rei1, we performed a high copy number suppressor screen with a strain deleted for REI1 grown at 20°C. In addition to REI1 itself, we selected the homologous gene REH1/YLR387C. The overexpression of this gene was able to partially complement the cold-sensitive phenotype of rei1Δ (Fig. 6 A) as previously shown (Iwase and Toh-e, 2004), yet we could not detect any defect in reh1Δ strains on polysome profiles nor on rRNA maturation (not depicted). Thus, we assume that although it shares partial functional overlap with Rei1, Reh1 is not limiting for the biogenesis of the large ribosomal subunit under physiological conditions.
Figure 6.
Kap121 is involved in the Rei1-dependent nuclear import of pre-60S factors. (A) KAP121 is a high copy number suppressor of rei1Δ. A rei1Δ strain was transformed with high copy number plasmids carrying distinct DNA inserts (REI1, REH1, KAP121, or no insert) and plated in 10−1 dilution series on synthetic medium without uracil at 23 or 30°C for 3 and 2 d, respectively. Generation times at 23°C were calculated from growth curves performed in liquid synthetic medium without uracil over a period of 30 h. The confidence intervals were calculated for a probability of 95%. (B) Overexpression of KAP121 is sufficient to overcome the import defect in rei1Δ strains. rei1Δ strains expressing ARX1-GFP, ALB1-GFP, or TIF6-GFP were transformed with vectors overexpressing REI1, KAP121, or nothing. The localization of Arx1-, Alb1-, or Tif6-GFP was detected after growth for 8 h in minimal medium without uracil at 23°C. (C) Kap121 copurifies with Arx1-TAP and Alb1-TAP. Tif6-, Rei1-, Arx1-, and Alb1-TAP–associated complexes were purified by TAP. The presence of Kap121 or Rlp24 in the eluates was assessed by Western blotting with specific antibodies against each of these proteins. The Ponceau red staining is shown on the bottom. (D) In the absence of Kap121, Arx1-GFP is blocked in the cytoplasm. KAP121 was placed under the control of the PGAL1 promoter in strains expressing ARX1-GFP or ALB1-GFP. The localization of Arx1-GFP or Alb1-GFP was detected in this mutant or in the corresponding wild-type strain after growth in glucose- containing medium for 16 h.
The third gene identified in our screen was KAP121/YMR308C (Fig. 6 A). It was selected as seven distinct DNA inserts containing the whole KAP121 ORF. The smallest insert constituted a DNA fragment extending from 589 bp upstream of the initiation codon to 762 bp downstream of the stop codon. Kap121, also known as Pse1, is a β-karyopherin, which was shown to be involved in the import of various ribosomal proteins (Rout et al., 1997) as well as pre-ribosomal factors such as Nop1 and Sof1 from the cytoplasm to the nucleus (Leslie et al., 2004). The mislocalization to the cytoplasm of Arx1-GFP, Alb1-GFP, or Tif6-GFP in a rei1Δ strain could be compensated by overexpression of either REI1 or KAP121 (Fig. 6 B). Both constructs were able to relocalize the three fusion proteins to the nuclear compartment compared with a strain transformed with an empty vector.
Because overexpression of the β-karyopherin Kap121 was sufficient to overcome the import defect of all three shuttling factors in the rei1Δ strain, one might wonder whether Kap121 was the physiological importin for these proteins. Such a hypothesis was supported by the presence of Kap121 in Arx1- and Alb1-TAP–associated complexes (Figs. 5 D and 6 C; Gavin et al., 2002; Nissan et al., 2002) and by two-hybrid data (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200510080/DC1). In contrast, Kap121 was not detected in Tif6-TAP–associated complexes; therefore, we did not further assess the possibility that Kap121 is the specific importin for this factor. As a control, Rlp24 was as efficiently copurified with Tif6-TAP as with Alb1-TAP or Arx1-TAP compared with Rei1-TAP, which was associated with neither Kap121 nor Rlp24 (Fig. 6 C).
Further evidence for Arx1 and Alb1 being Kap121 cargoes was provided by localization of the Arx1-GFP and Alb1-GFP fusion proteins in strains in which the expression of KAP121 was repressed (Fig. 6 D). Both fusions were redistributed to the cytoplasm compared with wild-type strains, where we observed bright nuclear–nucleolar signals. This result suggested that the karyopherin Kap121 might be the physiological importin for Arx1 and Alb1. Altogether, these results show that the nuclear import of Arx1 and Alb1 is Kap121 dependent. In the absence of Rei1, the return of Arx1–Alb1 to the nucleus as well as that of Tif6 can be restored by overexpressing this karyopherin.
The accumulation of Alb1–Arx1 in the cytoplasm prevents dissociation and recycling of Tif6 in rei1Δ strains
In the absence of Rei1, Arx1 and Alb1 accumulate in the form of a small complex, whereas Tif6 remains bound to the pre-60S particle. We hypothesized that the nonphysiological levels of the Arx1–Alb1 cytoplasmic subcomplex could prevent the dissociation of Tif6 from the premature large ribosomal subunit and subsequent return of this factor to the nucleus.
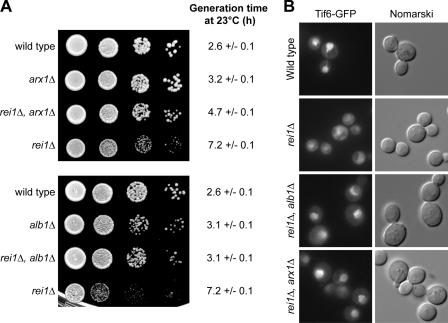
The deletion of either ARX1 or ALB1 in the rei1Δ strain restored growth at 23°C almost up to the levels of arx1Δ or alb1Δ single mutant strains, respectively (Fig. 7 A). Therefore, the cytoplasmic accumulation of Arx1–Alb1 accounts for the observed cold-sensitive phenotype of the rei1Δ strain.
Figure 7.
In the absence of Rei1, the cytoplasmic Arx1–Alb1 complex prevents the recycling of Tif6. (A) Disruptions of ARX1 or ALB1 compensate the cold-sensitive phenotype of a rei1Δ strain. Wild-type, rei1Δ, arx1Δ, or rei1Δ, arx1Δ strains were spotted on YPD medium in 10−1 dilution series (top); the same was performed for wild-type, rei1Δ, alb1Δ, or rei1Δ, alb1Δ strains (bottom). Cold-sensitive phenotypes were tested by incubating the plates for 3 d at 23°C. Generation times at 23°C were calculated from growth curves performed in liquid synthetic medium over a period of 30 h. The confidence intervals were calculated for a probability of 95%. (B) Disruptions of ARX1 or ALB1 compensate the Tif6 shuttling defect in a rei1Δ strain. The localization of Tif6-GFP was detected by epifluorescence microscopy in a wild-type, rei1Δ, and rei1Δ, alb1Δ or rei1Δ, arx1Δ double mutant strains.
According to our hypothesis, Arx1–Alb1 might also explain the cytoplasmic retention of Tif6. Thus, we monitored the localization of Tif6-GFP in rei1Δ alb1Δ or rei1Δ arx1Δ double mutant strains (Fig. 7 B). In these mutants, Tif6 was mainly nuclear, as in the wild-type strain, compared with the mostly cytoplasmic signal in the rei1Δ strain. Thus, we conclude that in the absence of Rei1, Arx1 and Alb1 participate in a cytoplasmic subcomplex that prevents the adequate release of Tif6 from pre-60S particles and its correct return to the nucleus for a new round of biogenesis.
Discussion
End of ribosome biogenesis and recycling of shuttling pre-60S factors are intertwined processes
At the end of ribosome biogenesis, all remaining pre-ribosomal factors on the particles are released into the cytoplasm. Recycling of these factors for a new round of biogenesis, instead of synthesizing new ones, constitutes a logical spare of resources for yeast cells. As far as shuttling factors such as Rlp24, Nmd3, Arx1, and Tif6 are concerned, this recycling involves reimport into the nucleus.
Previous studies have shown that some of the latest pre-60S factors can be involved in recycling shuttling factors. Indeed, the GTPase Lsg1 was shown to participate in the release and recycling of Nmd3 (Hedges et al., 2005). An additional GTPase, Efl1, is required for release from the particles and recycling of the pre-60S factor Tif6 (Senger et al., 2001). In this study, we characterized Rei1 as a new pre-60S–associated cytoplasmic factor at the center of maturation events taking place in the nucleus. We provide evidence for the role of this factor in the recycling of Arx1, Tif6, and a novel pre-60S shuttling factor, Alb1.
Arx1 and Alb1 are interacting pre-60S factors
Arx1 was first characterized as a component of pre-60S complexes by TAP experiments (Bassler et al., 2001; Nissan et al., 2002). This nonessential protein, which is present both in the nucleus and cytoplasm of yeast cells, contains a domain that is characteristic of methionine amino peptidases.
This study sheds light on an additional shuttling pre-60S factor, Alb1. Our data suggest that Arx1 and Alb1 can form a dimer because (1) they interact in a two hybrid (Fig. 4 A), (2) Arx1 is overrepresented in Alb1-TAP–associated complexes (Fig. 4 B), (3) they interact directly in vitro (Fig. 4 D), and (4) they can be copurified as a nonribosomal subcomplex (Fig. 5 D).
In physiological conditions, both Arx1 and Alb1 participate in pre-60S complexes from the nucleus to the cytoplasm. Nevertheless, neither of them seems to be essential for the maturation of the large ribosomal subunit. Indeed, in the absence of either factor, we failed to detect any strong rRNA maturation, export, or polysome profile defect (Fig. S3 and not depicted). In the absence of Rei1, both factors are retained in the cytoplasm (Figs. 3 A and 5 A) in the form of a small complex (Figs. 3 B and 5, B and D); they are not imported into the nucleus and, consequently, are not incorporated into new pre-60S complexes.
Recycling of the pre-60S factors Arx1 and Alb1 is Kap121 dependent
Ribosome biogenesis needs the import into the nucleus of many proteins through the nuclear pore complexes. In addition to ribosomal proteins, all of the factors that participate in the transcription of rRNA or in ribosome biogenesis must be imported into the nucleolus. Two main β-karyopherins have been shown to be involved in this directional transport: Kap123 and Kap121/Pse1 (Rout et al., 1997). Each of these importins is responsible for the nuclear import of specific cargoes: Kap123 imports most of the ribosomal proteins, and Kap121 also carries nucleolar proteins such as Nop1 and Sof1 (Leslie et al., 2004).
Although we did not detect any consensus Kap121-specific NLS in the sequence of Arx1 or Alb1, three independent pieces of evidence indicate that both factors are imported into the nucleus by a mechanism involving Kap121: (1) the cytoplasmic accumulation of Arx1 and Alb1, released from pre-60S complexes in the absence of Rei1, can be overcome by the overexpression of Kap121 (Fig. 6 B); (2) Arx1 and Alb1 remain in the cytoplasm in the absence of Kap121 (Fig. 6 D); and (3) Kap121 can be found in Arx1- and Alb1-associated complexes (Fig. 5 C; Nissan et al., 2002) and was found as a two-hybrid partner of Alb1 in a matrix assay (Fig. S4). Thus, one might assume that Arx1 and Alb1 are additional Kap121 cargoes. Nevertheless, we cannot exclude the possibility that the observed effects could be indirect.
So far, the nature of the nuclear import signal for Arx1–Alb1, which is affected when they dissociate from pre-60S particles in the absence of Rei1, is still unclear. However, the binding with Kap121 is not affected because Kap121 is still copurified with Arx1 and Alb1 in such conditions (Fig. 5 D).
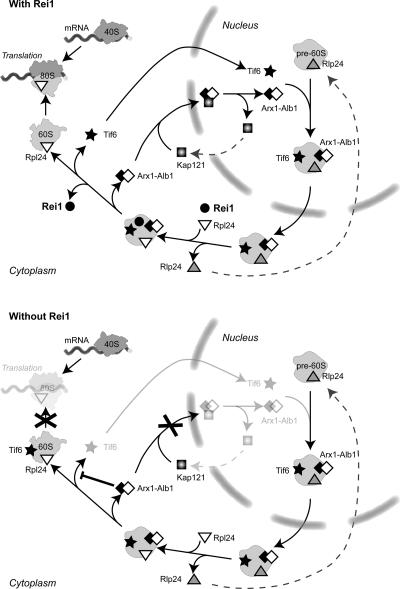
Rei1 controls the exit from 60S biogenesis
Fig. 8 recapitulates the conclusions of this study concerning the cytoplasmic events that take place at the end of the large subunit biogenesis in wild-type conditions or in the absence of Rei1. In the model that we propose, we only took into account the factors that had shown physical or functional links in our study. At the exit of the nucleus, pre-60S particles still contain shuttling pre-ribosomal factors such as Rlp24, Tif6, Arx1, and Alb1. At this stage in the pathway, the cytoplasmic pre-60S factor Rei1 and the late-associating ribosomal protein Rpl24 associate on the particle. So far, we could not determine which of the factors loads first; however, our data suggest that they associate with the particle after the dissociation of Rlp24 (Fig. 1 B). Arx1, Alb1, and Tif6 then leave the particle and return to the nucleus, where they can enter a new round of biogenesis.
Figure 8.
Model for the function of Rei1 at the exit of large ribosomal subunit biogenesis. Arx1, Alb1, and Tif6 are shuttling pre-60S factors that load on the precursors of the large ribosomal subunit in the nucleus and dissociate from the particles in the cytoplasm. At this last stage in biogenesis, the late ribosomal protein Rpl24 and the pre-60S factor Rei1 load on the particle. Rei1 triggers the Kap121-dependent recycling of Arx1 and Alb1 and subsequent dissociation and return of Tif6 to the nucleus. In the absence of Rei1, an Arx1, Alb1-containing subcomplex accumulates in the cytoplasm, which inhibits the dissociation of Tif6 from pre-60S particles. Thus, Tif6 antiassociation activity prevents the participation of novel large ribosomal subunits to translation initiation, and the lack of nuclear Tif6 results in pre-60S maturation defects.
In the absence of Rei1, we showed that the Kap121-dependent recycling of Arx1–Alb1 was impaired and that the cytoplasmic accumulation of this duplex inhibited the release of Tif6 from pre-60S particles and subsequent return of this factor to the nucleus. Because Tif6 and its homologues were described as putative antiassociation factors in several eukaryotic species (Valenzuela et al., 1982; Si and Maitra, 1999), we believe that stalling of Tif6 on the particles at the end of the 60S biogenesis could result in the observed rei1Δ phenotypes. Because Efl1 was previously characterized as the GTPase that triggers the dissociation of Tif6 from the particle (Senger et al., 2001), it was a good candidate for explaining the recycling defect observed for Tif6. However, Efl1 was always found as a free protein in sucrose gradients, whatever the presence of Rei1 (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200510080/DC1). Additionally, it was never found to be associated with Arx1 or with Alb1 in TAP experiments (unpublished data). Lastly, the overexpression of wild-type or mutant forms of Efl1 did not complement a rei1Δ strain (Fig. S2). Consequently, the retention of Tif6 on pre-60S particles in the absence of Rei1 is not likely caused by a lack of Efl1. Possibly, the Arx1–Alb1 cytoplasmic complex might sequester another, as yet unknown, factor required for the release of Tif6.
The late pre-60S network could be linked with the cell cycle control
Rei1 (required for isotropic bud growth) was previously described as a member of the mitotic signaling network (Iwase and Toh-e, 2004). This factor was proposed to be involved in the regulatory network around the Swe1 kinase by way of Nis1. The Swe1 kinase is the converging platform for several events that result in the negative regulation of cyclin-dependent kinases, which, in turn, control the switch between polar growth and isotropic growth at the bud neck. These data are not in contradiction with the present results and even offer the possibility that Rei1 might be involved in coupling ribosome biogenesis with cell cycle control.
Indeed, other factors appear to be implicated in both pathways. For instance, Nap1, which belongs to this mitotic signaling network, also displays interactions with proteins involved in ribosome biogenesis and, in particular, with Rei1 (Krogan et al., 2004). According to these data, Rei1 could also act at the intersection between these two major cellular pathways.
One way to address the question would be to confirm the existence of an E-MAP-115 domain in Rei1. This domain was characterized in ensconsin, a microtubule-associated protein of 115 kD. In higher eukaryotes, this protein was shown to bind microtubules in cell lines of epithelial origin (Masson and Kreis, 1993). To further speculate, we could imagine that Rei1, if it also interacted with microtubules, could orient newly synthesized large ribosomal subunits to the bud neck, where a high rate of protein synthesis is required for bud growth. An appealing hypothesis would be that Rei1 triggers the dissociation of Tif6 only once the 60S particles have reached active translation sites.
Materials and methods
Yeast strains, plasmids, and oligonucleotides
The yeast strains used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200510080/DC1). They were generated by homologous recombination using PCR products to transform either MGD353-13D or BY4741/4742 (S288C background) strains (Baudin et al., 1993). The pBS1479 vector was obtained from B. Seraphin (Centre National de la Recherche Scientifique [CNRS], Gif-sur-Yvette, France). A pFA6a-TAP plasmid was obtained from F. Stutz (University of Geneva, Geneva, Switzerland). The SC0708 and SC0406 strains were obtained from Euroscarf and have been constructed by CellZome AG. Disrupted strains in the BY4741/4742 background with the KanMX4 marker came from the Euroscarf collection of deletion strains (Brachmann et al., 1998). Plasmids for two-hybrid screens were obtained by Gateway cloning in pAS2ΔΔ and pACTIIst destination vectors (gift from E. Bertrand, CNRS). The sequence of oligonucleotides used for Northern hybridization and primer extension analysis were previously described (Saveanu et al., 2003).
Yeast two-hybrid genomic screen
The yeast two-hybrid screens were performed using a cell-to-cell mating strategy as previously described (Fromont-Racine et al., 2002). The strain CG1945, transformed with either pAS2ΔΔ-RPL24 or pAS2ΔΔ-ARX1 baits, was mated with the strain Y187 transformed with an S. cerevisiae DNA library cloned in pACTIIst. Diploids were spread on minimal medium without leucine, tryptophan, and histidine. 55 and 36 positive clones were recovered in the Rpl24b and Arx1 screens, respectively.
High copy number suppressor genetic screen
The BY4741 reiΔ strain was transformed with a yeast genomic high copy number vector library constructed in pFL44L (provided by F. Lacroute, CNRS). The transformants were grown on solid synthetic minimal medium lacking uracil at 20°C. Colonies that had lost their cold-sensitive phenotype when compared with the same strain transformed with an empty vector were selected. Plasmidic DNA was recovered, and DNA inserts were sequenced. The suppressor plasmids were checked by retransformation of the rei1Δ strain; their growth phenotypes were compared by spotting transformants in 10−1 dilution steps on minimal medium without uracil at 23 and 30°C.
RNA extraction, Northern blotting, and primer extension
Cells were broken with glass beads, and total RNAs were subjected to phenol-chloroform extraction. RNAs were resolved on 6% polyacrylamide-urea gels or on 1% agarose gels, transferred to Hybond-N+ membranes, and probed with various 32P-labeled oligonucleotides. Primer extensions were performed with 32P-labeled oligonucleotides; the products were then resolved on 5% polyacrylamide-urea gels. Quantifications were performed with ImageQuant software (GE Healthcare).
Sucrose gradients and Western blotting
Total protein extracts were prepared from exponentially growing yeast cells as previously described (Saveanu et al., 2001) and were separated on 10–50% sucrose gradients by centrifugation for 3 h at 190,000 g (SW41 rotor; Beckman Coulter). In each fraction of the gradient, the proteins were precipitated with 10% TCA, separated on 10% polyacrylamide-SDS gels, and transferred to nitrocellulose membranes. TAP-tagged proteins were detected with a 1:10,000 dilution of the peroxidase–antiperoxidase (PAP) soluble complex (Sigma-Aldrich). Native, 13Myc-tagged, and GFP-tagged proteins were detected by indirect immunoblotting using specific polyclonal rabbit antibodies as primary antibodies at a 1:5,000 dilution, anti-cMyc mouse monoclonal IgG1 (Santa Cruz Biotechnology, Inc.) at a 1:2,000 dilution, or anti-GFP rabbit polyclonal IgG (Santa Cruz Biotechnology, Inc.) at a 1:400 dilution. TAP-tagged proteins were detected with PAP (Sigma-Aldrich) at a 1:10,000 dilution. Anti-Nog1 antibodies were described previously (Saveanu et al., 2003). Antibodies against Kap121 and Tif6/Efl1 were obtained from J.D. Aitchison (Institute for Systems Biology, Seattle, WA) and F. Fasiolo (CNRS), respectively. Anti-Arx1 and anti-Rei1 antibodies were produced by the immunization of rabbits with specific immunogenic peptides and were affinity purified (Covalab). Secondary antibodies (goat anti–rabbit or goat anti–mouse HRP conjugate; Bio-Rad Laboratories) were used at a 1:10,000 dilutions. Visualization of the peroxidase activity was performed with the ECL+ chemiluminescence kit (GE Healthcare).
Purification of complexes
Complexes were purified according to the standard TAP protocol (Rigaut et al., 1999), starting from 4 liters of yeast culture. The TEV eluate was precipitated with 10% TCA and analyzed by Western blotting as described above.
In vitro binding assay
Interactions between the GST-tagged baits and the His6-Arx1 fusion prey were assessed as previously described (Saveanu et al., 2003).
Fluorescence microscopy
Cells transformed with a centromeric plasmid expressing RPL25-eGFP (gift from E. Hurt, University of Heidelberg, Heidelberg, Germany) or cells expressing chromosomal reporter constructs were cultured as indicated and mounted in liquid minimal culture medium on glass slides. Immunofluorescence detection of the TAP-tagged fusion proteins was performed as previously described (Pringle et al., 1991). The fluorescence of GFP or Cy3 was detected with an epifluorescence microscope (model DMRB; Leica) at room temperature with 100× NA 1.30–0.60 oil immersion objective lenses (Leica). Images were acquired with a dual mode cooled CCD camera (C4880; Hamamatsu) and the HiPic32 acquisition software (Hamamatsu). The pAJ368/544 vectors were provided by A.W. Johnson (University of Texas, Austin, TX).
Online supplemental material
Table S1 recapitulates all yeast strains used in this study. Fig. S1 shows the results of TIF6 overexpression. Fig. S2 presents the data showing that a lack of Efl1 is likely not the cause of the recycling defect observed for Tif6 in the absence of Rei1. Fig. S3 displays the phenotypes on growth and polysome profiles of arx1Δ or alb1Δ single or double mutants. Fig. S4 shows the two-hybrid interactions between Arx1 and Alb1 and between Alb1 and Kap121. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200510080/DC1.
Supplementary Material
Acknowledgments
We would like to thank Dr. J.D. Aitchison and Dr. F. Fasiolo for providing antibodies against Kap121 and Tif6/Efl1, respectively. We are grateful to Professor F. Lacroute, Dr. B. Seraphin, Dr. Edouard Bertrand, Dr. F. Stutz, Dr. E. Hurt, and Dr. A.W. Johnson, who provided the genomic bank in pFL44L for high copy number suppressor screens, the pBS1479 vector, the two-hybrid Gateway plasmids, the pFA6a-TAP-Tag-His3MX6 vector, the centromeric plasmid expressing RPL25-eGFP, and pAJ368/544, respectively.
This work was supported by grants from the European Union, the RNomics network (QLG2-CT-2001-01554), and the Ministère délégué à l'Enseignement Supérieur et à la Recherche (ACI-BCM0089-2003). A. Lebreton received a fellowship from the Association pour la Recherche sur le Cancer.
Abbreviations used in this paper: PAP, peroxidase–antiperoxidase complex; rRNA, ribosomal RNA; TAP, tandem affinity purification; YPD, yeast extract–peptone–d-glucose.
References
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 8:517–529. [DOI] [PubMed] [Google Scholar]
- Basu, U., K. Si, J.R. Warner, and U. Maitra. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21:1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C.B., A. Davies, G.J. Cost, E. Caputo, J. Li, P. Hieter, and J.D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Dez, C., and D. Tollervey. 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7:631–637. [DOI] [PubMed] [Google Scholar]
- Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313–318. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., J.C. Rain, and P. Legrain. 2002. Building protein-protein networks by two-hybrid mating strategy. Methods Enzymol. 350:513–524. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene. 313:17–42. [DOI] [PubMed] [Google Scholar]
- Gadal, O., B.D. Strau, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A.C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J.M. Rick, A.M. Michon, C.M. Cruciat, et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415:141–147. [DOI] [PubMed] [Google Scholar]
- Hedges, J., M. West, and A.W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, J.H., G. Kallstrom, and A.W. Johnson. 2000. a. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA. 6:1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, J.H., G. Kallstrom, and A.W. Johnson. 2000. b. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G.D. Bader, L. Moore, S.L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415:180–183. [DOI] [PubMed] [Google Scholar]
- Hurt, E., S. Hannus, B. Schmelzl, D. Lau, D. Tollervey, and G. Simos. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase, M., and A. Toh-e. 2004. Ybr267w is a new cytoplasmic protein belonging to the mitotic signaling network of Saccharomyces cerevisiae. Cell Struct. Funct. 29:1–15. [DOI] [PubMed] [Google Scholar]
- Johnson, A.W., E. Lund, and J. Dahlberg. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27:580–585. [DOI] [PubMed] [Google Scholar]
- Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., W.T. Peng, G. Cagney, M.D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D.P. Richards, et al. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 13:225–239. [DOI] [PubMed] [Google Scholar]
- Kruiswijk, T., R.J. Planta, and J.M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta. 517:378–389. [DOI] [PubMed] [Google Scholar]
- Leslie, D.M., W. Zhang, B.L. Timney, B.T. Chait, M.P. Rout, R.W. Wozniak, and J.D. Aitchison. 2004. Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol. Cell. Biol. 24:8487–8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, D., and T.E. Kreis. 1993. Identification and molecular characterization of E-MAP-115, a novel microtubule-associated protein predominantly expressed in epithelial cells. J. Cell Biol. 123:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy, T.I., and P.A. Silver. 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 13:2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T.A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R., A.E. Adams, D.G. Drubin, and B.K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565–602. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., G. Blobel, J.D. Aitchison, and M. Tyers. 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell. 89:715–725. [DOI] [PubMed] [Google Scholar]
- Saveanu, C., D. Bienvenu, A. Namane, P.E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu, C., A. Namane, P.E. Gleizes, A. Lebreton, J.C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger, B., D.L. Lafontaine, J.S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J.M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 8:1363–1373. [DOI] [PubMed] [Google Scholar]
- Si, K., and U. Maitra. 1999. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol. 19:1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydorskyy, Y., D.J. Dilworth, E.C. Yi, D.R. Goodlett, R.W. Wozniak, and J.D. Aitchison. 2003. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 23:2042–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255–263. [DOI] [PubMed] [Google Scholar]
- Valenzuela, D.M., A. Chaudhuri, and U. Maitra. 1982. Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6). J. Biol. Chem. 257:7712–7719. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.