Abstract
The significance of cadherin superfamily proteins in vascular smooth muscle cell (VSMC) biology is undefined. Here we describe recent studies of the Fat1 protocadherin. Fat1 expression in VSMCs increases significantly after arterial injury or growth factor stimulation. Fat1 knockdown decreases VSMC migration in vitro, but surprisingly, enhances cyclin D1 expression and proliferation. Despite limited similarity to classical cadherins, the Fat1 intracellular domain (Fat1IC) interacts with β-catenin, inhibiting both its nuclear localization and transcriptional activity. Fat1 undergoes cleavage and Fat1IC species localize to the nucleus; however, inhibition of the cyclin D1 promoter by truncated Fat1IC proteins corresponds to their presence outside the nucleus, which argues against repression of β-catenin–dependent transcription by nuclear Fat1IC. These findings extend recent observations about Fat1 and migration in other cell types, and demonstrate for the first time its anti-proliferative activity and interaction with β-catenin. Because it is induced after arterial injury, Fat1 may control VSMC functions central to vascular remodeling by facilitating migration and limiting proliferation.
Introduction
Vascular remodeling is a critical part of the pathogenesis of clinically important vascular disorders such as atherosclerosis, restenosis after angioplasty, and saphenous vein graft disease (Shanahan and Weissberg, 1998; Owens et al., 2004). Despite considerable study, the molecular mechanisms that control vascular smooth muscle cell (VSMC) activities during vascular remodeling are not fully understood. Recent reports linking cadherins to VSMC regulation (Jones et al., 2002; Uglow et al., 2003; Slater et al., 2004) suggest that these transmembrane adhesion proteins, characterized extensively as major mediators of epithelial cell homeostasis, may also be important in vascular remodeling.
Cadherins are involved in Ca2+-dependent cell–cell adhesion, intracellular junction assembly, and tissue morphogenesis during development (Yap et al., 1997; Angst et al., 2001; Wheelock and Johnson, 2003b). Major subdivisions of the large cadherin superfamily include the classical cadherins and the protocadherins (Gallin, 1998; Yagi and Takeichi, 2000; Angst et al., 2001). The extracellular domains of these proteins share a unique structure, the cadherin motif, which is repeated in tandem in variable numbers. Classical cadherins function as homophilic adhesive molecules, and both extracellular and cytoplasmic domains contribute to this function. Classical cadherin cytoplasmic domains interact with β-catenin and plakoglobin (Takeichi, 1995; Huber and Weis, 2001), members of the armadillo gene family of transcription factors. This interaction effectively sequesters β-catenin away from the nucleus, limits its transcriptional activity (Sadot et al., 1998; Kaplan et al., 2001; Simcha et al., 2001), and thus links cadherins to the canonical Wnt signaling pathway, a major determinant of cellular activity during development (Bhanot et al., 1999; Jamora et al., 2003; Nelson and Nusse, 2004).
We identified the protocadherin Fat1 in a screen for molecules expressed differentially after balloon injury of rat carotid arteries. Like classical cadherins, protocadherins have extracellular domains capable of Ca2+-dependent, homophilic interaction (Suzuki, 2000). Protocadherin cytoplasmic domains, on the other hand, are structurally divergent from those of the classical cadherins, and less is known about their function. Sequestration and inhibition of β-catenin by protocadherins has not been described.
Although mammalian Fat1 genes (Dunne et al., 1995; Ponassi et al., 1999; Cox et al., 2000) were initially characterized as homologues of the Drosophila protein Fat (Mahoney et al., 1991), recent bioinformatics analysis indicates that Fat1 is more closely related to Drosophila Fat-like (Ftl) (Castillejo-Lopez et al., 2004). In Drosophila, Ftl is expressed apically in luminal tissues such as trachea, salivary glands, proventriculus, and hindgut (Castillejo-Lopez et al., 2004). Silencing of ftl results in the collapse of tracheal epithelia, and it has been suggested that Ftl is required for morphogenesis and maintenance of tubular structures of ectodermal origin.
Like Drosophila Fat and Ftl, mammalian Fat1 is remarkable for its very large size (∼4,600 aa). It has a huge extracellular domain that contains 34 cadherin repeats, 5 EGF-like repeats and l laminin A-G motif, a single transmembrane region, and a cytoplasmic tail of ∼400 aa (Dunne et al., 1995). Sequences within the Fat1 intracellular domain (Fat1IC) show limited similarity to β-catenin binding regions of classical cadherins (Dunne et al., 1995).
Our studies show that Fat1 expression increases after injury of the rat carotid artery, and is positively regulated in cultured VSMCs by several factors that promote cell proliferation and migration. Interestingly, knockdown of Fat1 expression limits VSMC migration, but enhances VSMC growth. This anti-proliferative effect of Fat1 appears to be mediated by Fat1IC sequences because expression of a fusion protein containing the Fat1IC inhibits cyclin D1 expression and cell growth. Moreover, the Fat1IC can interact with β-catenin, prevent its nuclear translocation, and limit its transcriptional activity on both synthetic and native β-catenin–responsive promoters, including that of cyclin D1, a known target of canonical Wnt signaling. These findings point to an integrative role for Fat1 in regulation of critical VSMC activities in which it promotes migration and limits both canonical Wnt signaling and VSMC growth in the remodeling artery.
Results
Expression of Fat1 increases after arterial injury
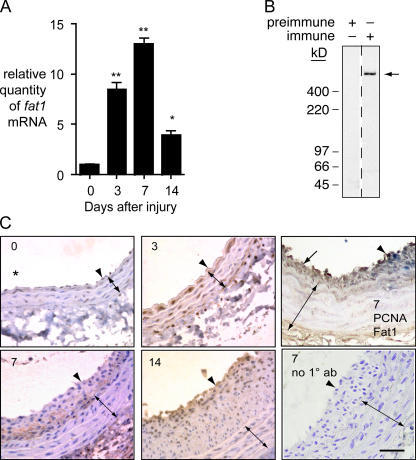
We quantitated fat1 mRNA expression by quantitative PCR (qPCR) of cDNA samples from normal and injured rat carotid arteries. Compared with uninjured arteries, fat1 mRNA expression was ∼8.5-, 13.0-, and 3.9-fold higher than control at 3, 7, and 14 d after injury, respectively (Fig. 1 A).
Figure 1.
Expression of Fat1 in normal and balloon-injured rat carotid arteries. (A) qPCR analysis of fat1 mRNA expression. Fat1 mRNA levels were corrected relative to gapdh mRNA levels, with day 0 (no injury) set = 1. *, P < 0.05, **, P < 0.01 vs. day 0. Data show the means ± SEM. (B) specificity of anti-Fat1 antiserum. Preimmune and anti-Fat1 immune rabbit sera (1:5,000 dilutions) were tested by immunoblotting of replicate total RASMC protein extracts (20 μg/lane). Arrow indicates Fat1 signal. (C) Immunohistochemical analysis of Fat1 expression in arteries 0, 3, 7, and 14 d after injury, as indicated. Fat1 immunoreactivity appears brown, with blue hematoxylin counterstain except in top right panel. Arrowheads indicate the internal elastic lamina, and double-ended arrows, the extent of the media. The neointima is the space between arrowhead and arrow. The orientation of all samples is similar, with an asterisk in the day 0 panel indicating the vessel lumen. The top right panel shows a day 7 sample costained for Fat1 (brown) and PCNA (blue); areas of increased Fat1 (arrow) and increased PCNA (arrowhead) staining are indicated. The bottom right panel shows a day 7 sample in which Fat1 antiserum was omitted (no 1° ab). Scale bar, 100 μm.
To localize Fat1 protein expression in injured arteries, we characterized rabbit antisera raised against a GST-Fat1IC immunogen. Immunoblotting of VSMC lysates with one such antiserum, but not preimmune serum, yielded a single high molecular weight band of ∼500 kD, in accord with the predicted size of full-length Fat1 (Fig. 1 B). Further specificity was demonstrated in RNAi experiments directed against multiple separate targets in the mouse Fat1 sequence (see Figs. 3 and 4). We then used the antiserum for immunohistochemical studies. As shown in Fig. 1 C, prominent Fat1 staining appeared in the media 3 d after injury, while at 7 and 14 d after injury Fat1 staining was less evident in the media, but clearly present in the developing neointima. Western analysis of Fat1 expression in the carotid artery injury model, like our qPCR findings, showed a clear induction after injury (unpublished data). To correlate Fat1 expression with the proliferative status of specific cells, we co-stained sections for Fat1 and the proliferation marker PCNA. Although some cells appeared positive for both, we also noted some spatial separation of the signals, particularly evident in areas with limited neointimal formation, which showed prominent Fat1 staining without PCNA (Fig. 1 C, top right). The latter observation raised the possibility that, despite its overall induction after injury, increased Fat1 expression might have negative effects on VSMC growth in vivo.
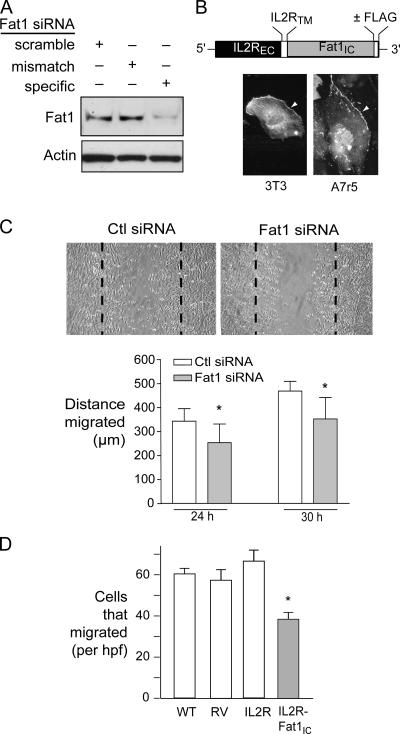
Figure 3.
Effect of decreased Fat1 expression on VSMC migration. (A) Western analysis of Fat1 specific and control siRNA efficacy, 48 h after transfection. (B) Schematic diagram of IL2R-Fat1IC. EC, extracellular; TM, transmembrane; IC, intracellular, with photomicrograph of subcellular localization of IL2R-Fat1IC-3XFLAG in transfected cells stained with FITC-conjugated anti-FLAG antibody. Arrowheads indicate cell surface staining. (C) MASMC migration 30 h after wounding of monolayer. Dashed lines indicate extent of initial denudation. Bottom, quantitative analysis of MASMC migration after control (Ctl) and specific Fat1 siRNA transfection. For 10 matched fields, the area of the wounded monolayer covered by cells at the indicated time points was determined by planimetry using NIH Image, and distance migrated calculated according to the difference from time 0. *, P < 0.05 vs. Ctl siRNA. (D) Transwell migration assay of A7r5 cells transduced with the indicated retroviruses. Six fields were counted per condition, and the values were averaged for each filter. Data show the means ± SEM. *, P < 0.05 vs. other groups.
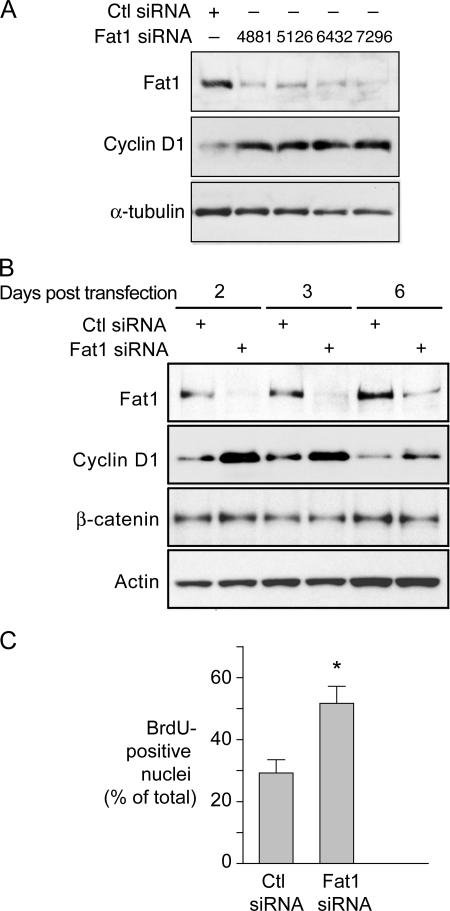
Figure 4.
Effect of decreased Fat1 expression on VSMC cell cycle progression. (A and B) Western analyses of Fat1 and cyclin D1 expression with control (Ctl) or Fat1-specific siRNAs. (A) Four distinct Fat1-specific siRNAs were transfected 48 h before protein extraction. Loading reference, α-tubulin. (B) Efficacy of Ctl or Fat1-specific siRNA 7296 over time. Actin and β-catenin expression were also tested. (C) Effect of Fat1 inhibition on DNA synthesis assessed by BrdU incorporation. Cells were transfected with Ctl or Fat1 siRNA, serum deprived for 48 h, and stimulated with 10% FBS. BrdU incorporation was assessed as described in Materials and methods. The graph depicts the means ± SEM of three independent experiments in which a total of 219–874 cells were counted each time for each group. *, P < 0.05 vs. Ctl siRNA.
Serum and growth factors induce Fat1 expression in VSMCs
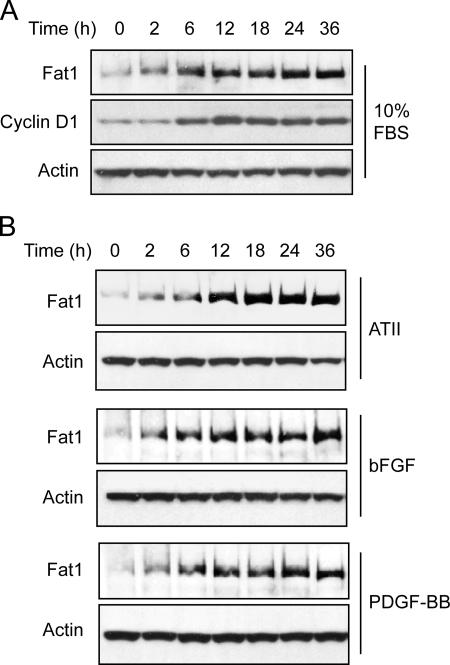
To identify factors that might contribute to Fat1 induction after arterial injury, we characterized its expression in primary cultured VSMCs. Quiescent rat aortic smooth muscle cells (RASMCs) (time 0 h) were treated with 10% FBS for 2, 6, 12, 18, 24, and 36 h, and the level of Fat1 protein was determined by Western analysis. The Fat1 signal increased strongly between 2 and 12 h and remained elevated through 36 h (Fig. 2 A). To assess cell cycle status, we also checked cyclin D1 expression in these lysates. Interestingly, Fat1 induction preceded the increase of cyclin D1, a mediator of progression through the G1 phase of the cell cycle (Fig. 2 A).
Figure 2.
Western analysis of Fat1 expression in RASMCs. Cells were serum deprived, stimulated as described, and harvested for protein extraction at the indicated time points. Actin expression is shown as a loading reference. Data are representative of three independent experiments. (A) Induction by 10% FBS for 0–36 h before protein extraction. The blot was also probed for cyclin D1. (B) Induction by specific factors. Cells were stimulated with 10−6 mol/L ATII, 20 ng/ml bFGF, or 20 ng/ml PDGF-BB.
We then assessed Fat1 expression in response to several factors known to affect the vascular response to injury. Western analyses showed that expression of Fat1 increased in response to Angiotensin II (ATII), basic FGF (bFGF), and PDGF-BB (Fig. 2 B). Increased Fat1 expression was apparent by 2 h and sustained at high levels from 12 to 36 h after stimulation with each of these factors. Thus, Fat1 expression is regulated consistently and strongly by multiple factors known to promote VSMC growth and migration.
Inhibition of Fat1 expression limits VSMC migration
Two recent studies have described a role for Fat1 in regulation of epithelial cytoskeletal actin dynamics, planar polarity, and migration, mediated through interactions of the Fat1 cytoplasmic domain with proteins of the Ena/VASP family (Moeller et al., 2004; Tanoue and Takeichi, 2004). Fat1 induction by known VSMC chemotactic factors (Fig. 2) suggested that Fat1 might also be involved in VSMC migration. To test this and other potential Fat1 functions, we developed reagents to effectively manipulate Fat1 expression. Transfection of mouse aortic smooth muscle cells (MASMCs) with Fat1 specific small interfering RNAs (siRNAs), but not scrambled or mismatch derivatives, resulted in significantly decreased levels of Fat1 protein (Fig. 3 A). To isolate and augment signals mediated by the Fat1IC, we generated a cDNA construct, IL2R-Fat1IC, in which the entire Fat1 cytoplasmic domain was fused to the extracellular domain and transmembrane region of the interleukin 2 receptor α-chain (IL2R), with or without a COOH-terminal FLAG epitope tag (Fig. 3 B). Subcellular localization of this fusion protein was tested in 3T3 cells, which do not express detectable Fat1, and A7r5 VSMCs, which express moderate amounts of endogenous Fat1; both transfected 3T3 and A7r5 cells showed an appropriate cell surface signal when stained with anti-FLAG epitope antibody (Fig. 3 B and unpublished data). Cell migration in monolayers treated with specific Fat1 siRNA was modestly but significantly decreased compared with control siRNA (Fig. 3 C), which indicates that Fat1 expression is required for optimal VSMC migration. Surprisingly, we also found decreased migration of VSMCs expressing the IL2R-Fat1IC protein in a Transwell assay using FBS as a stimulant in the lower chamber (Fig. 3 D). We confirmed both expression of Ena/VASP proteins in VSMCs and the ability of the IL2R-Fat1IC protein to interact with these signaling intermediates (unpublished data). We surmise that although the IL2R-Fat1IC construct may increase intracellular Fat1 signaling, it also dissociates Fat1 extracellular interactions from this intracellular signaling, and thus interferes with directional migration. Altogether, these findings indicate that Fat1 promotes VSMC migration; it is likely that, as described in epithelial cells, interactions with Ena/VASP proteins link Fat1 expression to VSMC cytoskeletal actin reorganization, polarization, and migration.
Inhibition of Fat1 expression promotes VSMC growth
In addition to increased migration, the VSMC response to injury is characterized by cell cycle entry and increased proliferation (Clowes et al., 1983a). To evaluate how Fat1 induction after injury might affect VSMC growth, we tested the effect of Fat1 knockdown on expression of cyclin D1, a marker of cell cycle activation. Four distinct mouse Fat1 siRNA duplexes attenuated endogenous Fat1 levels in MASMCs; with each duplex, we also observed a significant increase in cyclin D1 expression over control levels (Fig. 4 A and unpublished). The similarity of effect achieved by multiple distinct siRNAs argues strongly that increased cyclin D1 expression results from decreased Fat1, and not an off-target effect. The duration of Fat1 inhibition was more than 90% at 2 and 3 d after transfection, with persistent and strong inhibition still apparent after 6 d (Fig. 4 B). Decreased Fat1 expression corresponded to increased cyclin D1 signal at each time point (2.0–2.5-fold increase of cyclin D1/actin ratio vs. control), suggesting that endogenous Fat1 exerts a tonic inhibitory effect on cyclin D1 expression (Fig. 4 B). The level of total β-catenin in these cells, by contrast, showed little change.
We then examined the effect of Fat1 knockdown on DNA synthesis. Cells were transfected with Fat1 or control siRNA, and then were serum deprived for 48 h before stimulation with 10% FBS and evaluation of BrdU incorporation. In Fat1 knockdown cultures, the fraction of BrdU-positive cells was significantly higher than in control siRNA cells (52 ± 7% vs. 30 ± 8%, P < 0.05) (Fig. 4 C). These findings indicate that decreased Fat1 expression promotes cell cycle progression and DNA synthesis in VSMCs.
The Fat1IC is sufficient to inhibit VSMC growth
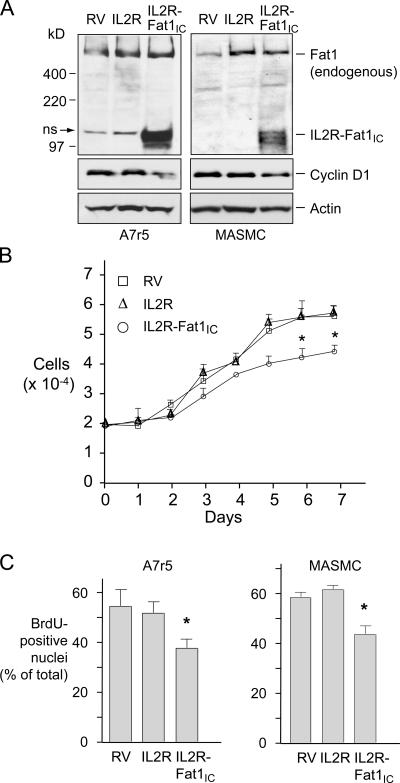
Classical cadherins interact with intracellular signaling pathways through their cytoplasmic domains (Wheelock and Johnson, 2003a). To establish cell populations differing primarily in their expression of the Fat1IC, we transferred the IL2R (without cytoplasmic domain) and IL2R-Fat1IC constructs into the GFP-RV retroviral vector (Ranganath et al., 1998), produced viral supernatants, and transduced A7r5 and primary MASMCs. Additional control cells, denoted RV, were produced using the unmodified GFP-RV vector. Western analysis confirmed IL2R-Fat1IC expression in A7r5 and MASMCs (Fig. 5 A). Interestingly, endogenous cyclin D1 levels were lower in both A7r5 and MASMCs expressing IL2R-Fat1IC (Fig. 5 A). In cell growth assays over 7 d, A7r5 cells expressing IL2R showed no significant change from control RV cells, but decreased cell numbers were evident in the IL2R-Fat1IC at all time points after 3 d (Fig. 5 B). In addition, both A7r5 and MASMCs expressing the IL2R-Fat1IC construct showed significantly lower fractions of BrdU-positive nuclei, indicating that this decrease in cell number reflected growth inhibition rather than decreased survival (Fig. 5 C).
Figure 5.
Effect of IL2R-Fat1IC expression on VSMC growth. RV, IL2R, and IL2R-Fat1IC designate A7r5 or MASMCs transduced with the corresponding retroviral constructs. (A) Western analyses of IL2R-Fat1IC expression in A7r5 and MASMC stable transfectants. A nonspecific band (ns) near the IL2R-Fat1IC protein is indicated. The blots were also probed for cyclin D1 and actin. (B) Effect of IL2R-Fat1IC on A7r5 cell growth. Cell number was calculated by CyQuant fluorescence assay by reference to a standard curve. (C) Effect of IL2R-Fat1IC on DNA synthesis in A7r5 and MASMCs, evaluated by BrdU incorporation. Data show the means ± SEM. *, P < 0.05 vs. control.
Fat1 and β-catenin colocalize and interact in VSMCs
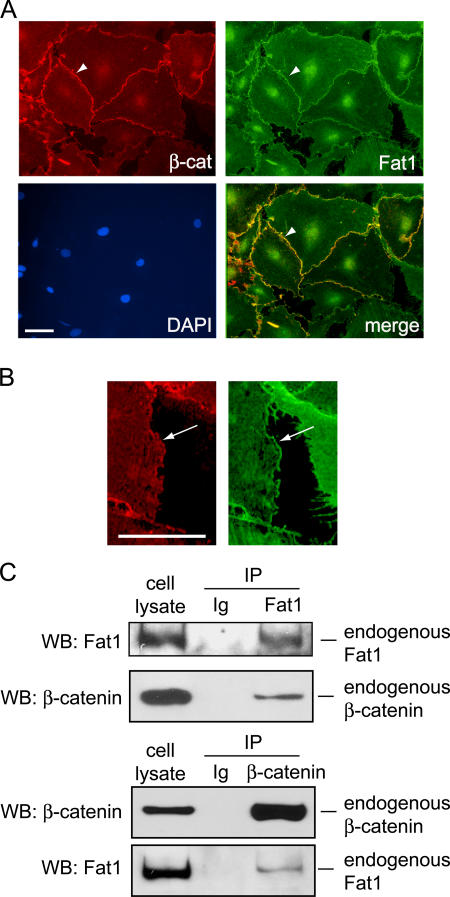
In epithelial cells, classical cadherins such as E-cadherin regulate Wnt signaling activity by physically associating with β-catenin at points of cell–cell contact (Nathke et al., 1994). The sequences, interacting proteins, and functions of protocadherin cytoplasmic domains are typically thought to be divergent from those of the classical cadherins (Yagi and Takeichi, 2000), and Fat1 is not regarded as part of the classical cadherin system (Tanoue and Takeichi, 2005). Nevertheless, we found that the Fat1IC has growth inhibitory activity, and that expression of cyclin D1, a known target of the canonical Wnt signaling pathway, correlated negatively with Fat1IC expression. Together, these findings suggested that growth inhibition by Fat1 might involve β-catenin. In our immunofluorescent analyses of RASMCs (Fig. 6), Fat1 localized to both cell–cell junctions and cellular free edges, while β-catenin was concentrated at sites of cell–cell contact. By two color immunofluorescence analysis, we found areas along cell–cell junctions where the two signals overlapped (Fig. 6 A). This overlap did not include the cellular free edges, where Fat1 alone was seen (Fig. 6 B).
Figure 6.
Co-localization and interaction of endogenous β-catenin and Fat1 in VSMCs. (A) Immunofluorescence analysis of β-catenin (β-cat, red), Fat1 (green), and areas of colocalization (merge, yellow). Nuclei were stained with DAPI (blue), as indicated. β-Catenin and Fat1 colocalization at cell–cell junctions is indicated by an arrowhead. Bar (10 μm) applies to all panels. (B) Detail from panels in A, showing staining for Fat1 (green), but not β-catenin (red), at the cellular free edge. Bar, 10 μm. (C) Co-immunoprecipitation of endogenous β-catenin and Fat1. Cell lysates were incubated with antibodies specific for Fat1 (top) or β-catenin (bottom) or normal rabbit or mouse IgG, and the immunoprecipitated complexes were analyzed by Western blot for Fat1 and β-catenin, as indicated.
Junctional β-catenin and Fat1 have been identified in epithelial cells that display apical–basal polarity, but it is thought that the two proteins occupy distinct domains, with β-catenin at apical adherens junctions and Fat1 at basolateral points of cell–cell contact (Tanoue and Takeichi, 2004, 2005). VSMCs are nonpolarized (Muller and Gimbrone, 1986), so this model of apical–basal domain specialization may not apply. To test directly if Fat1 and β-catenin can interact at physiologic levels of expression in VSMCs, we immunoprecipitated endogenous Fat1 and tested for recovery of β-catenin. Both this assay and reciprocal coimmunoprecipitations of β-catenin followed by immunoblotting for Fat1 demonstrated interaction of the two proteins (Fig. 6 C). This finding suggests that the nonpolarized nature of VSMCs allows for protein–protein interactions not found in polarized cell types such as epithelial cells. Further immunoblotting of Fat1 immunoprecipitates with a pan-cadherin antibody did not reveal associated (classical) cadherins that might associate with both Fat1 and β-catenin (unpublished data).
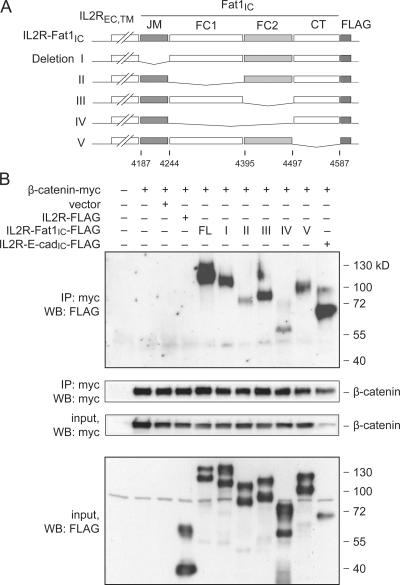
To characterize the Fat1–β-catenin interaction further, we used coimmunoprecipitation assays in cotransfected 293T cells to map the sequences required for interaction. We generated a series of constructs bearing deletions within the Fat1IC portion of the IL2R-Fat1IC-3XFLAG (Fig. 7 A). IL2R-E-cadherinIC-3XFLAG and IL2R-3XFLAG (containing no Fat1 sequences) constructs served as positive and negative controls, respectively. We confirmed the expression of Myc-tagged β-catenin and FLAG-tagged fusion proteins, and immunoprecipitation of transfected Myc-tagged β-catenin (Fig. 7 B, bottom panels). Interaction of β-catenin with the IL2R-Fat1IC-3XFLAG derivatives was assessed by immunoblotting with FLAG antibody (Fig. 7 B, top). A robust FLAG signal was obtained with the IL2R-Fat1IC-3XFLAG construct containing the complete Fat1IC domain and with derivatives I, III, and V. Weaker signals were seen with constructs II and IV, which lack the FC1 and both FC1 and FC2 domains, respectively. Although these findings based on overexpressed proteins must be interpreted with caution, they suggest that β-catenin interacts with the Fat1IC principally through the FC1 domain, but leave open the possibility that the FC2 domain or additional sequences also contribute to the interaction. Interestingly, the E-cadherin–based positive control yielded a comparatively strong band, despite input of substantially less protein.
Figure 7.
Identification of β-catenin-interacting residues in the Fat1IC domain by coimmunoprecipitation of epitope-tagged proteins. (A) Schematic depiction of FLAG-tagged IL2R-Fat1IC deletion constructs. Fat1IC domains indicated: juxtamembrane (JM), FC1, FC2, carboxy-terminus (CT). (B) Western analysis of immunoprecipitated protein complexes. The indicated constructs were transfected into 293T cells. Top: after 24 h, total cellular protein (400 μg) was harvested and analyzed by immunoprecipitation and Western blotting (WB) with antibodies against the epitope tags, as indicated. Bottom: protein input (7.5%). An analogous E-cadherin–derived construct (IL2R-E-cadIC-FLAG) was used as a positive control.
Expression of the Fat1IC affects β-catenin cellular distribution and transcriptional activity
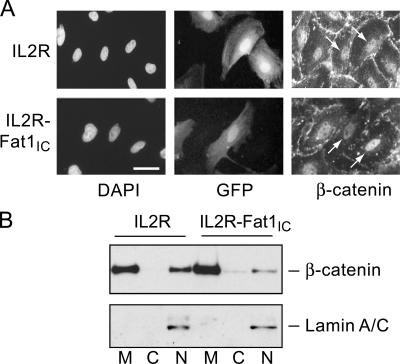
Thus, changes in Fat1 or Fat1IC expression affected expression of a β-catenin target gene, cyclin D1, but had little effect on overall β-catenin levels (Figs. 4 and 5). Having found evidence for colocalization and interaction of β-catenin and Fat1 in VSMCs, we postulated that Fat1 might be acting like a classical cadherin to affect the subcellular localization and activity of β-catenin. We examined this first using immunocytochemistry. Expression plasmids encoding IL2R or IL2R-Fat1IC were introduced into VSMCs, which were subsequently treated with 20 mM LiCl for 6 h to activate Wnt signaling and promote nuclear translocation of β-catenin (Hedgepeth et al., 1997). The intensity of nuclear β-catenin staining did not appear to be affected by expression of IL2R (Fig. 8 A, top, arrows). In contrast, nuclear accumulation of β-catenin appeared decreased in the IL2R-Fat1IC–expressing cells (Fig. 8 A, bottom, arrows) as compared with untransfected cells. To assess this effect in a more quantitative way, we determined the distribution of β-catenin in the membrane, cytoplasmic, and nuclear fractions of IL2R-GFP-RV– and IL2R-Fat1IC-GFP-RV–transduced VSMC cultures treated with LiCl. As shown in Fig. 8 B, immunoblotting showed a relative decrease in nuclear β-catenin accumulation in cells expressing IL2R-Fat1IC as compared with those expressing IL2R (respective nuclear β-catenin/lamin A/C ratios 0.8 [IL2R-Fat1IC] vs. 1.65 [IL2R]).
Figure 8.
Effect of Fat1IC overexpression on β-catenin nuclear localization in VSMCs. (A) Immunofluorescence analysis of β-catenin subcellular localization in IL2R-GFP-RV (top) and IL2R-Fat1IC-GFP-RV (bottom) transduced RASMCs. Cells were treated with LiCl (20 mmol/L) for 6 h, and then stained with anti-β-catenin antibody and DAPI. Transduced cells were identified by coexpressed GFP. Arrows indicate nuclear β-catenin signal of untransduced and transduced cells within each panel (see text). Bar, 10 μm. (B) Western analysis of β-catenin in membrane (M), cytoplasmic (C), and nuclear (N) fractions extracted from IL2R-GFP-RV and IL2R-Fat1IC-GFP-RV transduced A7r5 cells treated with LiCl, as above. The blot was probed for lamin A/C to assess fractionation and loading.
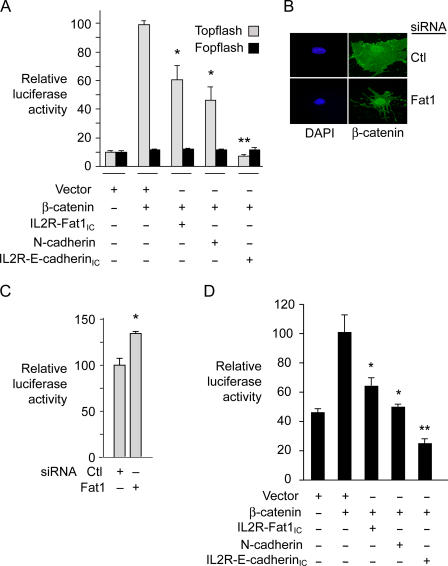
To assess further the functional significance of the Fat1–β-catenin interaction in VSMCs, we tested the effect of Fat1IC overexpression on β-catenin–mediated transcription. A7r5 cells were cotransfected with β-catenin and/or IL2R-Fat1IC, along with the TCF-luciferase reporter construct Topflash or its negative control, Fopflash (Fig. 9 A). Topflash reporter activity reflects activation of the canonical Wnt signaling pathway, β-catenin nuclear translocation, and formation of TCF/β-catenin heterodimers; Fopflash contains mutated TCF binding sites and serves as a control for nonspecific activation (Korinek et al., 1997). A full-length N-cadherin cDNA and the IL2R–E-cadherinIC construct were also tested as controls. Specific activation of Topflash by β-catenin was ∼10-fold above basal levels, and the three test constructs all inhibited this activation significantly. Interestingly, the inhibition due to both IL2R-Fat1IC (40%) and N-cadherin (55%) was less complete than that resulting from cotransfection of IL2R-E-cadherinIC, which abolished all β-catenin–mediated transactivation. We also evaluated the effect of decreased Fat1 expression. Immunocytochemistry of LiCl-stimulated MASMCs suggested a relative enhancement of nuclear β-catenin staining in Fat1-depleted cells (Fig. 9 B). To assess this observation more quantitatively, we transfected MASMCs first with control or Fat1-specific siRNA, and then with the Topflash reporter. As shown in Fig. 9 C, LiCl-stimulated TCF/β-catenin transcriptional activation was ∼30% higher in Fat1 knockdown cells compared with control.
Figure 9.
Effect of altered Fat1 expression on β-catenin transcriptional activity in VSMCs. (A) Topflash (TCF-luciferase reporter) activation. Topflash or Fopflash control was transfected into A7r5 cells along with expression constructs for β-catenin, IL2R-Fat1IC, N-cadherin, and/or IL2R-E-cadherinIC. The maximal reporter activity was set to 100. *, P < 0.05, **, P < 0.01, vs. activity with β-catenin alone. (B) β-Catenin localization in MASMCs transfected with control (Ctl, scrambled) or Fat1-specific (7296) siRNAs and stimulated with LiCl (20 mM) for 12 h. (C) Topflash activity with decreased Fat1 expression. MASMCs transfected with the indicated siRNAs and the Topflash reporter were stimulated with LiCl (20 mM) for 12 h before assay for luciferase activity. *, P < 0.05, vs. activity with Ctl siRNA. (D) Cyclin D1 promoter activation. The cyclin D1 promoter-luciferase construct was transfected into A7r5 cells along with test constructs, as in A. Data show the means ± SEM. *, P < 0.05, **, P < 0.01, vs. activity with β-catenin alone.
As shown in Figs. 4 and 5, cyclin D1 levels varied inversely with the level of Fat1IC expression. The cyclin D1 promoter is a known transcriptional target of Wnt signaling and activated TCF/β-catenin complexes (Shtutman et al., 1999; Tetsu and McCormick, 1999), so we postulated that Fat1IC might also inhibit the native cyclin D1 promoter. VSMCs were cotransfected with β-catenin and/or IL2R-Fat1IC, along with the cyclin D1 promoter–luciferase reporter construct (Herber et al., 1994). N-cadherin and the IL2R–E-cadherinIC fusion protein were also tested. Most of the β-catenin–mediated activation of the cyclin D1 promoter reporter was eliminated by IL2R-Fat1IC or N-cadherin expression (Fig. 9 D). Consistent with the Topflash results, IL2R–E-cadherinIC was more effective, as it decreased promoter activity to a level below baseline.
Inhibition of β-catenin activity depends on extranuclear localization of the Fat1IC
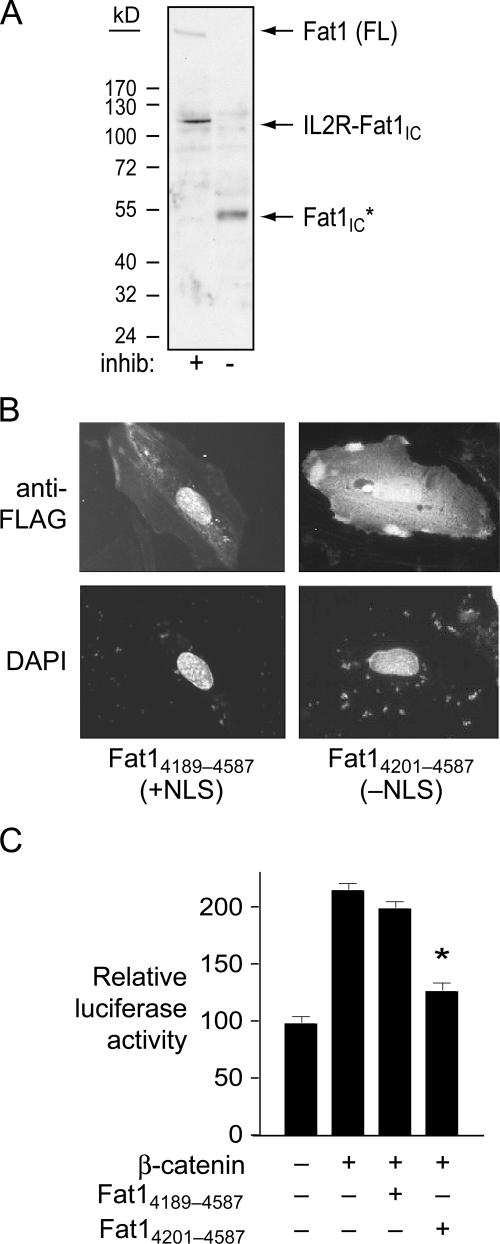
Fat1 is a type I transmembrane protein, and immunofluorescent studies with antiserum specific for Fat1IC sequences showed expression at the cell surface, as expected (Fig. 6). We also noted consistent signals in the cell nucleus with this antiserum. This observation, together with a recent report of localization of Fat1 cytoplasmic sequences to the nucleus (Magg et al., 2005), raised the possibility that inhibition of β-catenin by Fat1 might result from a nuclear (transcriptional repressor) function of a cleaved Fat1IC fragment, rather than sequestration of β-catenin outside the nucleus. Indeed, incubation without proteinase inhibitors of extracts of A7r5 cells expressing both native Fat1 and the IL2R-Fat1IC fusion protein showed the disappearance of these full-length proteins and rapid appearance of a single, relatively stable species of ∼50 kD (Fig. 10 A). Because the NH2 terminus of this cleaved product is not yet defined, we designate it as Fat1IC*; its apparent size in SDS-PAGE suggests that it contains most (if not all) of the ∼400 aa Fat1IC domain.
Figure 10.
Cleavage, localization, and activity of Fat1 cytoplasmic sequences in VSMC protein extracts. (A) Western analysis of A7r5 extracts transduced with IL2R-Fat1IC retrovirus. Total cellular protein was incubated at 37°C for 15 min with or without proteinase inhibitors (inhib). Both full-length (FL) Fat1 and the fusion protein (IL2R-Fat1IC) are apparent with proteinase inhibition; only a single band (Fat1IC*) of ∼50 kD is seen without inhibition. (B) Subcellular localization of the FLAG-tagged Fat1 cytoplasmic domain with (Fat14189–4587) or without (Fat14201–4587) the putative NLS in transfected A7r5 cells. Anti-FLAG immunofluorescence and DAPI nuclear stains are shown. (C) Effect of the NLS on Fat1IC-mediated inhibition of β-catenin activation of the cyclin D1 promoter. Luciferase activity was assessed 24 h after transfection of A7r5 cells with the indicated expression constructs and the cyclin D1 promoter reporter. Data show the means ± SEM. *, P < 0.01 vs. activity with β-catenin alone.
Like human Fat1IC (Magg et al., 2005), the mouse Fat1IC contains a potential NLS (RKMISRKKKR) near its NH2 terminus. We tested the effect of this sequence on Fat1IC localization by immunocytochemical analysis of A7r5 cells transfected with FLAG-tagged expression constructs that retain (Fat14189–4587) or exclude (Fat14201–4587) the NLS motif. Fat14189–4587 localized almost exclusively to the nucleus, whereas Fat14201–4587 was apparent in the nucleus and prominent throughout the cytoplasm (Fig. 10 B).
To evaluate these findings in the context of Fat1-mediated VSMC growth inhibition, we tested these Fat1IC derivatives for effects on cyclin D1 promoter activity. The IL2R-Fat1IC fusion protein yielded significant inhibition of β-catenin–mediated cyclin D1 promoter activation (Fig. 9 D); Fat14201–4587, but not Fat14189–4587, retained this inhibitory effect (Fig. 10 C). Both Fat14201–4587 and Fat14189–4587 are present in the nucleus, but the former has a cytoplasmic distribution not shared by Fat14189–4587; hence, we attribute this inhibitory effect on β-catenin to the extranuclear presence of Fat14201–4587.
Discussion
Fat1 is expressed widely during mouse and rat development (Ponassi et al., 1999; Cox et al., 2000), notably in areas with high levels of cellular proliferation. Although in situ hybridization of rat embryos demonstrated expression of fat1 mRNA in the developing aortic outflow tract (Ponassi et al., 1999), the significance of Fat1 in vascular tissues has not been previously explored.
We found relatively low expression of Fat1 in normal adult rat carotid arteries, and substantially increased levels during the first few days after injury (Fig. 1). Immunohistochemical analyses (Fig. 1 C) showed prominent Fat1 staining first in the injured arterial media, and subsequently in the neointima, a pattern of expression similar to that of VSMC proliferation in this model (Clowes et al., 1983b). Interestingly, areas of attenuated neointimal formation showed prominent Fat1 and decreased PCNA staining, providing an initial suggestion that Fat1 might act to limit VSMC proliferation in vivo (Fig. 1 C). Nevertheless, Fat1 levels in cultured VSMCs increased in response to serum and several factors known to promote VSMC activation and neointimal formation, including ATII (Powell et al., 1990), PDGF-BB (Ferns et al., 1991), and bFGF (Lindner and Reidy, 1991) (Fig. 2). This expression pattern contrasts with that described for N-cadherin, which decreases after stimulation of VSMC with serum or PDGF-BB (Uglow et al., 2003), and that of R-cadherin, which decreases substantially in the first few days after injury (Slater et al., 2004).
To evaluate how induction of this very large protocadherin might affect the response to vascular injury, we tested the effect of Fat1 on VSMC migration and proliferation, two of the key cellular functions activated in this setting. Both loss of Fat1 expression and expression of the IL2R-Fat1IC fusion protein attenuated VSMC migration (Fig. 3). In the context of recent reports regarding Fat1 function in epithelial cells (Moeller et al., 2004; Tanoue and Takeichi, 2004), these findings suggest that increased Fat1 expression facilitates VSMC migration by providing directional cues and stimulating actin cytoskeletal remodeling through its interactions with proteins of the Ena/VASP family. Together with the Fat1 knockdown results, inhibition of migration by the IL2R-Fat1IC fusion protein suggests that dissociation of Fat1 extracellular interactions from Fat1IC-mediated intracellular signaling interferes with directional migration.
Despite the induction of Fat1 in the proliferative phase after injury and in response to growth factor stimulation of cultured cells, our results in both loss- and gain-of-function studies (Figs. 4 and 5) suggest that Fat1 opposes VSMC proliferation. Loss of growth suppression resulting in imaginal disc overgrowth in Drosophila led to identification of Fat (Mahoney et al., 1991), the founding member of the cadherin subfamily that includes mammalian Fat1. Although recent analyses indicate that mammalian Fat1 is more closely related to Drosophila Ftl (Castillejo-Lopez et al., 2004) than to Fat, a growth regulatory function has yet to be described for Ftl. Altered growth characteristics were also not identified in mouse Fat1−/− neural progenitors and embryonic skin (Ciani et al., 2003). Thus, our findings in VSMCs may reflect cell type–specific differences in the expression of cadherins or other protocadherins functionally redundant with Fat1, or differences in the level of β-catenin expression. In either case, the results of Fat1 knockdown studies indicate that in VSMCs, endogenous levels of Fat1 expression are sufficient to limit cyclin D1 expression (Fig. 4) and β-catenin–mediated transcription (Fig. 9), whereas our gain-of-function studies (Fig. 5) suggest that decreased cyclin D1 expression and cell growth are likely physiologic consequences of Fat1 induction. Cyclin D1, a known TCF/β-catenin target gene (Shtutman et al., 1999; Tetsu and McCormick, 1999), plays a critical role in regulation of G1 phase progression and G1/S cell cycle transition (Jiang et al., 1993; Resnitzky et al., 1994), and the level of its expression is closely controlled. Increased Fat1 expression in response to injury probably acts to slow VSMC proliferation, at least in part by decreasing cyclin D1 expression.
Signaling by classical cadherins has been studied extensively, but the mechanisms of protocadherin signaling are not well understood. The intracellular portion of Fat1 shows limited similarity to classical cadherin cytoplasmic domains, with 30 of 137 (22%) residues matching consensus in the FC1 domain and 28 of 84 (33%) residues matching consensus in the FC2 domain (Dunne et al., 1995). Although Tanoue and Takeichi described partial colocalization of Fat1 and β-catenin in immortalized epithelial cell lines, they found more β-catenin in apical lateral cell contacts and more Fat1 in basal lateral cell contacts (Tanoue and Takeichi, 2004), and concluded that Fat1 does not participate in the classical cadherin system (Tanoue and Takeichi, 2005). Interestingly, these findings are consistent with the observation that in polarized epithelial cells, complexes forming between adjacent cells vary in composition according to their apical vs. basal position (Johnston and Gallant, 2002). Thus, our findings in VSMCs, which are morphologically and biochemically nonpolarized (Muller and Gimbrone, 1986), may differ because of the lack of apical–basal specialization in this cell type. In immunocytochemical studies, we found that β-catenin and Fat1 colocalized in a junctional pattern at points of contact between VSMCs (Fig. 6); Fat1 staining was also observed at cellular free edges, while β-catenin was not.
To our knowledge, a physical interaction between endogenous Fat1 and β-catenin has not been previously demonstrated. We found clear evidence that these proteins interact at physiologic levels of expression. Transfection studies with the IL2R-Fat1IC fusion protein indicated that, despite limited similarity to the β-catenin–interacting domains of classical cadherins, the Fat1IC domain was sufficient for this interaction (Fig. 7). Although mapping studies suggested that the Fat1 FC1 domain was most important for the β-catenin–Fat1 interaction, deletion of other domains within the Fat1IC also decreased the amount of protein coimmunoprecipitation, indicating that sequences both within and outside of the relatively conserved FC1 and FC2 domains may contribute to β-catenin–Fat1 interaction. Interestingly, the FC1 domain corresponds to the area of greatest similarity (54/196 aa identity [27%]) with the Drosophila Ftl cytoplasmic domain; its role in the β-catenin–Fat1 interaction described here suggests that Ftl may be capable of interaction with armadillo, the Drosophila homologue of β-catenin.
The IL2R-Fat1IC chimera allowed us to perform functional analyses without confounding effects attributable to increased expression of the Fat1 extracellular domain. Expression of IL2R-Fat1IC, but not a control protein lacking the Fat1IC domain, decreased nuclear translocation of β-catenin (Fig. 8), and inhibited β-catenin transactivation of both synthetic (Topflash) and native (cyclin D1) TCF-dependent promoters (Fig. 9). Although we found evidence of Fat1 cleavage resulting in a Fat1IC* fragment that may localize to the nucleus (Fig. 10), only a defined Fat1IC fragment lacking the NLS (aa 4189–4198) reproduced the inhibitory effect of the IL2R-Fat1IC fusion protein. This result suggests that inhibition of β-catenin transcriptional activity is mediated by Fat1IC outside the nucleus, and is not due to Fat1IC peptides in the nucleus. Thus, it remains to be determined if cleavage and nuclear translocation of Fat1IC underlies a specific function, perhaps as a chaperone or transcriptional regulator, or if it is important as a means to inactivate Fat1-mediated inhibition of β-catenin. Our studies to date suggest that the interaction of Fat1 cytoplasmic sequences with β-catenin has consequences for overall regulation of VSMC growth. The underlying mechanism appears similar to that described for classical cadherin-mediated sequestration of β-catenin in epithelial cells (Orsulic et al., 1999), but in the case of the protocadherin Fat1, this mechanism may be operative only in nonpolarized cells such as VSMCs.
Our findings suggest that increased expression of Fat1 after vascular injury facilitates migration and opposes proliferation of VSMCs. The former effect likely involves Fat1 interaction with Ena/VASP proteins, as described in other cell types (Moeller et al., 2004; Tanoue and Takeichi, 2004), whereas the latter effect relies in part on decreased nuclear accumulation of β-catenin (this paper). Interestingly, we found that the Fat1IC interaction with and inhibition of β-catenin both appeared less robust than that observed with classical cadherin sequences (Figs. 7 and 9), suggesting that Fat1 may be less efficient than the classical cadherins at sequestering β-catenin. Fat1 induction after injury and by growth factors contrasts with the expression pattern of other cadherins found in VSMCs. Together, these observations suggest that Fat1 may guide VSMC migration while remaining relatively permissive of growth in settings when VSMC proliferation is necessary for vascular repair. Drosophila Ftl is thought to use its exceptionally large extracellular domain to promote epithelial cell separation during formation of tubular organs in embryogenesis (Castillejo-Lopez et al., 2004); we speculate that mammalian Fat1, by virtue of its similar structure, may expedite circumferential distribution of VSMCs around the injured artery. Altogether, it is tempting to speculate that Fat1 limits VSMC proliferation while providing directional migration cues important during vascular remodeling, providing an integrative function that may oppose the formation of hyperproliferative cellular clusters. Finally, though expression of Fat1 in human vascular disease has not yet been evaluated, it is possible that loss of Fat1-mediated negative regulation could contribute to VSMC hyperplastic syndromes such as restenosis, transplant arteriopathy, or vein graft disease.
Materials and methods
Rat carotid artery balloon injury
All procedures were in accordance with institutional guidelines. The rat carotid artery balloon injury model was implemented as described previously (Sibinga et al., 1997). In brief, male Sprague-Dawley rats (20 in total, Zivic-Miller) weighing 350–400 g were anesthetized with 40 mg/kg ketamine and 5 mg/kg xylazine. The left common carotid artery was denuded of endothelium and stretched by three passages of a 2F embolectomy catheter according to standard protocols. At 3, 7, and 14 d after injury, animals were reanesthetized and killed, and carotid arteries were harvested and snap-frozen in liquid nitrogen for RNA and protein extraction, or fixed with 4% PFA and processed for paraffin embedding for immunohistochemical analysis.
qPCR
A cDNA fragment identified in differential mRNA display analysis of the rat carotid artery injury model (Sibinga et al., 1997) was cloned, sequenced, and subjected to BLAST analysis, which revealed homology of the sequence fragment with the 3′ end of the rat Fat1 ORF (GenBank/EMBL/DDBJ accession no.NM_031819). Total RNA was extracted from vascular tissues by homogenization in TRIzol (Invitrogen), treated with DNase I (1 U/μlPromega), and used for first-strand cDNA synthesis. The mRNA levels were quantified in triplicate by qPCR in the Mx3000P Real-Time PCR System with the Brilliant SYBR Green qPCR kit (Stratagene). Rat Fat1 specific primers for qPCR were 5′-CCCCTTCCAACTCTCCCTCA-3′ (forward) and 5′-CAGGCTCTCCCGGGCACTGT-3′ (reverse). PCR cycling conditions included 10 min at 95°C for 1 cycle followed by 45 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 60 s. Dissociation curve analysis confirmed that signals corresponded to unique amplicons. Expression levels were normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels for each sample, obtained from parallel assays and analyzed using the comparative ΔΔCt method (Bustin, 2000).
Western analysis
Fat1-specific antisera were raised in rabbits. A cDNA fragment encoding mouse Fat1 aa 4434–4587 was generated by PCR and cloned in frame with GST in the pGEX-2T plasmid. The resultant fusion protein was expressed in bacteria, purified by GST-Sepharose affinity chromatography (GE Healthcare), and used as an immunogen in a standard rabbit injection protocol (Cocalico Labs). Fat1-specific antiserum was purified by affinity chromatography performed sequentially on a GST column and a GST-Fat1 column. Antiserum specificity was evaluated by Western analysis of GST-Fat1 fusion protein and whole cell lysates from RASMCs (1:5,000 dilution). Other mouse antibodies used were anti-β-catenin (1:100; Santa Cruz Biotechnology, Inc.), anti-cyclin D1 (1:100; NeoMarkers), anti-FLAG M2 (1:5,000; Sigma-Aldrich), and anti-c-myc (1:250).
For protein analyses, cells or vascular tissue samples were homogenized and extracted in RIPA buffer with or without protease inhibitors. Whole cell lysate (30 μg) was separated by electrophoresis through 3–8% Novex Tris-acetate or 4–12% Bis-Tris polyacrylamide gels (Invitrogen) and transferred to Immobilon-P membrane (Millipore). After blocking in TBST (Tris pH 8.0, NaCl 150 mmol/L, and 0.1% Tween 20) plus 4% (wt/vol) nonfat milk, blots were incubated overnight at 4°C with primary antibodies. The blots were then incubated with HRP-conjugated secondary antibody and activity was visualized by ECL (GE Healthcare). Equivalent protein loading was evaluated with anti-α-tubulin (1:500; NeoMarkers), anti-lamin A/C (1:100; Santa Cruz Biotechnology, Inc.) or anti-actin (1:100, Santa Cruz Biotechnology, Inc.) antibodies.
Immunohistochemistry
Rat carotid arterial sections (5 μm) were incubated overnight with anti-Fat1 antiserum (1:2,000), washed extensively, and incubated with a 1:500 dilution of secondary antibody (biotinylated goat anti–rabbit IgG, DakoCytomation). Slides were incubated with avidin and biotinylated HRP, developed with a peroxidase substrate solution (DakoCytomation), and counterstained with hematoxylin (Fisher Schientific). Specificity of staining was confirmed by omission of the primary antibody. PCNA staining was performed with anti-PCNA (1:100; LabVision), alkaline phosphatase–conjugated goat anti–mouse secondary antibody (1:200), and visualization with BM Purple substrate (Roche). Images were obtained using a microscope (Eclipse E600; Nikon), 40×/NA 0.75 Plan objective, and Coolpix 5400 camera (Nikon).
Cell culture
Primary culture RASMCs were prepared as described previously (Sibinga et al., 1997) and maintained in DME (Invitrogen) containing 10% FBS (HyClone), 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 mmol/L Hepes (pH 7.4; Sigma-Aldrich). RASMCs were passaged every 3 to 5 d, and used between 4 and 8 passages from harvest. Primary culture MASMCs were harvested from the aortas of 12-wk-old male Friend virus B mice by enzymatic dissociation, evaluated by immunocytochemical analysis by using α-smooth muscle actin antibody (1:400, Clone 1A4; NeoMarkers), and maintained in DME containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. MASMCs were passaged every 2 to 4 d, and used between 4 and 8 passages from harvest. The A7r5 embryonic RASMC, 3T3, and 293T cell lines (American Tissue Type Collection) were cultured in DME containing 10% FBS. ATII was obtained from Sigma-Aldrich, and bFGF and PDGF-BB from Collaborative Biomedical. In stimulation experiments, the cells were made quiescent by incubation in medium containing 0.4% horse serum for 72 h before addition of the FBS or growth factor. Control cultures received an equivalent amount of vehicle. Whole cellular protein was extracted at designed time points.
RNAi
The mouse Fat1 siRNA templates were comprised of 19-bp sense sequences derived from GenBank/EMBL/DDBJ accession no. AJ250768 (position 4881, 5′-GGACCGAAGTCACCAAGTA-3′; position 5126, 5′-GCGACGCATTTAACATTAA-3′; position 6432, 5′-GCATGACACTTTAAATAAA-3′; position 7296; 5′-GTCTGGCAATGATCATAAA-3′) followed by a 9-bp loop sequence, a 19-bp antisense sequence, and a T7 promoter sequence. Control siRNAs included scrambled (GTAACCATAAACAGGCATT) and mismatched (underlined) (GTCTGATAATGCGCATAAA) derivatives of the 7296 sequence, and an unrelated siRNA based on the Renilla luciferase sequence. siRNA was transcribed in vitro using the T7-MEGAshortscript kit (Ambion), and transfected with X-tremeGENE Reagent (Roche) according to manufacturer's recommendations. Fat1 knockdown efficiency was assessed by Western analysis.
cDNA constructs
The mouse Fat1IC cDNA was generated by RT-PCR with primers containing HindIII and XbaI sites (underlined) to facilitate cloning: forward 5′-AAGCTTCTCTGCCGGAAGATGATCAGTCGG-3′ and reverse 5′-TCTAGACACTTCCGTATGCTGCTGGGA. The product was subcloned into the p3XFLAG-CMV-14 expression vector (Aldrich). The IL2R expression construct (a gift of S. LaFlamme, Albany Medical College, Albany, NY; LaFlamme et al., 1994) was used to construct a chimeric cDNA encoding the IL2R extracellular and transmembrane domains and the Fat1IC, with or without an in frame 3XFLAG tag (IL2R-Fat1IC-3XFLAG and IL2R-Fat1IC, respectively). The IL2R-E-cadherinIC-3XFLAG construct was produced using a similar strategy. The truncated FLAG-tagged Fat1IC constructs, Fat4189–4587 and Fat14201–4587, were generated by PCR from the IL2R-Fat1IC-3XFLAG template using forward primers 5′-CCATGGgcctctgccggaagatgatcagt-3'and 5′-CCATGGGCCAGGCTGAACCTGAAGACAAAC-3′ and the CMV24 reverse primer; the resulting fragments were cloned into pcDNA3.1v5 (Invitrogen). The FLAG-tagged N-cadherin and Myc-tagged β-catenin constructs were gifts from R. Hazan (Albert Einstein College of Medicine, Bronx, NY) and R. Kemler (Max Planck Institute of Immunobiology, Freiburg, Germany), respectively. All constructs were confirmed by sequencing.
Retrovirus preparation and transduction
The retrovirus system we used is based on the IRES-GFP-RV constructs developed by K. Murphy (Washington University, St. Louis, MO) and Phoenix ecotropic packing cells provided by G. Nolan (Stanford University, Stanford, CA). The IL2R-Fat1IC cDNA was inserted upstream of the encephalomyocarditis virus internal ribosomal entry sequence (IRES) and green fluorescent protein (GFP) ORF in the GFP-RV vector. A7r5 cells, MASMCs, or RASMCs (5 × 105) were infected with virus-containing supernatant in the presence of 8 μg/ml polybrene. Control cells transduced with virus encoding GFP alone or IL2R and GFP were generated in parallel, and FACS analysis of retroviral transduced cell lines indicated similar levels of GFP expression.
Cell migration assays
Cell migration was assessed by (1) scratch wounding of monolayers and (2) with Transwell 24-well cell culture inserts with 8-μm pores (Costar). For the former, MASMCs transfected with control or Fat1-specific siRNA were grown to confluence, and monolayers were denuded similarly using a 1,000-μl pipette tip. Photomicrographs of the same fields were obtained sequentially at 24 and 30 h after injury using a microscope (TMS; Nikon), Plan 4×/NA 0.13 DL objective, and camera (model 5400; Coolpix), and cellular progress was quantitated by planimetry of the denuded area and converted to distance migrated using NIH Image 1.63 software. For Transwell assays, quiescent cells were harvested, counted, and added (5 × 104/well) to the insert. Culture medium containing 10% FBS as chemotactic agent was added to the lower chamber. After 4 h, nonmigrating cells were removed from upper filter surfaces and the filter was washed, fixed, and stained. We then photographed six randomly selected 200× fields and counted cells that had migrated to the underside of the filter.
Cell proliferation assays
Cell number was evaluated with the CyQUANT Assay (Molecular Probes). Cells (2 × 104 per well) were plated in 6-well plates in DMEM containing 2% FBS, medium was replaced every other day, and at each time point triplicate wells were washed with PBS and frozen at −80°C. Net sample fluorescence was determined on a Victor 2 plate reader (Wallac) and enumerated by reference to a standard curve. For the BrdU incorporation assay, cells plated on chamber slides (Becton Dickinson) were serum starved (0.4% horse serum) for 48 h and then stimulated with 10% FBS. 10 μM BrdU (Sigma-Aldrich) was added to cells for 6 h before harvest at 24 h. Cells were washed in PBS, fixed in 4% PFA, treated with HCl, and stained sequentially with anti-BrdU antibody (1:200; Abcam) and AlexaFluor 555–conjugated secondary antibody (1:2,000; Molecular Probes). Cells were counterstained with DAPI (Molecular Probes). Signals were visualized by fluorescence microscopy, and the numbers of BrdU-positive and total nuclei per field calculated.
Immunocytochemistry
Cells were plated on chamber slides 24 h before staining, and then washed with PBS, fixed with PFA, blocked with 3% normal goat serum, and incubated with anti-β-catenin (1:100) and anti-Fat1 (1:1,000) antibodies. Specific staining was identified with goat anti–mouse and chicken anti–rabbit IgG (AlexaFluors, Molecular Probes). Expression of FLAG-tagged proteins was detected using FITC-conjugated anti-FLAG M2 antibody (8 μg/ml, Sigma-Aldrich). After counterstaining with DAPI, samples were mounted (Supermount medium; Biogenex) on glass slides and signals were visualized using an inverted fluorescent microscope (model IX70; Olympus) equipped with 20×/NA 0.4 and 40×/NA 0.6 LWD objectives and standard fluorescent filter sets, a CCD camera (SensiCam ; Cooke), and IPLab software (Scanalytics). Subsequent image processing was performed using Photoshop 7.0 and Illustrator 10.0 (Adobe Systems). Routine control experiments included omission of the primary antibodies. For Wnt pathway activation, cells were treated with LiCl (20 mmol/L) for 6–12 h, and then stained with anti-β-catenin antibody and DAPI nuclear stain.
Co-immunoprecipitation
Deletions within the Fat1IC portion of the IL2R-Fat1IC-3XFLAG construct were engineered using the vector XbaI site and introducing NheI restriction sites (QuikChange mutagenesis; Stratagene) in frame at the following positions in the mouse Fat1 aa sequence: 4187, 4244, 4395, and 4497. The sequences between selected pairs of restriction sites were excised, plasmids recircularized, and constructs confirmed by sequencing. Plasmids were introduced into 293T cells using Lipofectamine 2000 (Invitrogen). Whole cell lysates were harvested 24 h after transfection in lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.1% sodium deoxycholate, 1 mM Na3VO4, 1 mM NaF, with protease inhibitors. Myc-tagged β-catenin was immunoprecipitated by incubating 400 μg of precleared lysate with 2 μg of c-Myc antibody for 2 h at 4°C, followed by incubation with protein G–Agarose (Invitrogen) at 4°C overnight. For immunoprecipitation of endogenous proteins, RASMC whole cell lysates were precleared and then incubated with anti-Fat1 antiserum, anti-β-catenin antibody, or normal rabbit or mouse IgG for 2 h at 4°C, followed by incubation with protein G–Agarose overnight. The beads were washed and immune complexes recovered by boiling in sample buffer. Fat1 and β-catenin were detected by Western analysis, as described above.
Cell fractionation
Membrane, cytoplasmic, and nuclear fractions were prepared using the Compartment Protein Extraction Kit (Chemicon International, Inc.) according to the manufacturer's instructions. Fractionation and loading of proteins was evaluated by Western analysis with anti-lamin A/C antibody (Santa Cruz Biotechnology, Inc.).
Analysis of reporter gene activation
A7r5 cells growing in DMEM supplemented with 10% FBS were transfected transiently using Lipofectamine 2000 with β-catenin, IL2R-Fat1IC, Fat14189–4587, Fat14201–4587, or control expression constructs, along with the TCF wild-type (Topflash) and mutated control (Fopflash) luciferase reporter plasmids (Upstate Biotechnology), or cyclin D1 promoter luciferase reporter (a gift from R. Müller, Philipps-Universität, Marburg, Germany; Herber et al., 1994). MASMCs were transfected by Amaxa electroporation according to the manufacturer's instructions. The total amount of transfected DNA was kept constant. Cell lysates were harvested 24 h after transfection, and luciferase activity was determined using the Glo-lysis buffer system (Promega) and the Victor 2 plate reader. Luciferase activities were normalized to protein levels for each well. The data shown represent transfections repeated at least three times each.
Statistical analysis
Experiments were repeated at least three times. Data are presented as mean ± SEM. Comparisons between two groups were analyzed by t test, and comparisons between three or more groups were assessed by analysis of variance (ANOVA) with a Bonferroni/Dunn post hoc test. Significance was accepted for values of P < 0.05.
Acknowledgments
The authors thank Hanh Nguyen for critical reading of the manuscript, Rachel Hazan, Kenneth Murphy, Susan LaFlamme, Rolf Kemler, and Rolf Müller for providing DNA constructs, and Gary Nolan for the Phoenix cell line.
This work was supported by funds from the American Heart Association, Heritage Affiliate (Grant-in-Aid 0555803T) and from the National Heart, Lung, and Blood Institute of the National Institutes of Health (HL67944) to N.E.S. Sibinga.
Abbreviations used in this paper: Ftl, Fat-like; IC, intracellular; MASMC, mouse aortic smooth muscle cell; qPCR, quantitative PCR; RASMC, rat aortic smooth muscle cell; siRNA, small interfering RNA; VSMC, vascular smooth muscle cell.
References
- Angst, B.D., C. Marcozzi, and A.I. Magee. 2001. The cadherin superfamily: diversity in form and function. J. Cell Sci. 114:629–641. [DOI] [PubMed] [Google Scholar]
- Bhanot, P., M. Fish, J.A. Jemison, R. Nusse, J. Nathans, and K.M. Cadigan. 1999. Frizzled and Dfrizzled-2 function as redundant receptors for Wingless during Drosophila embryonic development. Development. 126:4175–4186. [DOI] [PubMed] [Google Scholar]
- Bustin, S.A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169–193. [DOI] [PubMed] [Google Scholar]
- Castillejo-Lopez, C., W.M. Arias, and S. Baumgartner. 2004. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J. Biol. Chem. 279:24034–24043. [DOI] [PubMed] [Google Scholar]
- Ciani, L., A. Patel, N.D. Allen, and C. ffrench-Constant. 2003. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol. Cell Biol. 23:3575–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes, A., M. Reidy, and M. Clowes. 1983. a. Mechanisms of stenosis after arterial injury. Lab. Invest. 49:208–215. [PubMed] [Google Scholar]
- Clowes, A.W., M.A. Reidy, and M.M. Clowes. 1983. b. Kinetics of cellular proliferation after arterial injury. I. Smooth muscle growth in the absence of endothelium. Lab. Invest. 49:327–333. [PubMed] [Google Scholar]
- Cox, B., A.K. Hadjantonakis, J.E. Collins, and A.I. Magee. 2000. Cloning and expression throughout mouse development of mfat1, a homologue of the Drosophila tumour suppressor gene fat. Dev. Dyn. 217:233–240. [DOI] [PubMed] [Google Scholar]
- Dunne, J., A.M. Hanby, R. Poulsom, T.A. Jones, D. Sheer, W.G. Chin, S.M. Da, Q. Zhao, P.C. Beverley, and M.J. Owen. 1995. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics. 30:207–223. [DOI] [PubMed] [Google Scholar]
- Ferns, G., E. Raines, K. Sprugel, A. Motani, M. Reidy, and R. Ross. 1991. Inhibition of neointimal smooth muscle accumulation after angioplasty byan antibody to PDGF. Science. 253:1129–1132. [DOI] [PubMed] [Google Scholar]
- Gallin, W.J. 1998. Evolution of the “classical” cadherin family of cell adhesion molecules in vertebrates. Mol. Biol. Evol. 15:1099–1107. [DOI] [PubMed] [Google Scholar]
- Hedgepeth, C.M., L.J. Conrad, J. Zhang, H.C. Huang, V.M. Lee, and P.S. Klein. 1997. Activation of the Wnt signaling pathway: a molecular mechanism for lithium action. Dev. Biol. 185:82–91. [DOI] [PubMed] [Google Scholar]
- Herber, B., M. Truss, M. Beato, and R. Muller. 1994. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 9:2105–2107. [PubMed] [Google Scholar]
- Huber, A.H., and W.I. Weis. 2001. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 105:391–402. [DOI] [PubMed] [Google Scholar]
- Jamora, C., R. DasGupta, P. Kocieniewski, and E. Fuchs. 2003. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 422:317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., S.M. Kahn, P. Zhou, Y.J. Zhang, A.M. Cacace, A.S. Infante, S. Doi, R.M. Santella, and I.B. Weinstein. 1993. Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene. 8:3447–3457. [PubMed] [Google Scholar]
- Johnston, L.A., and P. Gallant. 2002. Control of growth and organ size in Drosophila. Bioessays. 24:54–64. [DOI] [PubMed] [Google Scholar]
- Jones, M., P.J. Sabatini, F.S. Lee, M.P. Bendeck, and B.L. Langille. 2002. N-cadherin upregulation and function in response of smooth muscle cells to arterial injury. Arterioscler. Thromb. Vasc. Biol. 22:1972–1977. [DOI] [PubMed] [Google Scholar]
- Kaplan, D.D., T.E. Meigs, and P.J. Casey. 2001. Distinct regions of the cadherin cytoplasmic domain are essential for functional interaction with Galpha 12 and beta-catenin. J. Biol. Chem. 276:44037–44043. [DOI] [PubMed] [Google Scholar]
- Korinek, V., N. Barker, P.J. Morin, D. van Wichen, R. de Weger, K.W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 275:1784–1787. [DOI] [PubMed] [Google Scholar]
- LaFlamme, S.E., L.A. Thomas, S.S. Yamada, and K.M. Yamada. 1994. Single subunit chimeric integrins as mimics and inhibitors of endogenous integrin functions in receptor localization, cell spreading and migration, and matrix assembly. J. Cell Biol. 126:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner, V., and M. Reidy. 1991. Proliferation of smooth muscle cells after vascular injury is inhibited by an antibody against basic fibroblast growth factor. Proc. Natl. Acad. Sci. USA. 88:3739–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magg, T., D. Schreiner, G.P. Solis, E.G. Bade, and H.W. Hofer. 2005. Processing of the human protocadherin Fat1 and translocation of its cytoplasmic domain to the nucleus. Exp. Cell Res. 307:100–108. [DOI] [PubMed] [Google Scholar]
- Mahoney, P.A., U. Weber, P. Onofrechuk, H. Biessmann, P.J. Bryant, and C.S. Goodman. 1991. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 67:853–868. [DOI] [PubMed] [Google Scholar]
- Moeller, M.J., A. Soofi, G.S. Braun, X. Li, C. Watzl, W. Kriz, and L.B. Holzman. 2004. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 23:3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, W.A., and M.A. Gimbrone Jr. 1986. Plasmalemmal proteins of cultured vascular endothelial cells exhibit apical-basal polarity: analysis by surface-selective iodination. J. Cell Biol. 103:2389–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke, I.S., L. Hinck, J.R. Swedlow, J. Papkoff, and W.J. Nelson. 1994. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J. Cell Biol. 125:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, W.J., and R. Nusse. 2004. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 303:1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic, S., O. Huber, H. Aberle, S. Arnold, and R. Kemler. 1999. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1- mediated transactivation. J. Cell Sci. 112(Pt 8):1237–1245. [DOI] [PubMed] [Google Scholar]
- Owens, G.K., M.S. Kumar, and B.R. Wamhoff. 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84:767–801. [DOI] [PubMed] [Google Scholar]
- Ponassi, M., T.S. Jacques, L. Ciani, and C. ffrench Constant. 1999. Expression of the rat homologue of the Drosophila fat tumour suppressor gene. Mech. Dev. 80:207–212. [DOI] [PubMed] [Google Scholar]
- Powell, J.S., R.K. Muller, M. Rouge, H. Kuhn, F. Hefti, and H.R. Baumgartner. 1990. The proliferative response to vascular injury is suppressed by angiotensin-converting enzyme inhibition. J. Cardiovasc. Pharmacol. 16:S42–S49. [DOI] [PubMed] [Google Scholar]
- Ranganath, S., W. Ouyang, D. Bhattarcharya, W.C. Sha, A. Grupe, G. Peltz, and K.M. Murphy. 1998. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161:3822–3826. [PubMed] [Google Scholar]
- Resnitzky, D., M. Gossen, H. Bujard, and S.I. Reed. 1994. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14:1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadot, E., I. Simcha, M. Shtutman, A. Ben-Ze'ev, and B. Geiger. 1998. Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proc. Natl. Acad. Sci. USA. 95:15339–15344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan, C.M., and P.L. Weissberg. 1998. Smooth muscle cell heterogeneity: patterns of gene expression in vascular smooth muscle cells in vitro and in vivo. Arterioscler. Thromb. Vasc. Biol. 18:333–338. [DOI] [PubMed] [Google Scholar]
- Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 96:5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibinga, N.E., L.C. Foster, C.M. Hsieh, M.A. Perrella, W.S. Lee, W.O. Endege, E.H. Sage, M.E. Lee, and E. Haber. 1997. Collagen VIII is expressed by vascular smooth muscle cells in response to vascular injury. Circ. Res. 80:532–541. [DOI] [PubMed] [Google Scholar]
- Simcha, I., C. Kirkpatrick, E. Sadot, M. Shtutman, G. Polevoy, B. Geiger, M. Peifer, and A. Ben-Ze'ev. 2001. Cadherin sequences that inhibit beta-catenin signaling: a study in yeast and mammalian cells. Mol. Biol. Cell. 12:1177–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, S.C., E. Koutsouki, C.L. Jackson, R.C. Bush, G.D. Angelini, A.C. Newby, and S.J. George. 2004. R-cadherin:beta-catenin complex and its association with vascular smooth muscle cell proliferation. Arterioscler. Thromb. Vasc. Biol. 24:1204–1210. [DOI] [PubMed] [Google Scholar]
- Suzuki, S.T. 2000. Recent progress in protocadherin research. Exp. Cell Res. 261:13–18. [DOI] [PubMed] [Google Scholar]
- Takeichi, M. 1995. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 7:619–627. [DOI] [PubMed] [Google Scholar]
- Tanoue, T., and M. Takeichi. 2004. Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J. Cell Biol. 165:517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue, T., and M. Takeichi. 2005. New insights into Fat cadherins. J. Cell Sci. 118:2347–2353. [DOI] [PubMed] [Google Scholar]
- Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 398:422–426. [DOI] [PubMed] [Google Scholar]
- Uglow, E.B., S. Slater, G.B. Sala-Newby, C.M. Aguilera-Garcia, G.D. Angelini, A.C. Newby, and S.J. George. 2003. Dismantling of cadherin-mediated cell-cell contacts modulates smooth muscle cell proliferation. Circ. Res. 92:1314–1321. [DOI] [PubMed] [Google Scholar]
- Wheelock, M.J., and K.R. Johnson. 2003. a. Cadherin-mediated cellular signaling. Curr. Opin. Cell Biol. 15:509–514. [DOI] [PubMed] [Google Scholar]
- Wheelock, M.J., and K.R. Johnson. 2003. b. Cadherins as modulators of cellular phenotype. Annu. Rev. Cell Dev. Biol. 19:207–235. [DOI] [PubMed] [Google Scholar]
- Yagi, T., and M. Takeichi. 2000. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 14:1169–1180. [PubMed] [Google Scholar]
- Yap, A.S., W.M. Brieher, and B.M. Gumbiner. 1997. Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell Dev. Biol. 13:119–146. [DOI] [PubMed] [Google Scholar]