Abstract
The activity of the p53 gene product is regulated by a plethora of posttranslational modifications. An open question is whether such posttranslational changes act redundantly or dependently upon one another. We show that a functional interference between specific acetylated and phosphorylated residues of p53 influences cell fate. Acetylation of lysine 320 (K320) prevents phosphorylation of crucial serines in the NH2-terminal region of p53; only allows activation of genes containing high-affinity p53 binding sites, such as p21/WAF; and promotes cell survival after DNA damage. In contrast, acetylation of K373 leads to hyperphosphorylation of p53 NH2-terminal residues and enhances the interaction with promoters for which p53 possesses low DNA binding affinity, such as those contained in proapoptotic genes, leading to cell death. Further, acetylation of each of these two lysine clusters differentially regulates the interaction of p53 with coactivators and corepressors and produces distinct gene-expression profiles. By analogy with the “histone code” hypothesis, we propose that the multiple biological activities of p53 are orchestrated and deciphered by different “p53 cassettes,” each containing combination patterns of posttranslational modifications and protein–protein interactions.
Introduction
The product of the p53 gene is capable of inducing transcription activation or repression of a variety of genes, which in turn trigger cell cycle arrest and promote apoptosis, differentiation, or senescence (Vousden and Prives, 2005). p53 levels and activity are regulated by numerous stress-induced posttranslational modifications that converge on two distinct domains of the protein (Appella and Anderson, 2001). A kinase cascade phosphorylates several serine residues in the p53 NH2-terminal region. Among these, phosphorylation of serine 15, 20, and 46 and threonine 18 are of most significance, as they act by promoting p53 stabilization (Unger et al., 1999; Knights et al., 2003), by preventing nuclear export (Zhang and Xiong, 2001), or by favoring p53 recruitment on specific sets of promoters (Oda et al., 2000). A second pathway targets the p53 COOH terminus and involves the activity of at least two classes of acetyltransferases, p300/CBP and p300/CBP-associated factor (PCAF; Avantaggiati et al., 1997; Lill et al., 1997). p300/CBP acetylates lysines located in the last COOH-terminal portion of p53, specifically, lysines 370, 372, 373, and 382 (Gu and Roeder, 1997). PCAF has been linked to acetylation of a single residue, lysine 320, located within a flexible linker domain nearby the oligomerization domain, which also contains a nuclear localization signal (Sakaguchi et al., 1998; Liu et al., 1999). These initial studies demonstrated that all these acetylation events lead to enhancement of p53 DNA binding activity in vitro. More recently, acetylation has been shown to stimulate the p53–DNA interaction in vivo as well (Luo et al., 2004), in agreement with the original proposal that acetylation relieves the inhibitory role exerted by the COOH terminus on p53 DNA binding capacity (Prives and Manley, 2001; Luo et al., 2004). However, thus far, the biological significance of individual acetylation of each of these lysine clusters is unclear.
The proapoptotic activity is the most ancestral function of p53, but additional and more complex activities have clearly appeared during evolution. This is argued based on the properties of the p53 protein of Drosophila melanogaster, dp53, which differs from its human counterpart because of its exclusive role in promoting apoptosis. Similarly, the Caenorhabditis elegans homologue of p53, CEP-1, displays predominantly proapoptotic activity (Derry et al., 2001). The evolution of p53 toward cell cycle regulatory functions in complex multicellular organisms may reflect the need of cells to deal with various forms of stress in a more advantageous fashion and to mount adaptive responses that preserve the life of tissues with limited proliferative potential. In fact, in mammalian cells, the effects of p53 on cellular growth are pleiotropic and exhibit cell type specificities, and the p53-induced cell cycle arrest, although prolonged, is often reversible (Bates et al., 1999). This reversibility is presumably important in conditions of “repairable” cellular damage, for resumption of proliferation when apoptosis can be avoided. Yet, the molecular basis for p53's ability to commit cells toward these different outcomes is still an object of intense investigation. Given the large repertoire of p53-responsive genes, one possibility is that a particular combination of target genes, activated or repressed, may determine whether cells will survive or die after engagement of p53 activity. Because cells are continuously exposed to genotoxic signals of different nature and intensity, they must have elaborated ways that allow p53 to interact with specific classes of genes but not with others, ultimately leading to adaptation and differential susceptibility to stress.
How, then, does p53 select such combination patterns? Important determinants of selectivity are likely the accessibility of chromatin and the structural characteristics of the DNA consensus sequences within p53-responsive elements, which show a surprisingly high degree of variation (Resnick and Inga, 2003; Tomso et al., 2005). When several p53-response elements were analyzed for their ability to be activated in a yeast-based assay that eliminates the influence of chromatin on transcription, an unexpected 1,000-fold difference in p53 transactivation ability was detected depending on the central sequence present within each response element. In addition, tumor-derived p53 mutants that have altered conformation compared with the wild-type (WT) protein exhibit distinct promoter specificities and retain their ability to transactivate certain promoters but not others. Thus, the intrinsic DNA binding affinity of p53, together with conformational changes, may contribute to differential gene activation. Posttranslational modifications may play an important role in modifying both p53 conformation and p53 affinity for its downstream targets.
In this study, we have characterized the functional and biochemical properties of p53 mutants mimicking constitutive acetylation as well as of truly acetylated p53. We show that two distinct acetylable clusters at position 373 (K373) and 320 (K320) regulate p53 activity in quite a different fashion. A model is proposed to explain how site-specific acetylation operates and cross-talks with other events, such as additional posttranslational modifications and protein–protein interactions, to modify p53 affinity for different classes of genes during stress signals, leading to cell survival or death.
Results
Different kinetics of acetylation of K320 and K373 in response to DNA damage
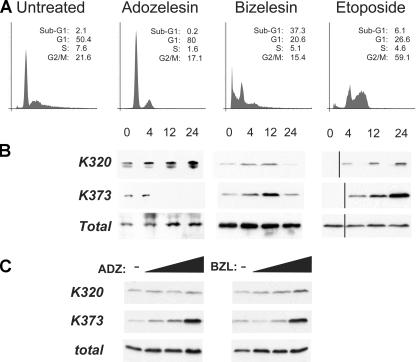
We began our studies by asking how acetylation of different sites is regulated during stress signals. For these experiments we used a p53-null, human lung carcinoma cell line, H1299, where the expression of p53 was reconstituted via a tetracycline-inducible vector. The kinetics of acetylation of K320 and K373 were then studied in cells exposed to the radiomimetic DNA-alkylating agents adozelesin and bizelesin or to the topoisomerase inhibitor etoposide. Adozelesin and bizelesin are members of the cyclopropylpyrroloindole (CPI) family of DNA minor groove alkylating agents endowed with anti-tumor activity (Lee et al., 1994). Adozelesin is a monofunctional CPI, whereas bizelesin is a CPI dimer that can alkylate adenines on one or both DNA strands, forming double-stranded DNA cross-links that render it more cytotoxic than adozelesin. Thus, these drugs allowed us to assess whether different types of DNA damage trigger distinct acetylation events. In cells treated with adozelesin, which arrested predominantly in G1 and did not undergo apoptosis (Fig. 1 A), acetylation of K373 was no longer visible after 4–12 h of treatment, whereas acetylation of K320 remained detectable for 24 h after the addition of the drug (Fig. 1 B). In contrast, treatment with bizelesin or etoposide resulted in robust and sustained levels of acetylation of K373 and induced an initial arrest in G2/M followed by apoptosis (Fig. 1 A and not depicted).
Figure 1.
The kinetics of K320 and K373 acetylation of p53 in response to DNA damage correlate with cell cycle arrest or apoptosis. (A) Cell cycle profile of H1299 cells expressing tetracycline-inducible p53. Cells were left untreated or treated with 5 μM adozelesin or bizelesin or with 100 μM etoposide after a 24-h induction of p53 with tetracycline. Flow cytometry was performed as described previously (Avantaggiati et al., 1997). The percentage of cells displaying Sub-G1, G1, S, or G2/M DNA content is provided as an inset for each histogram. (B) H1299 cells expressing p53 were treated as indicated at the top of each panel in A. At different times after treatment, cells were harvested and cell lysates were immunoprecipitated with a polyclonal antibody recognizing acetyl-p53K320 (Acp53K320) or acetyl-p53K373 (Acp53K373) or with an antibody recognizing total p53, as indicated on the left side of each panel. The products of these reactions were then subjected to immunoblot with a mixture of monoclonal antibodies directed against the p53 NH2 terminus (DO-1) and COOH terminus (PAb421). Black lines indicate that intervening lanes have been spliced out. (C) H1299 cells expressing WT p53 were left untreated or treated with increasing concentrations of adozelesin or bizelesin (1, 50, or 500 μM), harvested after 6 h, and analyzed as in B.
These results provided evidence that the kinetics of K320 and K373 acetylation can be dissected depending on the type of damage. To test whether the amount of DNA damage also influences the extent and sites of acetylation, we treated cells expressing p53 with increasing doses of adozelesin or bizelesin (Fig. 1 C). In this case, we observed a dose-dependent enrichment of the levels of K373 acetylation, whereas acetylation of K320 was only modestly enhanced. Thus, various acetylation sites act as a “sensor” system for the type and extent of DNA damage and may differentially regulate the ability of p53 to induce cell cycle arrest or apoptosis.
Opposing effects of K320 and K373 acetylation on the ability of p53 to induce apoptosis
To gain more insights into the function of acetylation, we created gain-of-function p53 acetylation mutants. Glutamine in place of lysine has been shown to mimic the effects of a constitutive acetylation in the case of histones and of p53 (Luo et al., 2004), probably because glutamine is a neutral amino acid, which, like N-acetyl-lysine, has an amide group that can function as a hydrogen bond donor or acceptor. Thus, we constructed mutants harboring lysine to glutamine substitutions at position 320 (Q320), a triple mutant at position 370/372/373 (Q373), or at all these positions (DM). Native p53 and its derivative acetylation mutants were expressed in the p53-null cell line, H1299, via a tetracycline-regulated promoter (see Fig. 4 F and Fig. S1 C, available at http://www.jcb.org/cgi/content/full/jcb.200512059/DC1, for typical expression levels of these p53 proteins). Cells were treated with different DNA-damaging agents, and their cell cycle distribution was assessed (Fig. 2 A). In untreated cells, expression of native p53 produced an arrest, especially at the G1 phase, accompanied by a reduction of cells transiting throughout S phase, as expected. The cell cycle profiles of cells harboring p53 acetylation mutants were different in several ways: a higher percentage of cells expressing p53DM arrested in G1, whereas cells harboring p53Q373 arrested markedly in G2/M, suggesting that these residues predominantly influence the activity of p53 on the G2 checkpoint. To rule out the possibility that these differences may be due to unique clonal characteristics of these p53 cell lines, similar experiments were performed by using either additional individual clones or polyclonal mixtures. The analysis of the growth characteristics and viability of these cells produced substantially similar results (Fig. S1, A and B).
Figure 4.
Characterization of the gene-expression pattern elicited by p53 acetylation mutants. (A) Clustering of genes regulated by p53 acetylation mimics. Probes corresponding to fold change >2 were hierarchically clustered. Data from each probe are in the columns, and each experiment is shown as a row. Red and green denote increased and decreased expression levels, respectively, with the intensity reflecting the magnitude of change. (B) Validation of microarray data by semiquantitative RT-PCR. To verify oligonucleotide microarray results, semiquantitative RT-PCR was used to estimate the amount of RNA from three altered genes in WT p53–, p53Q320-, and p53Q373-expressing cells (+). To ensure that clonal variability does not contribute to the different levels of gene expression, mRNA was collected from H1299-WT, -Q320, and -Q373 cells in the absence of tetracycline (−). Densitometry was performed using Fluor Chem 3.04A software (Alpha Innotech Corp.) and normalized to the β-actin signal. Fold changes represent the difference for each mRNA observed within each individual cell line. Each band detected in the absence of tetracycline was attributed an arbitrary value of 1 and compared with the band detected in the presence of tetracycline (lanes 1, 3, and 5 were compared with lanes 2, 4, and 6, respectively). (C) In vivo interaction of p53 with its DNA binding sites derived from different promoters. Untreated H1299 or H1299 treated with tetracycline were cross-linked with formaldehyde and immunoprecipitated with a polyclonal antibody recognizing p53 (FL393). The amount of p53 proteins contained in these immunoprecipitation reactions was normalized (E), such that similar amounts of DNA-bound p53 were used for sequence-specific amplification of the promoters of p21/WAF, PIG3, BAX, and p53AIP1. (D) H1299 cells were treated with tetracycline to induce WT p53 protein and then subjected to ChIP. Acetylated p53 was precipitated by using antibodies directed against Acp53K320 or Acp53K373/K382, whereas total p53 was precipitated with the polyclonal p53 antibody. Promoter binding was assessed as described in C. Input represents ∼2% of the total DNA content used for precipitation. (E) Equalization of p53 proteins used for the ChIP assays in C. p53 levels in each cell line were predetermined after DNA–protein cross-linking by direct immunoblot and before the immunoprecipitation reaction (not depicted). The amounts of cell extract were then adjusted based on this preliminary assessment, so that equivalent amounts of each acetylation mutant were subjected to immunoprecipitation and subsequent PCR amplification. The two panels show typical levels of p53 proteins after this equalization, derived from two different experiments. (F) H1299 cells were left untreated (time 0) or were treated with tetracycline for 12 and 24 h. The type of p53-expressing cell line is indicated at the top of the panels. Cell lysates were prepared and subjected to immunoblots with antibodies recognizing p53, p21/WAF, Bax, and actin.
Figure 2.
Different effects of p53 acetylation mutants on cell growth. (A) Cell cycle profile of H1299 cells expressing p53 proteins (indicated at the top of panels). Cells were left untreated or treated with 50 μM adozelesin, 50 μM bizelesin, or 100 μM etoposide. Drugs were added 24 h after tetracycline addition, and cells were grown for an additional 24–48 h and then harvested and stained with propidium iodide (shown in all panels) and with BrdU (not depicted), and their cell cycle profile was determined. (B and C) Tetracycline-induced H1299 cells were left untreated or treated with 1 μM adozelesin for 4 h. Cells were extensively washed and incubated in drug-free media but in the presence of tetracycline for an additional 24 h. Tetracycline was then removed to shut off p53 expression, and cells were allowed to recover for 6–8 d, after which one dish was stained with Coomassie brilliant blue (B) and the remaining were used for cell counting (C). Error bars indicate SEM.
To understand how site-specific acetylation of p53 affects sensitivity to anti-tumor agents, cells were treated with the DNA-damaging agents adozelesin, bizelesin, or etoposide. In drug-treated cells, expression of p53Q320 activated the G1 checkpoint and conferred significant protection from cell death compared with native p53-expressing cells, which underwent apoptosis when treated with bizelesin or etoposide. Quite in contrast, p53Q373 had a strong chemosensitizing effect, as the majority of cells bearing this mutant underwent apoptosis early after drug treatment. In cells harboring the mutant at all positions, the percentage of apoptosis was significantly lower than that observed in both native- and p53Q373-expressing cells, suggesting that the mutation of K320 has a dominant effect in preventing cell death.
K320 acetylation favors resumption of proliferation and cell survival
Results presented thus far are consistent with the possibility that acetylation of residues around position 373 activates the apoptotic pathway, whereas acetylation of K320 suppresses cell death. We then hypothesized that acetylation of K320 could function to disable or delay the apoptotic program elicited by p53, thus allowing cells to resume proliferation in conditions of moderate DNA damage once p53 signaling is extinguished. To test this, cells expressing native p53 or p53 acetylation mutants were pulsed with low concentrations of adozelesin for a short period of time in the presence of tetracycline. p53 expression was shut off by removing tetracycline from the media 24 h after drug treatment, and cell growth was monitored for several days thereafter (Fig. 2, B and C). In these conditions, untreated and drug-treated cells expressing p53Q373 displayed a significant loss of viability, whereas cells expressing p53Q320 were able to resume proliferation. In addition, cells expressing p53DM again displayed a phenotype intermediate between that exhibited by p53Q320 and -Q373. These results argue against a simple model in which the number of acetylated lysines controls p53-apoptotic activity in a dose-dependent manner. Rather, they suggest that the effects of acetylation are strictly position specific and place acetylation of K320 as a central event in favor of cell survival.
Site-specific acetylation differentially influences the interaction of p53 with transcriptional coactivators and corepressors
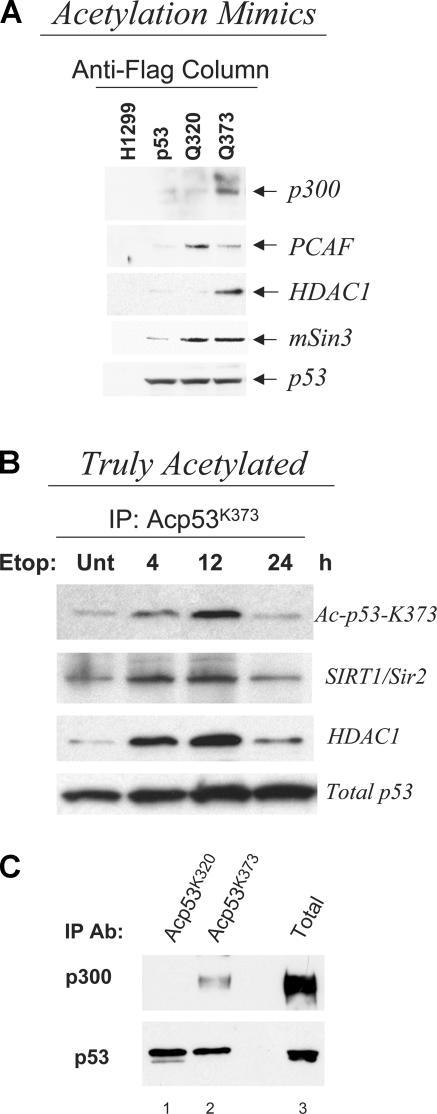
Posttranslational modifications regulate p53's interaction with other cellular proteins. We next asked whether acetylation influences the association with factors known to bind p53, particularly to coactivators and corepressors. p53 protein complexes isolated from native or p53 acetylation mutant–expressing cells were subjected to immunoblot analysis with antibodies recognizing p300, PCAF, HDAC1, mSin3, and p53 itself. HDAC1 and mSin3 are components of a corepressor complex, which plays an important role in the ability of p53 to silence transcription of critical antiapoptotic promoters (Murphy et al., 1999). As shown in Fig. 3 A, the p53Q373 mutant coprecipitated a significantly higher amount of p300 compared with native p53 and p53Q320 and, conversely, p53Q320 was more efficient in interacting with PCAF, indicating that each acetylation cluster specifically modifies the interaction of p53 with distinct types of histone acetyltransferases. Further, p53Q373 coprecipitated higher amounts of HDAC1 than native p53 or p53Q320 (Fig. 3 A). To further substantiate these findings, we studied the binding pattern of endogenously, “truly” acetylated p53 after treatment with etoposide. Consistent with data obtained with the acetylation mimics, acetylated p53 detected with the anti–acetyl-K373–specific antibody coprecipitated a significant amount of HDAC1 and SIRT1 (Fig. 3 B). Interestingly, these deacetylase–p53 complexes were detectable for at least 12 h after treatment, indicating that acetylation of K373 can stabilize the interaction of p53 with deacetylase–corepressor complexes. Similarly, in A549 cells treated with adozelesin, truly acetylated p53 at K373 displayed a significantly stronger affinity for p300 compared with acetyl-K320 p53 (Fig. 3 C, compare lanes 1 and 2). These data further support the notion that glutamine in place of lysine mimics the effects of constitutive acetylation and imply that acetylation of different sites selectively modifies the affinity of p53 for different types of chromatin-remodeling enzymes.
Figure 3.
Interaction of cofactors with acetylated forms of p53 protein. (A) Nuclear extracts prepared from H1299 cells treated with tetracycline were immunoprecipitated with an anti-Flag antibody, eluted with Flag peptide, and run on 4–20% gradient gels. The presence of p300, PCAF, HDAC1, and mSin3 was revealed by immunoblotting with specific antibodies. (bottom) Total p53 levels contained in the anti-Flag–specific immunoprecipitation. (B) H1299 cells expressing native p53 were treated with etoposide for the indicated times. Cell extracts were immunoprecipitated with the anti-Acp53K373–specific antibody. The presence of p53, SIRT1/Sir2, or HDAC1 in these reactions was revealed via immunoblots with specific antibodies, as indicated at the side of each panel. The bottom panel represents ∼20% of the total amount of precipitated p53 protein. (C) Analysis of the interaction between truly acetylated forms of p53 and p300. A549 cells expressing WT p53 were treated for 16 h with 50 μM adozelesin and immunoprecipitated with either Acp53K320 (lane 1) or Acp53K373 (lane 2). These reactions were subjected to p53- and p300-specific immunoblots. The total levels of p300 were determined with the C-20 (Santa Cruz Biotechnology, Inc.) antibody after immunoprecipitation, whereas p53 levels were detected from total extract (lane 3).
Delineation of the genetic program elicited by differently acetylated forms of p53
p53 conveys complex cellular responses that are due in part to its ability to function as a transactivator and as a transrepressor. To understand how acetylation operates, we performed microarray analysis on cells expressing native p53, p53Q320, and p53Q373 (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200512059/DC1). Approximately 40,000 probe sets were examined by using high-density oligonucleotides arrays at 12 h after addition of tetracycline to the media. For this analysis, we applied a present call noise filter (one in six arrays), fold-change thresholds (>2), and a p-value of <0.05 as our initial dataset for preliminary analysis and gene identification, leading to further consideration of ∼27,000 transcripts. To more specifically identify genes regulated in an acetylation-dependent manner, genes found in the array platform derived from cells expressing native p53 served as background, so that p53Q373 or -Q320 gene-expression patterns were directly compared with native p53. With this analysis, we determined that both p53Q320 and -Q373 lead to an increase of a similar number of transcripts, whereas p53Q320 repressed 955 genes, which was significantly less than the 1,576 transcripts repressed by p53Q373. Thus, p53Q373 is a stronger repressor than p53Q320, consistent with its ability to interact more efficiently with HDAC1 and SIRT1/mSir2 (Fig. 3). Further, in agreement with the observation that p53Q373 sensitizes cells to apoptosis, many of the genes found activated by this mutant promote cell death (i.e., APAF1, caspase 6, pig3, pig11, AMID, PCBP4, and IGFBP3), and a significant number of those repressed promote survival (i.e., survivin, API5, BIRC3, and IL31RA). The behavior of p53Q320 was, instead, opposite. For example, several of the proapoptotic genes activated by p53Q373 were repressed by p53Q320 (i.e., APAF1 and pig11) relatively to native p53 and, vice versa, some antiapoptotic genes repressed by p53Q373 were activated by p53Q320 (i.e., survivin, TRAF2, AATF, and BIRC4). Numerous other genes were also conversely expressed between p53Q320 and -Q373 (e.g., cyclin B1; Table S1, bold). To gain a further unbiased analysis of this phenomenon, a “heat map” was generated, which demonstrated differential, if not contrasting, gene-expression patterns between p53Q320 and -Q373 (Fig. 4 A). To validate the gene changes identified with the microarray platform, semiquantitative PCR was also performed on various relevant apoptotic- and growth arrest–related genes, some of which are shown in Fig. 4 B. This approach confirmed that the regulation of previously known p53 targets, such as pig11 and APAF1, or of new p53-regulated genes like Jagged2, was significantly different depending on the site of the mutation.
Although we cannot completely exclude the possibility that at least some of these changes are an epiphenomenon, reflecting events secondary to p53 activation, many of the relevant genes differentially modulated by p53Q320 and -Q373 are well-known primary targets of p53, for example, APAF1, pig11, survivin, caspase 6, mdm2, and cyclin B1. In addition, a large number of newly identified genes that we found up-regulated in the microarray platform contain p53 binding elements (Table I), thus suggesting that they might be direct p53 targets. In addition, this approach indicated that p53Q373, unlike p53Q320, predominantly controls the expression of genes that regulate apoptosis.
Table I.
Putative p53 binding sites in the genes up-regulated by p53 acetylation mimics
| Gene length | Site location | Score | Putative p53 binding site | |
|---|---|---|---|---|
| Q320 | ||||
| axl | 62851 | 47804 | 100 | GGGCTTGTCC ACATGGGCT GGGCATGTCT |
| cdk2 | 25017 | 9808 | 100 | AAACTTGTCT AGCCCCCAAGAACC AAACTTGCCT |
| rad9 | 27671 | 22729 | 93.66 | ACACAAGTCC AGATATAGACAG AAACTTGCCC |
| atm | 162402 | 89128 | 93.56 | AAGCATGTTT AAGA ACACTTGTTC |
| mad2l1 | 27376 | 25528 | 93.02 | AATCTAGCTC CTTATTTTC AAACATGTTT |
| mcm5 | 44646 | 23519 | 93.01 | AGGCAAGCTC TGTCCTCATTGCT AGGCAAGACC |
| mcm6 | 55966 | 41522 | 90.9 | AGGCATGCTT TAATACAAGTGATA TGACTTGTTA |
| birc5-survivin | 30454 | 15287 | 90.13 | AAGCATGCTG TGAGA GAGCTTGTCA |
| cdc6 | 34178 | 26674 | 90.89 | AGACTTGCCT G TGGCTAGCTA |
| tnfrsf6b-dcr3 | 64663 | 13579 | 88.94 | AAGCTTGTCT CAGGG CAGCTTGTGT |
| cdc25a | 51427 | 13156 | 93 | AGTCAAGTCT CACTATGTTGCCCC AGGCTTGTTT |
| cdc2 | 35523 | 1135 | 90.93 | TAACAAGCTC CTTTCATCAGTCCC AAGCAAGCCA |
| skp2 | 51917 | 41500 | 95.85 | AAACTAGTTT AAATTGA TGACTTGTTC |
| jun | 23255 | 3530 | 88.92 | TGGCTTGGCT GGGCTGGGCCATG AGACTAGTTT |
| cdkn2c | 25945 | 6870 | 87.82 | AGGCATGGTG . GAGCATGCCT |
| Q373 | ||||
| csnk1g1 | 148281 | 12563 | 100 | GGACTTGCCT GTAGTCAGAA GAACTTGTCT |
| pig11 | 40531 | 13335 | 93.59 | GCACAAGCCT TTTAAGTCAT GAGCTAGTCC |
| pml | 68790 | 1318 | 93 | AGACTTGCTT TA AGGCAAGATC |
| igfbp3 | 29032 | 25147 | 92.04 | TGACTAGCTT CTTTCACT TAGCATGTTT |
| jagged2 | 47019 | 18872 | 89.85 | GAGCATGTTG GGGTGTGTGGATG GAGCATGTTG |
| jag1 | 56265 | 29477 | 82.78 | ATACATGCTT GCTTA TTGCAAGCCT |
| pig3 | 27431 | 8069 | 83.82 | AGACATGTTT TTTGCCC TAACAAGTGA |
| ak1 | 28274 | 1838 | 83.45 | TGGCCAGCTC CAAACTTAC AGACTTGTCT |
| apaf1 | 110193 | 79794 | 88.99 | GGGCAAGCCC AGACA CCACTTGTTT |
| casp6 | 34733 | 18886 | 88.89 | TGGCTTGTTC AAAGG AGACAAGTGT |
| gas6 | 32013 | 11571 | 82.21 | CGTCTTGCCC TTCAGAAACATTGTAGACTTGTAT |
Genes that were activated by p53Q320 or -Q373 in the microarray platform were researched in a p53 database generated at The Rockefeller University (http://linkage.rockefeller.edu/p53). This database catalogues putative p53 DNA binding elements in 2,583 human genes and 1,713 mouse homologues on the basis of a newly developed p53MH algorithm. Genes containing putative p53 consensus sites and relative scores are shown. Gene length represents the combined base-pair length of the promoter and gene sequence. Site location represents the position within the gene length where the listed binding sequence begins. The consensus p53 binding sequence is PuPuPuC(A_T)(T_A)GPyPyPy.
Acetylation of various sites differentially influences the interaction of p53 with downstream promoters
Data based on the microarray analysis suggested that acetylation of K320 or K373 may lead to direct p53 binding onto different types of promoters. However, previous studies showed that simultaneous acetylation of multiple COOH-terminal lysines enhances the DNA binding activity of p53 on typical DNA binding elements (Gu and Roeder, 1997; Luo et al., 2004). In light of our results, we decided to investigate how each of the acetylation clusters individually influences p53–promoter interactions. The DNA binding properties of acetylation mutants and of truly acetylated p53 were studied in vivo in chromatin immunoprecipitation (ChIP) assays. To assess the influence of chromatin on the ability of p53 to interact with its DNA consensus sites, we also performed electrophoretic mobility shift assays (EMSAs) with in vitro–purified p53 (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200512059/DC1). A first observation arising from these experiments was that native p53 bound to the endogenous promoter of p21/WAF with significantly higher affinity compared with the PIG3, BAX, or p53AIP1 promoters (Fig. 4 C). Results obtained with EMSAs essentially recapitulated those seen with the ChIP assays and indicated that p53 possesses substantially different intrinsic affinity for its DNA binding elements. Noticeably, both p53Q320 (Fig. 4 C) and its truly acetylated counterpart immunoprecipitated with the anti–acetyl-320 antibody (Fig. 4 D), bound to the high-affinity p21/WAF promoter more efficiently than native p53 or p53Q373. Conversely, p53Q373, as well as endogenously acetylated p53 at position K373, interacted better with the low-affinity proapoptotic promoters of BAX (Fig. 4, C and D), PIG3, and p53AIP1 (Fig. 4 C), compared with other p53 proteins. It is important to note that both the EMSAs and ChIP assays were performed in conditions in which p53 levels were equalized (Fig. 4 E). Thus, these differences reflect acetylation-mediated changes in the intrinsic p53's DNA binding affinity. Furthermore, the different promoter binding pattern of p53 acetylation mutants resulted in coherent changes in the expression levels of corresponding encoded products, as demonstrated by the different protein levels of Bax and p21/WAF in these cell lines (Fig. 4 F). Based on these data, we infer that acetylation of K373 enhances the interaction of p53, particularly with low-affinity proapoptotic promoters.
Acetylation influences p53 nuclear retention and is coupled to site-specific phosphorylation
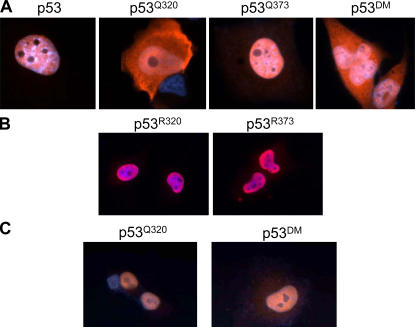
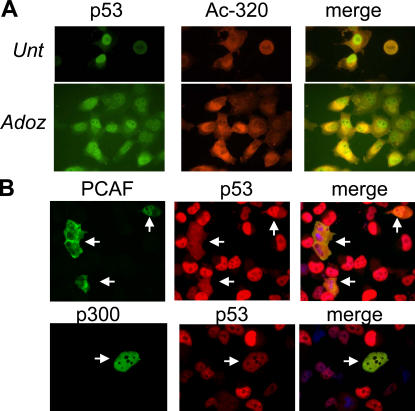
The aforementioned results offer a plausible molecular basis for the proapoptotic activity exhibited by p53Q373; however, they do not completely explain the biological effects of the mutant at position 320. Because K320 is located in a previously identified nuclear localization signal (Liang and Clarke, 2001), we suspected that acetylation of this residue might affect the subcellular localization of p53. In fact, as shown in Fig. 5, the p53 acetylation mimic at position K320 was localized predominantly in the cytoplasm, whereas p53Q373 was almost exclusively nuclear (Fig. 5 A). The presence of the mutation at position 320 also partially placed p53Q373 off the nucleus, as argued by the cytoplasmic localization of p53DM. In addition, when lysines were replaced with arginine rather than with glutamine, both R320 and R373 mutants localized in the nucleus (Fig. 5 B), indicating that neutralization of the positive charge of lysine interferes with nuclear accumulation of p53. Surprisingly, however, the increased cytoplasmic concentration of p53Q320 and -DM is not sustained by an increase in nuclear import but rather by an accelerated nuclear export. This is argued because treatment with leptomycin B, which blocks the activity of the exportin protein Crm1 (Fukuda et al., 1997), resulted in net nuclear accumulation of both mutants (Fig. 5 C). To further validate these results, two additional sets of experiments were performed. First, we studied the subcellular distribution of truly acetylated p53 in H1299 cells treated with adozelesin, which enhances the levels of K320 acetylation (Fig. 6 A). Treatment with adozelesin increased the cytoplasmic fraction of p53, and such fraction reacted with the anti–acetyl-320 antibody. Second, we determined the localization of p53 after overexpression of PCAF or of p300, which acetylate K320 and K370/372/373, respectively. Overexpression of PCAF resulted in increased cytoplasmic levels of p53 (Fig. 6 B), in contrast to overexpression of p300, where p53 remained substantially nuclear.
Figure 5.
Acetyl-mimic Q320 alters the nuclear-cytoplasmic shuttling of p53. (A and B) Tetracycline-induced H1299 cells were grown on glass coverslips, and 24 h later cells in A and B were stained with an anti-p53 polyclonal antibody (red) and with DAPI (blue). (C) Tetracycline-induced cells expressing p53Q320 and p53DM were grown in the presence of leptomycin B and subjected to indirect immunofluorescence.
Figure 6.
Acetylation of K320 increases the cytoplasmic localization of p53. (A) Tetracycline-induced WT p53–expressing H1299 cells were left untreated or were treated for 16 h with 50 μM adozelesin and then stained with a polyclonal antibody recognizing 320-acetylated p53 (Ac-320; red), a monoclonal antibody recognizing total p53 (green) and DAPI. (B) WT p53–H1299 cells were transfected with a PCAF or a p300 expression vector for 24 h and then stained with DAPI and with antibodies recognizing p300, p53, or PCAF, as indicated at the top of each panel.
The observation that K320 acetylation might place p53 off the nucleus by accelerating nuclear export led us to investigate the possible cross-talk between acetylation and phosphorylation of serine 15 (S15), which has been implicated in nuclear accumulation (Zhang and Xiong, 2001). As shown in Fig. 7 A, S15 phosphorylation was almost undetectable in cells expressing p53Q320, whereas the p53Q373 mutant was hyperphosphorylated at this residue compared with other p53 proteins, suggesting that acetylation and phosphorylation might be functionally linked. To assess whether other specific combination patterns of acetylation and phosphorylation do exist, we examined the phosphorylation state of S392 and S46. S392 phosphorylation increases the stability and the nuclear localization of p53 (Kim et al., 2004), whereas phosphorylation of S46 is specifically involved in mediating the interaction of p53 with the promoter of the proapoptotic gene p53AIP1 (Oda et al., 2000). We found that p53Q373, but not p53Q320, was hyperphosphorylated at both sites.
Figure 7.
Phosphorylation profile of p53 acetylation mutant proteins. (A) Cell extracts derived from tetracycline-induced H1299 cells expressing p53, p53Q320, or p53Q373 were subjected immunoblot with antibodies recognizing total or S15-phosphorylated p53. p53 levels were equalized among the different cell lines (bottom), so that similar amounts of each p53 acetylation mutant was probed with anti-phosphorylation–specific antibodies. S46 or S392 phosphorylation was detected via immunoprecipitation with a p53 polyclonal antibody, followed by immunoblot with anti-p53 antibodies directed against phospho-S46 or -S392. Fold induction for each phosphorylation site was determined by densitometry by using Fluor Chem 3.04A software, whereby cells expressing native p53 were attributed an arbitrary value of 1 and signals were normalized for total p53 levels. (B) WT p53, p53Q320, and p53Q373 were purified from baculovirus-infected cells and immunoprecipitated (IP) with different antibodies, each recognizing a different epitope, at the concentrations indicated at the top of each panel. The products of these immunoprecipitations were subjected to immunoblot (IB). The combination of antibodies used is indicated at the left side of each panel. (C) p53 mutants purified from baculovirus-infected cells were visualized via Coomassie blue staining (1) or by direct immunoblot with the DO-1 antibody (2).
Thus, K320 acetylation negatively interferes with the phosphorylation and the activity of the nuclear export domain of p53, whereas K373 acetylation enhances the phosphorylation of sites involved in regulation of retention and accumulation of p53 in the nucleus, as well as in p53 binding to proapoptotic promoters.
Modifications of K320 and K373 induce conformational changes in the NH2 terminus and DNA binding domain of p53
As a whole, our results imply that individual acetylation of lysines 320 and 373 of p53 has general, yet differential, effects on the intrinsic DNA binding activity and on the ability to interact with cellular proteins and influences other key posttranslational modifications, such as phosphorylation. We were interested in understanding, at a mechanistic level, how these acetylation events may affect so many important p53 functions. We hypothesized that modifications of K320 or K373 induce conformational changes that in turn influence multiple protein–protein interactions. To test this, we assayed the ability of p53Q320 and -Q373 to interact with antibodies recognizing epitopes located within spatially distant portions of p53 (Fig. 7, B and C). We used the p53 antibody DO-1, which binds to amino acids 20–25 in the NH2 terminus (B-1); the PAb421 antibody, which recognizes a COOH-terminal epitope located between amino acids 372–381 (B-2); and the PAb240 antibody, a typical conformational antibody that recognizes residues 213–217 within the DNA binding domain (B-3). This latter epitope becomes exposed when p53 is in an inactive mutant conformation or when WT p53 is bound to DNA (McLure and Lee, 1996). To assess the accessibility of these epitopes in the absence of other changes, native or acetylation mutant forms of p53 were purified in vitro to near homogeneity from insect cells infected with recombinant baculoviruses, and each protein was immunoprecipitated with limiting or saturating amounts of antibodies. The purity of these preparations is shown in Fig. S3 (available at http://www.jcb.org/cgi/content/full/jcb.200512059/DC1). The results of these experiments were striking. Indeed, all three epitopes were more accessible by their specific antibody in the case of p53Q320. The increased accessibility of the PAb240 epitope can particularly explain the lesser binding of this mutant to low-affinity promoters, as exposure of these residues indicates a partially denatured conformation. In contrast, p53Q373 completely disrupts reactivity with the PAb421, probably because the PAb421 epitope spans within this region. Importantly, no significant differences were observed when p53 proteins were immunoprecipitated with a goat polyclonal antibody (N-19; B-4) or were subjected to direct immunoblot after p53 had been denatured in SDS-PAGE (C-2).
Thus, in vitro, mutations of K320 and K373 impart conformational changes that modify the accessibility of epitopes located within the NH2 terminus, the central portion, or the COOH terminus of p53. Likewise, in vivo, acetylation-mediated changes in the availability of these sites might explain differential interactions with cellular factors, including coactivators and corepressors, or kinases and phosphatases that in turn influence transcription and phosphorylation, respectively.
Discussion
Why so many acetylation sites?
Our results are consistent with a model whereby acetylation of lysines 320 and 373 acts as a finely tuned sensor-effector system that enables p53 to coordinate gene-expression patterns in response to DNA damage (Fig. 8). We have demonstrated that various p53 acetylation mutants, as well as truly acetylated p53 (Fig. 4), possess different intrinsic affinities for their downstream promoters and that acetylation of residues around position K373 is necessary, particularly for binding to low-affinity proapoptotic promoters, such as BAX, both in vitro and in vivo (Fig. 4 and Fig. S2). It is important to note that although the expression levels of p53Q373 were consistently twofold higher than those of other p53 proteins (Fig. 4 and Fig. S1) in conditions in which p53 levels were predetermined and equalized, such as in EMSAs (Fig. S2), in ChIP assays (Fig. 4), and in the phosphorylation studies (Fig. 7), p53Q373 interacted more strongly with proapoptotic promoters and was hyperphosphorylated compared with WT p53 or p53Q320. Thus, these changes reflect qualitative differences in the behavior of this mutant. In light of these data, p53Q373-mediated activation of proapoptotic promoters might depend on a combination of effects, specifically, a direct enhancement of the intrinsic DNA binding affinity, hyperphosphorylation of strategic residues, and an increase in p53 nuclear retention that allows occupancy of “hard-to-reach” and low-affinity promoters. In contrast, we found that p53Q320 interacted efficiently with the high-affinity p21/WAF promoter but to a lesser extent with the promoters of BAX, p53AIP1, and PIG3. This correlated with a PAb240+ conformation, indicative of a partially denatured configuration, as well as with decreased phosphorylation of residues that favor nuclear retention of p53. We interpret these data to conclude that the effects of K320 acetylation rely, at least in part, on its accelerated nuclear export (Figs. 5 and 6) and different conformation (Fig. 7), both of which could prevent p53 from reaching nuclear concentrations and a configuration necessary to efficiently saturate low- affinity promoters. We have further shown that K373 acetylation stabilizes the interaction of p53 with p300, HDAC1, and SIRT1/Sir2 (Fig. 3). These molecular features were linked to an increase in the total number of repressed genes by p53Q373 compared with p53Q320 and with qualitative differences in the pattern of activated genes (Table S1). Expression of p53Q320, which interacts with PCAF but has significantly less affinity for p300 (Fig. 3), appeared instead to correlate with a lower threshold of activation and with a distinct pattern of gene expression. In addition, an unbiased “heat map” of genes regulated by the two acetylation mutants clearly demonstrated differential, if not contrasting, gene-expression patterns between p53Q320 and -Q373 (Fig. 4 A). Collectively, our results imply that each of the various clusters of acetylation regulates p53 distribution in different cell compartments and chromatin domains and acts as an interaction platform for various types of chromatin-remodeling enzymes to activate specific sets of genes.
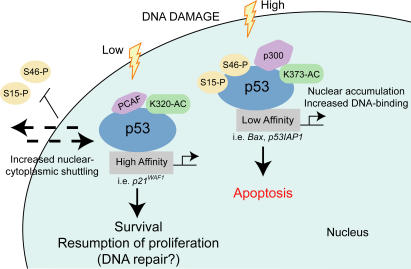
Figure 8.
p53 cassettes, created by combinations of posttranslational modifications and protein–protein interactions, control cell fate. By analogy with the “histone code” hypothesis elaborated by Strahl and Allis (2000) and based on data shown in this study, we propose that specific combinations of posttranslational modifications generate distinct p53 cassettes that direct p53 toward precise cellular functions. These p53 cassettes, in addition to directly influencing key biochemical properties of p53, such as its DNA binding affinity, also interact specifically with effector proteins that participate in deciphering and eliciting a particular response. We envision that these combination cassettes may exist for many p53 posttranslational modifications and establish a modular link between multiple upstream regulators of p53 and downstream events.
The aforementioned differences in the biochemical properties of p53 acetylation mutants are physiologically relevant. Indeed, cells expressing p53Q320 were capable of promoting cell survival and resumption of proliferation in conditions of moderate DNA damage, unlike those harboring p53Q373, which were irreversibly committed toward apoptosis (Fig. 2). Thus, in view of the DNA binding properties of the corresponding mutants, it seems legitimate to speculate that acetylation of K320 enables cells to tune the expression of various p53 target genes in a way that enhances the ability of cells to survive if the damage does not accumulate to catastrophic levels. While this work was in progress, a report showed that p53 activates a cell cycle checkpoint that responds to glucose availability and promotes survival (Jones et al., 2005). In keeping with our findings, these observations lead to the important conclusion that the cell cycle and death pathway are under the control of a subpopulations of p53 with very distinct biochemical properties.
As noted previously, the p53 protein of D. melanogaster (dp53) differs from its human counterpart because of its exclusive role in promoting apoptosis (Brodsky et al., 2000; Ollmann et al., 2000). Accordingly, unlike mammalian cells, D. melanogaster does not possess some of the pathways needed for recovery after p53 activation, as judged by the absence of genes encoding for key negative regulators of p53 (e.g., MDM2). Significantly, our analysis of the degree of conservation of the different p53 acetylation clusters between mammalian and D. melanogaster p53 (Fig. 9) showed that dp53 possesses a COOH-terminal domain with homology to the stretch of amino acids containing acetylated residues around position 373 of human p53, as noted by others (Brodsky et al., 2000), but no detectable K320 homology. Thus, regulation of p53 activity via K320 acetylation might have evolved in higher eukaryotes to suppress the apoptotic program and to allow recovery after damage. We postulate that this reflects the necessity of complex multicellular organisms to spare the life of tissues with limited proliferation potential. In strong support of this interpretation, we have now shown that K320 acetylation is particularly favored in neuronal cells after injury, where it regulates the expression of genes specifically involved in neuronal survival and regeneration (unpublished data).
Figure 9.
Conservation of K373 but not K320 between human and D. melanogaster p53. Human and D. melanogaster p53 were aligned using LALIGN from GeneStream (http://www.eng.uiowa.edu/∼tscheetz/sequence-analysis/examples/LALIGN/lalign-guess.html). Identical residues are shaded in gray, whereas conserved residues are colored in blue. K320 and K373 are indicated on human p53.
The COOH-terminal tail of p53 and a p53 acetylation code
It is becoming clear that the COOH-terminal region of p53 is a target of numerous additional posttranslational modifications, such as phosphorylation, ubiquitination, sumoylation, and methylation (Bode and Dong, 2004). The similarity between the posttranslational modifications that exist in the histone tail and in the p53 COOH-terminal domain is intriguing. In the case of histones, it has been proposed that different patterns of such modifications, acting alone or in combination on one or more tails, establish a “histone code” that is deciphered by other proteins to coordinate downstream events (Strahl and Allis, 2000). In this model, the recruitment of nucleosome-modifying enzymes and of effector molecules by such “histone cassettes” will execute the code by changing local chromatin structure. For p53, the challenge for further studies will be to understand whether specific combinations of posttranslational modifications do exist that modify and expand the ability of this protein to regulate gene expression in a manner similar to that proposed for the histone tail. Experiments presented here suggest that mutations of each of the two lysine clusters is associated at the very least with a different phosphorylation pattern in the NH2-terminal region of p53, with a different ability to interact with coactivators and corepressors, and with a different gene-expression profile. Moreover, in conditions whereby the activity of each acetylation mutant was studied independently, such as in the H1299 cell lines, we were able to trace an effect of each acetylation cluster on apoptosis or on the G1 and G2 checkpoints (Fig. 2). This finding, together with biochemical studies on the phosphorylation state and interaction profile of p53 acetylation mutants, supports the idea that the biological activities of p53 may be dictated by multiple cassettes, each interacting with specific effectors and containing well-defined combination patterns of posttranslational modifications. This interpretation might explain why so many posttranslational modifications target p53 (Fig. 8).
Implications for chemotherapy
It is well known that certain organs, such as the central nervous system, kidney, heart, liver, and lung, are naturally radio resistant, and there is good evidence that p53 is an important determinant of tissue-specific radio sensitivity (Gudkov and Komarova, 2003). We have shown that in a lung tumor cell line, the extent of K320 or K373 acetylation is qualitatively and quantitatively influenced by the type and extent of DNA damage. It is also possible that the extent of acetylation of each of these sites is determined in a tissue-specific manner by the local availability of acetylases or deacetylases. Based on the fact that expression of p53Q320 allows for resumption of proliferation after DNA damage, we predict that strategies aimed at enhancing acetylation of K320 could foster the development of compounds that protect peripheral tissues from toxicity during the course of chemotherapy.
Materials and methods
Cell lines, transfections, and the generation of the H1299 tetracycline-inducible cells
H1299 and A549 lung carcinoma cells were grown in DME supplemented with 10% FCS and 2 mM l-glutamine. The p53 mutants used for the tetracycline-inducible system were constructed by using a Pfu-polymerase–based, site-directed mutagenesis (Stratagene), followed by cloning of the amplified cDNAs into the pCDNA/TO4 vector (Invitrogen). The primers used for mutagenesis were as follows: p53Q320, 5′-CCCCAGCCAAAGCAGAAACCACTGGATGGAGAA and p53Q373, 5′-AGCCACCTGCAGTCCCAACAGGGTCAGTCTACC. p53DM was created with two consecutive rounds of site-directed mutagenesis. A Flag-encoding sequence was fused in frame to the NH2 terminus of p53 to facilitate immunodetection of the protein. Transfections were performed by using either Lipofectamine (Invitrogen) or FuGENE 6 (for coverslips; Roche). Full-length PCAF and HA-tagged p300 were expressed from pCDNA vectors. Cells were plated at 20–40% confluence 12–18 h before transfection. Cells were exposed to the transfection reagent for ∼16 h, washed twice with PBS, and refed with complete medium for an additional 24 h. Adozelesin and bizelesin were provided by T. Beerman (Rowell Park Cancer Institute, Buffalo, NY).
In vivo DNA binding assays
ChIP assays were performed as described elsewhere (Ogawa et al., 2002). The primers used for amplification of p53-responsive elements were as follows: p21, 5′-TCACCATTCCCCTACCCCATGCTGCTC and 3′-AAGTTTGCAACCATGCACTTGAATGTG; BAX, 5′-AGCTCATGCCTGTAATCCCAGCGCT and 3′-AAATAGCATGCTTCCAGGCAGGACGT; P53AIP1, 5′-AGCTGAGCTCAAATGCTGAC and 3′-CCAAGTTCTCTGCTTTC; and PIG3, 5′-CAGGACTGTCAGGAGGAGGCGAGTGATAAG and 3′-GTGCGATTCTAGCTCTCACTTCAAGGAGAG.
In vitro purification of p53 and EMSAs
p53 was purified from ∼10 dishes of SF21 cells infected with recombinant baculoviruses expressing native or acetylation mutant p53. Cells were harvested and lysed in extraction buffer (20 mM Hepes, pH 7.5, 0.5 M KCl, 0.4 mM EDTA, 0.2% NP-40, 10 mM β-mercaptoethanol, 0.1 mM PMSF, and 10 μg/ml pepstatin), incubated on ice for 20 min, and centrifuged at 14,000 g for 30 min at 4°C. p53 was purified by using a anti-Flag immunoaffinity column (Sigma-Aldrich) in the presence of 0.5 M KCl, extensively washed, and eluted with the Flag peptide. EMSAs were performed in a 30 μl total volume that contained buffer A (5× 100 mM Hepes, pH 7.9, 125 mM KCl, 0.5 mM EDTA, 50% glycerol, and 10 mM MgCl2); buffer B (10× 10 mM spermidine, 40 mM DTT, 1.2% NP-40, and 2 mg/ml BSA); 10 ng of double-stranded poly(d[I-C]); 50 ng of labeled oligonucleotide; and 50, 75, or 150 ng of p53. Reactions were incubated with the probe at room temperature for 20–40 min and run on native 6% polyacrylamide gels, which were run at room temperature until the xylen-cyanol blue reached 6 cm from the bottom of the gel. The oligonucleotides used were as follows: p21/WAF, 5′-TCTGGCCATCAGGAACATGTCCCAACATGTTGAGCTCTGG and 3′CCAGAGCTCAACATGTTGGGACATGTTCCTGATGGCCAGA; Gadd45, 5′TCTGTGGTACAGAACATGTCTAAGCATGCTGGGGACTGCC and 3′-GGCAGTCCCCAGCATGCTTAGACATGTTCTGTACCACAGA; and Bax, 5′AATTCGGCTACCTCACAAGTTAGAGACAAGCCTGGGCGTGGGCTATATTGTAGCGAAT and 3′-ATTCGCTACAATATAGCCCACGCCCAGGCTTGTCTCTAACTTGTGAGGTAGCCGAATT.
Immunoprecipitations and immunoblots
Preparation of cell extracts and immunoprecipitations were performed as previously described (Avantaggiati et al., 1997). Antibodies used in this study were for p53 (FL393 and N-19 [Santa Cruz Biotechnology, Inc.] and Ab-1 and Ab-6 [Calbiochem]), S15-, S46-, and S392-phospho-p53 (Cell Signaling Technology), Acetyl-320-, Acetyl-373-, and Acetyl-373/382-p53 (Upstate Biotechnology), PCAF (E8; Santa Cruz Biotechnology, Inc.), p300 (N-15; Santa Cruz Biotechnology, Inc.), p21 (WAF1/Ab-5; Calbiochem), mSin3 (AK-11; Santa Cruz Biotechnology, Inc.), Sir2 (7342; Abcam), Bax (anti-Bax/NT; Upstate Biotechnology), and actin (I19; Santa Cruz Biotechnology, Inc.). Proteins were detected by using a chemiluminescence-based system (Pierce Chemical Co.) according to the manufacturer's instructions.
Indirect immunofluorescence
The H1299 cells were plated on glass coverslips, fixed in 4% paraformaldehyde, and permeabilized by addition of 0.1% Triton X-100/PBS solution. Cells were stained using antibodies directed against p53 (FL393), PCAF (E-8), or p300 (N-15) and then stained for DNA content (DAPI; Invitrogen). The appropriate fluorescent secondary antibodies were obtained from Invitrogen. Where indicated, cells were treated with 5 ng/ml leptomycin B (Sigma-Aldrich) for 24 h. Cells were visualized using a microscope (Axiovert 200; Carl Zeiss MicroImaging, Inc.) with either a 40× or 60× objective. Images were acquired using a camera (Axio; Carl Zeiss MicroImaging, Inc.). Analysis of different Z stacks was performed using the AxioVision 3.0 software (Carl Zeiss MicroImaging, Inc.).
Microarrays
The cDNA microarrays were performed at the Children's National Medical Center. H1299 cells expressing WT p53, p53Q320, or p53Q373 were harvested for total RNA extraction 24 h after the addition of tetracycline. 7 μg total RNA was used for cDNA and biotinylated cRNA synthesis. Expression-profiling analysis was performed using the HG-U133 human high-density oligonucleotide microarray (Affymetrix, Inc.). Each GeneChip was used for a single hybridization with RNA isolated from one cell line from a single experiment. The total number of samples was six.
Expression profiling was performed as described previously and fulfilled all stringent quality-control measures as detailed previously (Di Giovanni et al., 2005). We used two normalization processes: one for chip–chip comparisons (scaling factors) and one for gene–gene comparisons (normalization to the mean of the naive signal intensities for each gene). The scaling factor determinations were done using default algorithms (MAS 5.0; Affymetrix, Inc.) with a target intensity of chip sector fluorescence to 800.
We have recently shown that the use of MAS 5.0 signal intensity values, together with a present call noise filter achieves an excellent signal/noise balance relative to other probe set analysis methods (dChip; robust multichip average [RMA]; Seo et al., 2004). Data analyses were limited to probe sets that showed one or more “present” (P “calls”) in the six GeneChip profiles in our complete dataset. Data were analyzed and visualized using the GeneSpring software (Silicon Genetics). Initial data analysis also included a fold-change filter of >2 increase or decrease relative to WT p53 (MAS 5.0). Functional classification was performed using DAVID software (http://apps1.niaid.nih.gov/david/upload.jsp).
For hierarchy and clustering analysis, the arrays were analyzed using the R statistics package (http://www.R-project.org) and the Affymetrix, Inc. library (Irizarry et al., 2003) of the Bioconductor software package. Expression values were determined using the RMA algorithm using the RMA function in the Affymetrix, Inc. library at its default settings. Genes that exhibited an expression change >2 relative to the control (WT p53) for at least one of replicates and one of the mutations were selected for further analysis. These genes were then clustered using hierarchical clustering with “complete” agglomeration, and each cluster was further analyzed based on the known function of the genes contained in the cluster.
Semiquantitative PCR was performed using random hexamer priming for first-strand synthesis (SuperScript III; Invitrogen), followed by amplification of specific genes with Taq polymerase and the following primer pairs: Jagged-2, 5′-GTTGACGCACCTGTGGTTGT-3′ and 5′-CTTCAATACTGCCGCCGTCC-3′ (32 cycles); pig11, 5′-GGCTGACAACTGGCTGTCTT-3′ and 5′-ACAGGCCATGTTGCTGTATC-3′ (35 cycles); APAF-1, 5′-TCTACCTCTGCTGACAAGAC-3′ and 5′-GTAGCAGCTCCTTCTTCTGA-3′ (35 cycles); and actin, 5′-GCTCGTCGTCGACAACGGCT-3′ and 5′-CAAACATGATCTGGGTCATCTTCTC-3′ (26 cycles).
Online supplemental material
Fig. S1 demonstrates that mixed populations of H1299–WT p53, p53Q320, p53Q373, or p53DM clones display similar cell cycle profiles and viability compared with a single clone population after p53 protein induction. In Fig. S2, EMSAs are used to study the DNA binding activity of p53 acetylation mimics in the absence of chromatin. Finally, Fig. S3 demonstrates that the p53 protein purified from baculovirus-infected incest cells has a high level of purity. Table S1 depicts apoptotic and cell cycle–related genes influenced in H1299 cells by the expression of the p53Q320 and -Q373 acetyl-mimics. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200512059/DC1.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 CA83979, R01 CA030716, and R01 CA102746 to M.L. Avantaggiati, and R01 CA70896, R01 CA75503, and R01 CA86072 to R.G. Pestell) and T32 Training Fellowship (CA009686 to C. Knights). Work conducted at the Lombardi Comprehensive Cancer Center was supported by the Comprehensive Cancer Center Core National Institutes of Health grant CA51008-14.
C.D. Knights and J. Catania contributed equally to this paper.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; CPI, cyclopropylpyrroloindole; EMSA, electrophoretic mobility shift assay; PCAF, p300/CBP-associated factor; WT, wild-type.
References
- Appella, E., and C.W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764–2772. [DOI] [PubMed] [Google Scholar]
- Avantaggiati, M.L., V. Ogryzko, K. Gardner, A. Giordano, A.S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 89:1175–1184. [DOI] [PubMed] [Google Scholar]
- Bates, S., E.S. Hickman, and K.H. Vousden. 1999. Reversal of p53-induced cell-cycle arrest. Mol. Carcinog. 24:7–14. [DOI] [PubMed] [Google Scholar]
- Bode, A.M., and Z. Dong. 2004. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer. 4:793–805. [DOI] [PubMed] [Google Scholar]
- Brodsky, M.H., W. Nordstrom, G. Tsang, E. Kwan, G.M. Rubin, and J.M. Abrams. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell. 101:103–113. [DOI] [PubMed] [Google Scholar]
- Derry, W.B., A.P. Putzke, and J.H. Rothman. 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 294:591–595. [DOI] [PubMed] [Google Scholar]
- Di Giovanni, S., A. De Biase, A. Yakovlev, T. Finn, J. Beers, E.P. Hoffman, and A.I. Faden. 2005. In vivo and in vitro characterization of novel neuronal plasticity factors identified following spinal cord injury. J. Biol. Chem. 280:2084–2091. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 390:308–311. [DOI] [PubMed] [Google Scholar]
- Gu, W., and R.G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 90:595–606. [DOI] [PubMed] [Google Scholar]
- Gudkov, A.V., and E.A. Komarova. 2003. The role of p53 in determining sensitivity to radiotherapy. Nat. Rev. Cancer. 3:117–129. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., B.M. Bolstad, F. Collin, L.M. Cope, B. Hobbs, and T.P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.G., D.R. Plas, S. Kubek, M. Buzzai, J. Mu, Y. Xu, M.J. Birnbaum, and C.B. Thompson. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 18:283–293. [DOI] [PubMed] [Google Scholar]
- Kim, Y.Y., B.J. Park, D.J. Kim, W.H. Kim, S. Kim, K.S. Oh, J.Y. Lim, J. Kim, C. Park, and S.I. Park. 2004. Modification of serine 392 is a critical event in the regulation of p53 nuclear export and stability. FEBS Lett. 572:92–98. [DOI] [PubMed] [Google Scholar]
- Knights, C.D., Y. Liu, E. Appella, and M. Kulesz-Martin. 2003. Defective p53 post-translational modification required for wild type p53 inactivation in malignant epithelial cells with mdm2 gene amplification. J. Biol. Chem. 278:52890–52900. [DOI] [PubMed] [Google Scholar]
- Lee, C.S., G.P. Pfeifer, and N.W. Gibson. 1994. Mapping of DNA alkylation sites induced by adozelesin and bizelesin in human cells by ligation-mediated polymerase chain reaction. Biochemistry. 33:6024–6030. [DOI] [PubMed] [Google Scholar]
- Liang, S.H., and M.F. Clarke. 2001. Regulation of p53 localization. Eur. J. Biochem. 268:2779–2783. [DOI] [PubMed] [Google Scholar]
- Lill, N.L., S.R. Grossman, D. Ginsberg, J. DeCaprio, and D.M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature. 387:823–827. [DOI] [PubMed] [Google Scholar]
- Liu, L., D. Scolnick, R. Trievel, H. Zhang, R. Marmorstein, T.D. Halazonetis, and S. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J., M. Li, Y. Tang, M. Laszkowska, R.G. Roeder, and W. Gu. 2004. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 101:2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLure, K.G., and P.W. Lee. 1996. A PAb240+ conformation of wild type p53 binds DNA. Oncogene. 13:1297–1303. [PubMed] [Google Scholar]
- Murphy, M., J. Ahn, K.K. Walker, W.H. Hoffman, R.M. Evans, A.J. Levine, and D.L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 102:849–862. [DOI] [PubMed] [Google Scholar]
- Ogawa, H., K. Ishiguro, S. Gaubatz, D.M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 296:1132–1136. [DOI] [PubMed] [Google Scholar]
- Ollmann, M., L.M. Young, C.J. Di Como, F. Karim, M. Belvin, S. Robertson, K. Whittaker, M. Demsky, W.W. Fisher, A. Buchman, et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 101:91–101. [DOI] [PubMed] [Google Scholar]
- Prives, C., and J.L. Manley. 2001. Why is p53 acetylated? Cell. 107:815–818. [DOI] [PubMed] [Google Scholar]
- Resnick, M.A., and A. Inga. 2003. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc. Natl. Acad. Sci. USA. 100:9934–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, K., J. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C.W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J., M. Bakay, Y.W. Chen, S. Hilmer, B. Shneiderman, and E.P. Hoffman. 2004. Interactively optimizing signal-to-noise ratios in expression profiling: project-specific algorithm selection and detection p-value weighting in Affymetrix microarrays. Bioinformatics. 20:2534–2544. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., and C.D. Allis. 2000. The language of covalent histone modifications. Nature. 403:41–45. [DOI] [PubMed] [Google Scholar]
- Tomso, D.J., A. Inga, D. Menendez, G.S. Pittman, M.R. Campbell, F. Storici, D.A. Bell, and M.A. Resnick. 2005. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc. Natl. Acad. Sci. USA. 102:6431–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger, T., T. Juven-Gershon, E. Moallem, M. Berger, R. Vogt Sionov, G. Lozano, M. Oren, and Y. Haupt. 1999. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 18:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden, K.H., and C. Prives. 2005. P53 and prognosis; new insights and further complexity. Cell. 120:7–10. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Y. Xiong. 2001. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 292:1910–1915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.