Abstract
NF-κB signaling is known to be critically regulated by the NF-κB–inducible inhibitor protein IκBα. The resulting negative feedback has been shown to produce a propensity for oscillations in NF-κB activity. We report integrated experimental and computational studies that demonstrate that another IκB isoform, IκBɛ, also provides negative feedback on NF-κB activity, but with distinct functional consequences. Upon stimulation, NF-κB–induced transcription of IκBɛ is delayed, relative to that of IκBα, rendering the two negative feedback loops to be in antiphase. As a result, IκBɛ has a role in dampening IκBα-mediated oscillations during long-lasting NF-κB activity. Furthermore, we demonstrate the requirement of both of these distinct negative feedback regulators for the termination of NF-κB activity and NF-κB–mediated gene expression in response to transient stimulation. Our findings extend the capabilities of a computational model of IκB–NF-κB signaling and reveal a novel regulatory module of two antiphase negative feedback loops that allows for the fine-tuning of the dynamics of a mammalian signaling pathway.
Introduction
The NF-κB family of transcription factors controls diverse mammalian signaling responses that mediate cell survival, inflammation, and immune response (Gerondakis et al., 1999; Li and Verma, 2002; Hoffmann and Baltimore, 2006). Functional NF-κB exists in a dimeric form that is composed of combinations of five proteins containing a Rel homology region, i.e., cRel, RelA, RelB, p50, and p52 (Ghosh et al., 1998; Hoffmann and Baltimore, 2006). In resting cells, the NF-κB dimer is bound to inhibitor IκB proteins, i.e., IκBα, -β, and -ɛ, which inhibit NF-κB DNA-binding activity and prevent its nuclear accumulation. Activation of the NF-κB signaling pathway relies upon signal-dependent phosphorylation and degradation of the IκB proteins that result in subsequent nuclear translocation of the NF-κB dimer (Ghosh et al., 1998).
Termination of NF-κB activity after cellular stimulation is critical, as deregulated inflammatory gene expression can be detrimental to the health of the organism, and several attenuation mechanisms have been described (Greten and Karin, 2004; Li et al., 2005). Importantly, IκBα, which is a target gene of NF-κB, is induced by numerous NF-κB–inducing stimuli, resulting in the termination of NF-κB DNA-binding activity and nuclear localization (Scott et al., 1993; Ghosh et al., 1998).
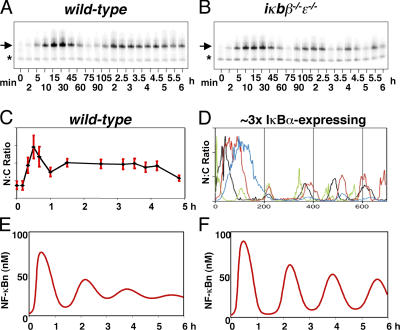
Temporal control of NF-κB activity has been shown to mediate stimulus-specific gene expression programs in response to different inflammatory stimuli (Werner et al., 2005), and understanding the dynamic regulation of NF-κB by IκB proteins is of critical importance. Negative feedback that is mediated by IκBα was shown to confer the propensity for oscillatory NF-κB nuclear activity, both when examined biochemically in gene knockout cells containing only the IκBα isoform (Fig. 1 B; Hoffmann et al., 2002) and when examined by microscopy using transiently transfected IκBα and RelA proteins fused to fluorescent moieties (Fig. 1 D; Nelson et al., 2004). The oscillations are not apparent in cells containing all three IκB proteins at normal expression levels (Fig. 1, A and C; Hoffmann et al., 2002; Barken et al., 2005).
Figure 1.
Oscillations in NF-κB nuclear localization. EMSAs show nuclear localization of NF-κB in wild-type (A) and iκbβ −/−ɛ−/− MEF cells (B) stimulated with TNF (adapted from Hoffmann et al., 2002). Arrows indicate specific nuclear NF-κB–binding activity. Asterisks indicate nonspecific DNA-binding complexes. (C) Quantitation of immunohistochemical analysis of individual cells stimulated with TNF (adapted from Barken et al., 2005). Error bars are the mean ± SD. (D) Recordings from live individual cells transduced with overexpressing RelA and IκBα fusion proteins and stimulated with TNF, where each colored line represents the recording from one cell (adapted from Nelson et al., 2004). NF-κB nuclear localization predicted by a computational model (Hoffmann et al., 2002) with (E) and without (F) the IκBβ-mediated protective mechanism described in the text and in Phillips and Ghosh (1997).
These observations suggested that IκBβ and/or -ɛ proteins play a role in dampening IκBα-mediated oscillations and determining the dynamics of NF-κB activity. The mechanism that confers dampening of oscillations in NF-κB activity was proposed to involve the nuclear accumulation of newly synthesized IκBβ that binds nuclear and promoter-bound NF-κB and shields it from IκBα-mediated nuclear export (Suyang et al., 1996; Phillips and Ghosh, 1997). This mechanism was included in our computational model that recapitulates NF-κB activation in response to TNF stimulation (Hoffmann et al., 2002), but our later studies were unable to observe the IκBβ effect in murine embryonic fibroblasts (MEFs), which are the cells for which the model was constructed (unpublished data). Removal of the mathematical term for this mechanism from the model resulted in highly oscillatory NF-κB responses (Fig. 1, E and F). Although genetic evidence points to important roles for IκBβ and -ɛ in regulating the dynamics of NF-κB signaling, the mechanisms by which they function remained unclear.
In this study, we investigated the dynamic behavior of all three canonical IκB isoforms, especially by contrasting IκBɛ and -β functions with that of IκBα. Our studies revealed that IκBɛ expression is, in fact, highly NF-κB inducible, and that it mediates functional negative feedback on NF-κB activity; however, it does so in antiphase to that of IκBα. Two antiphase negative feedbacks emerge as an important regulatory module that may be present for the dynamic control of signaling in other pathways as well.
Results and discussion
IκBɛ is TNF inducible via NF-κB
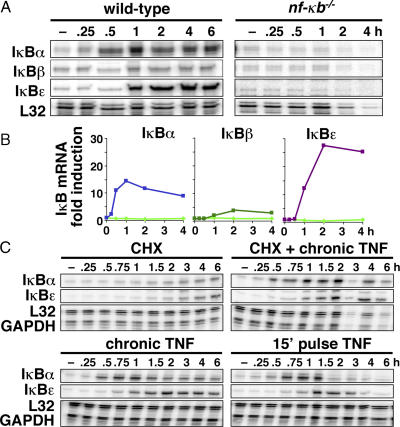
We constructed probes for RNase protection assays (RPAs) that allowed for the simultaneous quantitative monitoring of all three IκB mRNAs to characterize the regulation of IκBɛ and -β gene expression in response to stimulation. Analysis of the mRNA levels in MEFs that were stimulated with TNF confirmed that IκBα was strongly induced (Fig. 2 A). IκBβ showed only weak induction, which suggests that it is not a strong NF-κB–responsive gene in this cell type. Remarkably, IκBɛ transcription was highly induced by TNF stimulation, and quantitation of these results showed that IκBɛ was induced to a higher degree than IκBα (Fig. 2 B). Although the exact induction folds varied in replicate assays using separate MEF cell stocks (unpublished data), IκBɛ fold induction was consistently higher than that for IκBα. Furthermore, IκBɛ expression was also induced in response to LPS in MEFs, the macrophage cell line RAW264.7, and the B cell line 70Z (unpublished data), indicating that the dynamic control of its synthesis is not specific to TNF or to fibroblasts. Analysis of IκB mRNA levels in MEF cells deficient in NF-κB showed no induction of any of the three IκBs (Fig. 2 A).
Figure 2.
IκB gene transcription in response to inflammatory stimulation. (A) RPA revealing IκB mRNA levels in wild-type and NF-κB–deficient cells after chronic TNF stimulation. (B) IκB mRNA fold induction in wild-type (blue, dark green, and purple) and NF-κB–deficient (light green) stimulated cells that were normalized to L32 housekeeping gene expression. (C) IκB mRNA levels in wild-type cells in response to chronic treatment by cycloheximide and/or TNF and 15-min transient stimulation with TNF.
The temporal profile of IκBα transcript induction during chronic TNF stimulation is well described (Scott et al., 1993). It shows rapid activation as early as 15 min, a peak within 1 h, and a slow attenuation over many hours. We observed a similar activation profile for IκBɛ induction, but were surprised to note a distinct 45-min onset delay (Fig. 2, A and B). This suggests that the IκBɛ promoter may involve a delay mechanism, such as a requirement for the activation of an NF-κB–responsive transcription factor (a feed-forward regulation). However, the inhibition of protein synthesis by cycloheximide did not attenuate transcriptional activation of either IκBα or -ɛ (Fig. 2 D and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200510155/DC1) and, thus, does not support the notion of feed-forward regulation.
It has been previously shown that transient TNF stimulation leads to transient nuclear NF-κB activity lasting only ∼60 min (Hoffmann et al., 2002). To determine whether short stimulation would efficiently induce IκBɛ expression, an RPA was performed on MEF cells that were stimulated for 15 min with TNF. The results show that IκBɛ transcription is still activated with an onset delay and with an induction profile similar to that of chronically stimulated cells (Fig. 2 C and Fig. S1). Because NF-κB activity is diminished when IκBɛ mRNA levels are still rising, and because cycloheximide treatment precludes a feed-forward mechanism, we suggest that the time delay in the activation of IκBɛ transcription occurs after NF-κB recruitment to the promoter. Further study is required to elucidate the mechanism of this delay.
Computational modeling reveals dynamic control mechanisms
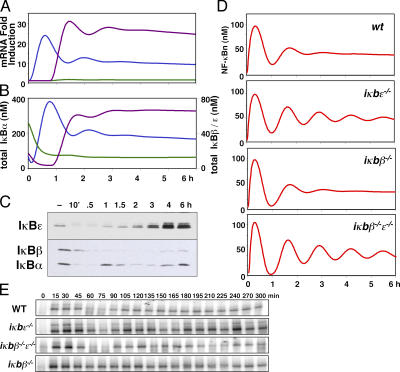
NF-κB–responsive syntheses of IκBɛ and -β were added to the mathematical model to explore the functional consequences imparted by these negative feedback regulators upon NF-κB activity. To determine the kinetic parameter values for the inducible transcription of each isoform, the temporal responses of IκB transcription in the model were constructed such that the mRNA induction profiles calculated by the model correlated with our RPA data (Fig. 3 A). This required the fitting of parameters defining transcription and translation rates and mRNA stability. The new model also includes revised IκB protein degradation parameters from earlier studies (unpublished data).
Figure 3.
Computational modeling of IκB mRNA and protein levels reveals a role for IκBɛ in regulating the dynamics of NF-κB activity in response stimulation. The results of computational simulations of the fold induction of mRNA synthesis (A) and of the protein levels for IκBα (blue), IκBβ (green), and IκBɛ (purple) in wild-type cells (B) in response to persistent stimulation with TNF. (C) Western blots of IκB proteins in wild-type cells in response to persistent stimulation with TNF. The results of computational simulations (D) and EMSAs of nuclear NF-κB activity (E) in wild-type, iκbɛ−/−, iκbβ−/−, and iκbβ−/−ɛ−/− cells in response to persistent TNF stimulation. Super-shift and oligonucleotide competition EMSAs are included in Fig. S2. Fig. S2 is available at http://www.jcb.org/cgi/content/full/jcb.200510155/DC1.
The revised model recapitulates IκBα protein degradation immediately after IκB kinase (IKK) activation and rapid synthesis in response to NF-κB nuclear localization (Fig. 3, B and C). Chronic stimulation results in repeated IκBα protein degradation and synthesis (Fig. 3 B). In addition, the model shows delayed induction of IκBɛ and -β protein syntheses. The low inducibility of IκBβ transcription results in very low IκBβ protein synthesis, whereas the high inducibility of IκBɛ transcription results in notable accumulation of IκBɛ protein (Fig. 3 B).
Earlier studies revealed oscillatory NF-κB activity in cells lacking IκBβ and -ɛ (Hoffmann et al., 2002) and in cells in which NF-κB–inducible IκBα was overexpressed (Nelson et al., 2004), whereas in wild-type cells late NF-κB activity (beyond 2 h) was remarkably steady (Hoffmann et al., 2002). However, the underlying dampening mechanism that results in steadied late activity remained obscure. Because induced synthesis of IκBɛ is delayed, we reasoned that IκBɛ may mediate an antiphase negative feedback that provides effective dampening of IκBα-mediated oscillations. Indeed, our simulations of signaling modules lacking IκBɛ revealed oscillations in nuclear NF-κB that persist with a higher amplitude than those that contain IκBɛ and represent wild-type cells (Fig. 3 D). In contrast, simulations of cells lacking IκBβ do not show such aberrant oscillations, whereas systems lacking both IκBɛ and -β do.
We set out to examine these predictions experimentally, using nuclear extracts prepared from TNF-treated MEFs harboring genetic deficiencies for IκBɛ and/or -β. We measured NF-κB DNA-binding activity by electrophoretic mobility shift assay (EMSA; Fig. 3 E) and nuclear localization of the NF-κB protein RelA by Western blot (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200510155/DC1). In both assays, all four cell types exhibited fast induction of nuclear NF-κB in response to TNF stimulation by 15 and 30 min, a transient trough at 60–75 min, and subsequent recovery at 90–120 min. However, a second trough at 135–150 min was most pronounced in iκbɛ−/− and iκbβ−/−ɛ−/− cells, as was a third trough at ∼225 min. These studies suggest that negative feedback, provided by IκBɛ in antiphase to that of IκBα, is the primary mechanism that dampens the propensity for oscillations in NF-κB activity.
The regulatory motif consisting of two antiphase negative feedback systems may be present in other signaling pathways to control the dynamics of signal transduction, and variations in the relative strength of the two systems may provide for altered response dynamics to the same stimulus. A similar model has recently been proposed for two NF-κB–inducing signaling pathways emanating from the TLR4 receptor (Covert et al., 2005). In contrast to the two interacting negative feedback mechanisms, this model depicts the coupling of two positive oscillatory signals in an antiphase relationship that produces stable NF-κB activity in response to LPS stimulation in wild-type cells. As the temporal control of NF-κB activity determines NF-κB–responsive gene expression (Hoffmann et al., 2002; Nelson et al., 2004; Werner et al., 2005), the interaction of antiphase regulation by IκBα and -ɛ may contribute to the regulation of stimulus-specific and cell type–specific gene expression programs by modulating the dynamics of this transcription factor.
IκBɛ mediates postinduction repression of NF-κB activity and inflammatory gene expression
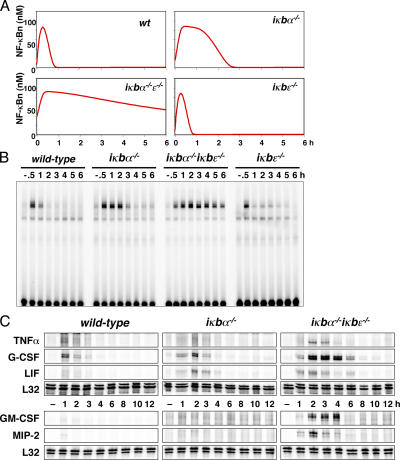
We used the computational model to identify conditions in which NF-κB–responsive IκBɛ expression would mediate negative feedback on stimulus-induced NF-κB activity and found that the most significant role for IκBɛ was in systems with reduced IκBα. To model such conditions, we used computational simulations to study the temporal profile of nuclear NF-κB in response to 15-min stimulation in systems lacking IκBα, -ɛ, or both (Fig. 4 A). Systems containing all three IκBs show rapid nuclear localization of NF-κB followed by removal from the nucleus within 1 h, as previously shown (Hoffmann et al., 2002). However, in systems lacking IκBα, we predicted effective down-regulation of NF-κB activity in the third hour and beyond. In this context, IκBɛ deficiency results in prolonged NF-κB activity, whereas in systems containing high IκBα expression it does not have an effect.
Figure 4.
IκBɛ mediates postinduction repression of NF-κB activity and inflammatory gene expression. (A) Nuclear NF-κB activity as predicted by computational modeling after 15 min of TNF stimulation. (B) EMSA of nuclear NF-κB activity in extracts prepared at the indicated time points from wild-type, iκbα−/− , iκbα−/− iκbɛ−/−, and iκbɛ−/− cells that were transiently stimulated for 15 min with 1 ng/ml TNF. (C) RPA reveals the mRNA levels over an extended time course of indicated NF-κB–responsive genes in wild-type, iκbα−/−, and iκbα−/− iκbɛ−/− cells that were transiently stimulated for 45 min with TNF.
We used IκBα-deficient MEFs as a model for cell types that have reduced IκBα expression. These MEFs showed NF-κB activity to last ∼3 h in response to 15-min transient TNF stimulation, after which it was dramatically attenuated (Fig. 4 B). In contrast, cells that were deficient in both IκBα and -ɛ showed a pronounced delay in attenuation, with NF-κB still present in the nucleus even at 6 h. Wild-type and IκBɛ-deficient cells are nearly indistinguishable, and both have strong NF-κB accumulation at 30 min and attenuation within 1 h. Collectively, these data strongly suggest that IκBɛ is responsible for the removal of NF-κB from the nucleus at late time points, allowing for dynamic functional interplay with the faster-acting feedback of IκBα.
Temporal control of NF-κB localization by IκBα was shown to control NF-κB–responsive gene expression not only quantitatively (Nelson et al., 2004) but also qualitatively (Hoffmann et al., 2002). To study the effects of IκBɛ-negative feedback on NF-κB–dependent gene expression, the transcription of five NF-κB–responsive genes was monitored by RPA after transient TNF stimulation in wild-type, iκbα−/−, and iκbα−/− iκbɛ−/− cells. The genes encoding TNF, G-CSF, and LIF are inducibly expressed in fibroblasts upon TNF stimulation, but mRNA levels return to baseline within 3 h in wild-type cells. In IκBα-deficient cells, these genes are attenuated within 4 h (Fig. 4 C). In this context, the loss of IκBɛ-negative feedback results in a further delay in attenuation and quantitative deregulation of TNF, G-CSF, and LIF expression. Interestingly, the loss of both IκBα- and -ɛ-negative feedback has a dramatic qualitative effect for GM-CSF and MIP-2. Although these genes are not induced in wild-type or IκBα-deficient cells, both are strongly responsive to NF-κB activation when both IκBα- and -ɛ-negative feedbacks are absent. The data presented here demonstrate that IκBɛ-dependent negative feedback regulates the termination of NF-κB–responsive gene expression in both a quantitative (in the cases of TNF, G-CSF, and LIF) and qualitative (in the cases of GM-CSF and MIP-2) manner.
The functional interplay between the antiphase IκBα- and -ɛ-negative feedback responses may explain differences in NF-κB–dependent gene expression profiles seen in various cell types. In MEFs, IκBɛ-mediated negative feedback appears to be secondary to that provided by IκBα in response to transient inflammatory stimuli, and it is therefore assumed that IκBα controls the bulk of the NF-κB–responsive gene expression (Ghosh et al., 1998). However, the ratio of the abundance of IκBɛ in relation to IκBα is cell type–specific (Memet et al., 1999; Spiecker et al., 2000; Emmerich et al., 2003; Doerre et al., 2005), suggesting that IκBɛ may play a predominant role in NF-κB–responsive gene expression in particular cell types. Indeed, in vivo studies have shown that a deficiency of functional IκBɛ has physiological consequences (Memet et al., 1999; Spiecker et al., 2000; Emmerich et al., 2003; Doerre et al., 2005) and, thus, emphasize the notion that no IκB isoform functions on its own. To understand regulation of NF-κB activity in different cell types and in response to diverse stimuli, the interplay of all IκB isoforms within the IKK–IκB–NF-κB signaling module must be considered.
Our studies aimed to quantitatively characterize the temporal expression profiles of the three IκB isoforms and to examine their functional consequences on NF-κB regulation. Earlier studies showed inducible IκBɛ expression (Simeonidis et al., 1997; Whiteside et al., 1997). We have demonstrated that induction of IκBɛ is NF-κB dependent and functions to attenuate NF-κB activity and terminate NF-κB–responsive gene expression. Based on these three criteria we conclude that IκBɛ mediates bona fide functional negative feedback regulation on NF-κB activity. Importantly, our studies reveal that inducible expression of IκBɛ is delayed by 45 min with respect to that of IκBα, thus, creating a two–negative feedback regulatory module that critically controls the dynamics of NF-κB activity. We suggest that the relative strength of the two feedback mechanisms and their temporal relationship to each other may account for cell type–specific dynamic regulation of NF-κB activity.
Materials and methods
Cell culture
The immortalized MEF cells used were previously described (Hoffmann et al., 2002). Cells were grown to confluency in DME containing 10% bovine calf serum and starved for 24 h in media containing 0.5% bovine calf serum. Stimulations were performed with 10 ng/ml TNF (Roche). Cells that were transiently stimulated with TNF were washed twice with 1× PBS after stimulation and returned to untreated media.
DNA-binding assays and Western blot
EMSAs were performed as previously described (Hoffmann et al., 2002). Western blots using whole-cell extracts were performed as previously described (Hoffmann et al., 2003). IκBα and -β antibodies were obtained from Santa Cruz Biotechnology, Inc. (SC-371 and SC-945, respectively). An antigen-purified polyclonal mouse antiserum raised against recombinant full-length mouse protein was used for IκBɛ.
RPA
Total cellular RNA was isolated from confluent and serum-starved cells with Trizol reagent (Invitrogen). Transcript levels were monitored with α-[32P]UTP–labeled probes using a RiboQuant kit (BD Biosciences) according to the manufacturer's instructions. Data was obtained using a storage phosphor screen (GE Healthcare) and a variable mode imager (Typhoon 9400; GE Healthcare). Data was quantitated using ImageQuant software version 5.2 (GE Healthcare) by normalization to L32 and/or glyceraldehyde- 3-phosphate dehydrogenase after local background subtraction. IκB probes were designed to select for mature mRNA species by spanning exon–exon junctions. The following primer pairs were used to amplify fragments from reverse-transcribed RNA: 5′-TCGCTCTTGTTGAAATGTGG-3′ and 5′TGGAGATTTTCCAGGGTCAG-3′ (IκBα); 5′-GCCCTTAGTCTTTGGCTACG-3′ and 5′-TCTCAGCCACCAACACTCCT-3′ (IκBβ); and 5′-GGCAGACAGCTTTCTCATCC-3′ and 5′-TGAGGTCGCAGTCTTCAATG-3′ (IκBɛ). G-CSF, LIF, MIP-2, TNF, L32, and glyceraldehyde-3-phosphate dehydrogenase probes were obtained from RiboQuant sets (BD Biosciences).
In silico studies
We previously constructed a computational model to describe NF-κB activation events in response to IKK activation by TNF (Hoffmann et al., 2002). This model comprises a singular NF-κB species, three IκB isoforms (IκBα, -β, and -ɛ), and IKK. Synthesis and degradation of the IκBs and cellular localization and interactions for all components were calculated using a system of ordinary differential equations. The model used in this study includes NF-κB–induced IκBɛ and -β transcription and was written in MatLab V7.0 (MathWorks) using previously described methods (Hoffmann et al., 2002). MatLab simulation files are available upon request.
Online supplemental material
Fig. S1 supports Fig. 2 C, with quantitation of IκBα and -ɛ gene induction profiles. Fig. S2 supports Fig. 3 E, with super-shift and oligonucleotide competition EMSAs, as well as nuclear westerns for RelA. Table S1 contains the computational model parameters and reactions.
Supplementary Material
Acknowledgments
We acknowledge Cristina Aguilera for experiments on IκBβ, Andre Levchenko and Gouri Ghosh for discussions and critical reading of the manuscript, and Santa Cruz Biotechnology, Inc. for antibodies.
This study was supported by National Institutes of Health grants GM071573 and GM08326.
Abbreviations used in this paper: EMSA, electrophoretic mobility shift assay; IKK, IκB kinase; MEF, murine embryonic fibroblast; RPA, RNase protection assay.
References
- Barken, D., C.J. Wang, J. Kearns, R. Cheong, A. Hoffmann, and A. Levchenko. 2005. Comment on “Oscillations in NF-kappaB signaling control the dynamics of gene expression.” Science. 308:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert, M.W., T.H. Leung, J.E. Gaston, and D. Baltimore. 2005. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 309:1854–1857. [DOI] [PubMed] [Google Scholar]
- Doerre, S., K.P. Mesires, K.M. Daley, T. McCarty, S. Knoetig, and R.B. Corley. 2005. Reductions in I kappa B epsilon and changes in NF-kappa B activity during B lymphocyte differentiation. J. Immunol. 174:983–991. [DOI] [PubMed] [Google Scholar]
- Emmerich, F., S. Theurich, M. Hummel, A. Haeffker, M.S. Vry, K. Dohner, K. Bommert, H. Stein, and B. Dorken. 2003. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J. Pathol. 201:413–420. [DOI] [PubMed] [Google Scholar]
- Gerondakis, S., M. Grossmann, Y. Nakamura, T. Pohl, and R. Grumont. 1999. Genetic approaches in mice to understand Rel/NF-kappaB and IkappaB function: transgenics and knockouts. Oncogene. 18:6888–6895. [DOI] [PubMed] [Google Scholar]
- Ghosh, S., M.J. May, and E.B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225–260. [DOI] [PubMed] [Google Scholar]
- Greten, F.R., and M. Karin. 2004. The IKK/NF-kappaB activation pathway- a target for prevention and treatment of cancer. Cancer Lett. 206:193–199. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A., and D. Baltimore. 2006. Circuitry of NF-kB signaling. Immunol. Rev. 210:171–186. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A., A. Levchenko, M.L. Scott, and D. Baltimore. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 298:1241–1245. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A., T.H. Leung, and D. Baltimore. 2003. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 22:5530–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., and I.M. Verma. 2002. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2:725–734. [DOI] [PubMed] [Google Scholar]
- Li, Q., S. Withoff, and I.M. Verma. 2005. Inflammation-associated cancer: NF-kappaB is the lynchpin. Trends Immunol. 26:318–325. [DOI] [PubMed] [Google Scholar]
- Memet, S., D. Laouini, J.C. Epinat, S.T. Whiteside, B. Goudeau, D. Philpott, S. Kayal, P.J. Sansonetti, P. Berche, J. Kanellopoulos, and A. Israel. 1999. I kappa B epsilon-deficient mice: reduction of one T cell precursor subspecies and enhanced Ig isotype switching and cytokine synthesis. J. Immunol. 163:5994–6005. [PubMed] [Google Scholar]
- Nelson, D.E., A.E. Ihekwaba, M. Elliott, J.R. Johnson, C.A. Gibney, B.E. Foreman, G. Nelson, V. See, C.A. Horton, D.G. Spiller, et al. 2004. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 306:704–708. [DOI] [PubMed] [Google Scholar]
- Phillips, R.J., and S. Ghosh. 1997. Regulation of IkappaB beta in WEHI 231 mature B cells. Mol. Cell. Biol. 17:4390–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, M.L., T. Fujita, H.C. Liou, G.P. Nolan, and D. Baltimore. 1993. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 7:1266–1276. [DOI] [PubMed] [Google Scholar]
- Simeonidis, S., S. Liang, G.Y. Chen, and D. Thanos. 1997. Cloning and functional characterization of mouse I kappa B epsilon. Proc. Natl. Acad. Sci. USA. 94:14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiecker, M., H. Darius, and J.K. Liao. 2000. A functional role of I kappa B-epsilon in endothelial cell activation. J. Immunol. 164:3316–3322. [DOI] [PubMed] [Google Scholar]
- Suyang, H., R. Phillips, I. Douglas, and S. Ghosh. 1996. Role of unphosphorylated, newly synthesized I kappa B beta in persistent activation of NF-kappa B. Mol. Cell. Biol. 16:5444–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, S., D. Barken, and A. Hoffmann. 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 309:1857–1861. [DOI] [PubMed] [Google Scholar]
- Whiteside, S.T., J.C. Epinat, N.R. Rice, and A. Israel. 1997. I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. EMBO J. 16:1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.