Abstract
Stp1 and Stp2 are homologous transcription factors in yeast that are synthesized as latent cytoplasmic precursors with NH2-terminal regulatory domains. In response to extracellular amino acids, the plasma membrane–localized Ssy1–Ptr3–Ssy5 (SPS) sensor endoproteolytically processes Stp1 and Stp2, an event that releases the regulatory domains. The processed forms of Stp1 and Stp2 efficiently target to the nucleus and bind promoters of amino acid permease genes. In this study, we report that Asi1 is an integral component of the inner nuclear membrane that maintains the latent characteristics of unprocessed Stp1 and Stp2. In cells lacking Asi1, full-length forms of Stp1 and Stp2 constitutively induce SPS sensor–regulated genes. The regulatory domains of Stp1 and Stp2 contain a conserved motif that confers Asi1-mediated control when fused to an unrelated DNA-binding protein. Our results indicate that latent precursor forms of Stp1 and Stp2 inefficiently enter the nucleus; however, once there, Asi1 restricts them from binding SPS sensor–regulated promoters. These findings reveal an unanticipated role of inner nuclear membrane proteins in controlling gene expression.
Introduction
Eukaryotic cells are able to control gene expression by regulating the movement of transcription factors across the nuclear envelope. Transcription factors can be synthesized as latent precursor forms that are excluded from the nucleus until proper environmental cues activate processes that trigger their targeting and entry into the nucleus (Brivanlou and Darnell, 2002). Several such regulated latent transcription factors have been described, including the extensively studied nuclear factor κB (NF-κB)/Relish, cubitus interruptus, and Notch proteins (Aza-Blanc and Kornberg, 1999; Gilmore, 1999; Baron, 2003). Notably, the activating signals that induce the translocation of these factors into the nucleus are initiated by receptors at the plasma membrane. Consequently, the movement of the regulated transcription factor provides the means to physically transmit signals from nonnuclear compartments to specific promoter sequences.
Fundamental to understanding regulated latent transcription factors is the elucidation of mechanisms that restrict their activity under noninducing conditions. A general mechanism appears to directly regulate nuclear targeting by physically tethering or anchoring latent precursor forms of transcription factors outside the nucleus. The first example of such a mechanism is the sterol regulatory element binding protein (Brown and Goldstein, 1997). Sterol regulatory element binding protein is an integral membrane protein that is anchored in the membranes of the early secretory pathway. The cytoplasmically oriented domain possessing transactivation activity is released from membranes in two successive rounds of proteolytic processing by site-specific membrane-bound proteases (Wang et al., 1994). An additional example is NF-κB/Relish signaling; IF-κB sequesters NF-κB in the cytoplasm and prevents nuclear translocation by binding the actin cytoskeleton via ankyrin repeats (Gilmore, 1999; Brivanlou and Darnell, 2002). However, with respect to nonmembrane factors like NF-κB, little is known regarding the efficiency of cytoplasmic retention. Given the nature and kinetics of protein–protein interactions, the retention of soluble latent transcription factors is expected to be incomplete even under noninducing conditions. Similarly, low level basal processing of soluble or membrane-bound transcription factors under noninducing conditions will generate active proteins that have the potential to inappropriately enter the nucleus. Consequently, to ensure the fidelity of signal transducing pathways, cells are likely to possess other modes of regulation in addition to cytoplasmic retention strategies to maintain the stringency of the inactive state under noninducing conditions.
Several of the recognized signaling pathways from the plasma membrane to the nucleus in Saccharomyces cerevisiae are involved in sensing nutrient availability and regulating nutrient uptake (Forsberg and Ljungdahl, 2001b; Van Belle and André, 2001). In the plasma membrane, the amino acid receptor Ssy1 (Wu et al., 2006) functions with two intracellular peripheral membrane proteins, Ptr3 and Ssy5, as the fundamental components of the Ssy1–Ptr3–Ssy5 (SPS)-sensing pathway (Forsberg and Ljungdahl, 2001a). This pathway induces the transcription of amino acid permease genes in response to extracellular amino acids. The homologous zinc finger transcription factors Stp1 and Stp2 are the downstream effectors of the SPS signaling pathway (de Boer et al., 2000; Nielsen et al., 2001). Stp1 and Stp2 bind to specific upstream activating sequences that are present within SPS sensor–regulated promoters (de Boer et al., 2000; Nielsen et al., 2001). Both Stp1 and Stp2 are synthesized as latent cytoplasmic factors that are mobilized by receptor-activated processing (Andréasson and Ljungdahl, 2002, 2004; Andréasson et al., 2006). In response to the addition of amino acids and in a strictly SPS sensor–dependent manner, Stp1 and Stp2 are endoproteolytically cleaved by the endoproteolytic activity of the Ssy5 protease (Abdel-Sater et al., 2004; Andréasson et al., 2006). This event liberates the DNA-binding and transactivation domains from an ∼10-kD NH2-terminal fragment. The shorter forms of Stp1 and Stp2 accumulate in the nucleus, where they function to transactivate SPS sensor–regulated genes.
Because of the inability to process Stp1 and Stp2, cells lacking a functional SPS sensor exhibit diminished capacities to take up amino acids. Recessive loss of function mutations in ASI1 (amino acid sensor independent) result in the constitutive expression of SPS sensor–regulated genes, bypass the requirement of a functional SPS sensor, and restore amino acid uptake in SPS sensor–deficient strains (Forsberg et al., 2001). ASI1 encodes a novel integral membrane protein. In this study, we examine the intracellular location and membrane structure of Asi1 and whether it is a constituent of the SPS signaling pathway. We report that Asi1 is a glycoprotein component of the inner nuclear membrane that restricts full-length unprocessed forms of Stp1 and Stp2 from binding SPS sensor–regulated promoters. Our findings indicate that Asi1 participates directly within the SPS signaling pathway and that its presence is required to maintain the repressed state of SPS sensor–regulated genes under noninducing conditions. These results reveal a novel role of inner nuclear membrane proteins and illuminate an additional layer of control that is required to establish proper levels of basal gene expression.
Results
Asi1 is a glycoprotein with five transmembrane segments and an essential RING domain
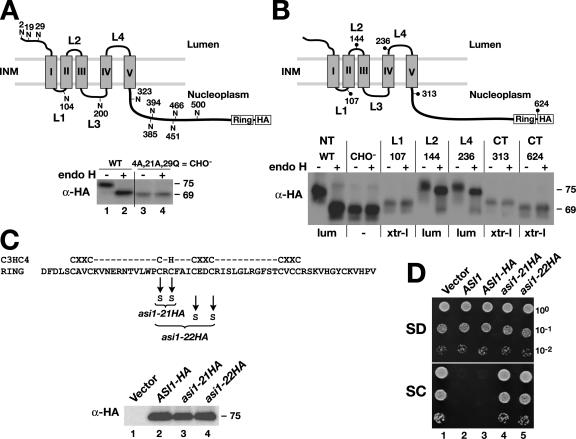
The observation that SPS sensor–regulated genes are constitutively expressed even under noninducing conditions in asi1 mutant cells prompted us to study Asi1 as a potential regulator of transcription factor latency. Primary sequence analysis of Asi1 reveals two equally sized domains: an NH2-terminal half with five hydrophobic segments predicted to be membrane spanning and a COOH-terminal hydrophilic half with a conserved Zn2+-binding RING motif (Fig. 1). To gain a more precise understanding of the significance of these domains, we determined the in vivo topology of Asi1. We created an epitope-tagged ASI1-HA allele by inserting a cloning cassette encoding an HA epitope tag just before the stop codon of the ASI1open reading frame. The function of the encoded Asi1-HA protein was assessed using a growth-based assay. In brief, leucine auxotrophic (leu2) cells lacking a functional SPS sensor (ssy1) are unable to grow on synthetic complex dextrose (SC) medium (Forsberg et al., 2001). Such ssy1 leu2 double mutant strains can only import leucine at sufficient rates to support growth in the absence of competing amino acids (synthetic minimal dextrose [SD]) or on SC when ASI1 is inactive (asi1; compare dilution series 1 with 2; Fig. 1 D). Thus, the lack of growth of the ssy1 leu2 asi1 triple mutant strain expressing the ASI1-HA allele (dilution series 3; Fig. 1 D) reflects the presence of a functional ASI1 allele. Initial experiments using the functional epitope-tagged allele supported the notion that ASI1 encodes an integral membrane protein. Asi1-HA pellets with membranes and is solubilized only in the presence of detergents (unpublished data).
Figure 1.
Structural and functional analysis of Asi1. (A) Asi1 is a glycoprotein. Schematic presentation of the predicted membrane topology of Asi1; the 11 possible NH2-linked glycosylation sites are shown. The gray boxes depict the five hydrophobic domains (I–V), and the COOH-terminal RING domain and HA epitope tag are indicated. Strain YMH349 (asi1Δ ssy1Δ leu2) carrying either plasmid pAZ002 (ASI1-HA) or pAZ020 (ASI1-HA-CHO −) was grown in SC. Extracts of total cell protein were prepared, treated with endoH as indicated, and resolved by SDS-PAGE and immunoblotted with monoclonal anti-HA antibody. The numbers to the right of immunoblots are the molecular mass (in kilodaltons) of protein bands calculated based on the migration of protein standards. INM, inner nuclear membrane. (B) Membrane topology of Asi1. Schematic representation of Asi1. Positions of the suc2 topological reporter cassette insertions are indicated by dots; the numbers refer to the amino acids to which the insertions were fused. Native Asi1-HA (pAZ014), glycosylation minus Asi1-HA-CHO− (pAZ020), and suc2 cassette gene fusion proteins Asi1-107 (pAZ034), Asi1-144 (pAZ035), Asi1-236 (pAZ037), Asi1-313 (pAZ039), and Asi1-624 (pAZ073) were expressed in YMH349. Cells were grown in SD, and extracts were prepared and analyzed as in A. The experimentally determined topological orientation of each reporter is indicated (lum, lumenal; xtr-l, extralumenal). NT, NH2 terminus; CT, COOH terminus. (C and D) The C3HC4-like RING motif (amino acids 568–608) is required for function. WT, wild type. (C) Alleles asi1-21HA (pAZ015) and asi1-22HA (pAZ016) encode mutant proteins with cysteine 583 and 585 and cysteine 583, 585, 589, and 592 replaced by serine, respectively. Immunoblot analysis of extracts from strain YMH349 carrying plasmids pRS202 (vector), pAZ014 (ASI1-HA), pAZ015 (asi1-21HA), or pAZ016 (asi1-22HA) grown in SD. Proteins were resolved on 10% SDS-PAGE gels and blotted with anti-HA antibody. (D) 10-fold dilution series of cell suspensions of strains as in C and YMH349 carrying pMH4 (ASI1) were spotted onto plates containing SD or SC, incubated for 3 d at 30°C, and photographed.
Asi1 has 11 possible NH2-linked (NXS/T) glycosylation sites (Fig. 1 A). We examined whether any of these sites were used in vivo by examining the electrophoretic mobility of Asi1 in extracts before and after treatment with endoglycosidase H (endoH). A distinct endoH-dependent gel mobility shift was observed (Fig. 1 A, bottom; lanes 1 and 2), indicating that Asi1 is indeed a glycoprotein. As a first step to experimentally determine the topology of Asi1, we constructed the ASI1-4A,21A,29Q allele, which encodes a mutant protein with three amino acid residue changes (S4 to A, T21 to A, and N29 to Q) that destroy the three potential NXS/T glycosylation sites in the hydrophilic NH2 terminus. This mutant protein is not glycosylated (Fig. 1 A, bottom; lanes 3 and 4), indicating that the other potential glycosylation sites are not used and that the NH2 terminus is oriented toward the ER lumen during biogenesis. The ASI1-4A,21A,29Q allele (CHO−) fully complements asi1-null mutant phenotypes (unpublished data); thus, glycosylation is not required for Asi1 function.
The availability of a functional but nonglycosylated CHO− form of Asi1 enabled us to use a glycosylation-dependent topological reporter cassette (Gilstring and Ljungdahl, 2000) to further assess the structure of Asi1. The topological reporter cassette was inserted in frame into the hydrophilic COOH-terminal domain and each of the four hydrophilic loops (L1–L4) that separate the five hydrophobic segments of Asi1 (I–IV; Fig. 1 B). The resulting gene sandwich fusions with the topological reporter inserted at positions 107, 144, 236, 313, and 624 encode functional proteins (unpublished data). We were unable to obtain functional proteins when the topological reporter was inserted into positions within loop L3. The glycosylation state of the fusion proteins was monitored. The results indicate that the topological reporter was efficiently glycosylated only when introduced into loops L2 and L4, indicating that these loops are oriented toward the lumenal side of the ER membrane during biogenesis (Fig. 1 B). The topological reporter was not glycosylated when placed in loop L1 or at various positions in the hydrophilic COOH-terminal domain, and, thus, these portions of Asi1 are in an extralumenal orientation (Fig. 1 B, xtr-l). In summary, the findings that the NH2 terminus of wild-type Asi1 is glycosylated and that topological reporter cassettes located in the COOH-terminal domain are not demonstrate that the NH2 and COOH termini are oriented toward opposite sides of a membrane. Consequently, Asi1 must have an odd number of hydrophobic segments that span the membrane. Although our experimental data are consistent with either three or five membrane-spanning segments (the inability to obtain a functional protein with a topological reporter in loop L3 introduces a degree of uncertainty), based on predictive computer algorithms (Persson and Argos, 1996), it is likely that Asi1 has five membrane-spanning segments. Notably, the data indicate that the conserved RING motif at the extreme COOH terminus of Asi1 (Fig. 1) is extralumenally oriented.
The RING motif has putative zinc atom–coordinating residues with a spacing typical of C3HC4-type (RING-HC) zinc fingers (i.e., C-x2-C-x(9–39)-C-x(1–3)-H-x(2–3)-C-x2-C-x(4–48)-C-x-C; Freemont, 2000). To investigate whether the RING domain is required for Asi1 function, we constructed two mutant alleles encoding proteins that lack the ability to bind zinc (Fig. 1 C, top). Plasmids expressing these mutant alleles (asi1-21HA and asi1-22HA; dilution series 4 and 5; Fig. 1 D) did not complement the asi1Δ mutation, and strains were able to grow on SC medium, indicating that the asi1-HA21 and -HA22 proteins are nonfunctional. The levels of Asi1-HA, asi1-HA21, and asi1-HA22 proteins were similar in extracts (Fig. 1 C), excluding the trivial possibility that the mutations merely affected protein stability. These results indicate that the RING domain is essential for Asi1 function.
Asi1 is an inner nuclear membrane protein
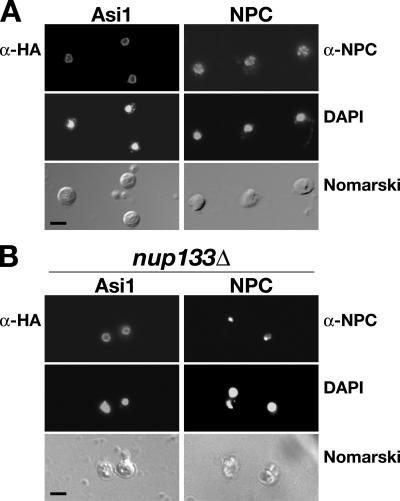
As Asi1 is an integral membrane protein, we anticipated that knowledge regarding its precise intracellular location would help us understand its role in modulating gene expression. The intracellular location of the functional Asi1-HA epitope-tagged protein was examined by immunofluorescence. The Asi1-HA–dependent fluorescence was restricted to and evenly distributed in the nuclear envelope (Fig. 2 A, left). The punctate fluorescence associated with nuclear pore complexes (NPCs; Fig. 2 A, right) was clearly distinct from the continuous rim staining of Asi1. To directly test whether Asi1 associates with NPCs, we expressed Asi1-HA in the nup133Δ strain (Doye et al., 1994; Pemberton et al., 1995). Loss of NUP133 causes the clustering of nuclear pores to distinct areas of the nuclear envelope (Fig. 2 B, right). In contrast to the single intense spot of NPC fluorescence at the nuclear periphery in nup133Δ mutant cells, Asi1 fluorescence remained evenly distributed in the nuclear membrane (Fig. 2 B, left). Our observations demonstrate that Asi1 is a component of the nuclear membrane that is not intimately associated with nuclear pores.
Figure 2.
Asi1 is a component of the nuclear envelope. Indirect immunolocalization of Asi1 (left) and nuclear pore complexes (NPCs; right) was performed with anti-HA (α-HA) and anti-NPC (α-NPC) monoclonal antibodies, respectively. (A) Strain PLY1314 (asi1Δ) carrying pAZ014 (ASI1-HA) was grown on SC (−ura). The pattern of NPC staining was examined in strain AZY262 (ASI1) grown in YPD. (B) Asi1 is not directly associated with NPCs. Strain nup133− (nup133Δ) carrying pAZ014 (ASI1-HA) was grown on SC. (top to bottom) Monoclonal α-HA (12CA5 or 3F10) or α-NPC (anti-rNup153) antibody-dependent AlexaFluor488 fluorescence; DAPI staining; cells viewed by Nomarski optics. Bars, 5 μm.
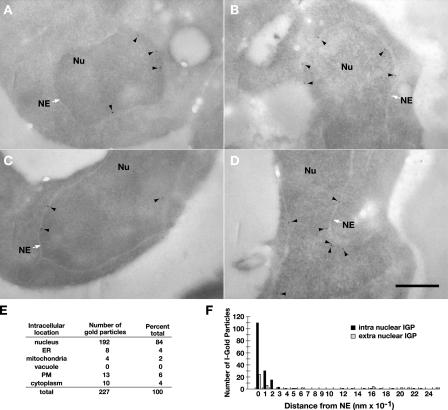
Next, we examined the localization of Asi1 by postembedding immunoelectron microscopy (Fig. 3). In this method, yeast cells are embedded in the acrylic resin LR White (Newman et al., 1983). Osmium fixation is omitted in these preparations; consequently, the nuclear envelope appears in negative contrast as a light band surrounding the nucleus. This light band includes the inner and outer nuclear membranes. In yeast, the intermembrane space does not have sufficient intrinsic contrast to be visualized. On sections through resin-embedded cells, gold-coupled antibodies label only antigens exposed at the surface; penetration into the depth of sections is limited to a few nanometers at most (Bendayan et al., 1987; Stierhof and Schwarz, 1991). We quantified the number of immunogold particles associated with different compartments in the cell (Fig. 3 E). The bulk of the gold particles (>80%) were associated with the nuclear compartment. Next, we measured the distance of gold particles from the nuclear membrane (Fig. 3 F). The distance from the inner or outer nuclear membrane was calculated by measuring the shortest distance between gold particles and the border between the negative contrast membrane and more contrast-rich cytoplasm and nucleoplasm. The data clearly demonstrate that immunolabeling was concentrated inside of the nucleus in close proximity (0–10 nm) to the inner nuclear membrane. These results are consistent with Asi1 being a component of the inner nuclear membrane. This finding coupled with our topology studies indicates that the essential RING domain of Asi1 is oriented into the nucleoplasm.
Figure 3.
Asi1 is a component of the inner nuclear membrane. Strain PLY1314 (asi1Δ) carrying pAZ014 (ASI1-HA) was grown in SC (−ura). Cells were fixed and prepared for analysis as described in Materials and methods. (A–D) Immunoelectron micrographs of yeast cell nuclei (Nu). The white arrows indicate the nuclear envelope (NE), and the black arrowheads show the locations of Asi1-HA–dependent labeling of gold particles. Quantification of immunogold labeling. Bar, 0.5 μm. (E) Intracellular distribution of immunogold particles in 44 cells decorated with at least two gold particles. PM, plasma membrane. (F) The distance of 165 immunogold particles (IGPs) from the nuclear membrane.
Asi1 is required to maintain the latent behavior of Stp1 and Stp2 under noninducing conditions
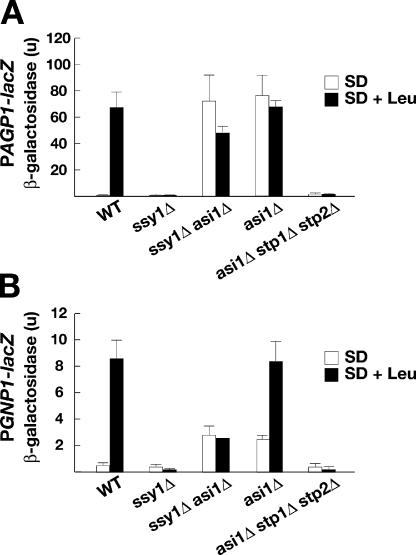
Mutations in ASI1 were identified in a genetic screen based on their ability to restore amino acid uptake in cells lacking a functional SPS sensor (Forsberg et al., 2001). Two possible explanations exist for the constitutive expression of SPS sensor–regulated genes in asi1 mutants. Either Asi1 negatively regulates the activity of the transcription factors Stp1 and Stp2, the only known effectors of the SPS-sensing pathway (Andréasson and Ljungdahl, 2002), or, alternatively, Asi1 functions independently of these factors. In the latter case, asi1 mutations could induce SPS sensor–regulated genes via an independent and perhaps parallel pathway. Accordingly, SPS sensor gene expression should remain constitutive in mutants lacking Stp1 and Stp2 (asi1Δ stp1Δ stp2Δ). To differentiate between these two possibilities, we monitored the expression of the SPS sensor–regulated genes AGP1 and GNP1 by measuring the levels of β-galactosidase in wild-type, ssy1Δ, asi1Δ, ssy1Δ asi1Δ, and asi1Δ stp1Δ stp2Δ strains carrying either a PAGP1-lacZ (Fig. 4 A) or PGNP1-lacZ (Fig. 4 B) reporter construct. Cells were grown in the presence or absence of the inducing amino acid leucine. In wild-type cells, the expression of AGP1 and GNP1 promoting β-galactosidase was dependent on amino acid availability; low levels of β-galactosidase were detected in uninduced cells (SD), whereas robust activity was detected upon induction (SD + leucine). The induced expression of PAGP1-lacZ and PGNP1-lacZ was strictly dependent on the SPS sensor, and no β-galactosidase was detected in the SPS sensor–deficient ssy1Δ mutant. The asi1Δ mutation bypassed the requirement of a functional SPS sensor and the presence of an inducing amino acid. Constitutive expression of both reporter constructs was observed in ssy1Δ asi1Δ and asi1Δ mutants. Importantly, β-galactosidase activity was not detected in the asi1Δ stp1Δ stp2Δ mutant; thus, the constitutive transcription of SPS sensor–regulated genes in asi1 mutants is strictly dependent on the presence of Stp1 or Stp2. These results indicate that Asi1 is a component of the SPS signaling pathway that negatively modulates the activity of Stp1 and Stp2 under noninducing conditions.
Figure 4.
Null alleles of ASI1 derepress SPS sensor–regulated genes in a strictly Stp1- and Stp2-dependent manner. The expression of two SPS sensor–dependent genes, AGP1 (A) and GNP1 (B), was monitored by measuring β-galactosidase activity (in units) of PAGP1-lacZ and PGNP1-lacZ reporter constructs. Wild-type (WT; CAY29), ssy1Δ (CAY91), ssy1Δ asi1Δ (CAY206), asi1Δ (PLY1313), and asi1Δ stp1Δ stp2Δ (CAY152) strains carrying either plasmid YCpAGP1-lacZ (PAGP1-lacZ) or pCA227 (PGNP1-lacZ) were grown in SD or SD supplemented with leucine (SD + Leu). β-galactosidase activity present in three independent transformants of each strain was quantified; error bars indicate one standard deviation.
asi1 mutations enable unprocessed full-length forms of Stp1 and Stp2 to activate transcription
The NH2-terminal regulatory domains of Stp1 and Stp2 possess two conserved sequence motifs that function independently and in parallel to confer amino acid–induced regulation of an artificial transcription factor (Andréasson and Ljungdahl, 2004). One motif (Region I) is required for cytoplasmic retention, and the other motif (Region II) mediates SPS sensor–dependent endoproteolytic processing. The finding that loss of Asi1 function constitutively activates the expression of SPS sensor genes in a Stp1- and Stp2-dependent manner raised the possibility that Asi1 somehow affected one or both of these two regulatory activities.
We initially considered that Asi1 prevents Stp1 and Stp2 processing under noninducing conditions. If this is so, Stp1 and Stp2 should be constitutively processed in an asi1Δ mutant. Stp1 and Stp2 processing was examined in wild-type (ASI1) and asi1Δ strains (Fig. 5 A). In the absence of amino acids, Stp1 and Stp2 were found exclusively in their unprocessed full-length forms. Both factors were processed normally in amino acid–induced cells, and, thus, the processing of Stp1 and Stp2 is Asi1 independent. We monitored the levels of β-galactosidase resulting from PAGP1-lacZ reporter gene expression in the same transformants (Fig. 5 B). Consistent with our previous results (Fig. 4), lacZ expression was constitutive in asi1Δ strains, whereas in ASI1 wild-type strains, expression was amino acid dependent. To conclusively address the possibility that asi1 mutations affect processing, perhaps at levels too low to detect by immunoblot analysis, we assessed the ability of asi1Δ mutations to activate SPS sensor–regulated genes in a strain lacking the Stp1 and Stp2 processing protease Ssy5. An asi1Δ ssy5Δ double mutant strain exhibited constitutive gene expression and exhibited growth phenotypes indicative of restored amino acid uptake capabilities (unpublished data). This finding formally rules out the possibility that Asi1 exerts regulatory effects by modulating Stp1 and Stp2 processing. Together, these results demonstrate that in the absence of Asi1, unprocessed full-length forms of Stp1 and Stp2 can enter the nucleus and induce transcription. Thus, the NH2-terminal regulatory domains do not appear to grossly interfere with the transactivation potential of Stp1 or Stp2.
Figure 5.
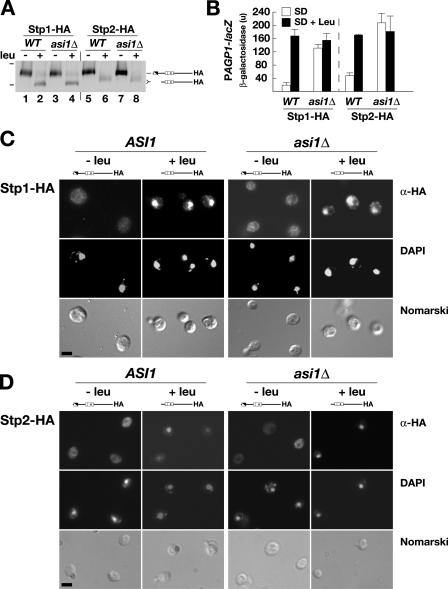
Characterization of the SPS sensor signaling pathway in asi1-null mutant strains. (A) Stp1 and Stp2 processing occurs independently of Asi1. ASI1 (PLY127) and asi1Δ (PLY1327) strains were transformed with pCA030 (PAGP1-lacZ) and pCA047 (Stp1-HA) or pCA111 (Stp2-HA). Cells were grown in SD medium (−leu), and, where indicated, leucine was added 30 min before harvest (+leu). Extracts were resolved by SDS-PAGE and immunoblotted with anti-HA. The immunoreactive forms of Stp1-HA and Stp2-HA present in the cell extracts are schematically represented (right), and the 83- and 62-kD protein standards (bars, left) are indicated at their corresponding positions of migration. WT, wild type. (B) The levels of β-galactosidase activity (in units) resulting from expression of the PAGP1-promoted β-galactosidase gene were assessed in strains as in A. The activity in three independent transformants was quantified; error bars indicate one standard deviation. (C and D) Indirect immunolocalization of Stp1-HA (C) and Stp2-HA (D) in ASI1 and asi1Δ cells was performed with anti-HA monoclonal antibodies. Cells were grown in SD to an OD600 of 0.7, the cultures were split into two equal aliquots, and leucine was added to one. The cultures were incubated by shaking at 30°C for an additional 30 min, and the cells were fixed. (top to bottom) α-HA monoclonal antibody-dependent AlexaFluor488 fluorescence; DAPI staining; and cells viewed by Nomarski optics. Strains CAY28 (ASI1) and PLY1314 (asi1Δ) carrying pCA078 (STP1-HA; C) and strains CAY29 (ASI1) and PLY1313 (asi1Δ) carrying pMB30 (STP2-HA; D) were used. The full-length (uninduced; −leu) and processed (induced; +leu) forms of Stp1-HA and Stp2-HA present in cells are schematically depicted above the appropriate panels. Bars, 5 μm.
Asi1 does not influence Stp1 and Stp2 localization
We have previously shown that Stp1 is efficiently targeted to the nucleus only upon induction by amino acids and in a strictly SPS sensor–dependent manner (Andréasson and Ljungdahl, 2002). Because asi1 mutations enable full-length forms of Stp1 and Stp2 to induce the gene expression of SPS sensor–regulated genes, we examined the possibility that the loss of Asi1 function interfered with the cytoplasmic retention of unprocessed Stp1 and Stp2. In this case, Stp1 and Stp2 should constitutively target to the nucleus in asi1Δ mutants. Using immunofluorescence microscopy, we determined the intracellular location of full-length (uninduced; −leu) and processed (induced; +leu) Stp1 and Stp2 in the wild-type (ASI1) and asi1Δ strains. In both uninduced ASI1 and asi1Δ cells, Stp1- and Stp2-dependent fluorescence was barely above background and diffused throughout cells (Fig. 5, C and D; −leu). In cells induced with leucine, intense and highly focused fluorescence that colocalized with DAPI-stained DNA was observed (Fig. 5, C and D; +leu). The finding that Stp1 and Stp2 do not constitutively accumulate in the nuclei of uninduced asi1Δ cells clearly demonstrates that Asi1 does not play a key role in retaining Stp1 and Stp2 in the cytoplasm.
Asi1 prevents access of unprocessed Stp1 and Stp2 to the promoters of SPS sensor–regulated genes
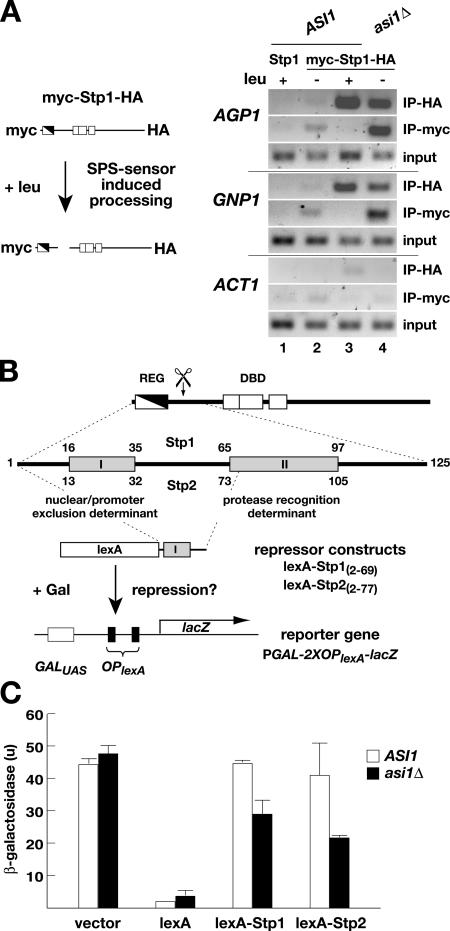
The apparent conflicting observations that asi1Δ mutant strains exhibit constitutive Stp1- and Stp2-dependent expression of SPS sensor–regulated genes (Fig. 4) without visible nuclear accumulations of Stp1 or Stp2 (Fig. 5, C and D) could be resolved if asi1Δ mutations enable only low levels of Stp1 and Stp2 to access SPS sensor–regulated promoters. We used chromatin immunoprecipitation (ChIP) to examine this possibility by analyzing the association of Stp1 with two SPS sensor–regulated promoters (i.e., AGP1 and GNP1). To facilitate the analysis, we used a Stp1 construct that carries a myc epitope at the NH2 terminus and an HA epitope at the COOH terminus (Fig. 6 A, schematic diagram); this construct complements stp1 stp2–null mutant phenotypes and thus is functional (unpublished data). The use of this doubly tagged Stp1 construct allowed the promoter association of full-length and processed forms of Stp1 to be experimentally distinguished. Anti-myc antibodies can only immunoprecipitate unprocessed Stp1, whereas anti-HA antibodies immunoprecipitate both full-length and processed forms of Stp1. A plasmid encoding native Stp1 without epitope tags was included in the experiment as a control for nonspecific immunoprecipitation. The ability to amplify the ACT1 promoter was used to control the quality of input DNA and the binding to nonspecific DNA sequences.
Figure 6.
Unprocessed Stp1 and Stp2 are excluded from SPS sensor–regulated promoters in an Asi1-dependent manner. (A) ChIP analysis of Stp1 association with AGP1 and GNP1 promoters before and after amino acid–induced processing. Schematic diagram of the double epitope-tagged myc-Stp1-HA protein and SPS sensor–dependent processing. Cultures of ASI1 (CAY60) and asi1Δ (CAY150) carrying plasmid pMB10 (myc-Stp1-HA) were grown in SD medium (−leu), and, where indicated, leucine was added 30 min before harvest (+leu). Cell lysates were prepared and split into two equal portions, and each portion was analyzed by ChIP using anti-myc or anti-HA antibody. The ability to amplify the ACT1 promoter was monitored to control nonspecific immunoprecipitation. The size of amplified fragments are as follows: AGP1, 246 bp; GNP1, 313 bp; and ACT1, 274 bp. (B) Diagram of Stp1 and Stp2 and the repression assay used to assess Asi1-dependent promoter access. Stp1 and Stp2 contain two distinct regions (I and II) of sequence conservation within their NH2-terminal regulatory domains (REG) and DNA-binding domains (DBD). Regions I and II are indicated in the enlargement of the NH2-terminal domain (amino acids 1–125). Region I is required to prevent unprocessed full-length forms from efficiently entering the nucleus and binding promoters. Region II is required for amino acid–induced endoproteolytic processing (scissors). Gene fusion constructs directing the expression of the DNA-binding protein lexA fused to NH2-terminal Stp1(2–69) (pMB16) and Stp2(2–77) (pMB17) Region I–containing sequences are shown. In the absence of lexA, the reporter gene PGAL1-2XOPlexA-lacZ (pMB18) is fully inducible by galactose; if lexA is present and able to localize to the nucleus, the induction is reduced. (C) The ability of lexA fusion proteins to enter the nucleus and bind to lexA operators (OPlexA) was assessed by measuring the levels of β-galactosidase activity (in units) in ASI1 (PLY127) and asi1Δ (PLY1327) strains carrying pMB18 (PGAL1-2XOPlexA-lacZ) and pCA160 (vector), pCA153 (lexA), pMB16 (lexA-Stp1), or pMB17 (lexA-Stp2). Strains were grown in SC and induced with galactose. The activity in three independent transformants was quantified; error bars indicate one standard deviation.
In ASI1 cells, we readily detected the association of Stp1 with AGP1 and GNP1 promoters, but only after induction with amino acids and only in lysates immunoprecipitated with anti-HA antibody (Fig. 6 A, compare lanes 2 with 3). The inability of anti-myc antibodies to immunoprecipitate these promoters indicates that only the shorter processed form of Stp1 is able to gain access to promoters in wild-type cells. In lysates prepared from asi1Δ cells, both anti-HA and anti-myc antibodies were able to immunoprecipitate the AGP1 and GNP1 promoters (Fig. 6 A, lane 4), even in cells grown in the absence of inducing amino acids. This finding indicates that in the absence of Asi1, full-length unprocessed Stp1 is indeed able to gain access to SPS sensor–regulated promoters. These results are entirely consistent with our phenotypic analysis of asi1 mutants and account for the constitutive expression of SPS sensor–regulated promoters observed in asi1Δ mutants.
Asi1-dependent promoter exclusion is mediated by a conserved sequence motif in the NH2-terminal regulatory domains of unprocessed Stp1 and Stp2
The NH2-terminal fragment of Stp1 comprised of amino acids 1–125 is modular and can be transferred to faithfully regulate the activity of an artificial transcription factor in an SPS sensor–dependent manner (Andréasson and Ljungdahl, 2004). As previously mentioned, this fragment contains two motifs that are also present in the NH2 terminus of Stp2 (Fig. 6 B). These motifs appear to have independent functions. Region I is required to prevent the unprocessed full-length forms of these factors from inducing SPS sensor–regulated genes, whereas Region II is required for amino acid–induced SPS sensor–mediated endoproteolysis. A smaller fragment spanning Region II of Stp1 (Stp163–125) interacts with Ssy5, the protease component of the SPS sensor (Andréasson et al., 2006). In contrast, a fragment spanning Region I (Stp11–62) does not interact with Ssy5. These findings prompted us to directly test whether the NH2-terminal regions of Stp1 and Stp2 containing Region I are sufficient to mediate Asi1-dependent promoter exclusion when fused to a nonrelated DNA-binding protein.
The NH2-terminal domains of Stp1 (amino acids 2–69) and Stp2 (amino acids 2–77) were fused to the COOH terminus of the bacterial DNA-binding protein lexA (Fig. 6 B). The lexA protein contains an intrinsic nuclear localization signal and, in the absence of additional sequences, is efficiently targeted to the nucleus (Rhee et al., 2000). Because of the lack of Region II sequences, the lexA-Stp1(aa 2–69) and lexA-Stp2(aa 2–77) fusion proteins are not subject to amino acid–induced SPS sensor–dependent processing (unpublished data). The ability of these fusion proteins to access DNA was tested using a reporter plasmid containing lexA operators (OPlexA; schematically depicted in Fig. 6 B). The expression of the β-galactosidase lacZ gene is regulated in a galactose-dependent manner as a result of the presence of the GAL1 promoter. However, when lexA is present, it binds to the two OPlexA that are placed between the GAL1 promoter and the lacZ open reading frame, and, consequently, the level of GAL1-driven lacZ expression is repressed. A similar repression assay is routinely used to test whether fusion proteins intended to serve as bait in two-hybrid approaches are able to enter the nucleus (Gyuris et al., 1993).
β-galactosidase activity was measured in ASI1 wild-type and asi1Δ mutant cells expressing the lexA-Stp1(aa 2–69) and lexA-Stp2(aa 2–77) constructs. Experimental controls included the empty vector and plasmid encoding only the lexA DNA- binding domain. As expected, cells carrying the empty vector and thus lacking lexA exhibited high levels of β-galactosidase activity. Cells expressing lexA lacking Stp1 or Stp2 sequences exhibited very low levels of β-galactosidase activity, clearly demonstrating the repressive effect of lexA. ASI1 cells expressing either lexA-Stp1(aa 2–69) or lexA-Stp2(aa 2–77) exhibited high β-galactosidase activity at levels similar to that observed in cells carrying the empty vector control. These results demonstrate that the presence of either the NH2-terminal fragments of Stp1 or Stp2 spanning the Region I motif prevent lexA from gaining access to the OPlexA sites. Importantly, the expression of lexA-Stp1(aa 2–69) or lexA-Stp2(aa 2–77) in asi1Δ mutant cells significantly lowered β-galactosidase activity, indicating that in the absence of Asi1, the fusion proteins are able to access the promoter and reduce the expression of the reporter gene. This repression assay provides independent evidence supporting the view that Asi1 normally functions to prevent unprocessed Stp1 and Stp2 from accessing SPS sensor–regulated promoters.
Discussion
The nuclear envelope is a highly specialized structure that delimits the nucleus of eukaryotic cells. The nuclear envelope is comprised of inner and outer membranes, each with a distinct inventory of resident proteins. Aside from the detailed description of NPCs that provide entry and exit routes through the nuclear envelope, remarkably little is known regarding the role of nonpore proteins (Hetzer et al., 2005). Importantly, the nuclear envelope is not only a physical barrier that restricts the movement of macromolecules between the cytoplasm and nucleoplasm but is thought to function as a scaffold for proteins required for diverse nuclear functions. Our finding that Asi1 is a component of the inner nuclear membrane that functions to maintain the latent properties of two related transcription factors by preventing them from binding DNA introduces an unanticipated and novel mechanism of gene regulation in eukaryotic cells.
We previously reported that loss of function mutations in ASI1 are recessive and lead to the constitutive expression of SPS sensor–regulated genes (Forsberg et al., 2001). In this study, we have experimentally addressed Asi1 function. We show that Asi1 is a polytopic membrane protein (Fig. 1) and an integral component of the inner nuclear membrane (Figs. 2 and 3). Furthermore, the latent precursor forms of transcription factors Stp1 and Stp2 were defined as the targets of Asi1-dependent regulation (Fig. 4), clearly demonstrating that Asi1 is a bona fide constituent of the SPS pathway. Our studies have revealed an additional layer of regulation in this nutrient- induced signaling pathway. The constitutive expression of SPS sensor–regulated genes in asi1Δ mutants was traced to the ability of full-length unprocessed forms of Stp1 and Stp2 to enter the nucleus, bind promoters, and induce transcription (Figs. 5 and 6). In contrast, unprocessed forms of Stp1 and Stp2 do not bind SPS sensor–regulated promoters in wild-type cells.
During the course of this study, we experimentally addressed and ruled out several possible mechanisms that could explain the constitutive expression of SPS-regulated genes in asi1 mutants. First, the loss of Asi1 could facilitate precocious Stp1 or Stp2 processing; amino acid induction and an intact SPS sensor were found to be required for the processing of both factors in asi1Δ mutant strains (Fig. 5 A). Thus, Asi1 does not negatively modulate the endoprotease activity of the SPS sensor. Second, although Asi1 is not intimately associated with nuclear pores (Fig. 2 B), Asi1 could indirectly affect transport across the nuclear membrane. The loss of Asi1 did not visibly perturb the intracellular distribution of unprocessed Stp1 or Stp2 (Fig. 5, C and D). Therefore, Asi1 appears not to be mechanistically linked with cytoplasmic retention. Third, the presence of the essential RING domain in the nucleoplasmically oriented COOH-terminal region of Asi1 (Fig. 1) raised the possibility that Asi1 is a ubiquitin ligase specifically involved in the degradation of nuclear-localized Stp1 and Stp2. Using standard ubiquitylation assays in which positive controls exhibited robust auto- and transubiquitylation activity, Asi1 was not autoubiquitylated, nor did it catalyze ubiquitylation of the purified NH2-terminal regulatory domain of Stp1(aa 1–125) (unpublished data). Consistent with these latter results, the turnover rates of Stp1 and Stp2 are similar in both wild-type and asi1Δ mutant strains (unpublished data).
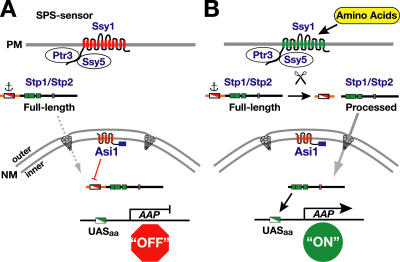
Our current model for the SPS sensor signaling pathway is schematically presented in Fig. 7. In the absence of inducing amino acids (Fig. 7 A), newly translated Stp1 and Stp2 are excluded from gaining access to SPS sensor–regulated genes by two parallel activities that converge on the NH2-terminal regulatory motif spanning Region I (Fig. 6 B). The primary mechanism functions to anchor the unprocessed full-length forms to a presently undefined cytoplasmic determinant, which prevents targeting of unprocessed factors to the nucleus. Our current studies reveal that the efficiency of anchoring is not absolute. A second Asi1-dependent mechanism prevents the low levels of full-length Stp1 and Stp2 that enter or “leak” into the nucleus from activating transcription. Therefore, Asi1 is required to maintain the basal uninduced level of SPS sensor gene expression. In the presence of inducing amino acids (Fig. 7 B), the NH2-terminal regulatory domains of Stp1 and Stp2 are proteolytically removed. The SPS sensor–catalyzed processing event depends on the conserved motif designated Region II (Andréasson and Ljungdahl, 2004). The processed forms of Stp1 and Stp2 target to the nucleus, and because they lack Region I, they bypass Asi1-dependent control and efficiently bind SPS sensor–regulated promoters.
Figure 7.
Model of the SPS-sensing pathway and the role of Asi1 in maintaining the latency of Stp1 and Stp2. (A) In the absence of inducing amino acids, the SPS sensor (Ssy1–Ptr3–Ssy5) of extracellular amino acids is present in the plasma membrane (PM) in its preactivation conformation (red). The transcription factors Stp1 and Stp2 are synthesized as inactive precursors that localize to the cytosol as a result of the presence of a cytoplasmic retention signal (anchor) that prevents the unprocessed full-length forms to efficiently enter the nucleus. In the absence of Asi1, full-length Stp1 and Stp2 enter the nucleus (dashed arrow) at rates sufficient to derepress amino acid permease (AAP) gene expression. The ability of Asi1 located in the inner nuclear membrane (NM) to prevent transcription is dependent on the nucleoplasmically oriented COOH-terminal RING motif (blue box) and on the presence of the sequences of Stp1 and Stp2 (Region I) in the NH2-terminal regulatory domain (red and white box). Consequently, there are low levels of amino acid permeases in the plasma membrane. (B) In the presence of inducing amino acids, the SPS sensor is activated (green), leading to the endoproteolytic processing of Stp1 and Stp2 (scissors). The shorter activated forms of Stp1 and Stp2 are efficiently targeted to the nucleus (solid arrow), where they bind SPS sensor–regulated promoters (DNA-binding domains; green boxes) and induce their transcription. The increased transcription of amino acid permease genes results in a concomitant increase in amino acid permeases in the plasma membrane, and cells exhibit induced rates of amino uptake.
Perhaps the simplest mechanism by which Asi1 could restrict Stp1 and Stp2 from accessing promoters would be by directly binding and sequestering them at the inner nuclear membrane. We tested this possibility using the NH2-terminal regulatory domain (amino acids 1–125) of Stp1 and the hydrophilic COOH-terminal domain of Asi1 in reciprocal and unbiased genome-wide yeast two-hybrid approaches but failed to detect any interactions. Our lack of success raises the possibility that other proteins may be involved. An obvious candidate is the membrane protein encoded by ASI3. Null mutations in this gene give rise to identical phenotypes as asi1 mutations (Forsberg et al., 2001). Asi3 is structurally related to Asi1, it has five putative membrane-spanning segments and a COOH-terminal localized RING domain (Forsberg et al., 2001), and Asi3 also localizes to the inner nuclear membrane (unpublished data). Using immunoprecipitation approaches, we have found that Asi1 but not Stp1 copurifies with Asi3 (unpublished data). We are currently pursuing alternative strategies to more definitively assess potential Stp1 and Asi protein interactions.
It is known that proteins that localize to or associate with the inner nuclear membrane can affect patterns of gene expression (for reviews see Taddei et al., 2004; Gruenbaum et al., 2005). Recruitment of chromatin to the nuclear periphery often correlates with reduced gene expression; however, recent studies in yeast have clearly demonstrated that these events are not obligatorily linked (Gartenberg et al., 2004; Taddei et al., 2005). Our data also provide a clear indication that inner nuclear membrane proteins can regulate gene expression independently of chromatin recruitment. In uninduced cells, Asi1 is required to maintain the repressed state of plasmid-encoded SPS sensor–regulated genes and, importantly, synthetic reporter gene constructs. Consequently, Asi1-mediated repression is independent of chromatin context. Furthermore, our repression and ChIP assays provide incontrovertible evidence that rule out the possibility that Asi1 recruits chromatin to the nuclear periphery indirectly via interactions with the NH2-terminal regulatory domains of Stp1 or Stp2. Asi1 negatively controls the repressive activity of lexA-Stp1(aa 2–69) or lexA-Stp2(aa 2–77), and Stp1 is not bound to promoters when gene expression is repressed.
In the nucleus of mammalian cells, lamins are involved in an extensive network of protein–protein interactions, including several known transcriptional regulators, some of which bind DNA (for reviews see Taddei et al., 2004; Gruenbaum et al., 2005). For example, it was recently shown that direct interaction of lamin A/C with c-Fos suppresses AP-1 DNA binding and transcriptional activation presumably by reducing c-Fos/c-Jun heterodimer formation (Ivorra et al., 2006). Additionally, several lamin-binding proteins (e.g., LBR, LAP2β, emerin, and MAN1) are integral components of the inner nuclear membrane. The ability to bind lamin is believed to be important because it provides the means to repress gene expression by recruiting lamin-associated regions of chromatin to the nuclear periphery. In addition to its ability to bind lamin, MAN1 can antagonize TGF-β and BMP2 signaling by binding Smads (Osada et al., 2003; Lin et al., 2005; Pan et al., 2005). Smad proteins are transcriptional regulators that become phosphorylated by activated TGF-β and BMP receptors at the plasma membrane. The phosphorylated forms of Smads target to the nucleus, where they interact with various transcription factors and induce gene expression. The binding of Smads to MAN1 appears to be independent of stimulation by TGF-β or BMP2. However, Smads bound to MAN1 are hypophosphorylated with respect to free Smads (Pan et al., 2005). These observations and the similarity in our findings regarding Asi1 raise the possibility that mechanisms that retain inactive Smads in the cytoplasm are not absolutely efficient, and, consequently, a small quantity of Smads enter the nucleus, where they are sequestered by MAN1 (Lin et al., 2005). Such a mechanism may contribute to restricting the ability of Smads to transactivate gene expression.
It seems that the biological regulation of gene expression is not limited to controlling cytoplasmic retention of latent factors. Cytoplasmic anchoring mechanisms that limit nuclear targeting are clearly not absolute, and transcription factors do inappropriately enter the nucleus, where they directly or indirectly affect gene expression. Consequently, eukaryotic cells have apparently evolved additional mechanisms to ensure the fidelity of signaling and to maintain the dormant, or repressed, state in the absence of inducing signals. The regulatory mechanism that we have described represents an additional layer of control that functions to establish the basal expression of genes controlled by a discrete signal transduction pathway. We believe that the insights gained from these studies can be extended to a more general and classic biochemical problem: how to generate a large difference between “off” and “on” catalytic states, which is an important and potentially difficult task in controlling multicomponent biological systems. The Asi1 story provides an intriguing example of how this can be achieved.
Materials and methods
Media, strains, and plasmids
Standard media, including YPD and ammonia-based SD supplemented as required to enable the growth of auxotrophic strains, were prepared as described previously (Burke et al., 2000). Ammonia-based SC was prepared as described previously (Andréasson and Ljungdahl, 2002). Where indicated, l-leucine was added at a concentration of 1.3 mM to induce the SPS sensor. When required, 5-fluoroortic acid was added to 1 g/l SC. Media were made solid with 2% (wt/vol) Bacto Agar (Difco). Antibiotic selections were made on solid YPD supplemented with 200 mg/l G418 (Invitrogen), 100 mg/l clonNAT (Werner Bioagents), or 300 mg/l hygromycin B (Duchefa). The yeast strains used in data collection are listed in Table I. All strains except nup133− are isogenic descendants of the S288c-derived strain AA255/PLY115 (Antebi and Fink, 1992). Plasmids used are listed in Table II. Details regarding strain and plasmid constructions are provided online in the supplemental material (available at http://www.jcb.org/cgi/content/full/jcb.200601011/DC1).
Table I.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| AZY262 | MATa ura3-52 ASI3∷13MYC | This study |
| CAY28 | MATα ura3-52 | Andréasson and Ljungdahl, 2002 |
| CAY29 | MATa ura3-52 | Andréasson and Ljungdahl, 2002 |
| CAY60 | MATα ura3-52 stp1Δ51∷Agleu2 | This study |
| CAY62 | MATα ura3-52 lys2Δ201 stp1Δ51∷Agleu2 | This study |
| CAY91 | MATa ura3-52 ssy1Δ13∷hisG | Andréasson and Ljungdahl, 2002 |
| CAY126 | MATa ura3-52 asi1Δ8∷kanMX stp2Δ50∷hphMX4 | This study |
| CAY150 | MATa ura3-52 asi1Δ8∷kanMX stp1Δ51∷Agleu2 | This study |
| CAY152 | MATa ura3-52 stp1Δ51∷Agleu2 stp2Δ50∷hphMX4 asi1Δ8∷kanMX | Andréasson and Ljungdahl, 2004 |
| CAY206 | MATa ura3-52 ssy1Δ13∷hisG asi1Δ8∷kanMX | Andréasson and Ljungdahl, 2004 |
| PLY127 | MATα ura3-52 lys2Δ201 | Kuehn et al., 1996 |
| PLY1313 | MATa ura3-52 asi1Δ80∷hphMX4 | This study |
| PLY1314 | MATα ura3-52 asi1Δ80∷hphMX4 | This study |
| PLY1327 | MATα ura3-52 lys2Δ201 asi1Δ80∷hphMX4 | This study |
| nup133− | MATα ura3 leu2 his3 trp1 ade2 nup133∷HIS3 | Doye et al., 1994 |
| YMH349 | MATa ade2 leu2-3, 112 lys2Δ201 ura3-52 ssy1Δ13 asi1Δ8∷kanMX | Forsberg et al., 2001 |
Table II.
Plasmids
| Plasmid | Description | Reference |
|---|---|---|
| pAZ002 | ASI1-3xHA in pRS316 | This study |
| pAZ011 | asi1-21-3xHA in pRS316 | This study |
| pAZ012 | asi1-22-3xHA in pRS316 | This study |
| pAZ014 | ASI1-3xHA in pRS202 | This study |
| pAZ015 | asi1-21-3xHA in pRS202 | This study |
| pAZ016 | asi1-22-3xHA in pRS202 | This study |
| pAZ020 | ASI1-3xHA-CHO − in pRS316 | This study |
| pAZ024 | asi1Δ8∷kanMX in pRS316 | This study |
| pAZ025/26/28/30/33 | BglII sites introduced into ASI1 in pAZ020 | This study |
| pAZ034/35/37/39/73 |
suc2 topology reporter inserted into BglII sites in pAZ025/26/28/30/33 |
This study |
| pCA030 | PAGP1-lacZ in pRS317 | Andréasson and Ljungdahl, 2004 |
| pCA047 | STP1-3xHA in pRS316 | Andréasson and Ljungdahl, 2002 |
| pCA078 | PHXT7-STP1-3xHA in pRS202 | Andréasson and Ljungdahl, 2002 |
| pCA088 | STP1-3xHA in pRS316 | This work |
| pCA111 | STP2-3xHA in pRS316 | Andréasson and Ljungdahl, 2002 |
| pCA145 | lexA in pRS202 | This study |
| pCA153 | lexA in 2 μm LYS2 | This study |
| pCA160 | 2 μm LYS2 PADH1 ΔSph1 | Andréasson and Ljungdahl, 2004 |
| pCA181 | STP1-6xHA in pRS316 | This study |
| pCA227 | PGNP1-lacZ in CEN URA3 | This study |
| pMH4 | ASI1 in pRS316 | Forsberg et al., 2001 |
| pMB09 | STP1-13xMYC in pRS316 | This study |
| pMB10 | 13xMYC-STP1-6xHA in pRS316 | This study |
| pMB12 | STP2-3xHA in pRS202 | This study |
| pMB13 | STP2-6xHA in pRS202 | This study |
| pMB16 | lexA-Stp1(aa 2–69) in 2 μm LYS2 | This study |
| pMB17 | lexA-Stp2(aa 2–77) in 2 μm LYS2 | This study |
| pMB18 | PGAL1-OPlexA-lacZ in CEN URA3 | This study |
| pMB27 | STP2-13xMYC in pRS316 | This study |
| pMB30 | 13xMYC-STP2-6xHA in pRS202 | This study |
| pRS202 | 2 μm URA3 | Connelly and Hieter, 1996 |
| pRS316 | CEN URA3 | Sikorski and Hieter, 1989 |
| pRS317 | CEN LYS2 | Sikorski and Hieter, 1989 |
| YCpAGP1-LacZ | PAGP1-lacZ in CEN URA3 | Iraqui et al., 1999 |
β-galactosidase activity assays
β-galactosidase activity was determined with N-lauroyl-sarcosine–permeabilized cells (Kippert, 1995). Cells grown in SD were harvested by centrifugation, resuspended in Z buffer, and the OD600 was measured. A 250-μl aliquot of cell suspension was mixed with 550 μl of 0.3% (wt/vol) Na N-lauroyl-sarcosine Z buffer and incubated at 30°C for 15 min. A 160-μl aliquot of 4 mg/ml ONPG (2-nitrophenyl β-D-galactopyranoside) solution was added, and tubes were mixed by vortexing. The reactions were stopped by the addition of 400 μl of 1 M Na2CO3, the tubes were centrifuged at 12,000 g for 5 min, and the absorbance of the supernatant was measured at 420 nm. Activity was calculated according to the following formula: 103 × A420/OD600/0.25 (volume of cell suspension added)/time (minutes).
Immunoblot analysis
Whole cell extracts were prepared under denaturing conditions with NaOH and TCA treatment according to Silve et al. (1991). Extracted proteins were resolved using SDS-PAGE and analyzed by immunoblotting. Immunoblots were incubated with primary antibody (12CA5 ascites fluid; anti-HA monoclonal) diluted 1:1,000 in blocking buffer. Immunoreactive bands were visualized by chemiluminescence detection (SuperSignal West Dura Substrate; Pierce Chemical Co.) of HRP conjugated to a secondary antibody (anti–mouse Ig from sheep and anti–rabbit Ig from donkey; GE Healthcare) and quantified by using a gel documentation system (LAS1000; Fuji).
Determination of Asi1 membrane topology
Whole cell protein extracts derived from 1 ml of cultures (OD600 of 1) were prepared as described previously (Silve et al., 1991). 25 μl of duplicate protein samples (equivalent to an OD600 of 0.2 cell suspension) were diluted with an equal volume of 100 mM sodium citrate, pH 5.5, and heated for 10 min at 37°C. Three milliunits of endoH (Roche) was added to half of the samples, and all samples were incubated overnight at 37°C (Gilstring and Ljungdahl, 2000). Proteins were resolved by 7% SDS-PAGE and immunoblotted with monoclonal anti-HA antibody (12CA5).
Microscopy
Cells were grown to an OD600 of 0.8 and processed for indirect immunofluorescence analysis essentially as described previously (Burke et al., 2000). Cells were fixed by the addition of an aliquot of 37% formaldehyde directly to the cultures to a final concentration of 4.5% and incubated for 45 min at 30°C. To detect HA-tagged proteins, the primary antibody used was the 12CA5 or 3F10 anti-HA monoclonal antibody diluted 1:300. To visualize nuclear pores, monoclonal antibody raised against rat Nup153 (monoclonal antibody PF190 × 7A8; gift of V. Cordes, University of Heidelberg, Heidelberg, Germany) was used in a 1:10 dilution (Cordes et al., 1993). The secondary antibody was AlexaFluor488 conjugated to goat anti–mouse or donkey anti–rat IgG (H + L; Invitrogen) diluted 1:500. Strain nup133− is temperature sensitive for growth and was grown at 23°C and fixed at RT. Cells were viewed using a microscope (Axiophot; Carl Zeiss MicroImaging, Inc.) with a plan-Apochromat 63× NA 1.40 objective. Digital images of cells examined using Nomarski optics and antibody-dependent and DAPI fluorescence (standard filter sets) were captured using a CCD camera (C4742-95; Hamamatsu) and QED Imaging software (Media Cybernetics). Image files were incorporated into figures using Photoshop CS (Adobe).
Immunoelectron microscopy
Cells were grown to an OD600 of 0.7 and were fixed in the presence of 2% formaldehyde/0.1% glutaraldehyde at 30°C for 1 h with gentle shaking. The fixed cells were washed in SP buffer (1.2 M sorbitol and 0.1 M potassium phosphate, pH 7.5), pelleted by centrifugation, dehydrated in graded ethanol (70–100%), and embedded in LR White Resin (London Resin) by UV polymerization as previously described (Krull et al., 2004). Thin sections were cut on an ultramicrotome (Ultracut; Leica) and picked up on formvar-coated nickel grids. For immunostaining, the sections were blocked with 5% BSA in PBS, pH 7.3, for 30 min and incubated for 120 min with rat anti-HA antibodies (clone 3F10; Roche) diluted 1:50 in PBS/5% BSA. The grids were then rinsed repeatedly with PBS/5% BSA and incubated for 60 min with goat anti–rat IgG conjugated to 12-nm gold particles (Jackson ImmunoResearch Laboratories) that were diluted 1:20 in PBS/0.5% BSA. After repeated washing with PBS/0.5% BSA and PBS alone, the grids were postfixed for 5 min with 2% glutaraldehyde in PBS, washed with PBS and water, and allowed to dry. Contrast staining was performed with saturated uranyl acetate for 5 min and lead citrate for 5 s. Finally, the sections were examined in an electron microscope (CM120; Philips) at 80 kV and photographed using a CCD camera (MegaPlus; Kodak). For quantitative evaluation of the immunogold labeling, successive cells were photographed at high magnification, and the distance of all gold particles from the midline of the nuclear envelope was measured using the analySIS system (Soft Imaging Software).
ChIP
ChIP analysis was performed according to Strahl-Bolsinger et al. (1997) with minor modifications. Cells were grown to an OD600 of 0.7 and were fixed for 30 min at RT in the presence of 1% formaldehyde. The formaldehyde was added directly to the cultures. Cells were harvested by centrifugation, resuspended in lysis buffer, and disrupted with glass beads by beating six times for 40 s in the beadbeater. The resulting lysate was sonicated twice for 10 s using a sonifier (Sonifier 250; Branson Ultrasonics Corp.) with output control set to 5 (average size of DNA fragments was 0.5 kb). Sonicated lysates were clarified by centrifugation (twice for 10 min at 15,000 g). The protein content was measured, the samples were adjusted to 10 mg/ml in 1,200 μl, and 10 μl of the total lysate was put aside to control input levels. The remaining lysates were split into two equal fractions. Magnetic beads with covalently attached sheep anti–mouse or sheep anti–rat IgG (Dynabeads M-450; Dynal) were incubated with mouse monoclonal anti-myc antibody (clone 9E10; Roche) or rat monoclonal anti–HA antibody (clone 3F10; Roche), respectively. 50 μl of coated beads were used in immunoprecipitation reactions. Immunoprecipitates were sequentially washed in lysis buffer, lysis buffer containing 500 mM NaCl, washing buffer (10 mM Tris-Cl, pH 8.0, 500 mM LiCl, 1% NP-40, 1% Na-deoxycholate, and 1 mM EDTA), and Tris-EDTA. Bound protein was eluated by incubating beads twice for 10 min at 65°C in 75 μl of eluation buffer (50 mM Tris-Cl, pH 8.0, 10 mM EDTA, and 1% SDS). Cross-linking of immunoprecipitates and input samples was reversed by an overnight incubation at 65°C, after which DNA was extracted (PCR Purification Kit; QIAGEN). PCR was performed with primers that amplify promoter regions of AGP1 (PrMB23/24), GNP1 (PrMB31/32), and ACT1 (PrMB41/42). Taq polymerase (Invitrogen) and the corresponding buffer system were used. Hot start was achieved by using TaqStart antibody (BD Biosciences). The appropriate dilution of template DNA and the number of cycles (25–30) were empirically determined. Samples were first incubated for 3 min at 94°C, and the amplification cycle was as follows: 45 s at 94°C, 45 s at 50°C, and 20 s at 72°C. The reactions were stopped in the logarithmic phase of amplification, and the PCR products were separated on 2.3% agarose gel and visualized by ethidium bromide.
Repression assay
Plasmid pMB18 is a centromeric version of pJK101 (Brent and Ptashne, 1984) that contains a GAL1-promoted lacZ reporter gene (PGAL1-lexAop-lacZ). Two lexA operators (OPlexA) have been placed between the GAL1 promoter and the lacZ gene; lexA fusion proteins that bind to these operators decrease the level of galactose-induced lacZ expression. Yeast strains were grown for 2 d in SC medium containing 4% galactose, 1% raffinose, and 0.2% glucose, and β-galactosidase activity was measured.
Online supplemental material
Supplemental material provides detailed descriptions of strain and plasmid constructions. Table S1 provides data on yeast strains. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200601011/DC1.
Supplementary Material
Acknowledgments
We thank the other members of the Ljungdahl laboratory for constructive comments throughout the course of this work and Volker Cordes for providing the anti–rat Nup153 monoclonal antibody.
This research was supported by the Ludwig Institute for Cancer Research.
C. Andréasson's present address is Zentrum fur Molekulare Biologie Heidelberg, D-69120 Heidelberg, Germany
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; endoH, endoglycosidase H; NF-κB, nuclear factor κB; NPC, nuclear pore complex; SC, synthetic complex dextrose; SD, synthetic minimal dextrose; SPS, Ssy1–Ptr3–Ssy5.
References
- Abdel-Sater, F., M. El Bakkoury, A. Urrestarazu, S. Vissers, and B. André. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 24:9771–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., and P.O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., and P.O. Ljungdahl. 2004. The N-terminal regulatory domain of Stp1p is modular and fused to an artificial transcription factor confers full SPS-sensor control. Mol. Cell. Biol. 24:7503–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., S. Heessen, and P.O. Ljungdahl. 2006. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. In press. [DOI] [PMC free article] [PubMed]
- Antebi, A., and G.R. Fink. 1992. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell. 3:633–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc, P., and T.B. Kornberg. 1999. Ci: a complex transducer of the hedgehog signal. Trends Genet. 15:458–462. [DOI] [PubMed] [Google Scholar]
- Baron, M. 2003. An overview of the Notch signalling pathway. Semin. Cell Dev. Biol. 14:113–119. [DOI] [PubMed] [Google Scholar]
- Bendayan, M., A. Nanci, and F.W. Kan. 1987. Effect of tissue processing on colloidal gold cytochemistry. J. Histochem. Cytochem. 35:983–996. [DOI] [PubMed] [Google Scholar]
- Brent, R., and M. Ptashne. 1984. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 312:612–615. [DOI] [PubMed] [Google Scholar]
- Brivanlou, A.H., and J.E. Darnell Jr. 2002. Signal transduction and the control of gene expression. Science. 295:813–818. [DOI] [PubMed] [Google Scholar]
- Brown, M.S., and J.L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89:331–340. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson, and T. Stearns. 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 205 pp.
- Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 86:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V.C., S. Reidenbach, A. Kohler, N. Stuurman, R. van Driel, and W.W. Franke. 1993. Intranuclear filaments containing a nuclear pore complex protein. J. Cell Biol. 123:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer, M., P.S. Nielsen, J.P. Bebelman, H. Heerikhuizen, H.A. Andersen, and R.J. Planta. 2000. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28:974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye, V., R. Wepf, and E.C. Hurt. 1994. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 13:6062–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, H., and P.O. Ljungdahl. 2001. a. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, H., and P.O. Ljungdahl. 2001. b. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91–109. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., M. Hammar, C. Andréasson, A. Molinér, and P.O. Ljungdahl. 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics. 158:973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont, P.S. 2000. RING for destruction? Curr. Biol. 10:R84–R87. [DOI] [PubMed] [Google Scholar]
- Gartenberg, M.R., F.R. Neumann, T. Laroche, M. Blaszczyk, and S.M. Gasser. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 119:955–967. [DOI] [PubMed] [Google Scholar]
- Gilmore, T.D. 1999. The Rel/NF-kappaB signal transduction pathway: introduction. Oncogene. 18:6842–6844. [DOI] [PubMed] [Google Scholar]
- Gilstring, C.F., and P.O. Ljungdahl. 2000. A method for determining the in vivo topology of yeast polytopic membrane proteins demonstrates that Gap1p fully integrates into the membrane independently of Shr3p. J. Biol. Chem. 275:31488–31495. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., A. Margalit, R.D. Goldman, D.K. Shumaker, and K.L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31. [DOI] [PubMed] [Google Scholar]
- Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 75:791–803. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., T.C. Walther, and I.W. Mattaj. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347–380. [DOI] [PubMed] [Google Scholar]
- Iraqui, I., S. Vissers, F. Bernard, J.O. de Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivorra, C., M. Kubicek, J.M. Gonzalez, S.M. Sanz-Gonzalez, A. Alvarez-Barrientos, J.E. O'Connor, B. Burke, and V. Andres. 2006. A mechanism of AP-1 suppression through interaction of c-Fos with lamin A/C. Genes Dev. 20:307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert, F. 1995. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128:201–206. [DOI] [PubMed] [Google Scholar]
- Krull, S., J. Thyberg, B. Björkroth, H.R. Rackwitz, and V.C. Cordes. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell. 15:4261–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn, M.J., R. Schekman, and P.O. Ljungdahl. 1996. Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J. Cell Biol. 135:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F., J.M. Morrison, W. Wu, and H.J. Worman. 2005. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum. Mol. Genet. 14:437–445. [DOI] [PubMed] [Google Scholar]
- Newman, G.R., B. Jasani, and E.D. Williams. 1983. A simple post-embedding system for the rapid demonstration of tissue antigens under the electron microscope. Histochem. J. 15:543–555. [DOI] [PubMed] [Google Scholar]
- Nielsen, P.S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen, R.J. Planta, M.C. Kielland-Brandt, and H.A. Andersen. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264:613–622. [DOI] [PubMed] [Google Scholar]
- Osada, S., S.Y. Ohmori, and M. Taira. 2003. XMAN1, an inner nuclear membrane protein, antagonizes BMP signaling by interacting with Smad1 in Xenopus embryos. Development. 130:1783–1794. [DOI] [PubMed] [Google Scholar]
- Pan, D., L.D. Estevez-Salmeron, S.L. Stroschein, X. Zhu, J. He, S. Zhou, and K. Luo. 2005. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-{beta} superfamily of cytokines. J. Biol. Chem. 280:15992–16001. [DOI] [PubMed] [Google Scholar]
- Pemberton, L.F., M.P. Rout, and G. Blobel. 1995. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 92:1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, B., and P. Argos. 1996. Topology prediction of membrane proteins. Protein Sci. 5:363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, Y., F. Gurel, Y. Gafni, C. Dingwall, and V. Citovsky. 2000. A genetic system for detection of protein nuclear import and export. Nat. Biotechnol. 18:433–437. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve, S., C. Volland, C. Garnier, R. Jund, M.R. Chevallier, and R. Haguenauer-Tsapis. 1991. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol. Cell. Biol. 11:1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierhof, Y.-D., and H. Schwarz. 1991. Yield of immunolabel compared to resin sections and thawed cryosections. In Colloidal Gold. Principles, Methods, and Applications. vol. 3. M.A. Hayat, editor. Academic Press, London. 87–115.
- Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83–93. [DOI] [PubMed] [Google Scholar]
- Taddei, A., F. Hediger, F.R. Neumann, and S.M. Gasser. 2004. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 38:305–345. [DOI] [PubMed] [Google Scholar]
- Taddei, A., M.R. Gartenberg, F.R. Neumann, F. Hediger, and S.M. Gasser. 2005. Multiple pathways tether telomeres and silent chromatin at the nuclear periphery: functional implications for sir-mediated repression. Novartis Found Symp. 264:140–156; discussion 156–165, 227–230. [PubMed] [Google Scholar]
- Van Belle, D., and B. André. 2001. A genomic view of yeast membrane transporters. Curr. Opin. Cell Biol. 13:389–398. [DOI] [PubMed] [Google Scholar]
- Wang, X., R. Sato, M.S. Brown, X. Hua, and J.L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 77:53–62. [DOI] [PubMed] [Google Scholar]
- Wu, B., K. Ottow, P. Poulsen, R.F. Gaber, E. Albers, and M.C. Kielland-Brandt. 2006. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 173:327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.