Abstract
Specific spatial arrangements of proteins and lipids are central to the coordination of many biological processes. Tetraspanins have been proposed to laterally organize cellular membranes via specific associations with each other and with distinct integrins. Here, we reveal the presence of tetraspanin-enriched microdomains (TEMs) containing the tetraspanins CD9, CD63, CD81, and CD82 at the plasma membrane. Fluorescence and immunoelectron microscopic analyses document that the surface of HeLa cells is covered by several hundred TEMs, each extending over a few hundred nanometers and containing predominantly two or more tetraspanins. Further, we reveal that the human immunodeficiency virus type 1 (HIV-1) Gag protein, which directs viral assembly and release, accumulates at surface TEMs together with the HIV-1 envelope glycoprotein. TSG101 and VPS28, components of the mammalian ESCRT1 (endosomal sorting complex required for transport), which is part of the cellular extravesiculation machinery critical for HIV-1 budding, are also recruited to cell surface TEMs upon virus expression, suggesting that HIV-1 egress can be gated through these newly mapped microdomains.
Introduction
Tetraspanins are medium-sized (∼250 amino acids) membrane proteins that contain cytoplasmic NH2 and COOH termini and two extracellular domains separated from each other by a short inner loop. The mammalian family of these evolutionarily conserved proteins contains 32 members. Tetraspanins are expressed in a wide range of tissues and cell types, and members of this protein family have been implicated in regulating various biological functions, including antigen presentation, cell adhesion and migration, cell–cell fusion, cell activation, and proliferation (for reviews see Berditchevski, 2001; Vogt et al., 2002; Hemler, 2003; Stipp et al., 2003; Tarrant et al., 2003; Hemler, 2005).
Tetraspanins associate specifically with distinct integrins, various Ig superfamily members, and other tetraspanins, thus establishing a scaffold for various cellular functions. Numerous biochemical analyses and functional studies predicted the existence of tetraspanin-enriched microdomains (TEMs) that together form the so-called tetraspanin web (Charrin et al., 2003; Levy and Shoham, 2005). TEMs are thought to organize the plasma membrane and intracellular membranes, where some tetraspanins are primarily located, by selectively concentrating specific membrane proteins and membrane-peripheral signaling molecules. Such TEM-based concentration/exclusion of proteins involved, for example, in adhesion or in intracellular signaling is thought to dynamically segregate molecules, similar to how lipid rafts are proposed to laterally organize cellular membranes. Although some studies have documented colocalization of individual tetraspanins with various membrane receptors and costimulatory molecules, e.g., in adhesion complexes (Berditchevski et al., 1997; Berditchevski and Odintsova, 1999), the concept that different members of the tetraspanin family associate at membranes, thus forming distinct microdomains, is based largely on coimmunoprecipitation and protein cross-linking data. Neither the mean size of TEMs nor their overall distribution at the plasma membrane of these microdomains has been determined.
Human immunodeficiency virus type 1 (HIV-1), like other enveloped viruses, exits from cells by budding through membranes, a process that does not lead to disintegration of the cell. For its budding, HIV-1 uses the host cell machinery that is responsible for the formation of intralumenal vesicles in multivesicular bodies (MVBs), components of the endosomal compartment (for review see Morita and Sundquist, 2004). Nevertheless, HIV-1 primarily buds through the plasma membrane of T lymphocytes and other cell types. Only in macrophages is HIV-1 known to bud exclusively into MVBs. Viruses sequestered in these late endosomes (LEs) are thought to exit from macrophages upon fusion of the limiting membrane of MVBs with the plasma membrane (Raposo et al., 2002; Pelchen-Matthews et al., 2003).
HIV-1 produced in macrophages specifically incorporates the tetraspanin CD63, compatible with the finding that this antigen largely resides at the limiting membrane and on intralumenal vesicles of LEs/MVBs (Escola et al., 1998; Kobayashi et al., 2000; Gruenberg, 2001; Rous et al., 2002; Pelchen-Matthews et al., 2003). However, despite its low abundance at the plasma membrane of cells, CD63 is also specifically incorporated into HIV-1 particles produced in primary and transformed T lymphocytes and in other nonmacrophages where this virus buds mainly through the cell cortex (Orentas and Hildreth, 1993; Ott, 2002). Furthermore, we previously reported that we occasionally observed colocalization of HIV-1 Gag and CD63 at the periphery of T lymphocytes and melanocytes, though it was difficult to distinguish with certainty between the small fraction of CD63 associated with the plasma membrane and the vast majority of this antigen residing on intracellular membranes (Nydegger et al., 2003).
Here, we tested the hypothesis that the small fraction of CD63 that resides at the plasma membrane is concentrated at distinct microdomains, i.e., in TEMs, together with other members of the tetraspanin family. Fluorescence microscopy analysis of cells in which we selectively stained the surface fractions of tetraspanins combined with EM of immunolabeled sheets of plasma membrane allowed us to visualize TEMs containing the tetraspanins CD9, CD81, CD82, and CD63. Our data also document the size and distribution of these TEMs in membranes. Furthermore, the data presented in this paper suggest that these newly visualized plasma membrane microdomains can function as exit gateways for HIV-1.
Results
Identification of CD63-enriched microdomains situated at the plasma membrane
Although the tetraspanin CD63 is known to cycle via the cell surface, at steady state, the vast majority (>98%) of this antigen resides in perinuclear LEs/MVBs of HeLa cells (Fig. 1 A). The overall distribution of CD63 is thus identical to what is seen in melanocytes and T lymphocytes (Nydegger et al., 2003) and other cell types (Stinchcombe and Griffiths, 1999; Kobayashi et al., 2000). To examine specifically the cell surface distribution of CD63, we incubated HeLa cells with the antibody H5C6, whose epitope is located either in the small or in the large extracellular loop (EC1 or EC2, respectively) of this tetraspanin (Azorsa et al., 1991). After fixation and incubation with a secondary, fluorophore-conjugated antibody, cells were analyzed by widefield fluorescence deconvolution microscopy (Gerlich and Ellenberg, 2003). Such selective staining of surface CD63 in live cells revealed that the small fraction of this antigen that resides at the plasma membrane, rather than being diffusely distributed, clusters in discrete microdomains. To exclude the possibility that CD63 clustering was due to antibody cross-linking, cells were also stained after fixation in some experiments. Fig. 1 B visualizes that discrete CD63 assemblages exist at the cell surface before antibody binding. These microdomains are not restricted to the adhesion structures but are present in the entire cell periphery, as shown in Fig. 1 B (bottom). Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200508165/DC1) shows a 3D-rendering of a cell surface stained for CD63 and visualizes that clusters of this antigen are equally abundant in those areas of the cell surface that are not contacting other cells or the plastic of the tissue culture dish. Fig. 1 C documents that the plasma membrane of an individual cell contains several hundred CD63-enriched microdomains (mean number of domains per 25 μm2 = 18 ± 4).
Figure 1.
Identification of CD63-enriched microdomains at the surface of HeLa cells. HeLa cells were stained with an anti-CD63 antibody after fixation and permeabilization (A; a middle section is shown) or after fixation without permeabilization (B [middle and bottom sections are shown] and C [a bottom section is shown]), followed by incubation with a fluorophore-conjugated secondary antibody. The selective staining of surface CD63 (B and C) allows visualization of discrete microdomains where this antigen clusters at the plasma membrane. Bar, 10 μm. (C, right) Sevenfold-magnified view of the boxed region in the cell shown in the left panel. Bar, 1 μm.
Partial colocalization of the tetraspanins CD9, CD81, CD82, and CD63: visualization of surface TEMs
Through biochemical studies, CD63 is known to associate with other members of the tetraspanin family (Rubinstein et al., 1996; Tarrant et al., 2003). We were interested in investigating the localization of CD9, CD81, and CD82, three tetraspanins that were previously shown to coprecipitate with CD63 in preB cells (Rubinstein et al., 1996) and two of which (CD81 and CD82) were shown to associate with HIV-1 produced in MVBs of macrophages (Pelchen-Matthews et al., 2003). First, we sought to compare the overall distribution of these tetraspanins. Cells were incubated with the respective antibodies after permeabilization. Compared with CD63, the three other members of the tetraspanin family showed significantly higher surface expression (Fig. 1 A and Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200508165/DC1). Although some colocalization at intracellular membranes was observed for these tetraspanins (Fig. S1 B), at steady state, substantial fractions of those antigens apparently reside at different intracellular membranes.
Next, cells were stained for the plasma membrane fraction of the selected tetraspanin, as described in Fig. 1 B, and the relative colocalization of surface CD63 with CD9, CD81, or CD82 was assessed. Substantial colocalization of CD63 with any one of the three tetraspanins was observed (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200508165/DC1), suggesting that relatively large fractions of CD9 and CD82 reside in plasma membrane TEMs together with CD63, whereas a small fraction of total CD63 contributes to these surface microdomains. The concept that various tetraspanins contribute dissimilarly to the formation of TEMs is based on biochemical studies only (for review see Hemler, 2003). The data presented in Fig. 2 and Fig. S2 support that concept and provide an unprecedented visualization of bona fide surface TEMs.
Figure 2.
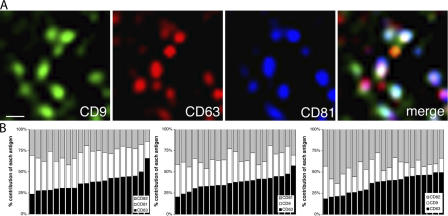
CD9, CD63, CD81, and CD82 cocluster in surface TEMs. To identify surface TEMs containing three of the tetraspanins analyzed here, HeLa cells were surface stained as described in Materials and methods. Insets show sixfold-magnified views of the boxed region in the cell. The percentages of TEMs containing one (30 ± 9), two (24 ± 7), or three (46 ± 14) of the tetraspanins visualized in these triple stainings were assessed as described in Materials and methods (n > 300). Bar, 10 μm.
Contribution of individual tetraspanins to the formation of surface TEMs
At the plasma membrane, CD63 typically colocalizes with one or more of the other three tetraspanins (Fig. S2). Also, CD9, CD81, and CD82 partially colocalize with each other (unpublished data). It follows that a subset of the TEMs visualized in Fig. S2 contains at least three of the analyzed tetraspanins. In Fig. 2, we demonstrate that this is indeed the case. A detailed visual inspection of the three triple stainings revealed that more than two thirds of TEMs contain at least two of the three analyzed tetraspanins and about half of them contain all three antigens.
Although biochemical analyses (i.e., coprecipitation experiments) allow determination of how much an individual tetraspanin contributes to an average TEM, such analyses of the bulk population do not permit determination of whether those relative contributions are uniform, i.e., whether tetraspanins accumulate at TEMs in defined ratios. The analysis of triple immunostainings, however, can provide such information. The magnifications in Fig. 2 (right) and Fig. 3 A visualize that the composition of TEMs at steady state varies widely, and the statistical analysis shown in Fig. 3 B documents that the tetraspanins analyzed in this study do not exist in TEMs in distinct, uniform ratios. Note that TEMs containing only one or two of the three analyzed tetraspanins were excluded from this quantification. TEM heterogeneity may thus be even larger than suggested by the graph shown in Fig. 3 B.
Figure 3.
Quantitative analysis of surface TEM composition. Cells were triple stained with anti-tetraspanin antibodies as described in the legend for Fig. 2, and the relative presence in surface TEMs of CD9, CD63, CD81, and CD82 was analyzed as described in Materials and methods. (A) A representative micrograph reveals that the relative contribution of the tetraspanins to the build-up of individual TEMs varies. (B) Statistical analyses confirming the stochastic nature of the tetraspanin contributions to TEM formation. Each bar represents a single TEM that showed staining for three of the above tetraspanins. The relative contribution of each tetraspanin was determined as described in Materials and methods. The bars were sorted in ascending order according to CD63 signal. Bar, 0.5 μm.
Ultrastructural analysis of surface TEMs
The overall presence of CD63 at the limiting membrane of MVB and on intraluminal vesicles of MVBs has been well documented previously by immuno-EM in various cell types (Escola et al., 1998; Pelchen-Matthews et al., 2003). We used immuno-EM to analyze CD63-enriched TEMs at the plasma membrane. To visualize the lateral arrangement of these domains with higher spatial resolution and to determine the approximate size of these domains, we used a technique that allows an en face view of the cytosolic face of the plasma membrane and that preserves submembrane structures such as clathrin lattices and cortical cytoskeleton (Sanan and Anderson, 1991; Foti et al., 1997). After incubation with the anti-CD63 antibody 1B5, HeLa cells, transfected or not with a CD63-YFP expressor plasmid, were allowed to sediment on poly-l-lysine–coated grids. Subsequently applied hypotonic shocks, followed by sonication, disrupted the cells and removed the cytoplasm. The plasma membrane, still adherent to the grid, was processed for visualization by EM. Fig. 4 A shows a representative micrograph. It confirms the fluorescence microscopy data (Figs. 1 and 2) by visualizing that CD63 expression at the plasma membrane is restricted to distinct small islands that are separated from each other by membrane that is virtually free of this antigen. The size and shape of the CD63-enriched domains, as well as the distance between these microdomains, vary considerably (mean area = 0.2 μm2; mean distance between CD63-enriched domains = 0.65 μm; see Table I). No significant differences were observed between CD63-enriched microdomains in cells expressing only the endogenous version of this tetraspanin (Fig. 4 A) and cells expressing also CD63-YFP (not depicted). Interestingly, many CD63 molecules appear to be situated along cytoskeletal elements and adjacent to clathrin lattices and/or clathrin-coated pits.
Figure 4.
Visualization of CD63- and CD9-enriched microdomains by immuno-EM. HeLa cells were incubated with either an anti-CD63 or an anti-CD9 antibody or a combination of both antibodies and processed as described in Materials and methods. Representative electron micrographs of tetraspanin immunogold labeling associated with discrete membrane microdomains on HeLa cell plasma membrane are shown for cells stained for CD63 (A), CD9 (B), or both tetraspanins (C and D). CD9- and CD63-associated gold particles (20 and 10 nm, respectively) are seen by transparency. CD63 clusters are encircled with dashed lines and are marked with arrows in D; CD9 clusters are encircled with dotted lines, and microdomains containing both antigens are encircled by dashed/dotted lines. Bars, 0.5 μm.
Table I.
Size and distribution of plasma membrane TEMs
| TEM size | Distance between individual TEMs |
|
|---|---|---|
| μm2 | μm | |
| Endogenous CD63 only | 0.19 ± 0.1 | 0.7 ± 0.3 |
| Cells expressing CD63-YFP | 0.22 ± 0.1 | 0.6 ± 0.4 |
Fields of plasma membrane sheets as shown in Fig. 4 A were photographed and analyzed for the presence of CD63 clusters, as described in Materials and methods (n > 50).
As shown in Fig. 2 by immunofluorescence, despite displaying an overall distribution that differs dramatically from the distribution of CD63, CD9 shows substantial colocalization with CD63 in surface TEMs. We thus sought to confirm the features of surface TEMs revealed in Fig. 4 A by analyzing CD9 clusters at the plasma membrane. Live cells were incubated with the anti-CD9 antibody KMC8.8, and CD9 localization was visualized by EM (Fig. 4 B) as described for CD63. CD9-containing TEMs display the same characteristics with regard to size and their 2D distribution within the plasma membrane as those enriched in CD63. Also, as expected given the fluorescence microscopy data displayed in Fig. 2 and Fig. S2, double labeling of cells with antibodies against CD9 and CD63 reveals microdomains containing one or both tetraspanins (Fig. 4, compare C and D, displaying separate CD9 and CD63 clusters and colocalizing tetraspanins, respectively). As for the analysis of TEM distance (see Materials and methods), the relative colocalization of the two antigens was quantified by positioning a grid consisting of squares with 200-nm length onto the images. CD9- and CD63-associated gold particles residing in the same or adjacent squares were scored as colocalizing. This analysis revealed that about a third (29.7 ± 4.7%; n = 20 micrographs) of the CD63-associated gold particles are situated proximal to CD9, in good agreement with the data shown in Fig. S2.
TEM positioning relative to HA-enriched microdomains
A recent ultrastructural study analyzed the surface distribution in HAb2 fibroblasts of the influenza viral envelope glycoprotein HA. HA, a membrane protein that is sometimes used as marker for fluid-ordered raft domains, was demonstrated to cluster at distinct plasma membrane microdomains if expressed independently or together with the other viral proteins (Takeda et al., 2003; Hess et al., 2005). To investigate the spatial relationship between HA and the tetraspanins CD9 and CD63, HeLa cells expressing HA were surface stained with the respective antibodies. Fig. 5 (left) documents the 2D distribution of HA at the plasma membrane of these cells. As in HAb2 cells, HA is restricted to discrete microdomains of variable size. Although the overall appearance (size and distribution) of HA clusters resembles that of CD9- and CD63-containing TEMs (Fig. 5, middle), very little colocalization (<10%) was detected between HA and either CD9 or CD63 (right).
Figure 5.
Distribution of surface TEMs relative to influenza virus HA-enriched microdomains. HeLa cells expressing HA were surface stained for HA and CD63 (A) or CD9 (B). Insets show sixfold-magnified views of the boxed region in the cell. Colocalization analysis (see Materials and methods) showed that 7.7 ± 5.3% of CD63 and 9.7 ± 5.8% of CD9 colocalized with HA. Bar, 10 μm.
HIV-1 Gag accumulates at surface TEMs
Despite its low abundance at the plasma membrane of cells, the LE marker CD63 is one of few cellular antigens that are specifically enriched in HIV-1 particles even if this virus is produced in cells where newly assembled particles bud primarily through the plasma membrane (Orentas and Hildreth, 1993; Gluschankof et al., 1997). We thus reasoned that HIV-1 Gag, the major structural protein of this virus, must assemble at areas of the plasma membrane that are enriched in CD63. The results shown in Fig. 6 B demonstrate that this is indeed the case. In HeLa cells transiently transfected with a plasmid carrying the HIV-1 provirus and giving rise to the expression of the complete set of viral proteins and subsequent particle release, a large fraction of viral Env at the plasma membrane colocalizes with surface CD63 (Fig. 6 A), supporting the hypothesis that HIV-1 Gag is specifically accumulating at these microdomains. We previously showed that, in a melanocyte cell line, about one third of viral Gag, the major viral structural protein, colocalizes with CD63 (Nydegger et al., 2003). However, unlike in the current analysis, in those studies we looked at total Gag and total CD63 and thus were visualizing primarily LE/MVB-hosted CD63. As documented in Fig. S3 (available at http://www.jcb.org/cgi/content/full/jcb.200508165/DC1), many of these CD63-enriched surface TEMs must also contain other tetraspanins because significant proportions of viral Gag also colocalize with the tetraspanins CD9, CD81, and CD82.
Figure 6.
HIV-1 Env and Gag colocalize with surface CD63. (A) HeLa cells expressing full-length HIV-1 were surface stained for CD63 and HIV-1 Env. (B) To visualize Gag, cells were subsequently permeabilized and treated with an anti-Gag antibody followed by incubation with a fluorophore-conjugated secondary antibody. Bottom sections and middle sections are shown for each cell. Bar, 10 μm.
To confirm that viral Gag is indeed colocalizing with CD9- or CD63-containing TEMs at the plasma membrane and not, for example, at tetraspanin-enriched endosomes or MVBs situated right beneath the plasma membrane, we used the same EM technique as in Fig. 4. To achieve high expression levels for the viral proteins, we cotransfected cells with Gag and Env expressor plasmids, which gives rise to the production of virus-like particles (VLPs). Fig. 7 confirms that tetraspanin surface expression is limited to distinct, often cytoskeleton-associated plasma membrane microdomains, as shown also in Fig. 4. Further, it documents that Gag can indeed associate with surface assemblages of either tetraspanin analyzed here. Colocalization analysis showed that 21.1 ± 5.0% of Gag colocalized with CD63-containing TEMs and 41.8 ± 4.8% of Gag colocalized with CD9-containing TEMs (n = 27/29 micrographs, respectively; TEMs were defined as in Table I).
Figure 7.
Immuno-EM analysis of HIV-1 Gag accumulation at surface TEMs containing CD9 or CD63. HeLa cells were incubated with either an anti-CD63 or an anti-CD9 antibody, and cells were treated as described for Fig. 4. CD9- and CD63-associated gold particles (10 nm) and Gag-associated gold particles (15 nm) are seen by transparency. Representative electron micrographs of HIV-1 Gag immunogold labeling associated with discrete plasma membrane microdomains are shown for cells stained for either CD63 or CD9. Bar, 0.2 mm.
Recruitment to surface TEMs of cellular proteins necessary for HIV-1 particle formation
To determine whether viral particle morphogenesis and egress are likely to take place at surface TEMs, we analyzed if TSG101 and VPS28, constituents of endosomal sorting complex required for transport (ESCRT1) and as such known to be essential components of the HIV-1 particle–forming machinery (Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001; Demirov et al., 2002; Martin-Serrano et al., 2003), are recruited to Gag-containing TEMs. Cells transfected with expressor plasmids for YFP-tagged TSG101 (TSG101-YFP) and CFP-tagged VPS28 (VPS28-CFP) and cotransfected with a HIV-1 Gag expressor plasmid, which leads to the formation and release VLPs even in the absence of Env expression, were analyzed for the localization of TSG101 and VPS28. A strong shift toward surface localization was immediately apparent for both ESCRT1 components (Fig. 8 A and Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200508165/DC1), similar to what was previously observed in 293T cells producing HIV-1 (Martin-Serrano et al., 2001). Upon closer examination, it became evident that TSG101 and VPS28 are recruited to small but distinct Gag-containing patches at the plasma membrane (Fig. 8 A). To evaluate whether these Gag-containing microdomains are surface TEMs, cells coexpressing HIV-1 Gag, TSG101-YFP, and FLAG-tagged VPS28 were double stained with anti-Gag and anti-CD63 antibodies and were analyzed for the localization of TSG101, Gag, and CD63. As demonstrated in Fig. 8 B, in cells expressing HIV-1 Gag and thus producing VLPs, a significant fraction of the ESCRT1 component TSG101 relocates to the surface, where it colocalizes with surface CD63, strongly suggesting that surface TEMs can serve as sites for the production and the release of viral particles. Finally, a similar recruitment to the plasma membrane and colocalization with viral Gag was found for VPS28, another ESCRT1 component required for HIV-1 budding (unpublished data), providing further evidence for the notion that surface TEMs can provide exit gateways for HIV-1.
Figure 8.
TSG101 and VPS28 are recruited to CD63-containing surface TEMs where Gag clusters. (A) HeLa cells expressing HIV-1 Gag, TSG101-YFP, and VPS28-CFP were fixed, permeabilized, and stained for Gag. (B) HeLa cells expressing HIV-1 Gag, TSG101-GFP, and FLAG-VPS28 were fixed, surface stained for CD63, permeabilized, and stained for Gag. Bottom sections are shown for each cell. Blow-ups show 11-fold–magnified views of the boxed region in the cell. Bar, 10 μm.
Association of individual viral components and TEMs
The data shown in Figs. 7 and 8 suggest that a considerable fraction of HIV-1 Gag, which can form VLPs if expressed in the absence of other viral components, is targeted to CD63-containing surface TEMs. However, as previously noted (Nydegger et al., 2003), Gag's colocalization with membranes carrying the late endosomal markers major histocompatibility antigen type II and/or CD63 is increased if it is expressed in the context of a full-length virus. We thus sought to quantify the colocalization of Gag and Env with tetraspanins if the viral proteins were expressed individually or together. Fig. 9 and Fig. S5 (available at http://www.jcb.org/cgi/content/full/jcb.200508165/DC1) document that very little Env and relatively modest amounts of Gag colocalize with CD9- and CD63-containing surface TEMs if either of the viral antigens is expressed alone. Colocalization of viral Gag with the two tetraspanins is barely augmented in cells that coexpress viral Env, whereas clearly more Env colocalizes with the two tetraspanins in cells cotransfected with plasmids carrying the gag gene. Note that TEM association of either viral component is most pronounced if Env and Gag are expressed from full-length provirus. Importantly, and in further support of the hypothesis that TEMs can function as egress sites, we found that if we restricted our analysis to the subpopulation of Gag that colocalizes with Env, an even higher fraction of Gag was found to accumulate at surface TEMs (almost 50% colocalization with CD63-containing TEMs and almost 70% colocalization with CD9-containing TEMs).
Figure 9.
Quantitative analysis of viral Env and Gag colocalization with tetraspanins. HeLa cells expressing HIV Gag and Env individually, together but expressed from different plasmids, or coexpressed from a proviral vector were incubated with anti-Env and anti-CD63 (A) or anti-CD9 (B) antibodies without permeabilization. After fixation, the cells were permeabilized and incubated with an anti-Gag antibody and with fluorophore-conjugated secondary antibodies. Fluorescent images were captured and analyzed for colocalization (see Materials and methods). At least 10 cells were analyzed for each condition. The bar diagrams summarize the data from that analysis. The percentage of Gag or Env that colocalizedwith either CD63 or CD9 is shown. White bars show Gag or Env colocalization with CD63 or CD9 if the viral antigens were expressed separately, gray bars show Gag or Env colocalization with CD63 or CD9 if the viral antigens were coexpressed from separate plasmids, and black bars show Gag or Env colocalization with CD63 or CD9 if the viral antigens were coexpressed from a proviral plasmid. Error bars indicate SD.
HIV-1 Env and Gag are sorted to surface TEMs in T lymphocytes
Though nonlymphocytes such as HeLa and 293T cells are model systems in which many of the cellular components of the HIV-1 assembly/release functions were identified and characterized, it was necessary to determine whether viral release is routed though surface TEMs in T lymphocytes. However, as in HeLa cells, the overwhelming majority of CD63 at steady state resides in LEs/MVBs of Jurkat T lymphocytes (Nydegger et al., 2003). Live, nonpermeabilized cells thus had to be stained with anti-CD63 antibodies to visualize the distribution of this antigen in the plasma membrane. Fig. 10 A (left) documents that CD63 clusters in plasma membrane TEMs that display sizes and overall distribution comparable to those analyzed in HeLa cells (Fig. 1). Also, as in HeLa cells, there is substantial colocalization of surface CD63 and CD9. Most important, Fig. 10 (B and C) demonstrates that virtually all HIV-1 Env expressed at the cell surface in the context of the full-length virus colocalizes with surface CD9 and surface CD63, signifying that plasma membrane TEMs can serve as exit gateways for HIV-1 in T lymphocytes. Judging by Env localization, both nonpolarized and polarized viral release was observed, the latter probably being triggered by cell–cell contact (Phillips, 1994; Jolly et al., 2004).
Figure 10.
HIV-1 at surface TEMs in Jurkat T lymphocytes. Jurkat T lymphocytes were surface stained for CD9 and CD63 (A). Surface TEMs containing CD9 and CD63 are comparable in size, and their distribution is similar to the one in HeLa cells (Fig. 2). (B and C) Jurkat T lymphocytes transfected with HIV-1 provirus–carrying plasmids and thus producing infectious virus were surface stained for Env and CD63 or CD9 to visualize virus and surface TEMs, respectively. Bar, 5 μm.
Discussion
Despite its very low abundance at the cell surface, the tetraspanin CD63 is enriched in HIV-1 particles produced in cells where this virus buds primarily through the plasma membrane. This prompted us to investigate whether CD63 is concentrated at those sites. We demonstrate that the surface fraction of CD63, together with other tetraspanins, clusters at discrete sites, thus forming surface TEMs. Besides providing a map of these surface TEMs, we also show that the structural components of HIV-1, together with elements of the host cell machinery responsible for viral budding, are recruited to precisely these microdomains. Thus, TEMs likely function as exit gateways for HIV-1.
Various biochemical and functional studies have linked specific cellular processes to the association of distinct proteins and tetraspanins. However, few studies have also analyzed the localization of tetraspanins involved in these processes, and physical aspects of TEMs, particularly size and distribution of these microdomains, have remained enigmatic. We hypothesized that CD63-enriched microdomains exist at the cell surface despite the fact that this tetraspanin is a resident primarily of LEs/MVBs. Fig. 1 documents for HeLa cells that this is indeed the case. These surface microdomains at first sight appear morphologically featureless; i.e., they are not restricted to pseudopods, for example, but they are distributed over the entire plasma membrane. The same holds for the other three tetraspanins analyzed in this study. Not only do they colocalize to a significant extent with CD63, surface TEMs containing these other tetraspanins are also spread over the entire cell surface and are not apparently engaged in any adhesion/migration-related functions (Fig. 2), though, as noted below, they clearly appear to be enriched in microvilli.
Size and 2D distribution of plasma membrane TEMs
To confirm the distribution pattern visualized by fluorescence microscopy and to get information about the size of surface TEMs, we used an immuno-EM technique that we had applied before and that allowed us to look perpendicularly with respect to the plane of the plasma membrane (Sanan and Anderson, 1991; Foti et al., 1997). Although the majority of the plasma membrane appeared devoid of CD63 and CD9, distinct clusters of these two antigens can be found dispersed over the entire cell surface. As integral membrane proteins, CD9 and CD63 arrive at the cell surface on vesicles that are inserted into the plasma membrane. The finding that all the TEMs analyzed in this study (Figs. 1–4 ) are separated from each other by areas that are virtually free of these antigens suggests that the four tetraspanins, rather than diffusing in the 2D plane, stay clustered in small areas that cover only a relatively small fraction of the membrane. Even though TEMs are immediately recognizable as distinct microdomains, size, shape, and the relative presence of the four tetraspanins in these domains varies substantially, as is evident in Figs. 2–4 and 7. Whether the larger domains visualized here represent clusters of smaller base units or whether TEMs grow continuously until they reach a maximal size remains to be analyzed. Finally, though these domains appear uniformly distributed within the plasma membrane, our ultrastructural analysis of plasma membrane sheets (Figs. 4 and 7) reveals that TEMs form preferentially at or adjacent to clathrin-coated areas and next to elements of the cytoskeleton. The former feature clearly sets them apart from HA-enriched microdomains, which are situated in membrane areas free of clathrin-coated pits (Hess et al., 2005).
TEM biogenesis
Recent studies in rodent cells have suggested that CD63, upon synthesis, is sorted directly to LEs/MVBs, where most of this antigen resides (Rous et al., 2002; Ihrke et al., 2004), though some of the newly synthesized protein may be sorted via the plasma membrane. Fluorescence microscopy and ultrastructural analyses of secretory lysosomes and MVBs, respectively (Murk et al., 2003; Jaiswal et al., 2004), suggested that proteins are not uniformly distributed within the limiting membrane of these LEs/MVBs. Data presented in one of these studies (Jaiswal et al., 2004) are compatible with the possibility that CD63 is clustered within distinct microdomains of the limiting membrane. It would thus be conceivable that TEMs nucleate in the limiting membrane of endosomes/secretory lysosomes and become part of the plasma membrane after these compartments have moved to the cell surface and the limiting membrane has been inserted into the plasma membrane. Alternatively, significant fractions of newly synthesized LE/lysosomal markers lamp1, lamp2, and CD63 appear to be sorted via the plasma membrane before reaching their final destination in HeLa cells (Janvier and Bonifacino, 2005). Insertion of post-Golgi vesicles into the plasma membrane may thus contribute to the formation of surface TEMs.
TEMs roles in viral egress
Two lines of evidence in our current study suggest that surface TEMs can serve as exit gateways for HIV-1. First, most cell surface punctae where either Gag or Env clusters in both HeLa cell and in Jurkat T lymphocytes are also occupied by one of the tetraspanins (Figs. 6–8 and 10). Second, cellular TSG101 and VPS28, the components of the cellular budding machinery responsible for viral egress (Morita and Sundquist, 2004), when recruited to the plasma membrane in cells producing HIV-1, accumulate at CD63-containing TEMs (Fig. 8). Furthermore, we find that distortion of TEM distribution in virus-producing cells by an anti-CD9 antibody (K41) correlates with inhibition of HIV-1 release (unpublished data).
In a study where we used the FlAsH technique for successive dual-color labeling (Gaietta et al., 2002) of Gag in various virus-secreting cell types, we observed localization of newly synthesized Gag at distinct areas on the plasma membrane (Rudner et al., 2005). We also documented that viral budding structures and CD63 colocalize at MVBs and at the surface of HeLa cells (Nydegger et al., 2003). Based on this latter finding and on the relatively large size (1–4 μm) of patches at the plasma membrane through which the virus appeared to bud in this preceding, lower resolution study, we hypothesized that the microdomains through which HIV-1 buds are derivatives of LE/MVB limiting membrane (see above). However, the results presented in Figs. 6–8 indicate that the large patches observed in our previous studies may indeed have been clusters of smaller, possibly post-Golgi vesicle-derived TEMs that form supraclusters in cells producing virus (Figs. 6 and 8 and Fig. S3).
Although it is well documented that HIV-1 exit from cells is restricted to distinct areas of the cell cortex, it remained unclear which of the viral components is the primary determinant that targets release to these sites. A fluorescence microscopy analysis demonstrated that Gag recruits Env to distinct areas at the plasma membrane (Hermida-Matsumoto and Resh, 2000). Consistent with this paper, we show that Gag has intrinsic TEM targeting information as it colocalizes with TEMs if expressed alone (Fig. 9) and thus codetermines where HIV-1 buds. Nevertheless, Gag colocalizes more extensively with surface tetraspanins if Env is coexpressed (Figs. 6 and 9 and Fig. S5), and Env thus plays at least a modulatory role in determining where HIV-1 exits from cells.
Biochemical studies (Rousso et al., 2000; Lindwasser and Resh, 2001; Ono and Freed, 2001) and a fluorescence microscopy analysis in T lymphocytes, where HIV-1 was shown to exit at the same pole where the raft marker GM1 accumulates (Nguyen and Hildreth, 2000), suggested that lipid rafts or barges are critical for HIV-1 exit. On the other hand, it was also reported that Gag complexes, defined by flotation studies, do not incorporate classical lipid raft markers and were not disrupted by cholesterol extraction (Ding et al., 2003). Further, as reviewed recently elsewhere, rafts probably have many different properties, and these lipid-based microdomains remain difficult to define (Edidin, 2003; Mukherjee and Maxfield, 2004). Nevertheless, the two hypotheses, i.e., that rafts are important during HIV-1 assembly and that TEMs serve as viral exit gateways, are not mutually exclusive. Rafts, for example, could potentially form and/or coalesce during viral budding at TEMs. However, as recently noted (Yang et al., 2004), though tetraspanins are palmitoylated, a proteomics analysis of lipid rafts did not reveal any tetraspanins or integrins, nor were typical raft proteins found in a mass spectrometry analysis to be partners for tetraspanins and palmitoylated integrins (Foster et al., 2003; Yang et al., 2004). Also, HA-containing rafts (Hess et al., 2005) for the most part do not colocalize with TEMs containing CD9 and/or CD63 in HeLa cells, as documented in Fig. 5.
Although this study provides evidence for the concept that surface TEMs can serve as HIV-1 exit sites, we are currently investigating whether the viral components can also be endocytosed via these domains. The concept that viral components may be sequestered in the endosomal compartment to be mobilized for virion formation later, e.g., upon attachment to a target cell, was first discussed by Rodriguez-Boulan et al. (1983). The positioning of TEMs adjacent to clathrin-coated areas (Figs. 4 and 7), together with various reports about the itineraries of CD63 as well as HIV-1 Gag and Env (Blot et al., 2003; Lindwasser and Resh, 2004; Murray et al., 2005), are compatible with such a scenario.
In summary, data presented in this paper provide a map of surface TEMs and demonstrate that these domains constitute sites at which HIV-1 proteins gather for potential subsequent particle exit. TEMs have been implicated in coordinating functions at the cell–cell interface. It is thus conceivable that HIV-1 evolved to egress through these microdomains to ensure efficient release toward adjacent target cells.
Materials and methods
Cell culture, plasmids, and transfections
HeLa and Jurkat cells were grown in DME or RPMI (Invitrogen), respectively, supplemented with 10% FBS (Invitrogen). Transfections were performed using Lipofectamine 2000 (Invitrogen) or by electroporation (Bio-Rad Laboratories) according to the manufacturers' protocol.
The following plasmids were used: pNL4-3, pGagopt (a gift from D. Ott, National Cancer Institute at Frederick, Frederick, MD), pSRalpha-Env, pVPS28-CFP, pTSG101-YFP (a gift from P. Bienasz, The Rockefeller University, New York, NY), pTSG101-GFP, pFLAG-VPS28 (a gift from U. von Schwedler and W. Sundquist, University of Utah, Salt Lake City, UT), pCD82-YFP (a gift from W. Mothes, Yale University School of Medicine, New Haven, CT), and pCD63-YFP (a gift from J. Bonafacino, National Institutes of Health, Bethesda, MD). Analysis of cells was performed 24 h after transfection with pGagopt or pSRalpha-Env alone or when cotransfected with pGagopt, pSRalpha-Env, pVPS28-CFP, pFLAG-VPS28, or pTSG101-GFP/YFP, and after 30–48 h when transfected with pNL4-3.
Immunocytochemistry and fixed cell imaging analysis by fluorescence microscopy
Adherent cells were plated in chambered coverglasses (Lab-Tek; Nunc) or in 35-mm glass-bottomed culture dishes (MatTek). Transfection and staining were performed directly in these chambers.
The following antibodies were used: anti-CD9 (K41; BMA Biomedicals AG); anti-CD63 (H5C6; Developmental Studies Hybridoma Bank); anti-CD63 clone 1B5 (a gift from J. Gruenberg [University of Geneva, Geneva, Switzerland] and M. Marsh [University College London, London, UK]); anti-CD81 (JS-81; BD Biosciences); anti-CD82 (B-L2; Diaclone); anti-Env (B12; NIH AIDS Research & Reference Reagent Program); anti-p6 (AIDS Vaccine Program, National Cancer Institute); anti-VPS28 serum; anti-HA rabbit serum (gift from M. Roth, The University of Texas Southwestern Medical Center, Dallas, TX); anti-mouse Alexa Fluor 488, 594, and 647; anti-rabbit Alexa Fluor 488, 594, and 647; and anti-human Alexa Fluor 488 and 594 (Invitrogen).
The slides were examined on a Delta Vision (DV) Workstation (DV base 3/3.5; epifluorescence microscope [Eclipse TE200; Nikon] fitted with an automated stage [Applied Precision, Inc.]) using a Plan Apo 60× objective (Nikon), 1.40 NA. Images were captured in z series with a charge-coupled device digital camera (CH350E or CoolSnap HQ [Roper Scientific]) using the DV Softworx software. Out-of-focus light was digitally removed using Softworx deconvolution software (Applied Precision Inc.), using the standard DV iterative deconvolution algorithm. Levels on each channel were adjusted in the DV Softworx software, and z sections of interest were exported as Tiff files. Tiffs were assembled into figures using Photoshop (Adobe).
Quantification of colocalization was computed using the Classifier Module of the Volocity Software 3.1 (Improvision). The bottom sections of representative cells were analyzed. Fluorescence thresholds for individual channels were set to two times the standard deviation of the signal, and the fluorescence intensity of each pixel was measured. Colocalization percentages were calculated by dividing the total number of pixels with intensity above the threshold that overlapped between two channels by the total number of pixels with intensity above the threshold for a given channel.
The relative contributions of tetraspanins to the formation of TEMs was assessed by selecting fluorescence thresholds for each channel followed by counting the individual clusters of each tetraspanin to gauge the presence or absence of one, two, or all three channels in a given cluster. The relative contributions of tetraspanins to the formation of triple TEMs (Fig. 3) was calculated by selecting TEMs containing signal for three tetraspanins and quantifying the relative intensity and area of each channel in a given TEM.
Overall staining
Cells were washed with PBS and fixed with 3.7% paraformaldehyde (Electron Microscopy Sciences) for 10 min at room temperature. After washing with PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min, washed again, and blocked with 1% BSA in PBS for 10 min, followed by a 1-h incubation with primary antibody at 37°C. After extensive washing with PBS, cells were blocked again with 1% BSA in PBS for 10 min and then incubated with the appropriate fluorophore-conjugated secondary antibody for 30 min. After extensive washing with PBS, cells were overlaid with 1% BSA in PBS and examined by fluorescence microscopy.
In case multiple antibodies of the same species were used, the antibodies were conjugated with a fluorophore using the Zenon technology according to the manufacturer's protocol (Invitrogen) before incubation with cells. Also, we routinely incubate cells with no, or only one (in dual stainings) or two (in triple stainings), primary antibodies, to control for nonspecific binding of the secondary antibodies.
Surface staining
Cells were washed with PBS and fixed with 3.7% paraformaldehyde for 10 min. Without permeabilization, cells were incubated for 1 h with primary antibody at 37°C in 1% BSA in PBS. Alternatively, live cells were precooled on ice and incubated for 1 h on ice at 4°C with primary antibody in complete medium supplemented with 1% Ca2+. Cells were then washed extensively with PBS and fixed for 10 min with 3.7% paraformaldehyde. After washing with PBS, cells were blocked for 10 min with 1% BSA in PBS and incubated with the appropriate fluorophore-conjugated secondary antibody for 30 min at room temperature. After extensive washing with PBS, cells were overlaid with 1% BSA in PBS and examined by fluorescence microscopy.
If two different surface stainings were performed on the same cells, both primary antibodies were added at the same time, followed by fluorophore-conjugated secondary antibody incubation. In case surface staining was followed by an overall staining, cells were permeabilized subsequently and treated with primary antibody followed by fluorophore-conjugated secondary antibody as described above.
Surface staining of Jurkat T lymphocytes
Jurkat cells were washed with PBS and incubated in suspension for 1 h on ice at 4°C with primary antibody in complete medium supplemented with 1% Ca2+. The cells were then washed with PBS and fixed with 3.7% paraformaldehyde for 10 min. The cells were washed, blocked for 10 min in 1% BSA in PBS, and transferred to chambered coverglasses precoated with CellTak (BD Biosciences) and incubated with the secondary antibodies at 37°C, simultaneously immobilizing the cells on the coverglass. In case multiple antibodies of the same species were used, the antibodies were conjugated with a fluorophore using the Zenon technology, and the stainings were done in series.
EM analysis
CD63- or CD9-enriched microdomains were visualized at the EM ultrastructural level on the inner face of the plasma membrane of HeLa cells as previously described (Sanan and Anderson, 1991; Foti et al., 1997). In brief, HeLa cells were incubated for 1 h at 4°C with either anti-CD63 antibodies (clone 1B5 of mouse anti-CD63 IgG2b [Fraile-Ramos et al., 2001] and antibody H5C6) or anti-CD9 antibodies (clone KMC8.8 of rat anti-CD9 [Santa Cruz Biotechnotolgy, Inc.] or K41 [Bachem]) in cold 1% BSA in PBS. After antibody binding, cells were washed twice in cold PBS to remove excess antibody and incubated with secondary species-specific gold-conjugated antibodies (10- or 20-nm colloidal gold) for 1 h at 4°C. Unbound secondary antibody was removed by washing twice with cold PBS. Cells were detached from the Petri dishes and allowed to sediment on formvar-coated EM nickel grids previously coated with poly-l-lysine. Adherent isolated plasma membranes were obtained by incubating the cells attached to the grids with hypotonic (0.65×) PBS for 30 s and then sonicating the cells at a weak power. This procedure disrupts the cells but allows a large portion of plasma membranes with conserved internal structures, such as clathrin-coated membranes and cytoskeleton elements, to stay adherent to the poly-l-lysine–coated grids. Adherent membranes were next washed with cold PBS and prefixed for 15 min at 4°C with 1% N- (3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride/0.2% glutaraldehyde, followed by 15 min at room temperature with 4% glutaraldehyde in PBS. Adherent membranes were subsequently fixed in 2% osmium tetroxide in PBS for 8 min and 30 s at room temperature, washed three times for 5 min with PBS, incubated with 1% aqueous tannic acid for 10 min, washed twice for 5 min in distilled water, incubated with 1% uranyl acetate for 10 min, and washed twice for 1 min with distilled water before air-drying. For CD63–CD9 double-labeling experiments, CD63 gold labeling was performed as described above, but CD9 was localized using a rabbit polyclonal anti-CD9 antibody (clone H-110; Santa Cruz Biotechnology, Inc.). Membranes considered well conserved were then photographed on an electron microscope (CM10; Philips). The mean size of CD63-enriched microdomains, as well as the mean distance separating these domains, was determined by analyzing 23 EM micrographs illustrating >50 well-delimited CD63-enriched microdomains. For this analysis, a grid consisting of squares with 200-nm length was positioned onto the images, and the numbers of gold particles per square were evaluated. Adjacent squares containing a sum of at least five particles were considered one TEM, and the mean size of these domains was calculated. The mean distance between TEMs was calculated by measuring the distances between the outermost squares defining the individual TEMs. To quantify either colocalization of CD63 and CD9 (Fig. 4) or Gag and tetraspanins (Fig. 7), the numbers of tetraspanin-associated gold particles situated either within the same square or in adjacent squares were determined (Fig. 4), or the numbers of Gag-associated gold particles overlapping with or directly adjacent to TEMs (as defined above) were determined. The percentages ± SEM are listed in the Results section.
For immunogold labeling of ultrathin cryosections, cells were detached and fixed for 1 h in phosphate buffer (100 mM NaPO4, pH 7.4) containing 4% paraformaldehyde (EMS) and 0.1% glutaraldehyde (EMS). Thereafter, the fixative was rinsed out three times with phosphate buffer and the cells were processed for cryosectioning as described previously (Liou et al., 1996). In brief, the cell pellet was infiltrated with sucrose and frozen in liquid nitrogen. Frozen sections (45-nm thickness) were cut with a cryotome (FCS; Leica), transferred to grids, and incubated with antibodies against CD63 (clone 1B5 of mouse anti-CD63 IgG2b). Grids were examined with a transmission electron microscope (Tecnai G-12; FEI Company).
Online supplemental material
Fig. S1 shows the overall distribution of the tetraspanins CD9, CD81, CD82, and CD63 when the cells were stained for only one of the antigens or dual labeled for two tetraspanins. Fig. S2 identifies surface TEMs containing two tetraspanins. Fig. S3 documents accumulation of viral Gag at plasma membrane TEMs containing CD9, CD81, or CD82. Fig. S4 demonstrates that TSG101 and VPS28, rather than accumulating at surface TEMs, are concentrated at intracellular compartments throughout the cell in the absence of viral Gag expression. Fig. S5 provides an immunofluorescence analysis of viral Env and Gag colocalization with tetraspanins representative of some of the data shown in Fig. 9. Video 1 shows untransfected HeLa cells that were surface stained for CD63, as described in Fig. 1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200508165/DC1.
Supplementary Material
Acknowledgments
We thank Geneviève Porcheron-Berthet for excellent technical assistance in the EM analysis, Marc Johnson and Volker Vogt for sharing and discussing unpublished data, and the Pôle Facultaire de Microscopie Électronique at the Geneva Medical Faculty for access to electron microscope and ancillary equipments. Alan Howe and members of the Thali laboratory are acknowledged for discussions and for critical reading of the manuscript.
This work was supported by National Institutes of Health grants R01 AI 47727 and R03 AI 060679 to M. Thali and Swiss National Science Foundation grant 3100A0-104489/1 to M. Foti.
Note added in proof. A recent article (Booth, A.M., Y. Fang, J.K. Fallon, J.-M. Yang, J.E.K. Hildreth, and S.J. Gould. 2006. J Cell Biol. 172:923–935) showed data compatible with the hypothesis that HIV-1 can exit through TEMs.
S. Nydegger and S. Khurana contributed equally to this paper.
Abbreviations used in this paper: ESCRT, endosomal sorting complex required for transport; HIV-1, human immunodeficiency virus type 1; LE, late endosome; MVB, multivesicular body; TEM, tetraspanin-enriched microdomain; VLP, virus-like particle.
References
- Azorsa, D.O., J.A. Hyman, and J.E. Hildreth. 1991. CD63/Pltgp40: a platelet activation antigen identical to the stage-specific, melanoma-associated antigen ME491. Blood. 78:280–284. [PubMed] [Google Scholar]
- Berditchevski, F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114:4143–4151. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., and E. Odintsova. 1999. Characterization of integrin-tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J. Cell Biol. 146:477–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berditchevski, F., K.F. Tolias, K. Wong, C.L. Carpenter, and M.E. Hemler. 1997. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272:2595–2598. [DOI] [PubMed] [Google Scholar]
- Blot, G., K. Janvier, S. Le Panse, R. Benarous, and C. Berlioz-Torrent. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for Env incorporation into virions and infectivity. J. Virol. 77:6931–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin, S., S. Manie, M. Billard, L. Ashman, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304:107–112. [DOI] [PubMed] [Google Scholar]
- Demirov, D.G., A. Ono, J.M. Orenstein, and E.O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA. 99:955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., A. Derdowski, J.J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edidin, M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257–283. [DOI] [PubMed] [Google Scholar]
- Escola, J.M., M.J. Kleijmeer, W. Stoorvogel, J.M. Griffith, O. Yoshie, and H.J. Geuze. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273:20121–20127. [DOI] [PubMed] [Google Scholar]
- Foster, L.J., C.L. De Hoog, and M. Mann. 2003. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. USA. 100:5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti, M., A. Mangasarian, V. Piguet, D.P. Lew, K.H. Krause, D. Trono, and J.L. Carpentier. 1997. Nef-mediated clathrin-coated pit formation. J. Cell Biol. 139:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile-Ramos, A., T.N. Kledal, A. Pelchen-Matthews, K. Bowers, T.W. Schwartz, and M. Marsh. 2001. The human cytomegalovirus US28 protein is located in endocytic vesicles and undergoes constitutive endocytosis and recycling. Mol. Biol. Cell. 12:1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta, G., T.J. Deerinck, S.R. Adams, J. Bouwer, O. Tour, D.W. Laird, G.E. Sosinsky, R.Y. Tsien, and M.H. Ellisman. 2002. Multicolor and electron microscopic imaging of connexin trafficking. Science. 296:503–507. [DOI] [PubMed] [Google Scholar]
- Garrus, J.E., U.K. von Schwedler, O.W. Pornillos, S.G. Morham, K.H. Zavitz, H.E. Wang, D.A. Wettstein, K.M. Stray, M. Cote, R.L. Rich, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 107:55–65. [DOI] [PubMed] [Google Scholar]
- Gerlich, D., and J. Ellenberg. 2003. 4D imaging to assay complex dynamics in live specimens. Nat. Cell Biol. 5(Suppl.):S14–S19. [PubMed] [Google Scholar]
- Gluschankof, P., I. Mondor, H.R. Gelderblom, and Q.J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology. 230:125–133. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. 2001. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Biol. 2:721–730. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397–422. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6:801–811. [DOI] [PubMed] [Google Scholar]
- Hermida-Matsumoto, L., and M.D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, S.T., M. Kumar, A. Verma, J. Farrington, A. Kenworthy, and J. Zimmerberg. 2005. Quantitative electron microscopy and fluorescence spectroscopy of the membrane distribution of influenza hemagglutinin. J. Cell Biol. 169:965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrke, G., A. Kyttala, M.R. Russell, B.A. Rous, and J.P. Luzio. 2004. Differential use of two AP-3-mediated pathways by lysosomal membrane proteins. Traffic. 5:946–962. [DOI] [PubMed] [Google Scholar]
- Jaiswal, J.K., S. Chakrabarti, N.W. Andrews, and S.M. Simon. 2004. Synaptotagmin VII restricts fusion pore expansion during lysosomal exocytosis. PLoS Biol. 2:E233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier, K., and J.S. Bonifacino. 2005. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell. 16:4231–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly, C., K. Kashefi, M. Hollinshead, and Q.J. Sattentau. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., U.M. Vischer, C. Rosnoblet, C. Lebrand, M. Lindsay, R.G. Parton, E.K. Kruithof, and J. Gruenberg. 2000. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol. Biol. Cell. 11:1829–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5:136–148. [DOI] [PubMed] [Google Scholar]
- Lindwasser, O.W., and M.D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser, O.W., and M.D. Resh. 2004. Human immunodeficiency virus type 1 Gag contains a dileucine-like motif that regulates association with multivesicular bodies. J. Virol. 78:6013–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, W., H.J. Geuze, and J.W. Slot. 1996. Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 106:41–58. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano, J., T. Zang, and P.D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano, J., T. Zang, and P.D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, E., and W.I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S., and F.R. Maxfield. 2004. Membrane domains. Annu. Rev. Cell Dev. Biol. 20:839–866. [DOI] [PubMed] [Google Scholar]
- Murk, J.L., B.M. Humbel, U. Ziese, J.M. Griffith, G. Posthuma, J.W. Slot, A.J. Koster, A.J. Verkleij, H.J. Geuze, and M.J. Kleijmeer. 2003. Endosomal compartmentalization in three dimensions: implications for membrane fusion. Proc. Natl. Acad. Sci. USA. 100:13332–13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J.L., M. Mavrakis, N.J. McDonald, M. Yilla, J. Sheng, W.J. Bellini, L. Zhao, J.M. Le Doux, M.W. Shaw, C.C. Luo, et al. 2005. Rab9 GTPase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus. J. Virol. 79:11742–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, D.H., and J.E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 Egress is gated through late endosomal membranes. Traffic. 4:902–910. [DOI] [PubMed] [Google Scholar]
- Ono, A., and E.O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA. 98:13925–13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orentas, R.J., and J.E. Hildreth. 1993. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res. Hum. Retroviruses. 9:1157–1165. [DOI] [PubMed] [Google Scholar]
- Ott, D.E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12:359–374. [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, D.M. 1994. The role of cell-to-cell transmission in HIV infection. AIDS. 8:719–731. [DOI] [PubMed] [Google Scholar]
- Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 3:718–729. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan, E., K.T. Paskiet, and D.D. Sabatini. 1983. Assembly of enveloped viruses in Madin-Darby canine kidney cells: polarized budding from single attached cells and from clusters of cells in suspension. J. Cell Biol. 96:866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rous, B.A., B.J. Reaves, G. Ihrke, J.A. Briggs, S.R. Gray, D.J. Stephens, G. Banting, and J.P. Luzio. 2002. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell. 13:1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso, I., M.B. Mixon, B.K. Chen, and P.S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA. 97:13523–13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, E., F. Le Naour, C. Lagaudriere-Gesbert, M. Billard, H. Conjeaud, and C. Boucheix. 1996. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur. J. Immunol. 26:2657–2665. [DOI] [PubMed] [Google Scholar]
- Rudner, L., S. Nydegger, L.V. Coren, K. Nagashima, M. Thali, and D.E. Ott. 2005. Dynamic fluorescent imaging of human immunodeficiency virus type 1 Gag in live cells by biarsenical labeling. J. Virol. 79:4055–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanan, D.A., and R.G. Anderson. 1991. Simultaneous visualization of LDL receptor distribution and clathrin lattices on membranes torn from the upper surface of cultured cells. J. Histochem. Cytochem. 39:1017–1024. [DOI] [PubMed] [Google Scholar]
- Stinchcombe, J.C., and G.M. Griffiths. 1999. Regulated secretion from hemopoietic cells. J. Cell Biol. 147:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp, C.S., T.V. Kolesnikova, and M.E. Hemler. 2003. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28:106–112. [DOI] [PubMed] [Google Scholar]
- Takeda, M., G.P. Leser, C.J. Russell, and R.A. Lamb. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA. 100:14610–14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant, J.M., L. Robb, A.B. van Spriel, and M.D. Wright. 2003. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 24:610–617. [DOI] [PubMed] [Google Scholar]
- VerPlank, L., F. Bouamr, T.J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C.A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA. 98:7724–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, A.B., S. Spindeldreher, and H. Kropshofer. 2002. Clustering of MHC-peptide complexes prior to their engagement in the immunological synapse: lipid raft and tetraspan microdomains. Immunol. Rev. 189:136–151. [DOI] [PubMed] [Google Scholar]
- Yang, X., O.V. Kovalenko, W. Tang, C. Claas, C.S. Stipp, and M.E. Hemler. 2004. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J. Cell Biol. 167:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.