Abstract
Misfolded proteins in the endoplasmic reticulum (ER) are retained in the organelle or retrotranslocated to the cytosol for proteasomal degradation. ER chaperones that guide these opposing processes are largely unknown. We developed a semipermeabilized cell system to study the retrotranslocation of cholera toxin (CT), a toxic agent that crosses the ER membrane to reach the cytosol during intoxication. We found that protein disulfide isomerase (PDI) facilitates CT retrotranslocation, whereas ERp72, a PDI-like protein, mediates its ER retention. In vitro analysis revealed that PDI and ERp72 alter CT's conformation in a manner consistent with their roles in retrotranslocation and ER retention. Moreover, we found that PDI's and ERp72's opposing functions operate on endogenous ER misfolded proteins. Thus, our data identify PDI family proteins that play opposing roles in ER quality control and establish an assay to further delineate the mechanism of CT retrotranslocation.
Introduction
The lumen of the ER enables proteins to fold properly before they are transported along the secretory pathway (Ellgaard and Helenius, 2003). When proteins misfold, the ER quality control system ensures that they are retained in the ER to prevent them from reaching their final destination and/or to allow for their refolding. Irreversibly misfolded proteins are eliminated by retrotranslocation to the cytosol, where they are ubiquitinated and degraded by the proteasome (for review see Tsai et al., 2002). The ER factors facilitating these opposing reactions are largely unknown.
We used cholera toxin (CT), which is secreted by the bacterium Vibrio cholerae, to study retrotranslocation. CT consists of a receptor-binding homopentameric B subunit that is noncovalently associated with a single catalytic A subunit. Once CT is secreted from V. cholerae, the A subunit is cleaved into the A1 toxic domain and the A2 domain, which are connected by a disulfide bond and other noncovalent interactions. To intoxicate cells, the holotoxin is endocytosed and travels from the plasma membrane to the ER lumen (Fujinaga et al., 2003). In the ER, the A subunit is disguised as a misfolded protein and hijacks the retrotranslocation machinery so that the A1 chain reaches the cytosol, where it is resistant to proteasomal degradation (Rodighiero et al., 2002), whereas the B subunit remains in the ER (Fujinaga et al., 2003). In the cytosol, the A1 peptide activates a cAMP-dependent signal cascade that results in chloride and water secretion, leading to diarrhea (Sears and Kaper, 1996). Elucidating the ER–cytosol transport mechanism of CT will not only clarify a decisive step in toxin trafficking but will also clarify the retrotranslocation mechanism of misfolded proteins.
Previous in vitro analysis found that the ER oxidoreductase protein disulfide isomerase (PDI) unfolds the A and A1 chains of CT (Tsai et al., 2001), a reaction we believe prepares the toxin for retrotranslocation. The PDI-like protein ERp29 has also been implicated in protein unfolding reactions (Magnuson et al., 2005). However, PDI family proteins have also been shown to facilitate protein folding (for review see Wilkinson and Gilbert, 2004). Thus, it is possible that certain PDI-like proteins are dedicated to the retention and refolding of misfolded polypeptides, whereas other PDI family members function to unfold misfolded proteins in preparation for their retrotranslocation.
In this study, we developed a semipermeabilized cell system that monitors the ER–cytosol transport of CT and found that PDI facilitates the toxin's retrotranslocation, whereas ERp72, a PDI-like protein, mediates its ER retention. Furthermore, these activities were found to operate on endogenous ER misfolded proteins, indicating the generality of this mechanism. These results identify PDI family members as playing opposite roles in ER quality control and establish an assay to elucidate the retrotranslocation process of CT.
Results and discussion
Retrotranslocation of CT
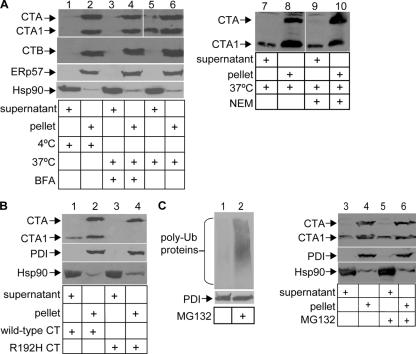
To study CT retrotranslocation, we developed an assay that monitors the transport of the A and A1 subunits from the ER into the cytosol, taking advantage of a semipermeabilized cell assay that efficiently separates cytosolic from ER proteins (Le Gall et al., 2004). CT-intoxicated HeLa cells were treated with 0.04% digitonin to permeabilize the plasma membrane and were fractionated by centrifugation. The supernatant should contain cytosolic proteins as well as ER–cytosol-transported CT, whereas the pellet should contain the plasma membrane, intracellular organelles (including the ER), and toxin that did not undergo retrotranslocation. We tested the purity of these fractions and found the ER resident protein ERp57 to be entirely in the pellet (Fig. 1 A, second panel from bottom; lanes 2, 4, and 6) and the cytosolic protein Hsp90 to be mostly in the supernatant (Fig. 1 A, bottom; lanes 1, 3, and 5). When cells were intoxicated with CT at 37°C, a portion of the A1 subunit was found in the supernatant (Fig. 1 A, top; compare lane 6 with 5), whereas the B subunit was absent in this fraction (Fig. 1 A, second panel from top; compare odd with even lanes) as expected. Typically, 15–30% of toxin and <0.01% of ER resident proteins was detected in the supernatant (calculation not depicted), indicating that the presence of the toxin in this fraction is not caused by ER leakage. This range is likely caused by the variable efficiency of the low level of detergent used in this study in permeabilizing the plasma membrane.
Figure 1.
Retrotranslocation of the CTA1 subunit. (A) HeLa cells were incubated with CT for 90 min at 4 or 37°C with or without BFA or NEM. Cells were permeabilized, centrifuged, and the supernatant and pellet fractions were separated, subjected to nonreducing SDS-PAGE, and immunoblotted with the indicated antibodies. CTA, CTA1, and CTB are 28, 22, and 11 kD, respectively. (B) As in A except a CT mutant, R192H, was used. PDI is 58 kD. (C) As in A except where indicated, cells were treated with MG132. Polyubiquitinated proteins range from 100 to 300 kD. White lines indicate that intervening lanes have been spliced out.
When cells were incubated with CT at 4 or at 37°C in the presence of brefeldin A (BFA), which are conditions shown previously to block the arrival of CT to the ER (Fujinaga et al., 2003), the A1 chain did not appear in the supernatant (Fig. 1 A, top; compare lane 5 with 1 and 3). A fraction of the A1 peptide in the pellet was generated after lysis, as cell permeabilization in the presence of the alkylating reagent N-ethylmaleimide (NEM) diminished the appearance of the A1 chain (Fig. 1 A, compare lane 10 with 8). However, the level of A1 peptide in the supernatant is similar regardless of whether NEM is present in the lysis buffer (Fig. 1 A, compare lane 9 with 7). Thus, postlysis reduction of CT in the pellet does not trigger the release of the A1 chain to the supernatant.
To further verify the assay, we tested the fractionation pattern of a CT mutant whose A chain cannot be cleaved because of a mutation at the cleavage site (R192H). Chloride secretion triggered by the R192H mutant is attenuated dramatically (Lencer et al., 1997); thus, we anticipated that less of this toxin would arrive to the cytosol when compared with the wild-type toxin. Indeed, when cells were incubated with the R192H toxin, no toxin was found in the supernatant (Fig. 1 B, compare lane 3 with 1).
CT-stimulated chloride secretion was previously shown to be resistant to proteasome inactivation (Rodighiero et al., 2002), suggesting that the toxin escapes proteasomal degradation in the cytosol. As expected, proteasome inactivation with MG132, which was confirmed by the accumulation of polyubiquitinated proteins (Fig. 1 C, compare lane 2 with 1), did not significantly alter the toxin level in the cytosol (Fig. 1 C, top; compare lane 5 with 3). The consistency of these results (Fig. 1) with previous chloride secretion studies (Lencer et al., 1997; Rodighiero et al., 2002) validates the use of this semipermeabilized assay as a tool to study CT retrotranslocation directly.
Down-regulation of PDI-like proteins
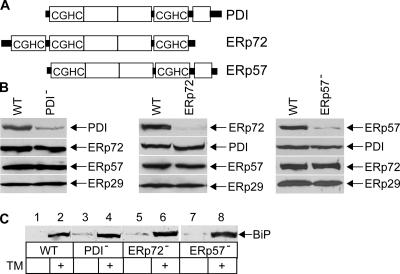
We next tested whether PDI, an ER chaperone that unfolds the A and A1 chains in vitro (Tsai et al., 2001), plays a role in the ER–cytosol transport of CT. Our approach was to down-regulate PDI and other PDI-like proteins in cells using RNAi and test their effect on CT retrotranslocation. Of the 14 known human PDI-like proteins in the ER, we chose to down-regulate ERp72 and ERp57 as controls because they are expressed at similar levels as PDI (unpublished data). PDI, ERp72, and ERp57 are characterized by the presence of a CxxC sequence within their thioredoxin domain (Fig. 2 A). PDI (PDI−), ERp72 (ERp72−), and ERp57 (ERp57−) were down-regulated in cells separately (Fig. 2 B, top); in each case, the expression of other PDI-like proteins (Fig. 2 B, bottom three panels) was unaffected. These results indicate that PDI, ERp72, and ERp57 expression can be reduced efficiently and specifically.
Figure 2.
Down-regulation of PDI-like proteins. (A) Structural organization of PDI, ERp72, and ERp57. (B) PDI, ERp72, ERp57, and ERp29 protein levels were examined in wild-type (WT), PDI−, ERp72−, and ERp57− cells. (C) Wild-type, PDI−, ERp72−, and ERp57− cells were incubated with 2 μg/ml tunicamycin (TM) for 8 h, and BiP expression was assessed by SDS-PAGE and immunoblot analysis. BiP is 78 kD.
We asked whether down-regulation of the PDI-like proteins elicited the unfolded protein response, a cellular stress response in which ER misfolded protein accumulation triggers the expression of ER chaperones such as BiP to alleviate protein misfolding. We found that BiP expression was only up-regulated marginally in PDI−, ERp72−, and ERp57− cells compared with wild-type cells (Fig. 2 C, compare lanes 3, 5, and 7 with 1), indicating that the lack of these proteins did not significantly induce the unfolded protein response. Moreover, the addition of tunicamycin, a drug that blocks N-glycosylation and, thus, causes the accumulation of misfolded proteins, induced BiP expression in wild-type, PDI−, ERp72−, and ERp57− cells (Fig. 2 C, compare lane 2 with 4, 6, and 8). We conclude that down-regulation of the PDI family proteins did not globally disrupt ER function.
PDI and ERp72 play opposing roles during CT retrotranslocation
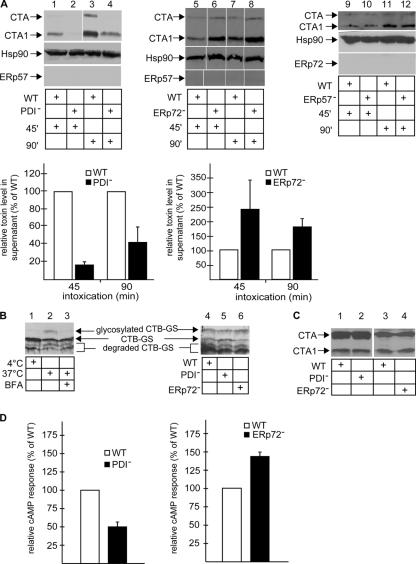
We assessed the effect of PDI down-regulation on the transport of CT to the cytosol by examining the toxin level in the supernatant after cell fractionation. When cells were incubated with CT for 45 or 90 min, we found a decreased level of both the A and A1 chains from PDI− cells when compared with wild-type cells (Fig. 3 A, top; compare lane 2 with 1 and lane 4 with 3; quantified as shown in the graphs). These data indicate that PDI facilitates toxin transport from the ER to the cytosol, which is consistent with our hypothesis that PDI-dependent unfolding of CT mediates the toxin's retrotranslocation (Tsai et al., 2001). We note that a low A subunit level also appeared in the supernatant. This is unlikely to be caused by ER leakage, as ER markers and CTB (CT B subunit) were absent from this fraction. Thus, it is possible that although the A1 subunit is the preferred substrate for retrotranslocation, the A chain is also transported, albeit with lower efficiency.
Figure 3.
PDI and ERp72 exert opposing effects on CT retrotranslocation. (A, top) Wild-type (WT), PDI−, ERp72−, and ERp57− cells were incubated with CT for 45 or 90 min, permeabilized, and the supernatant fraction was analyzed as in Fig. 1. (bottom) The intensities of the CTA and CTA1 bands in the supernatant were quantified. Graphs show the mean ± SD (error bars) of two to four experiments. (B) Wild-type, PDI−, and ERp72− cells were incubated with CT-GS, and the cell lysate was subjected to SDS-PAGE followed by immunoblotting with an anti-CTB antibody. Nonglycosylated and glycosylated CTB are 17 and 21 kD, respectively. (C) Whole cell lysates from CT-intoxicated wild-type, PDI−, and ERp72− cells were prepared in the presence of NEM and subjected to immunoblotting with CTA antibody. (D) Wild-type, PDI−, and ERp72− cells were incubated with CT for 45 or 90 min, and the cAMP level was measured by a cAMP Biotrak Enzyme Immunoassay System (GE Healthcare). Means ± SD of two to four experiments are shown. White lines indicate that intervening lanes have been spliced out.
In contrast, the A and A1 peptide level in the supernatant increased in ERp72− cells when compared with wild-type cells (Fig. 3 A, top; compare lane 6 with lane 5 and lane 8 with 7; quantified as shown in the graphs), suggesting that ERp72 acts to retain CT in the ER, a reaction that opposes PDI's function. The effects of down-regulating PDI and ERp72 on toxin transport were also observed when NEM was present in the lysis buffer (unpublished data). In ERp57− and wild-type cells, the toxin level in the supernatant was similar (Fig. 3 A, top; compare lane 10 with 9 and lane 12 with 11). We conclude that the opposing effects of PDI and ERp72 on CT transport are specific and are unlikely the result of a general disruption of ER chaperone functions.
To show that PDI and ERp72 did not affect CT trafficking to the ER, we used a variant of CT harboring consensus motifs for N-glycosylation on the B subunit (glycosylated CT [CT-GS]). Modification by N-glycosylation, detected by a molecular mass increase in the B subunit, indicates the toxin's arrival to the ER. This toxin was previously used to show that CT is transported as a holotoxin from the plasma membrane to the ER (Fujinaga et al., 2003). Thus, N-glycosylation of the B subunit indicates the arrival of the A subunit to the ER. When cells were incubated with CT-GS at 37°C and the cell lysate was analyzed by SDS-PAGE followed by immunoblotting with an antibody against the B subunit, a band larger than the nonglycosylated B subunit was observed (Fig. 3 B, lane 2). This band was shown previously to be N-glycanase sensitive (Fujinaga et al., 2003) and, thus, represents glycosylated B subunits. Glycosylated B subunits were not detected in cells incubated with the toxin at 4 or 37°C in the presence of BFA (Fig. 3 B, lanes 1 and 3). These experiments validate the use of CT-GS in monitoring A-subunit arrival to the ER. Significantly, the similar level of glycosylated B subunits found in wild-type, PDI−, and ERp72− cells (Fig. 3 B, lanes 4–6) indicate that PDI and ERp72 act on the toxin only after it reaches the ER. Furthermore, PDI and ERp72 down-regulation does not affect CT reduction (Fig. 3 C, compare lane 2 with 1 and lane 4 with 3).
Upon reaching the cytosol, the catalytic A1 subunit ADP ribosylates the Gαs protein, activating adenylate cyclase that, in turn, generates cAMP. Therefore, we measured the CT-induced cAMP response in PDI− and ERp72− cells. The CT-triggered cAMP level in PDI− cells was 50% lower than in wild-type cells (Fig. 3 D, left). Forskolin, which stimulates adenylate cyclase directly, elicited a similar cAMP level in wild-type and PDI− cells (unpublished data), indicating that PDI down-regulation did not affect adenylate cyclase. In contrast, the CT-induced cAMP level in ERp72− cells was 40% higher than in wild-type cells (Fig. 3 D, right). This result was normalized against the forskolin-induced cAMP response, as forskolin also triggered a higher cAMP response in ERp72− cells. The higher forskolin-induced cAMP response in these cells is likely caused by a concomitant increase in cell surface expression of adenylate cyclase (unpublished data). Thus, the cAMP data are consistent with the ER–cytosol transport assay—namely that PDI retrotranslocates CT, whereas ERp72 retains the toxin in the ER.
Opposing effects of PDI and ERp72 on CT conformation
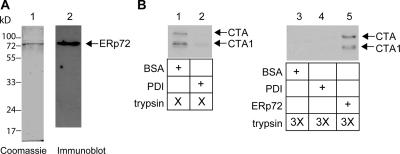
PDI's ability to unfold CT (Tsai et al., 2001) is consistent with its role in toxin retrotranslocation. ERp72's role in facilitating ER retention suggests that it may recognize CT as a misfolded protein and attempt to “refold” the toxin's structure. Incubation of purified PDI but not the control protein BSA with CTA (CT A subunit) was shown previously to render the A and A1 chains sensitive to trypsin digestion (Fig. 4 B, compare lane 2 with 1; Tsai et al., 2001), indicating that PDI unfolded the toxin. Purified ERp72 (Fig. 4 A) did not cause the toxin to become protease sensitive (not depicted). We now find that regardless of PDI's presence, a higher trypsin concentration rendered the A and A1 chains sensitive to degradation (Fig. 4 B, compare lanes 3 and 4 with 1). However, under this condition, ERp72 protected the toxin from degradation (Fig. 4 B, compare lane 5 with 3 and 4), indicating that ERp72 alters CT's conformation to render it more folded and compact. Alternatively, it is possible that ERp72 protects the toxin against protease digestion by either stabilizing the native conformation of the toxin or interacting with CT in a similar manner as PDI but with reduced efficiency. Nonetheless, PDI and ERp72's opposing effects on CT's conformation likely reflect their roles in facilitating retrotranslocation and ER retention during ER quality control.
Figure 4.
Opposing effects of PDI and ERp72 on the conformation of CT. (A) His-tagged ERp72 protein was purified from bacteria and analyzed by SDS-PAGE followed by Coomassie staining or immunoblotting with an antibody against ERp72. (B) CTA was incubated with BSA, PDI, or ERp72 followed by the addition of trypsin. Samples were subjected to SDS-PAGE followed by immunoblotting with an anti-CTA antibody.
A general role of PDI and ERp72 in quality control of ER misfolded proteins
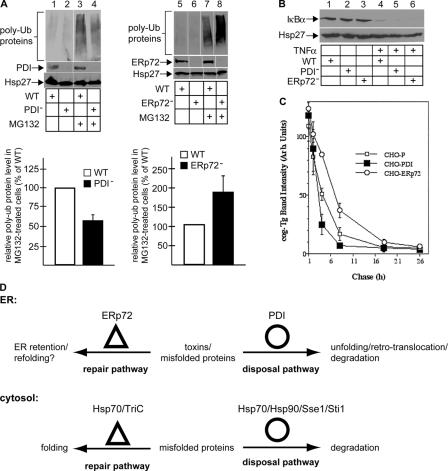
Do the opposing functions of PDI and ERp72 represent a general mechanism operating in the ER? We developed a method to measure the bulk arrival of misfolded proteins from the ER to the cytosol. Upon reaching the cytosolic surface of the ER membrane, ER misfolded polypeptides are polyubiquitinated before being targeted for proteasomal degradation (for review see Tsai et al., 2002). Consequently, proteasome inactivation ought to lead to the accumulation of polyubiquitinated ER misfolded proteins as well as cytosolic proteins. The effect of ER resident protein down-regulation on the accumulation of total polyubiquitinated proteins should, therefore, reflect their role in the ER–cytosol transport process. We measured the level of polyubiquitinated proteins and found that incubation of cells with the proteasome inhibitor MG132 resulted in an increase of polyubiquitinated proteins (Fig. 5 A, compare lane 3 with 1 and lane 7 with 5). However, in PDI− cells, less MG132-induced polyubiquitinated proteins appeared when compared with wild-type cells (Fig. 5 A, compare lane 4 with 3; quantified as shown in the graphs), whereas in ERp72− cells, more polyubiquitinated proteins were present (Fig. 5 A, compare lane 8 with 7; quantified as shown in the graphs). Although the source of the polyubiquitinated proteins is unclear, they likely originated from the ER. Quantification showed that although PDI down-regulation reduced the total level of polyubiquitinated proteins by ∼40%, a significant portion (∼60%) was unaffected; the unaffected polyubiquitinated proteins are presumably cytosolic proteins.
Figure 5.
A general role of PDI and ERp72 in the retrotranslocation of ER misfolded proteins. (A, top) Wild-type (WT), PDI−, or ERp72− cells were treated with MG132, and the cell lysate was subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. (bottom) The total ubiquitin signal intensity was measured. Graphs show the mean ± SD (error bars) of three to five experiments. (B) Wild-type, PDI−, or ERp72− cells were treated with TNFα, and the lysate was subjected to SDS-PAGE followed by immunoblotting with an antibody against IκBα. IκBα is 39 kD. (C) CHO cells stably expressing cogTg (CHO-P) and PDI (CHO-PDI) or ERp72 (CHO-ERp72) were pulse labeled with [35S]methionine and chased for the times indicated. CogTg was immunoprecipitated from the cell lysate with anti-Tg antibody and analyzed by SDS-PAGE. Data show quantification of the radioactive cogTg band intensity. (D) Diagram depicting the similarity between the quality control system in the ER and cytosol. See last paragraph of Results and discussion. White lines indicate that intervening lanes have been spliced out.
We then asked whether cytosolic ubiquitination reactions are affected nonspecifically by PDI and ERp72 down-regulation. IκBα is a cytosolic protein that is polyubiquitinated in response to TNFα stimulation before being degraded by the proteasome (for review see Karin and Ben-Neriah, 2000). Indeed, we found that IκBα was degraded with similar efficiency upon TNFα treatment in wild-type, PDI−, and ERp72− cells (Fig. 5 B, compare lanes 4–6 with 1–3), suggesting that IκBα ubiquitination was not disrupted by PDI and ERp72 down-regulation. These findings indicate that PDI facilitates the retrotranslocation of some ER misfolded proteins, whereas ERp72 mediates their ER retention.
We further characterized this effect on a specific ER misfolded substrate. Thyroglobulin (Tg), the thyroid prohormone, is synthesized and folded in the ER before being secreted. A mutant form of Tg, cogTg, was shown previously to undergo retrotranslocation and proteasomal degradation (Tokunaga et al., 2000). PDI overexpression in CHO cells stably expressing cogTg (CHO-PDI) stimulated the rate of cogTg degradation when compared with parental CHO cells expressing cogTg (CHO-P; Fig. 5 C, compare open with closed squares). In contrast, ERp72 overexpression in CHO cells (CHO-ERp72) decreased the rate of cogTg degradation (Fig. 5 C, compare open squares with circles). These results suggest that PDI stimulates the ER–cytosol transport of cogTg, whereas ERp72 retains it in the ER. We conclude that PDI and ERp72's opposing roles operate not only on CT retrotranslocation but more generally for ER misfolded proteins.
The role of PDI and ERp72 in retrotranslocation and ER retention
How do chaperones that belong to the same family serve opposite functions? PDI possesses two thioredoxin domains containing the redox-active CxxC motif (Fig. 2 A, CxxC box) and two thioredoxin-like domains without the CxxC motif (Fig. 2 A, white rectangles), whereas ERp72 contains three redox-active and two redox-inactive domains (Fig. 2 B). Their major difference is that PDI contains an additional c domain that is absent in ERp72. It is possible that this domain participates in the unfolding of polypeptides. Interestingly, PDI but not ERp72 was implicated in the unfolding of the nonnative structure of bovine pancreatic trypsin inhibitor during disulfide bond rearrangement (Weissman and Kim, 1993; Satoh et al., 2005), supporting our conclusion that PDI specifically unfolds misfolded substrates. PDI has also been shown to facilitate the proteasomal degradation of ER misfolded proteins in yeast (Gillece et al., 1999), a process that presumably requires substrate unfolding and retrotranslocation. Our findings that ERp72 mediates the ER retention of CT and misfolded proteins are consistent with recent data demonstrating that ER retention of a misfolded secretory (Cotterill et al., 2005) and a transmembrane protein (Sorensen et al., 2005) is coincident with binding to ERp72.
ERp72 and PDI's dedicated roles in ER quality control appear analogous to the cytosolic chaperone system that functions in protein folding and degradation (Fig. 5 D; McClellan et al., 2005). In this case, the chaperones Hsp70 and TriC assist in protein folding, whereas the Hsp70–Hsp90–Sti1–Sse1 complex mediates protein degradation. As PDI can promote protein folding and unfolding reactions, it is possible that PDI cofactors exist to control these opposing processes, which is similar to Hsp70 regulation.
Materials and methods
Materials
Antibodies against PDI, BiP, and Hsp90 were purchased from Santa Cruz Biotechnology, Inc. Antibodies against ERp72 were obtained from StressGen Biotechnologies, antibodies against ubiquitin were purchased from Zymed Laboratories, and antibodies against IκBα were obtained from Biolegend. CT A and B antibodies and CT-GS were provided by the Lencer laboratory, the Tg antibodies were obtained from the Arvan laboratory, and the CT R192H mutant was provided by R. Holmes (University of Colorado, Boulder, CO). Hsp27, ERp57, and ERp29 antibodies were gifts from M. Welsh (University of Michigan, Ann Arbor, MI), S. High (University of Manchester, Manchester, England), and S. Mkrtchian (Karolinska Institutet, Stockholm, Sweden), respectively. CT, CTA, and PDI were purchased from Calbiochem. Mouse ERp72 cDNA was subcloned into pQE30, and the protein was purified using a Ni–nitrilotriacetic acid agarose column.
Protein down-regulation
PDI-specific (5′-GACCTCCCCTTCAAAGTTGTT-3′) siRNA was synthesized by Ambion, and ERp72- (5′-CAAGCGUUCUCCUCCAAUUTT-3′) and ERp57-specific (5′-UGAAGGUGGCCGUGAAUUATT-3′) siRNAs were synthesized by Invitrogen. 10 nM duplexed siRNA was transfected into HeLa cells using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Protein expression was assessed by SDS-PAGE and immunoblot analysis. Experiments were initiated 48 (ERp57) or 72 h (PDI and ERp72) after transfection.
Retrotranslocation assay
Cells were incubated with 10 nM CT in HBSS at 37°C for 45 or 90 min. For permeabilization, 2 × 106 cells were resuspended in 100 μl of 0.04% digitonin in HCN buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 2 mM CaCl2, and protease inhibitors) with or without 10 mM NEM, incubated on ice for 10 min, and centrifuged at 16,000 g for 10 min. The supernatant was removed, and the pellet was washed with PBS and resuspended in 100 μl of the original buffer. Fractions were analyzed by nonreducing SDS-PAGE and immunoblot analysis. Where indicated, cells were treated with 2 μM MG132 for 90 min.
N-glycosylation of CTB
HeLa cells were incubated with 50 nM CT-GS in HBSS for 3 h at 4 or 37°C in the presence or absence of 5 μg/ml BFA. Cells were harvested in TN lysis buffer (1% Triton X-100, 1.75% n-octyl-B-/d-glucopyranoside, 10 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, and protease inhibitors), and the lysates were analyzed for the presence of glycosylated CTB.
cAMP assay
A cAMP enzyme immunoassay system (GE Healthcare) was used to quantify cAMP synthesis induced by 10 nM CT or 50 μM forskolin in HBSS. Samples were assayed in duplicate, and the mean optical density was used to calculate the cAMP level/well. The cAMP response was determined by dividing the cAMP level in CT- or forskolin -treated cells by the cAMP level in unstimulated cells. Forskolin induced a sevenfold higher cAMP response in ERp72− cells than in wild-type cells. Results are reported as a percentage of the wild-type CT-induced cAMP response normalized to the forskolin-induced cAMP response.
Trypsin sensitivity assay
Purified CTA was incubated with 1 mM glutathione, 2 μg/ml BSA, 2 μg/ml PDI, or 0.2 μg/ml ERp72 for 30 min at 37°C. 100 or 300 μg/ml trypsin was added to the samples for 30 min at 4°C. Samples were analyzed by nonreducing SDS-PAGE followed by immunoblot analysis.
Accumulation of polyubiquitinated proteins
Cells were incubated with or without 20 μM MG132 for 30 min and lysed in a buffer containing 1% Triton-X, 10 mM NEM, 30 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM EDTA, and protease inhibitors. Cleared lysates were analyzed by 4–20% SDS-PAGE followed by immunoblot analysis. To monitor ubiquitin-dependent degradation of IκBα, cells were treated with 10 ng/ml TNFα for 15 min, and the cell lysate was analyzed for the presence of IκBα.
Pulse-chase analysis
The cogTg degradation rate was analyzed as previously described (Tokunaga et al., 2000).
Acknowledgments
We thank Tom Rapoport for critical review of the manuscript.
B. Tsai is a Biological Scholar at the University of Michigan.
Abbreviations used in this paper: BFA, brefeldin A; CT, cholera toxin; NEM, N-ethylmaleimide; PDI, protein disulfide isomerase; Tg, thyroglobulin.
References
- Cotterill, S.L., G.C. Jackson, M.P. Leighton, R. Wagener, O. Makitie, W.G. Cole, and M.D. Briggs. 2005. Multiple epiphyseal dysplasia mutations in MATN3 cause misfolding of the A-domain and prevent secretion of mutant matrilin-3. Hum. Mutat. 26:557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191. [DOI] [PubMed] [Google Scholar]
- Fujinaga, Y., A.A. Wolf, C. Rodighiero, H. Wheeler, B. Tsai, L. Allen, M.G. Jobling, T. Rapoport, R.K. Holmes, and W.I. Lencer. 2003. Gangliosides that associate with lipid rafts mediate transport of cholera and related toxins from the plasma membrane to endoplasmic reticulum. Mol. Biol. Cell. 14:4783–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillece, P., J.M. Luz, W.J. Lennarz, F.J. de La Cruz, and K. Romisch. 1999. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J. Cell Biol. 147:1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621–663. [DOI] [PubMed] [Google Scholar]
- Le Gall, S., A. Neuhof, and T. Rapoport. 2004. The endoplasmic reticulum membrane is permeable to small molecules. Mol. Biol. Cell. 15:447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer, W.I., C. Constable, S. Moe, P.A. Rufo, A. Wolf, M.G. Jobling, S.P. Ruston, J.L. Madara, R.K. Holmes, and T.R. Hirst. 1997. Proteolytic activation of cholera toxin and Escherichia coli labile toxin by entry into host epithelial cells. Signal transduction by a protease-resistant toxin variant. J. Biol. Chem. 272:15562–15568. [DOI] [PubMed] [Google Scholar]
- Magnuson, B., E.K. Rainey, T. Benjamin, M. Baryshev, S. Mkrtchian, and B. Tsai. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell. 20:289–300. [DOI] [PubMed] [Google Scholar]
- McClellan, A.J., M.D. Scott, and J. Frydman. 2005. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 121:739–748. [DOI] [PubMed] [Google Scholar]
- Rodighiero, C., B. Tsai, T.A. Rapoport, and W.I. Lencer. 2002. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 3:1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, M., A. Shimada, A. Kashiwai, S. Saga, and M. Hosokawa. 2005. Differential cooperative enzymatic activities of protein disulfide isomerase family in protein folding. Cell Stress Chaperones. 10:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, C.L., and J.B. Kaper. 1996. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol. Rev. 60:167–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, S., T. Ranheim, K.S. Bakken, T.P. Leren, and M.A. Kulseth. 2005. Retention of mutant low density lipoprotein receptor in ER leads to ER stress. J. Biol. Chem. 281:468–476. [DOI] [PubMed] [Google Scholar]
- Tokunaga, F., C. Brostrom, T. Koide, and P. Arvan. 2000. Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase I. J. Biol. Chem. 275:40757–40764. [DOI] [PubMed] [Google Scholar]
- Tsai, B., C. Rodighiero, W.I. Lencer, and T.A. Rapoport. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 104:937–948. [DOI] [PubMed] [Google Scholar]
- Tsai, B., Y. Ye, and T.A. Rapoport. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 3:246–255. [DOI] [PubMed] [Google Scholar]
- Weissman, J.S., and P.S. Kim. 1993. Efficient catalysis of disulphide bond rearrangements by protein disulphide isomerase. Nature. 365:185–188. [DOI] [PubMed] [Google Scholar]
- Wilkinson, B., and H.F. Gilbert. 2004. Protein disulfide isomerase. Biochim. Biophys. Acta. 1699:35–44. [DOI] [PubMed] [Google Scholar]