Abstract
Centrins are calmodulin-like proteins present in centrosomes and yeast spindle pole bodies (SPBs) and have essential functions in their duplication. The Saccharomyces cerevisiae centrin, Cdc31p, binds Sfi1p on multiple conserved repeats; both proteins localize to the SPB half-bridge, where the new SPB is assembled. The crystal structures of Sfi1p–centrin complexes containing several repeats show Sfi1p as an α helix with centrins wrapped around each repeat and similar centrin–centrin contacts between each repeat. Electron microscopy (EM) shadowing of an Sfi1p–centrin complex with 15 Sfi1 repeats and 15 centrins bound showed filaments 60 nm long, compatible with all the Sfi1 repeats as a continuous α helix. Immuno-EM localization of the Sfi1p N and C termini showed Sfi1p–centrin filaments spanning the length of the half-bridge with the Sfi1p N terminus at the SPB. This suggests a model for SPB duplication where the half-bridge doubles in length by association of the Sfi1p C termini, thereby providing a new Sfi1p N terminus to initiate SPB assembly.
Introduction
Centrins are a family of EF-hand–containing proteins most closely related to calmodulins and, like calmodulin, probably have multiple unrelated functions in association with other proteins. At present, the best-characterized functions of centrins are in microtubule organizing centers (MTOCs), such as centrosomes and basal bodies (Salisbury, 1995), where they are often present in different parts of the MTOC (Levy et al., 1996; Stemm-Wolf et al., 2005) and probably have specialized functions associated with these various locations. Two clear functions in MTOCs have been established: one is in the duplication of the MTOC (Byers, 1981b; Middendorp et al., 2000; Salisbury et al., 2002) and the other is as constituents of filaments within and attached to the MTOC. Some of these filaments can contract in response to changes in Ca2+ concentration (Salisbury et al., 1984). However, the molecular basis for these centrin-based functions in MTOCs has not yet been established.
Budding yeast has a single simple MTOC, the spindle pole body (SPB), responsible for the organization of both the spindle and cytoplasmic microtubules (see diagram in Fig. 7 A). The SPB is a multilayered structure embedded in the nuclear envelope, which remains intact during yeast mitosis. Attached to one side of each SPB is a specialized area of the nuclear envelope called the half-bridge, which has a critical role during SPB duplication (Adams and Kilmartin, 1999) and is where the single centrin in budding yeast, Cdc31p, is localized (Spang et al., 1993). The half-bridge consists of a densely stained rectangular area of the nuclear membrane together with a cytoplasmic outer layer. The assembly of cytoplasmic components of the daughter SPB initiates from a satellite structure at the distal end of the bridge (Byers, 1981a) and continues to form a duplication plaque on the cytoplasmic side of the bridge (Adams and Kilmartin, 1999). The SPB is then inserted into the nuclear envelope, nuclear SPB components are added, and the two SPBs separate by dissociation or cleavage of the bridge, leaving a half-bridge with each SPB.
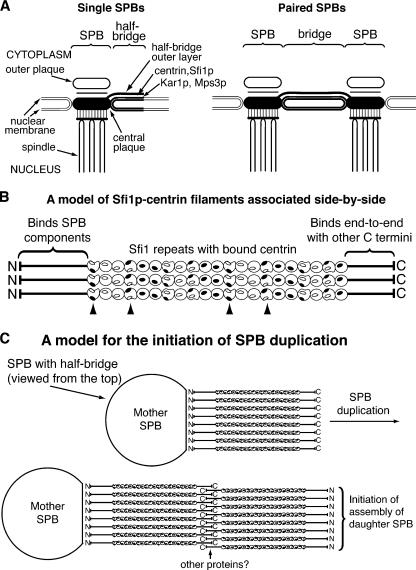
Figure 7.
Diagram of single and paired SPBs and a model for the initiation of SPB duplication. (A) Diagram of single and paired SPBs showing the location of half-bridge and bridge components. (B) A model of Sfi1p–centrin filaments associated side-by-side. Centrins have a 65° twist along Sfi1p and are shown as spheres or semispheres with filled and unfilled ovals as interaction patches, where filled patches interact with unfilled patches. Arrowheads show centrins interacting between filaments, assuming interactions occur within 15° of a total twist of 0° or 180°. (C) A model for the initial step in SPB duplication, viewed from above the SPB. We propose that the Sfi1p N terminus binds SPB components, whereas the C termini can associate end-to-end in an antiparallel way with or without other proteins. This provides a fresh Sfi1p N terminus capable of binding SPB components and thereby initiating SPB assembly.
Centrin has an essential function during SPB duplication, and temperature-sensitive (ts) mutants arrest with a single large SPB (Byers, 1981b). Centrin binds to three proteins in the half-bridge, Kar1p (Biggins and Rose, 1994; Spang et al., 1995), Mps3p (Jaspersen et al., 2002), and Sfi1p (Kilmartin, 2003). Kar1p and Mps3p have transmembrane domains and are probably associated with the lipid bilayers of the half-bridge. Sfi1p, which has an essential function during SPB duplication, does not have a transmembrane domain; however, it has ∼20 continuous conserved repeats in the center of the protein. Five of these repeats were tested, and four were found to bind centrin in pull-down assays (Kilmartin, 2003). This suggests a model for the Sfi1p–centrin complex where ∼20 molecules of centrin bind continuously to the repeats on a molecule of Sfi1p, possibly producing a filamentous structure. This paper describes structural studies on the Sfi1p–centrin complex, its arrangement on the half-bridge and bridge, and a model for its role during SPB duplication.
Results
Centrin binding Sfi1 repeats
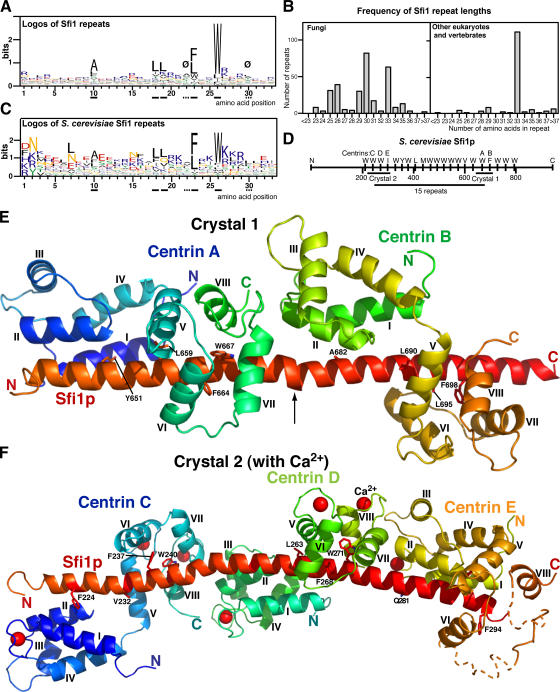
Sfi1 repeats that bind centrin have a consensus AX7LLX3F/LX2W (Kilmartin, 2003). There were only a small number of sequences available when Sfi1 repeats were first analyzed, but since then more sequences have become available and these show a similar consensus (Fig. 1 A), and there are also biases in sequence at other positions in the repeat (see Fig. 1 legend and the supplemental text, available at http://www.jcb.org/cgi/content/full/jcb.200603153/DC1). Fungi have a variable repeat length, whereas most repeats in other eukaryotes are 33 amino acids long (Fig. 1 B). All the complete sequences analyzed in Fig. 1 have between 20 and 24 continuous repeats. Saccharomyces cerevisiae Sfi1p was originally given only 17 repeats with gaps (Kilmartin, 2003). These gaps were close to exact multiples of repeats, and inspection of the gap sequence, particularly when aligned with other fungal species, has identified potential repeats in the gaps (Fig. 1, C and D).
Figure 1.
Sequence analysis of Sfi1 repeats and structure of the Sfi1p–centrin complex. (A) Logos of 454 Sfi1 repeats from 16 fungi, Chlamydomonas, Giardia lamblia, Ciona intestinalis, chicken, dog, mouse, and human (see the supplemental text, available at http://www.jcb.org/cgi/content/full/jcb.200603153/DC1). Ø marks positions 22 and 30, where there is an usually low content of charged aliphatic amino acids (DEKR; 2.9% at position 22 and 7.0% at position 30 compared with 29.5% for all positions). Underlined full and dashed lines are for orientation in Figs. 1 and 2. (B) Frequency of Sfi1-repeat lengths. Lengths were measured between the tryptophan positions (position 26). (C) Logos of 21 S. cerevisiae Sfi1 repeats. (D) Diagram of S. cerevisiae Sfi1p, showing 21 repeats and the positions of constructs used to prepare crystals 1 and 2 and the 15-repeat construct. (E) Crystal 1. Ribbon diagram of the Sfi1p–centrin complex containing two Sfi1 repeats and two centrins at low Ca2+ concentration. Conserved residues in the repeats are shown (positions 10, 18, 23, and 26; panel A). The arrow indicates a bulge and bend in the α helix. Centrin helices are marked. (F) Crystal 2. Sfi1p–centrin complex containing three Sfi1 repeats (N218-H306) and three centrins with Ca2+ bound. Notations are as explained in E, and Ca2+ ions are indicated by red spheres. The end of the third Sfi1 repeat and parts of the C-terminal domain of centrin E were indistinct (dotted lines). Coordinates for the two structures have been deposited with the RCSB Protein Data Bank under accession numbers 2GV5 (crystal 1) and 2DOQ (crystal 2).
An obvious structural model for the Sfi1 repeats is that centrin binds to each repeat and, therefore, ∼20 centrins are bound continuously along the repeat region. So, how does the highly conserved centrin bind to this heterogeneous Sfi1 sequence, particularly in the less conserved N-terminal region and, in the case of fungi, how does it bind to Sfi1 sequences of different length?
Structure of Sfi1p–centrin complexes containing two and three Sfi1 repeats
To determine the structure of the Sfi1p–centrin complex, we coexpressed fragments of Sfi1p containing two to three repeats from S. cerevisiae as GST fusion proteins together with yeast centrin (Cdc31p) in bacteria. All constructs tested gave stable complexes that could be purified by gel filtration and Q Sepharose chromatography in 1 mM EGTA–containing buffers, confirming that the Sfi1p–centrin interaction is stable at low Ca2+ concentrations (Kilmartin, 2003). One construct (crystal 1; Fig. 1 D) containing two repeats (K643-E710) gave crystals without Ca2+ (see Materials and methods) that could be solved at 3.0 Å (Fig. 1 E, Table S1, and the supplemental text, available at http://www.jcb.org/cgi/content/full/jcb.200603153/DC1). Another construct (N218-H306; crystal 2; Fig. 1 D) had three repeats, was crystallized with 0.1 M calcium acetate, and was solved at 3.2 Å (Fig. 1 F, Table S1, and the supplemental text). High Ca2+ concentrations were necessary because concentrations <50 mM did not give suitable crystals. In these crystals, Ca2+ was not present at positions other than EF-hands, as an anomalous difference map (see Materials and methods) showed Ca2+ only in EF-hands 1, 3, and 4; EF-hand 2 is probably unable to bind Ca2+ because it lacks essential ligands (Falke et al., 1994).
Both of these crystal structures show the Sfi1p fragments as α helices with centrin in an extended conformation bound to each repeat. The centrin N-terminal domains bind to the N-terminal half of the Sfi1 repeat containing the conserved alanine, whereas the centrin C-terminal domains bind to the more conserved C-terminal half of the Sfi1 repeat (Fig. 2). Parts of the C-terminal domain of centrin E, and that part of the Sfi1 repeat it probably binds to (R295-W304), were not visible in the electron density map.
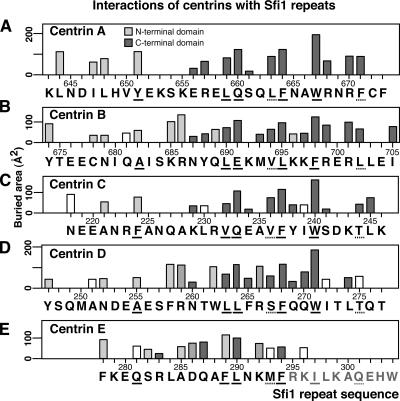
Figure 2.
Interactions of centrins with Sfi1 repeats. (A–E) Interactions with centrins A–E plotted as buried area (Collaborative Computational Project, Number 4, 1994). Light gray bars show interactions with the centrin N-terminal domain, dark gray with the C-terminal, and intermediate gray with both; interacting amino acids (within 4.0 Å) were identified by Ligplot (Wallace et al., 1995). Unshaded bars represent buried areas where Ligplot fails to show residues within 4.0 Å. Underlined amino acids are the conserved positions in the Sfi1 repeat.
One surprising feature of the electron density map was that the N-terminal ends of all the centrins (S2-P12) were not visible; the electron density for all the centrins started close to L13. This extra N-terminal sequence, which is variable in length and sequence and is before the helical EF-hand domains, is one of the defining characteristics of centrins that calmodulins lack. We tested whether this sequence was necessary for yeast centrin function and found that a construct with a deletion of 14 amino acids from the N terminus (S2-S15) was able to replace the wild-type protein in yeast, showing that this N-terminal end sequence has no essential function in vivo.
Surprisingly, despite the absence of Ca2+ in crystal 1 and the presence of Ca2+ in crystal 2, the conformations of the N- and C-terminal domains of centrins A–E are very similar (Fig. 3, A and B). The root mean square deviations (rmsd's) for a comparison of the main chain of the N-terminal domain (Cα L18-K91) of centrin A with the same domains of centrins B–E are 0.5, 0.7, 0.7, and 1.0 Å, respectively, and for the C-terminal domains (Cα L95-C158) the rmsd's are 1.2, 0.5, and 0.3 Å (centrin E was excluded from this comparison). The N-terminal domain is in the closed conformation, whereas the C-terminal is open (see the supplemental text). EF-hand proteins such as calmodulin in the apo form have closed or semi-open conformations of their N- and C-terminal domains, respectively. On binding Ca2+, the domains open up, exposing hydrophobic surfaces that are able to bind other proteins (Swindells and Ikura, 1996). However, in the case of yeast centrin bound to Sfi1p, and despite the rather high Ca2+ concentration in the crystal, there is little influence of Ca2+ on the conformation.
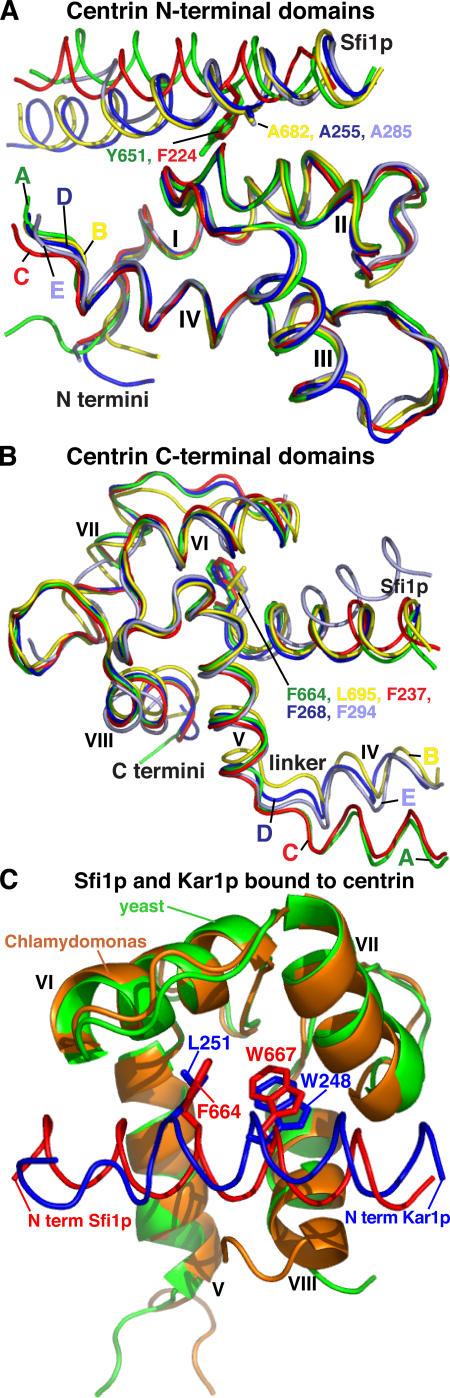
Figure 3.
Comparison of centrin N- and C-terminal domains and centrin binding sequences. (A) Comparison of N-terminal domains. Centrin A is green, B is yellow, C is red, D is blue, and E is gray. The bound Sfi1ps are in the corresponding colors, and centrin helices I–IV are marked. Also shown are the amino acids in the conserved position 10 of the Sfi1 repeat (Fig. 1), except for A285, which is at position 14 (Fig. 2 E). Here, because of the short repeat, centrin E binds to Sfi1p one turn of the helix later and binds to A285 in the same way as A682 and A255. (B) Comparison of C-terminal domains with notation as in A. Helices IV–VIII are marked, and the amino acids shown are in position 23 of the Sfi1 repeat. (C) Comparison of Sfi1p (red) and Kar1p (purple) bound to the C-terminal domains of yeast centrin A (green) and Chlamydomonas centrin (brown). Helices V–VIII are marked, together with amino acids L251 and W248 of Kar1p (Hu and Chazin, 2003) and F664 and W667 of Sfi1p.
The conformationally near identical N-terminal domains interact with the N-terminal part of the Sfi1p repeat through parts of a hydrophobic patch between centrin helices I and II containing L31, F32, V47, the side chain of K50, and A51. Centrins A and C interact with bulky residues Y651 and F224 in the alanine position 10 of the Sfi1 repeat (Fig. 1 A), and these push the Sfi1p helix away from the N-terminal domain (Fig. 3 A). In contrast, centrins B and D interact with the less bulky alanines A682 and A255. These allow the Sfi1p helix closer to the N-terminal domains (Fig. 3 A) so that the carbon atoms of the side chains of residues K685 and R686 (Fig. 2 B) and F258 and R259 (Fig. 2 D) can interact with a second overlapping hydrophobic patch containing A28, L31, A51, and L52. In addition, R686 and R259 can form a potential salt bridge with E97. These extra interactions may explain the preference for alanine at position 10 of the Sfi1 repeat.
The N-terminal domain of centrin E also interacts with Sfi1p similarly to centrins B and D, but, probably because of the shorter Sfi1 repeat, this occurs one turn of the helix further down. The alanine interaction is now with A285 (Fig. 2 E), which is in the equivalent interaction position to A682 and A255 (Fig. 3 A), and allows F289 to interact with the second hydrophobic patch, similar to the side chain of R686. Unfortunately, part of the C-terminal domain of centrin E is not visible, but that part that is visible does make normal interactions with F294 (Fig. 3 B). This centrin has to be in a less extended conformation to make these interactions, which seems to necessitate a bend in the Sfi1p helix between the N- and C-terminal domains of centrin E (Fig. 3 B).
The centrin C-terminal domains also have a similar conformation and have mainly hydrophobic interactions with Sfi1p (Fig. 2). They interact mainly with the more conserved C-terminal part of the Sfi1p repeat and, as expected, these amino acids are involved in a large part of the interactions. These interactions are not described in detail, as they are similar to interactions of Kar1p with the C-terminal domain of Chlamydomonas centrin (Hu and Chazin, 2003). This domain is very similar to the centrin A C-terminal domain (rmsd 0.9 Å) and to the open form of the calmodulin C-terminal domain (Hu and Chazin, 2003). However, although the interactions of centrin with Kar1p and Sfi1p are similar, the two binding sequences are reversed (Fig. 3 C). This ability to bind the same motif in reverse has previously been found for calmodulin (Osawa et al., 1999) and other helical motifs (Teo et al., 2004).
In conclusion, these results show the N- and C-terminal domains of centrin as relatively rigid structures, unaffected by Ca2+ when they are bound to Sfi1p but able to recognize heterogeneous sequences of variable lengths as α helices.
Interactions between the centrins and the structure of an Sfi1p–centrin filament
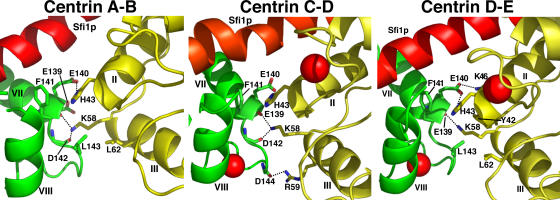
One of the most interesting features of the two crystal structures is that similar centrin–centrin interactions are made (Fig. 4). These are between the C-terminal domain of one centrin and the N-terminal domain of the next, with the second centrin rotated ∼65° clockwise around the Sfi1p helix. These interactions are mainly potential hydrogen bonds between K58 and the main chain carbonyls of E139 and D142, a potential salt bridge between H43 and E140, and hydrophobic interactions between F141 and H43 and also L62 and L143 or a potential salt bridge between R59 and D144. All of these amino acids were mutated to alanine to determine whether any phenotype was associated with the loss of these interactions. Earlier studies showed that the mutation F141A is lethal (Ivanovska and Rose, 2001) and D142A gave a ts phenotype (Geier et al., 1996). We found H43A also gave a ts phenotype, but single mutations in the other amino acids gave no clear phenotype, although the double mutations H43A, K46A and H43A, K58A were lethal (not all combinations were tested). These results suggest that some of these interactions are important, though we cannot exclude an indirect conformational effect. Several of the amino acids involved—H43, L62, and F141—are specific to the centrin 3 family (Azimzadeh and Bornens, 2004), suggesting that only this subfamily can form this particular type of filament in association with Sfi1p.
Figure 4.
Interactions between centrins A and B, C and D, and D and E when bound to Sfi1p. In all the interfaces, H43 forms a possible hydrogen bond with the main chain carbonyl of E139, and in the A–B and C–D interfaces, also with the main chain carbonyl of D142. H43 forms a possible salt bridge with E140 in the A–B and D–E interfaces, and there are additional possible salt bridges between R59 and D144 in the C–D and K46 and E140 in the D–E interfaces. There are potential hydrophobic interactions between the side chains of H43 and F141 in all interfaces and between L62 and L143 in the A–B and D–E interfaces. There are additional potential hydrophobic interactions between the side chains of K58 and E139 in the A–B interface, H43 and E140 in the C–D interface, and Y42 and L143 in the D–E interface. When interactions are with the carbonyl only (E139 and D142), the side chain is omitted.
The most obvious model for the structure of the Sfi1p-repeat region based on the two crystal structures solved here is a filament, with the 20 or so repeats as a single continuous α helix wrapped with centrins, also in a semiordered helical array. We assume that all the centrins within Sfi1p make the centrin–centrin interactions described in the previous paragraph, which sets the angle between adjacent centrins at ∼65°. This leads to a problem because of the geometry of the α helix, where each amino acid is rotated by 100° along the helix. Only one repeat length, 33 amino acids, will rotate the corresponding amino acids in the next repeat by 60°, close to the correct angle, which presumably accounts for why this repeat length is favored in higher eukaryotes (Fig. 1 B). Other repeat lengths spread the corresponding amino acids at 20° intervals through the circumference of the helix. An example of how other repeat lengths are accommodated can be seen between centrins A and B and centrins C and D, where the repeats are both 31 amino acids (measured between the conserved tryptophan positions), predicting a rotation of 220°. This large angle is compensated for by partially unwinding the Sfi1p helix between the repeats in both cases and probably also by bending this helix and variations in the linker angles between the centrin N- and C-terminal domains. There is little unwinding in the Sfi1p helix between centrins D and E, probably because the repeat length here is 26 amino acids, predicting a rotation of 80°, close to the 65° needed. This shorter repeat is accommodated by the N-terminal domain interacting with the Sfi1p helix one turn later (Fig. 3 A), putting centrin E into a less extended form and probably bending the Sfi1p helix so that the C-terminal domain can make normal interactions (Fig. 3 B). This ability of the N-terminal domain to interact with different α helix turns probably accounts for the peak distribution of repeat lengths in fungi (Fig. 1 B), which are separated by distances corresponding to helical turns.
In conclusion, the two crystal structures solved here suggest how a filament of centrin is formed by centrin–centrin interactions stabilized by an Sfi1p α helix, which is subject to local distortions to allow specific centrin–centrin interactions.
Structure of an Sfi1p–centrin complex containing 15 repeats
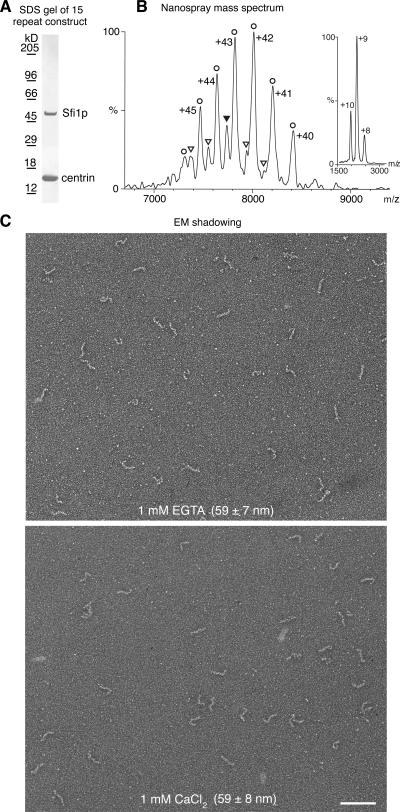
The model for the Sfi1p–centrin complex as a single Sfi1p α helix with centrins wrapped continuously around in a helical arrangement was tested by coexpression of ∼15 Sfi1 repeats (K246-E677; Fig. 1 D) with yeast centrin (Fig. 5 A). The molecular mass of this complex was measured by nanospray mass spectrometry under nondenaturing conditions and gave a value of 336,290 D (Fig. 5 B). This is reasonably close to a theoretical value of 332,713 D for a 1:15 Sfi1p–centrin complex, with retention of ∼3.6 kD of buffer molecules in the gas phase ions, as is often found for large complexes analyzed under conditions gentle enough to preserve quaternary interactions (Ilag et al., 2004). A minor component with a mass of 317,308 D was also observed. This is consistent with a 1:14 stoichiometry (theoretical mass = 314,093 D), suggesting that partial dissociation is able to occur in solution.
Figure 5.
Analysis of an Sfi1p–centrin complex containing 15 repeats. (A) Coomassie-stained SDS gel of the 15-repeat Sfi1p–centrin complex (Fig. 1 D). (B) Nanospray mass spectrum of the 15-repeat Sfi1p–centrin complex under nondenaturing conditions. The observed charge series correspond to centrin monomers at low m/z (inset: observed mass, 18,619.3 D; theoretical mass, 18,619.9 D, with N-terminal methionine removed) and to 1:15 (circles) and 1:14 (triangles) Sf1p–centrin complexes at high m/z (main panel: observed masses, 336,290 ± 191 and 317,308 ± 206 D; theoretical masses, 332,713 and 314,093 D, with excess mass due to solvent/buffer adduct retention under nondenaturing conditions). The errors in the observed masses are the standard deviations of the mass calculated for individual peaks in a series and are mainly due to variation in the degree of residual solvation of different charge states. The charge states for the 1:14 complex are omitted for clarity. The ion indicated by the filled triangle carries a charge of +41. (C) EM shadowing of the 15-repeat Sfi1p–centrin complex. Images were obtained with either 1 mM EGTA or 1 mM CaCl2, and the observed average length of the filaments is shown. Bar, 100 nm.
This complex was examined by EM shadowing (Fig. 5 C) and showed filaments 59 ± 7 nm (n = 74) in 1 mM EGTA and 59 ± 8 nm (n = 82) in 2 mM CaCl2. Thus, there was no discernable difference in filament length between low and high Ca2+. This is in agreement with the structures (Fig. 1, E and F) solved from crystal 1 (no Ca2+) and crystal 2 (0.1 M Ca2+), where Ca2+ has little effect on the conformations. If the Sfi1p fragment examined here was a single continuous α helix covered by centrins, then the 432 amino acids in this construct would predict a length of 65 nm, close to the 59 nm observed.
Arrangement of Sfi1p in the bridge
The bridge structure of the SPB, where Sfi1p is localized (Kilmartin, 2003), has been reported to change in length during the cell cycle from 90 nm in single SPBs to 150 nm in satellite-bearing SPBs (Winey et al., 1991); however, the numbers of cells examined in this study were small because the appropriate images occur rarely in thin sections. To confirm this change in length and measure the bridge length in paired SPBs, we reexamined all of our EM data collected over the years (see Materials and methods). We found the following lengths: for half-bridges from single SPBs in haploid (K699) mitotic cells, 57 ± 5 nm (n = 20), and for bridges from satellite-bearing and paired SPBs, 117 ± 9 nm (n = 53). The same numbers for a diploid strain (K842) were 60 ± 5 (n = 6) and 112 ± 9 nm (n = 9). There was no significant difference in bridge lengths between paired SPBs and satellite or duplication plaque-bearing SPBs. Another diploid strain (NCYC74) gave 109 ± 11 nm (n = 12) for paired SPBs, and a tetraploid strain (Adams and Kilmartin, 1999) showed 118 ± 9 nm (n = 9) for satellite-bearing SPBs. These relative distances confirm the differences found earlier (Winey et al., 1991), though there is a discrepancy in the absolute value of the numbers that we cannot explain. There may be some ambiguity in the absolute value of the numbers because of different fixation conditions; however, it seems probable that differences between the numbers under the same fixation conditions are valid. The numbers suggest an approximate doubling in bridge length between single SPBs in mitotic cells and cells undergoing SPB duplication and indicate that, in contrast to SPB size, which increases with ploidy (Byers and Goetsch, 1974), bridge length remains constant as ploidy increases.
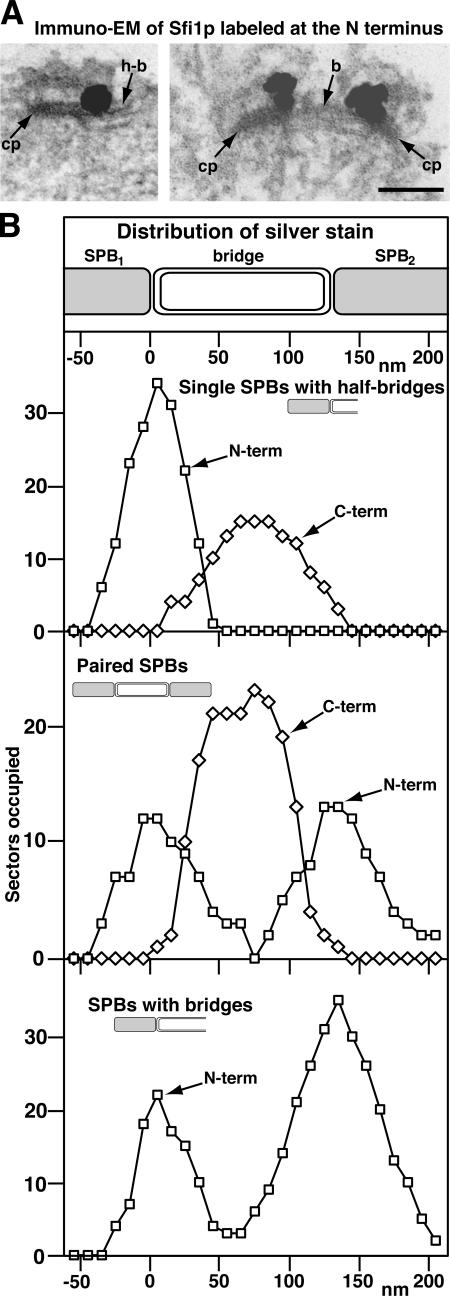
When the localization of Sfi1p was examined by immuno-EM in yeast, it was noticed that it appeared to have a restricted distribution on the bridge: at the distal end of the half-bridge in single SPBs and in the center of the bridge in paired SPBs (Kilmartin, 2003). This staining used anti-GFP with Sfi1p labeled with GFP at the C terminus; thus, the staining pattern may reflect the vicinity of the C-terminal region of Sfi1p. We have shown that a fragment of the Sfi1p–centrin complex containing ∼15 repeats forms a filament 60 nm long, which suggests that the full complex containing 21 repeats could be 90 nm long. This is long enough to span the 60-nm half-bridge with the rest possibly in the central plaque, and it agrees with the staining of centrin spread along the mainly cytoplasmic side of the bridge (Spang et al., 1993). To determine whether the N and C termini of Sfi1p are distant from each other, with the N terminus associated with the edge of the central plaque, a yeast strain containing Sfi1p labeled with GFP at the N terminus was stained with anti-GFP (Adams and Kilmartin, 1999). To quantify these observations, these data, together with the earlier results ( Kilmartin, 2003), were plotted as the presence or absence of silver deposition for 10-nm sectors along the SPBs and bridge (note that this would broaden the actual distribution, as it does not allow for the volume of deposition). The label itself before silver deposition, rabbit IgG with Fab anti–rabbit-Nanogold, would span ∼20 nm (Ribrioux et al., 1996). The data (Fig. 6) show a different distribution for the GFP on the N terminus compared with the C terminus. The N-terminal label is always close to the junction between the SPB and the proximal end of the half-bridge or bridge and in paired SPBs shows two separate sites of localization close to each SPB. In contrast, the C-terminal label, as found previously (Kilmartin, 2003), is either close to the distal end of the half-bridge or close to the center of the bridge in paired SPBs (Fig. 6 B). The N-terminal label also showed another class of labeling for single SPBs that appeared to have full bridges (Fig. 6 B, bottom). These are either SPBs at a very early stage of duplication, when daughter SPB components start to assemble (Adams and Kilmartin, 1999), or satellite-bearing SPBs or paired SPBs where it was not possible to locate a satellite or second SPB in the serial sections. Here, the mother SPB often had reduced accessibility to the antibody, hence the difference in peak heights at the N and C termini. Again, these SPBs show a bimodal distribution of stain, peaking close to the edge of the mother SPB and the expected position of new SPB assembly. The staining with both labels was always cytoplasmic. Our interpretation of these data is that the N terminus of Sfi1p lies close to the edge of the central plaque and the filamentous Sfi1p–centrin repeats span the length of the half-bridge, ending with the C terminus of Sfi1p at the distal end of the half-bridge. During duplication, the doubling in the length of the bridge takes place by the end-to-end addition of another molecule of Sfi1p by association of their C termini, and the N terminus of this second Sfi1p molecule is eventually associated with the edge of the central plaque of the daughter SPB.
Figure 6.
Immuno-EM of Sfi1p labeled at the N and C termini with GFP. (A) Images of a single (left) and paired SPBs (right) of cells containing Sfi1p labeled with GFP at the N terminus and stained with anti-GFP, anti-rabbit Fab-Nanogold, and silver intensification. cp, central plaque of the SPB; h-b, half-bridge; b, bridge. Bar, 100 nm. (B) Distribution of silver stain for stained cells labeled with GFP at the N terminus of Sfi1p as above and at the C terminus as described previously (Kilmartin, 2003). For each image, the region of staining at the SPB was divided into 10-nm sectors perpendicular to the long axis of the central plaque and along the bridge, starting from the edge of the central plaque. The presence or absence of staining in each of these sectors along the SPB and bridge was recorded. The numbers of SPBs examined in each category was slightly less than the maximum numbers of sectors occupied.
Discussion
Effect of Ca2+
Earlier work showed that, in contrast to the binding of yeast centrin to Kar1p (Geier et al., 1996), Ca2+ did not appear to be necessary for the binding between centrin and Sfi1p as assayed by pull downs (Kilmartin, 2003), and this was confirmed here, as all complexes were prepared in the presence of EGTA. Our structures provide a clear reason for this: Ca2+ does not appear to change the conformation of centrin when bound to Sfi1p. Very recently, the structure of the N-terminal domain of Chlamydomonas centrin in the presence of Ca2+ was solved by nuclear magnetic resonance (Sheehan et al., 2006). This has a different conformation from our five structures (the rmsd against the N-terminal domain of centrin A is 2.6 Å). Probably the constraints caused by binding Sfi1p and the centrin–centrin interactions prevent the N-terminal domain taking up such a conformation, or this may reflect the different members of the centrin family (the Chlamydomonas centrin is a centrin 2, whereas yeast centrin is a centrin 3). It would be very interesting to examine the recently discovered Chlamydomonas centrin 3 (Ruiz et al., 2005).
The lack of a significant effect of Ca2+ on the yeast centrin–Sfi1p interaction may reflect the fact that there is so far little evidence for a role of Ca2+ in the yeast cell cycle (Davis, 1995). The only increase in cytosolic Ca2+ concentration that has been detected in budding yeast under laboratory conditions is after mating pheromone treatment (Iida et al., 1990), and indeed yeast calmodulin is able to perform all its essential functions in the absence of Ca2+ binding (Geiser et al., 1991). In higher eukaryotes, Ca2+ transients have been detected during the cell cycle (Kahl and Means, 2003), and we found that stabilization of several human Sfi1–centrin 3 complexes containing three repeats required the presence of Ca2+ to remain intact on ion exchange chromatography (unpublished data). Thus, Ca2+ may play more of a role in Sfi1–centrin interactions in higher eukaryotes.
Mutations in budding yeast centrin
An extensive study of yeast centrin has been performed to identify mutations that cause a ts phenotype or lethality (Ivanovska and Rose, 2001) and thus may destabilize the structure or an interaction. A total of 40 mutants were isolated in this screen and in an earlier study (Vallen et al., 1994). For 33 of these, we can explain their effect by either altering a particularly conserved position in the EF-hand (Falke et al., 1994) or changing a core residue in centrin or a residue involved in the interaction between centrin and Sfi1p (E97G, A101T, F105L or Y, and C158F). Of the remainder, four (K112E, D144N, E148A, and E148Q) are within the EF-hands and should be acceptable substitutions (Falke et al., 1994), though they may perturb the relationships between the adjacent helices within an EF-hand. One mutation (R73W) places a hydrophobic tryptophan on the surface, and the final two (K91R and P94S) are in the linker region. P94 is conserved in all centrin 3 sequences (Azimzadeh and Bornens, 2004), whereas in calmodulins and other centrins it is replaced by S or T. With S or T in place of P94, helices IV and V can be continuous, as they are in Ca2+-calmodulin. The presence of the helix-breaker P94 between these two helices ensures they are always separate and may give additional flexibility to the linker. We cannot explain structurally why K91R gives a ts phenotype, as this is a surface residue with no interactions; however, this is one of the substitutions that causes a large decrease in Kic1p kinase activity (Ivanovska and Rose, 2001) and thus may perturb the binding between centrin and Kic1p (Sullivan et al., 1998).
Comparison with IQ repeats
Perhaps the most similar structure to the Sfi1p–centrin filament are the IQ repeats in myosin, which bind either calmodulin or myosin light chains, sometimes in the absence of Ca2+ (Bahler and Rhoads, 2002). These might be a lever arm that amplifies the movement of the myosin heavy chain along the actin filament. The structures of several of these have been solved and, like Sfi1p, all show the IQ repeats as an α helix but also with some distortions. In scallop myosin, the helix is bent between the two light chains, and toward the C terminus it is distorted into a right angle (Houdusse and Cohen, 1996). The light chains are less extended along the helix than centrin, they stabilize ∼20 residues, compared with 25–33 for centrin, and are in the opposite orientation to centrin. Both light chains make direct interactions with each other, but these are different from the centrin–centrin interactions found here. Myosin V has up to six continuous IQ repeats, and the structure of two of these has recently been solved. The structure is similar to that of scallop myosin but shows no direct interactions between the light chains (Terrak et al., 2005).
These structures and the two in this paper show that by imposition of kinks, bends, and bulges in an α helix, EF-hand proteins build a filament using a long α helix as a scaffold. It is not yet clear what the particular properties of these different types of filament are, though this type of arrangement is an attractive model (Kilmartin, 2003; Salisbury, 2004) for Ca2+-dependent contractile organelles such as the spasmoneme (Amos, 1975) and centrin-containing striated flagellar roots (Salisbury et al., 1984).
Structural role of Sfi1p in SPB duplication
The immuno-EM staining of Sfi1p at both the N terminus (Fig. 6 A) and C terminus (Kilmartin, 2003) shows only cytoplasmic staining, so it seems likely that Sfi1p, which has no transmembrane domain, either is positioned just above the densely stained outer nuclear membrane of the bridge or is a constituent of the cytoplasmic half-bridge outer layer (Fig. 7 A). This location suggests that a potential role in the elasticity of the bridge during assembly of the daughter SPB (Adams and Kilmartin, 1999; Kilmartin, 2003) is less likely and, indeed, if Sfi1p is part of the outer layer, then its length would not change during SPB duplication (Adams and Kilmartin, 1999), and this would be more consistent with the structural work presented here. The cytoplasmic part of the bridge is a flat rectangular structure (see plate 5 in Byers [1981a]), and our immuno-EM results suggest that Sfi1p is positioned across this structure with the N-terminal domain associated with the side of the SPB, the filamentous Sfi1 repeats and centrin laid in rows across the rectangle with the C-terminal domain at the distal end of the half-bridge (Fig. 7 C). The bridge, which is twice the length, would have the Sfi1p C-terminal domains associated, thus connecting the two SPBs.
The flat shape of the bridge suggests that there is only one or very few layers of Sfi1p–centrin filaments. If this is the case, our crystal structures suggest that there are unlikely to be extensive similar side-to-side interactions between the centrin-coated Sfi1p filaments because of the 65° twist in the centrin positions along the Sfi1p α helix (Fig. 7 B). This arrangement would resemble a Venetian blind with the slats held together at wide intervals, and the positions of the intervals would depend on the twist between each centrin. This relative lack of side-to-side interactions could explain how both microtubules and Sfi1p filaments are accommodated at the bridge at the same time. Microtubule assembly is initiated at the bridge during G1/S (Byers and Goetsch, 1975) and involves Kar1p, a transmembrane protein localized to the cytoplasmic side of the bridge (Spang et al., 1995), which binds the γ-tubulin–containing Tub4p complex via Spc72p (Pereira et al., 1999) and initiates microtubule assembly. Thus, these microtubules are anchored close to the outer nuclear membrane and might have to pass through cytoplasmic bridge components. If the Sfi1p filaments were organized like a venetian blind with widely spaced linkages between the filaments, then with some adjustments in filament position, microtubules could be accommodated between the filaments.
The suggested arrangement of Sfi1p on the half-bridge and bridge (Fig. 7 C) extends the earlier model for SPB duplication (Adams and Kilmartin, 1999), which proposed that the bridge had the property of binding cytoplasmic SPB components at either end. We extend this and propose that it is the N terminus of Sfi1p that either directly or indirectly binds the SPB components. An early step in SPB duplication would be the end-to-end association of Sfi1p at the C terminus, which might have to be activated at the appropriate point in the cell cycle or bind other proteins. This association would double the length of the bridge and provide a new N terminus of Sfi1p able to bind SPB components and thus start the assembly of the new SPB. The specificity of the association of the two C termini and the binding of the N terminus to SPB components would ensure that only a single copy of the SPB is produced. After assembly, the two SPBs would separate to form a spindle by dissociation of the connection between the C termini of Sfi1p.
Materials and methods
Protein expression and crystallization
Fragments of Sfi1p containing between 2 and 15 repeats were cloned as GST fusion proteins into the dicistronic vector pGEX-6p-2rbs (a gift from A. Musacchio, European Institute of Oncology, Milan, Italy) with yeast centrin (Cdc31p) in the non-GST site. All constructs produced soluble protein at 25°C, in contrast to expression of Sfi1p alone (Kilmartin, 2003), which only gave soluble GST fusion proteins in the presence of sarkosyl. These bound to glutathione beads but could not be eluted with glutathione. Complexes were isolated with glutathione beads, GST was removed, and the complex was released by PreScission cleavage. Further purification was by gel filtration on Superdex 200 in 10 mM Tris-Cl, pH 8.0, 0.15 M NaCl, 1 mM DTT, and 1 mM EGTA and then ion exchange chromatography on Q Sepharose using a 0.3–0.43 M NaCl gradient in the same buffer.
Crystals of first wild type and then selenomethionine-substituted protein (Teo et al., 2004) were obtained by sitting drop vapor diffusion at 4°C. The reservoir solutions contained 0.2 M sodium acetate, 18% polyethylene glycol 3350 (crystal 1), or 9% isopropanol, 0.1 M MES, pH 6.2, and 0.2 M calcium acetate (crystal 2). Drops contained 5 μl protein (20 mg/ml in 20 mM Tris-Cl, pH 8.0, 1 mM DTT, and 1 mM EGTA) mixed with 5 μl of the reservoir buffer.
Ca2+ measurements
Some electron density was present in the Ca2+ position in EF-hand 1, 3, and 4 of both centrins in crystal 1. This is probably water and not Ca2+ because refinement as Ca2+ resulted in a much higher average B factor than the average residue and the Ca2+ content of the protein and crystals is very low. This was measured by inductively coupled optical emission spectrometry and gave an EF-hand occupancy of around 1%, assuming three active EF-hands/centrin. In addition, to detect Ca2+ ions directly in both crystals 1 and 2, diffraction data were collected from crystals at a longer wavelength (λ = 1.74 Å) for crystal 1 at Beamline ID23 ESRF (Grenoble) or in-house at CuKα (λ = 1.54 Å) for crystal 2. An anomalous difference map was calculated using phases from the refined model. This showed no detectable Ca2+ in the EF-hands or elsewhere in crystal 1 and after refinement showed Ca2+ only at EF-hands 1, 3, and 4 in crystal 2 grown at high Ca2+ concentration.
S. cerevisiae strains
S. cerevisiae strains and yeast vectors were used as before (Kilmartin, 2003). Mutants in yeast centrin, CDC31, were prepared by QuikChange mutagenesis, confirmed by sequencing, and transferred to pRS314. These replaced the wild-type gene by plasmid shuffle, and if the strains were viable, PCR and sequencing were used to check that the mutation was retained. A strain containing Sfi1p labeled with GFP at the N terminus was prepared by insertion of an NcoI site and then GFP into SFI1, integration of a single copy at the TRP1 locus, and plasmid shuffle to remove the wild-type gene.
Nanospray mass spectrometry
Nanospray mass spectrometry was performed using modified instrument technology, involving increased pressures and reduced quadrupole frequencies to assist in the analysis of high m/z ions (Sobott et al., 2002), incorporated into a Q-Star XL instrument (MDS Sciex; Chernushevich and Thomson, 2004). Protein samples were transferred into 0.1 M ammonium acetate, pH 7.0, using three successive Bio-spin 6 columns (Bio-Rad Laboratories). Nanospray capillaries were prepared as described previously (Sobott and Robinson, 2004). Desolvation of high m/z ions was assisted by “collisional cleaning” using argon gas in the instrument's collision cell. The observed ion series were assigned manually and were consistent with a 1:15 (major component) and 1:14 (minor component) Sfi1p–centrin complex. Higher energy collision-induced dissociation experiments, either in “normal” mass spectrometry mode or by tandem mass spectrometry, caused the ejection of a single centrin monomer from the complex, to give 1:14 and 1:13 “stripped oligomers,” supporting the assigned parent ion stoichiometry.
EM
Immuno-EM and EM shadowing were done as before (Kilmartin et al., 1993; Adams and Kilmartin, 1999). In thin-section EM, bridge length was measured as the distance between the edge of the central plaques to the edge of the second central plaque or satellite or duplication plaque. Half-bridge length was measured from the edge of the central plaque to the end of the electron-dense nuclear membrane or cytoplasmic outer layer. Half-bridge and bridge length between paired SPBs was measured from log phase cells, and other measurements were from α factor–blocked or –released cells. EM photographic negatives were digitized and transferred to Photoshop 7.0 (Adobe).
Online supplemental material
Table S1 shows the MAD data collection, phasing, and refinement statistics. The supplemental text gives the GenBank identifiers of the sequences used in the analysis of Sfi1 repeats, describes the methods used in the refinement of the crystal structure, and gives the interhelical angles for the closed and open N- and C-terminal domains of centrin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200603153/DC1.
Supplementary Material
Acknowledgments
We thank Ingar Leiros and Didier Nurizzo for assistance with Beamline ID23-1 and Laurent Terradot for help with Beamline ID14-4 at the European Synchrotron Radiation Facility, John Finch for help with the EM shadowing, Mervyn Greaves for the inductively coupled optical emission spectrometry measurements, and M.S. Robinson for discussion.
Abbreviations used in this paper: MTOC, microtubule organizing center; rmsd, root mean square deviation; SPB, spindle pole body; ts, temperature-sensitive.
References
- Adams, I.R., and J.V. Kilmartin. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145:809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos, W.B. 1975. Contraction and calcium binding in the vorticellid ciliates. Soc. Gen. Physiol. Ser. 30:411–436. [PubMed] [Google Scholar]
- Azimzadeh, J., and M. Bornens. 2004. The centrosome in evolution. In Centrosomes in Development and Disease. E.A. Nigg, editor. Wiley-VCH, Weinheim, Germany. 93–122.
- Bahler, M., and A. Rhoads. 2002. Calmodulin signaling via the IQ motif. FEBS Lett. 513:107–113. [DOI] [PubMed] [Google Scholar]
- Biggins, S., and M.D. Rose. 1994. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 125:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. a. Cytology of the yeast cell cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 59–96.
- Byers, B. 1981. b. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In Molecular genetics in yeast, Alfred Benzon Symposium 16. D. von Wettstein, J. Friis, M. Kielland-Brand, and A. Stenderup, editors. Munksgaard, Copenhagen, Denmark. 119–131.
- Byers, B., and L. Goetsch. 1974. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 38:123–131. [DOI] [PubMed] [Google Scholar]
- Byers, B., and L. Goetsch. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernushevich, I.V., and B.A. Thomson. 2004. Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 76:1754–1760. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. [DOI] [PubMed] [Google Scholar]
- Davis, T.N. 1995. Calcium in Saccharomyces cerevisiae. Adv. Second Messenger Phosphoprotein Res. 30:339–358. [DOI] [PubMed] [Google Scholar]
- Falke, J.J., S.K. Drake, A.L. Hazard, and O.B. Peersen. 1994. Molecular tuning of ion binding to calcium signaling proteins. Q. Rev. Biophys. 27:219–290. [DOI] [PubMed] [Google Scholar]
- Geier, B.M., H. Wiech, and E. Schiebel. 1996. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J. Biol. Chem. 271:28366–28374. [DOI] [PubMed] [Google Scholar]
- Geiser, J.R., D. van Tuinen, S. Brockerhoff, M.M. Neff, and T.N. Davis. 1991. Can calmodulin function without binding calcium? Cell. 65:949–959. [DOI] [PubMed] [Google Scholar]
- Houdusse, A., and C. Cohen. 1996. Structure of the regulatory domain of scallop myosin at 2 Å resolution: implications for regulation. Structure. 4:21–32. [DOI] [PubMed] [Google Scholar]
- Hu, H., and W.J. Chazin. 2003. Unique features in the C-terminal domain provide caltractin with target specificity. J. Mol. Biol. 330:473–484. [DOI] [PubMed] [Google Scholar]
- Iida, H., Y. Yagawa, and Y. Anraku. 1990. Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway. J. Biol. Chem. 265:13391–13399. [PubMed] [Google Scholar]
- Ilag, L.L., L.F. Westblade, C. Deshayes, A. Kolb, S.J. Busby, and C.V. Robinson. 2004. Mass spectrometry of Escherichia coli RNA polymerase: interactions of the core enzyme with sigma70 and Rsd protein. Structure. 12:269–275. [DOI] [PubMed] [Google Scholar]
- Ivanovska, I., and M.D. Rose. 2001. Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics. 157:503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S.L., T.H. Giddings Jr., and M. Winey. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl, C.R., and A.R. Means. 2003. Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr. Rev. 24:719–736. [DOI] [PubMed] [Google Scholar]
- Kilmartin, J.V. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162:1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin, J.V., S.L. Dyos, D. Kershaw, and J.T. Finch. 1993. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell cycle-regulated. J. Cell Biol. 123:1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, Y.Y., E.Y. Lai, S.P. Remillard, M.B. Heintzelman, and C. Fulton. 1996. Centrin is a conserved protein that forms diverse associations with centrioles and MTOCs in Naegleria and other organisms. Cell Motil. Cytoskeleton. 33:298–323. [DOI] [PubMed] [Google Scholar]
- Middendorp, S., T. Kuntziger, Y. Abraham, S. Holmes, N. Bordes, M. Paintrand, A. Paoletti, and M. Bornens. 2000. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, M., H. Tokumitsu, M.B. Swindells, H. Kurihara, M. Orita, T. Shibanuma, T. Furuya, and M. Ikura. 1999. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat. Struct. Biol. 6:819–824. [DOI] [PubMed] [Google Scholar]
- Pereira, G., U. Grueneberg, M. Knop, and E. Scheibel. 1999. Interaction of the yeast γ-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J. 18:4180–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribrioux, S., G. Kleymann, W. Haase, K. Heitmann, C. Ostermeier, and H. Michel. 1996. Use of nanogold- and fluorescent-labeled antibody Fv fragments in immunocytochemistry. J. Histochem. Cytochem. 44:207–213. [DOI] [PubMed] [Google Scholar]
- Ruiz, F., N.G. De Loubresse, C. Klotz, J. Beisson, and F. Koll. 2005. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr. Biol. 15:2097–2106. [DOI] [PubMed] [Google Scholar]
- Salisbury, J.L. 1995. Centrin, centrosomes and mitotic spindle poles. Curr. Opin. Cell Biol. 7:39–45. [DOI] [PubMed] [Google Scholar]
- Salisbury, J.L. 2004. Centrosomes: Sfi1p and centrin unravel a structural riddle. Curr. Biol. 14:R27–R29. [DOI] [PubMed] [Google Scholar]
- Salisbury, J.L., A. Baron, B. Surek, and M. Melkonian. 1984. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J. Cell Biol. 99:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury, J., K. Suino, R. Busby, and M. Springett. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12:1287–1292. [DOI] [PubMed] [Google Scholar]
- Sheehan, J.H., C.G. Bunick, H. Hu, P.A. Fagan, S.M. Meyn, and W.J. Chazin. 2006. Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function. J. Biol. Chem. 281:2876–2881. [DOI] [PubMed] [Google Scholar]
- Sobott, F., and C.V. Robinson. 2004. Characterising electrosprayed biomolecules using tandem-MS—the noncovalent GroEL chaperonin assembly. International Journal of Mass Spectrometry. 236:25–32. [Google Scholar]
- Sobott, F., H. Hernandez, M.G. McCammon, M.A. Tito, and C.V. Robinson. 2002. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74:1402–1407. [DOI] [PubMed] [Google Scholar]
- Spang, A., I. Courtney, U. Fackler, M. Matzner, and E. Schiebel. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., I. Courtney, K. Grein, M. Matzner, and E. Schiebel. 1995. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128:863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf, A.J., G. Morgan, T.H. Giddings Jr., E.A. White, R. Marchione, H.B. McDonald, and M. Winey. 2005. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 16:3606–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, D.S., S. Biggins, and M.D. Rose. 1998. The yeast centrin, Cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143:751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindells, M.B., and M. Ikura. 1996. Pre-formation of the semi-open conformation by the apo-calmodulin C-terminal domain and implications binding IQ-motifs. Nat. Struct. Biol. 3:501–504. [DOI] [PubMed] [Google Scholar]
- Teo, H., O. Perisic, B. Gonzalez, and R.L. Williams. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 7:559–569. [DOI] [PubMed] [Google Scholar]
- Terrak, M., G. Rebowski, R.C. Lu, Z. Grabarek, and R. Dominguez. 2005. Structure of the light chain-binding domain of myosin V. Proc. Natl. Acad. Sci. USA. 102:12718–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen, E.A., W. Ho, M. Winey, and M.D. Rose. 1994. Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae. Genetics. 137:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, A.C., R.A. Laskowski, and J.M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134. [DOI] [PubMed] [Google Scholar]
- Winey, M., L. Goetsch, P. Baum, and B. Byers. 1991. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114:745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.