Abstract
Through a functional genomic screen for mitotic regulators, we identified hepatoma up-regulated protein (HURP) as a protein that is required for chromosome congression and alignment. In HURP-depleted cells, the persistence of unaligned chromosomes and the reduction of tension across sister kinetochores on aligned chromosomes resulted in the activation of the spindle checkpoint. Although these defects transiently delayed mitotic progression, HeLa cells initiated anaphase without resolution of these deficiencies. This bypass of the checkpoint arrest provides a tumor-specific mechanism for chromosome missegregation and genomic instability. Mechanistically, HURP colocalized with the mitotic spindle in a concentration gradient increasing toward the chromosomes. HURP binds directly to microtubules in vitro and enhances their polymerization. In vivo, HURP stabilizes mitotic microtubules, promotes microtubule polymerization and bipolar spindle formation, and decreases the turnover rate of the mitotic spindle. Thus, HURP controls spindle stability and dynamics to achieve efficient kinetochore capture at prometaphase, timely chromosome congression to the metaphase plate, and proper interkinetochore tension for anaphase initiation.
Introduction
Proper chromosome alignment at the metaphase plate ensures the fidelity of chromosome segregation in mitosis (Kops et al., 2005). This process of bipolar spindle formation and chromosome congression is mediated by spindle poles, microtubules, and kinetochores through a combination of centrosome- and chromatin-mediated pathways (Compton, 2000; Gadde and Heald, 2004). Through a search and capture mechanism, microtubules that nucleated from the centrosomes attach to kinetochores. In a parallel pathway, chromatin promotes the nucleation of microtubules that extend toward the centrosomes to link kinetochores to the mitotic spindle poles.
To ensure the equal segregation of chromosomes during mitosis, the spindle checkpoint monitors both the attachment of microtubules to kinetochores and the tension generated across bioriented sister kinetochores (Shah and Cleveland, 2000; Cleveland et al., 2003). Satisfaction of these two requirements abrogates signaling at the kinetochores through the checkpoint proteins Mad2 and BubR1, thereby allowing activation of the anaphase-promoting complex/cyclosome (APC/C). This ubiquitin ligase targets the anaphase inhibitor securin for degradation, leading to anaphase onset. Thus, the spindle checkpoint controls the fidelity of chromosome separation by preventing inappropriate anaphase initiation.
Disruption of mitotic events such as spindle formation and checkpoint response can promote genomic instability (Wassmann and Benezra, 2001; Weaver and Cleveland, 2005). Therefore, an understanding of mitotic regulation is central to dissecting the basic mechanisms of tumorigenesis. In a functional genomic screen, we identified hepatoma up-regulated protein (HURP) as a mitotic regulator. Upon the depletion of HURP, we found that HeLa cells at metaphase had a high frequency of unaligned chromosomes lacking microtubule attachment. Even chromosomes aligned at the metaphase plate were only under partial tension. Although Mad2 and BubR1 were recruited to kinetochores, these cells bypassed the spindle checkpoint and initiated anaphase prematurely. Biochemically, HURP is a microtubule-associated protein (MAP) that colocalizes with the mitotic spindle in a gradient that increases toward the chromosomes. HURP enhances microtubule polymerization and stabilizes the mitotic spindle by reducing the turnover rate of α/β-tubulin subunits on the spindle. We concluded that HURP promotes the efficient capture of kinetochores, timely chromosome congression, and proper interkinetochore tension by controlling the stability and dynamics of spindle microtubules.
Results
A genomic screen identifies novel mitotic regulators
Genes with mitotic functions such as cyclin B, Aurora A, and TPX2 have similar transcriptional expression profiles during the cell cycle, as they are all induced in G2/M (de Lichtenberg et al., 2005). Whitfield et al. (2002) reported that 566 genes in the human transcriptome are induced in G2 or G2/M, and we expected that a subset of these genes would have mitotic functions. In our initial analysis, we focused on 30 novel genes with the best induction in G2/M (see Materials and methods for the gene list).
To refine this list of potential mitotic regulators, we applied a second genomics filter based on the fact that in tumor cells, known mitotic regulators are transcriptionally up- or down-regulated in concert, thereby resulting in stereotypical covariation profiles (Rhodes et al., 2004). By surveying the analysis of transcriptional array data from 944 cancer tissue samples covering 22 different tumor types that were reported by Segal et al. (2004), we found that 12 of the 30 aforementioned genes covaried with known mitotic regulators such as Plk1, Cenp-A, and Nek2.
The functions of these 12 potential mitotic regulators were analyzed in two assays. First, the cellular localization of these genes was determined by expression of the respective GFP fusion protein in HeLa cells. Second, the expression of endogenous gene products was reduced by RNA interference, and spindle and DNA morphology was analyzed in mitotic cells by immunofluorescence staining. These analyses led to our identification of HURP as a potential mitotic regulator (Tsou et al., 2003).
To determine its potential role in mitosis, we ectopically expressed GFP-HURP in HeLa cells and found that although GFP-HURP was diffusely cytoplasmic during interphase, it localized predominantly to the chromatin-proximal regions of the mitotic spindle. Thus, HURP may regulate microtubule function specifically in mitosis (Fig. 1 B).
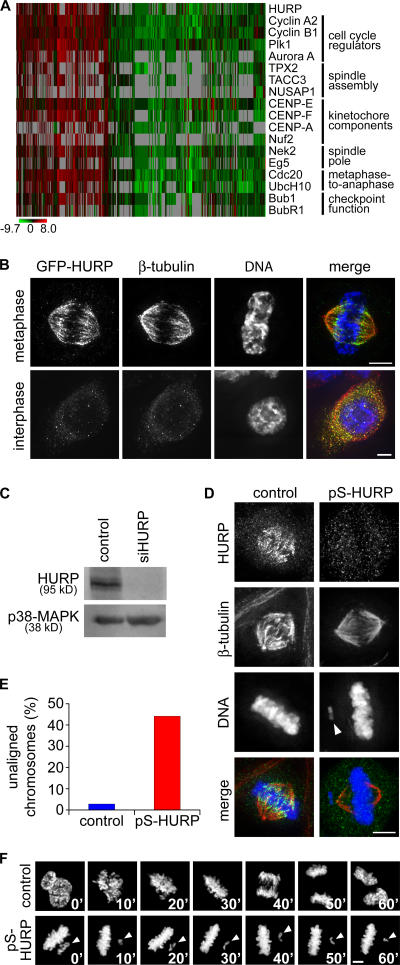
Figure 1.
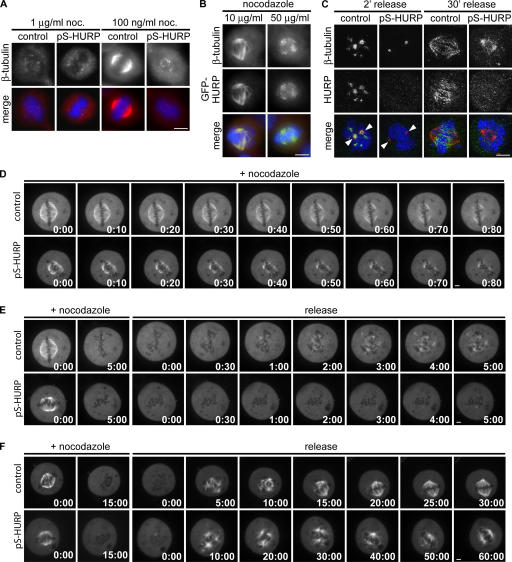
Functional genomic analysis identified HURP as a spindle- associated protein required for complete chromosome congression. (A) Coexpression of HURP with known mitotic regulators in tumor microarrays. Each of the 944 columns represents a tumor microarray experiment. The expression data were extracted, normalized, and calculated as described by Segal et al. (2004). The change in expression (in log scale) of each gene in a given array is relative to the mean expression of the gene across all arrays. Gray pixels represent missing values. (B) Maximum projections from deconvolved z stacks of HeLa cells expressing GFP-HURP stained for GFP (green), β-tubulin (red), and DNA (blue). (C) HeLa cells transfected with control or siRNAs against HURP (siHURP) were collected 48 h after transfection and analyzed by Western blotting. p38 MAP kinase (p38-MAPK) served as a loading control. (D) Maximum projections from deconvolved z stacks of control or HURP-depleted HeLa cells stained for HURP (green), β-tubulin (red), and DNA (blue). (E) The percentage of unaligned chromosomes at metaphase was quantified in control (n = 107) or HURP-depleted (n = 111) HeLa cells. (F) Still frames from confocal time-lapse microscopy of control or HURP-depleted HeLa/GFP-H2B cells (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). Time is given in minutes. Arrowheads indicate unaligned chromosomes. Bars, 5 μm.
Next, we depleted endogenous HURP from HeLa cells using pSUPER constructs and siRNAs targeting five different sequences, all of which generated the same phenotypes. siRNA-mediated knockdown reduced the protein level of HURP to <5% of that in control cells (Fig. 1 C and not depicted) and abolished the localization pattern of endogenous HURP on the spindle (Fig. 1 D). We found that 44% of HURP-depleted metaphase cells contained at least one unaligned chromosome compared with only 3% in control cells (Fig. 1 E). HURP depletion in Hct116 cells also generated a similar phenotype, with 8.9% of HURP-depleted metaphase cells containing at least one unaligned chromosome compared with only 2.8% of control cells (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). Based on confocal time-lapse microscopy of HURP-depleted HeLa cells stably expressing GFP–histone H2B (HeLa/GFP-H2B; n = 41), these unaligned chromosomes at metaphase persisted for up to 60 min (Fig. 1 F, bottom; and Video 1). In contrast, control cells (n = 37) progressed from prophase to mitotic exit within 40 min. Thus, HURP is a mitotic regulator that is required for efficient chromosome congression.
HURP depletion activates the spindle checkpoint
To determine the cellular basis for the lack of congression of unaligned chromosomes, we analyzed HeLa cells for indications of kinetochore attachment to microtubules. In control and HURP-depleted cells, the kinetochore marker CREST and spindle microtubules were juxtaposed for chromosomes aligned at the metaphase plate (Fig. 2 A). However, in HURP-depleted cells, the kinetochores of unaligned chromosomes were not attached to any microtubules.
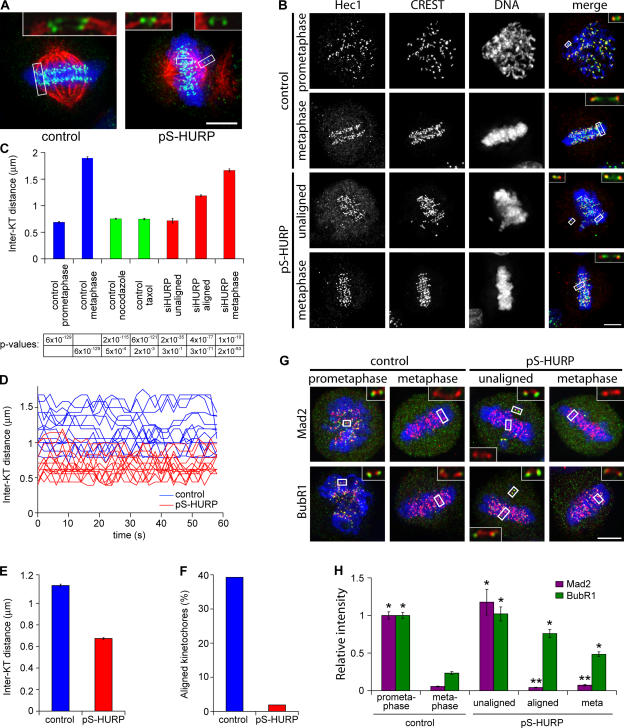
Figure 2.
HURP controls both microtubule attachment to kinetochores and tension across sister kinetochores, the lack of which activates the spindle checkpoint. (A) Maximum projections from deconvolved z stacks of control or HURP-depleted HeLa cells stained for CREST (green), β-tubulin (red), and DNA (blue). (B) Maximum projections from deconvolved z stacks of control or HURP-depleted HeLa cells stained for Hec1 (green), CREST (red), and DNA (blue). (C) Interkinetochore (Inter-KT) distance was quantified based on Hec1 and CREST staining in five HeLa cells (n > 150 kinetochore pairs) for each category except for unaligned chromosomes (n = 42 kinetochore pairs from 19 cells). P values were generated from one-tailed t tests. (D and E) Interkinetochore distances (n = 10 kinetochore pairs) across the time course quantified and averaged (from three cells) by time-lapse confocal microscopy of HeLa/GFP–Cenp-A cells (Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). P = 1.6 × 10−149 (one-tailed t test). (F) The presence of an aligned kinetochore track as shown in B (control metaphase) was quantified in control (n = 112) or HURP-depleted (n = 106) HeLa cells. (G) Maximum projections from deconvolved z stacks of control or HURP-depleted HeLa cells stained for Mad2 or BubR1 (green), Hec1 (red), and DNA (blue). (H) Mad2 and BubR1 signals were quantified and normalized against Hec1 signals in five control (n = 60 kinetochores) or HURP-depleted (n = 60 kinetochores) HeLa cells; in the case of unaligned chromosomes, n = 12 kinetochores from six cells. Insets show a single z slice of boxed regions. *, P < 3 × 10−5; **, P < 0.02 (one-tailed t test). Error bars represent SEM. Bars, 5 μm.
The lack of attachment to microtubules should abolish tension across sister kinetochores. We measured interkinetochore distance between pairs of Hec1 dots as an indication of the level of tension. In control cells, metaphase chromosomes under full tension had longer interkinetochore distances (1.90 ± 0.02 μm; Fig. 2, B and C) compared with prometaphase chromosomes that were unattached and under no tension (0.69 ± 0.02 μm) and mitotic cells treated with 1 μg/ml nocodazole (0.75 ± 0.01 μm) or with 1 μM taxol (0.75 ± 0.01 μm). As expected, the interkinetochore distance for unaligned chromosomes in HURP-depleted cells (0.71 ± 0.04 μm) was similar to that of control prometaphase chromosomes, indicating that the unaligned chromosomes are not under any tension.
Surprisingly, in HURP-depleted cells with unaligned chromosomes, the interkinetochore distance of aligned chromosomes (1.19 ± 0.02 μm) was shorter than that of control metaphase chromosomes. In fact, even in HURP-depleted cells with all chromosomes aligned at the metaphase plate, sister kinetochores still exhibited an intermediate interkinetochore distance (1.67 ± 0.03 μm), which is consistent with partial tension. This decrease in interkinetochore distance was confirmed by confocal time-lapse microscopy in HeLa cells stably expressing GFP–Cenp-A (HeLa/GFP–Cenp-A; Fig. 2, D and E; and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). For chromosomes aligned at the metaphase plate, the mean interkinetochore distance in control cells (1.16 ± 0.01 μm), as measured by the distance between pairs of GFP–Cenp-A dots, was longer compared with HURP-depleted cells (0.67 ± 0.01 μm). Finally, HURP depletion resulted in an irregular alignment of kinetochores along the metaphase plate, possibly as a result of a reduction in tension. Only 2% of HURP-depleted metaphase cells exhibited a pattern of two parallel tracks of kinetochores compared with 39% in control cells (Fig. 2, B and F). Thus, HURP is essential for the attachment of microtubules to kinetochores and is required for generating full tension to precisely align chromosomes along the metaphase plate.
Because HURP-depleted cells at metaphase contained kinetochores that were unattached or under partial tension, we investigated the state of the spindle checkpoint. In control cells, the checkpoint proteins Mad2, which monitors attachment (Chen et al., 1996; Li and Benezra, 1996), and BubR1, which monitors attachment and tension (Chan et al., 1999; Skoufias et al., 2001), localized to kinetochores at prometaphase but disappeared at metaphase (Fig. 2, G and H). As expected, kinetochores on unaligned chromosomes in HURP-depleted cells exhibited high Mad2 and BubR1 levels. Furthermore, in HURP-depleted cells, BubR1 also remained on kinetochores of aligned chromosomes that were under partial tension. These data indicate that in the absence of HURP, checkpoint signaling on kinetochores is activated because of a lack of attachment and insufficient tension.
The spindle checkpoint transiently arrests HURP-depleted cells
Although HURP-depleted HeLa cells activated the spindle checkpoint, the mitotic population did not accumulate significantly. Mitotic indices for HURP-depleted cells were slightly increased compared with control cells when quantified by flow cytometry (control, 2.08%; HURP depleted, 2.75%; Fig. 3 A) or by immunofluorescence staining (control, 2.8%; HURP depleted, 3.4%; Fig. 3 B). A similar increase in mitotic index was also found by flow cytometry in HURP-depleted Hct116 cells (control, 1.96%; HURP depleted, 3.01%; Fig. S1 B). Furthermore, within the mitotic population itself, HURP-depleted cells were only slightly enriched at metaphase (Fig. 3 C). Thus, the spindle checkpoint only transiently arrests HURP-depleted cells at metaphase.
Figure 3.
HURP depletion delays mitotic progression. (A) Mitotic index of control or HURP-depleted cells determined by flow cytometry. Cells were fixed 48 h after transfection and stained for phosphohistone H3 (H3P). Boxed areas indicate the mitotic populations. PI, propidium iodide. (B) Mitotic index determined by DNA and spindle morphology in control or HURP-depleted HeLa cells (n > 1,800 cells). Cells were treated with or without 1 μg/ml nocodazole for 14 h and fixed for immunofluorescence. (C) Mitotic stages were quantified by DNA and spindle morphology (n > 100 mitotic cells) in the mitotic population of untreated cells in B. (D) Mitotic stages were quantified by DNA and spindle morphology in control or HURP-depleted HeLa cells treated with nocodazole for 14 h, released, and fixed at the indicated time points for immunofluorescence (n > 100 mitotic cells). (E) Control (n = 184) or HURP-depleted (n = 73) HeLa/GFP-H2B cells were monitored by time-lapse microscopy using a very low level of light intensity to determine the duration from NEB to anaphase, NEB to the formation of a bipolar spindle with some unaligned chromosomes (NEB to unaligned), the unaligned state to metaphase (unaligned to metaphase), and metaphase to anaphase (Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). Error bars represent SEM. *, P < 5 × 10−3; **, P < 10−6; ***, P < 3 × 10−9 (one-tailed t test).
To investigate the role of spindle checkpoint response in HURP-depleted cells, we examined the kinetics of mitotic progression. Control or HURP-depleted HeLa cells were treated for 14 h with 100 ng/ml nocodazole to stably arrest cells in prometaphase. In the presence of nocodazole, control and HURP-depleted cells had similar mitotic indices of 47.5 and 46.9%, respectively (Fig. 3 B). This indicates that HURP itself is not a checkpoint protein required for the establishment or maintenance of the spindle checkpoint. Upon release from nocodazole arrest, the fraction of cells progressing beyond prometaphase was lower in HURP-depleted samples compared with control samples at both 1 h (HURP depleted, 24.9%; control, 44.0%) and 2 h after release (HURP depleted, 38.4%; control, 61.6%; Fig. 3 D). We concluded that the spindle checkpoint in HURP-depleted cells delays mitotic progression but does not maintain a stable mitotic arrest.
To determine the defects in HURP-depleted cells that delayed mitotic progression, we analyzed the kinetics of mitotic progression by time-lapse imaging using HeLa/GFP-H2B cells. Among the cells that progressed through mitosis, we found that the duration from nuclear envelope breakdown (NEB) to anaphase was substantially longer in HURP-depleted cells (59.1 ± 3.0 min) compared with control cells (42.8 ± 1.1 min; Fig. 3 E and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). To further define the mitotic stages that were delayed in HURP-depleted cells, we characterized the duration between four mitotic transitions: (1) NEB; (2) an unaligned state in which the metaphase plate and bipolar spindle had formed but before complete chromosome congression; (3) complete chromosome congression at metaphase; and (4) anaphase onset. The duration from NEB to the unaligned state was only slightly increased in HURP-depleted cells (9.5 ± 0.2 min) compared with control cells (8.2 ± 0.1 min), indicating that the kinetics of bipolar spindle formation and of the initial capture of chromosomes were only slightly affected by HURP. On the other hand, the duration from the unaligned state to metaphase was substantially longer in HURP-depleted cells (15.0 ± 1.0 min) compared with control cells (8.1 + 0.4 min), suggesting that the capture of the remaining unaligned chromosomes was much less efficient in the absence of HURP. Finally, the duration from metaphase to anaphase onset was longer in HURP-depleted cells (34.7 ± 2.9 min) compared with that in control cells (26.4 ± 1.0 min), indicating that the metaphase to anaphase transition was also delayed in the absence of HURP, likely because of insufficient tension. Thus, to varying degrees, HURP is required for timely bipolar spindle formation, chromosome congression, and anaphase onset.
HURP-depleted HeLa cells bypass the spindle checkpoint arrest
To investigate the fate of HURP-depleted cells in greater detail, we tracked their mitotic progression by confocal time-lapse microscopy in HeLa/GFP-H2B cells. Surprisingly, despite a delay during metaphase, 18 of 32 HURP-depleted cells containing one to two unaligned chromosomes initiated anaphase without complete chromosome congression, resulting in an unequal segregation of chromosomes (Fig. 4 A and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). To determine whether this checkpoint bypass phenotype is an intrinsic property of HeLa cells or a result of HURP depletion, we partially knocked down ch-TOG (Fig. 4 D) using siRNAs targeted to four different sequences, all of which gave the same phenotypes. Although the complete depletion of ch-TOG generated the previously reported phenotype of multipolar spindles (Cassimeris and Morabito, 2004), the partial depletion of ch-TOG using a limiting amount (1%) of specific siRNA generated only one to eight unaligned chromosomes in metaphase cells (Fig. 4 E). Among these ch-TOG–depleted HeLa/GFP-H2B metaphase cells analyzed during the 90-min time lapse, we observed that 6 of 30 metaphase cells containing one to four unaligned chromosomes initiated anaphase without complete chromosome congression (Fig. 4 B and Video 4). Unaligned chromosomes at metaphase were also generated in wild-type HeLa/GFP-H2B cells by treatment with a very low concentration of nocodazole (10 ng/ml) for 1 h, which neither depolymerized microtubules nor stably arrested HeLa cells at metaphase. Under these conditions, 3 of 20 cells that contained one to four unaligned chromosomes initiated anaphase without complete chromosome congression within the duration of the time lapse (Fig. 4 C and Video 4). Thus, HeLa cells have the intrinsic ability to initiate anaphase in the presence of a few unaligned chromosomes, which is a potential mechanism for genomic instability.
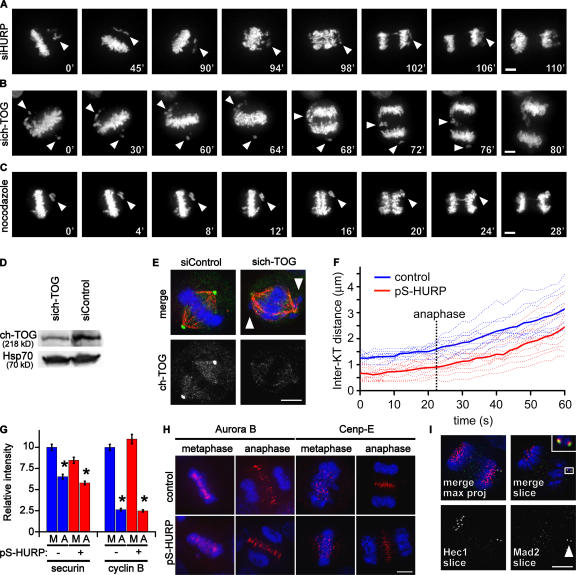
Figure 4.
HeLa cells initiate anaphase in the presence of unattached chromosomes or kinetochores under partial tension. (A–C) Still frames from confocal time-lapse microscopy of a HeLa/GFP-H2B cell depleted of HURP (A), partially depleted of ch-TOG (B), or treated with 10 ng/ml nocodazole (C; Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). Anaphase onset was between 90 and 94 min in A, 60 and 64 min in B, and 12 and 16 min in C. Arrowheads indicate unaligned chromosomes. Time is given in minutes. (D) HeLa cells transfected with control or siRNAs against ch-TOG (sich-TOG) were collected 24 h after transfection and analyzed by Western blotting. Hsp70 served as a loading control. (E) Maximum projections from deconvolved z stacks of control or ch-TOG–depleted HeLa cells stained for ch-TOG (green), β-tubulin (red), and DNA (blue). Arrowheads indicate unaligned chromosomes. (F) Interkinetochore distance (n = 10 kinetochore pairs) was quantified by time-lapse confocal microscopy in three control or HURP-depleted HeLa/GFP–Cenp-A cells (Video 5). Dotted traces indicate individual kinetochore pairs, and thick traces indicate an averaged distance. Traces were aligned temporally for anaphase onset, which was visually identified. (G) Levels of cytoplasmic securin or cyclin B were quantified by immunofluorescence in control or HURP-depleted cells at metaphase (M) or anaphase (A; n = 10 cells). Error bars represent SEM. *, P < 2 × 10−5 (one-tailed t test). (H) Maximum projections from deconvolved z stacks of control or HURP-depleted HeLa cells stained for Aurora B or Cenp-E (red) and DNA (blue). (I) HURP-depleted anaphase cells were stained for Mad2 (green), Hec1 (red), and DNA (blue). A maximum projection of the deconvolved z stack for an anaphase cell is presented in the top left panel. The remaining panels are taken from a single z slice to show the colocalization of Mad2 (arrowhead) and Hec1 on one pair of kinetochores (inset) in this anaphase cell. Bars, 5 μm.
Although only 44% of HURP-depleted cells contained unaligned chromosomes at metaphase (Fig. 1 E), all HURP-depleted cells exhibited a decrease in tension across sister kinetochores (Fig. 2, B–D). We measured the interkinetochore distance as an indication of tension at the metaphase to anaphase transition by confocal time-lapse microscopy of HeLa/GFP–Cenp-A cells. In control cells, the mean interkinetochore distance at anaphase onset was 1.7 ± 0.1 μm (Fig. 4 F and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). However, HURP-depleted cells with all chromosomes aligned at the metaphase plate initiated chromosome segregation at a shorter mean interkinetochore distance of 0.9 ± 0.1 μm, indicating anaphase onset with only partial tension. Because metaphase HeLa cells initiated anaphase in the presence of unaligned chromosomes or of sister kinetochores under partial tension, HeLa cells have an inherently less sensitive response to the spindle checkpoint.
To determine whether anaphase onset in HURP-depleted cells is initiated through APC/C activation, we assayed for the degradation of the APC/C substrates securin and cyclin B. By quantitative analysis of cytoplasmic fluorescence intensities, we found that the levels of these proteins were lower in anaphase compared with metaphase in both control and HURP-depleted cells (n = 10; Fig. 4 G). The anaphase markers Aurora B and Cenp-E were also assayed to determine whether other anaphase events occurred normally. In control and HURP-depleted cells, both proteins correctly localized to the centromeres/kinetochores at metaphase and redistributed to the central spindle at anaphase (Fig. 4 H). Thus, HURP-depleted cells initiated global anaphase events through the activation of APC/C even in the presence of unaligned chromosomes and kinetochores under partial tension.
These observations then raised questions concerning the status of the spindle checkpoint and the mechanism of its bypass at the onset of anaphase. Through extensive immunofluorescence analysis, we observed Mad2 staining on one to three kinetochores per cell in a subset of HURP-depleted anaphase A cells (Fig. 4 I), indicating that a subset of anaphase chromosomes were unattached in these cells. Because the signaling pathway up to the kinetochore association of Mad2 remained intact, HURP-depleted cells bypassed or adapted to the spindle checkpoint at a point downstream of this signal.
HURP is a MAP that controls spindle morphology
We next determined the localization of endogenous HURP across the cell cycle. Although HURP did not associate with interphase microtubules, it did colocalize with spindle microtubules during mitosis and cytokinesis (Fig. 5 A). Specifically, HURP is localized in a gradient along the pole-to-pole axis with higher concentrations at chromatin-proximal regions that decline toward the poles (Fig. 5 B). This localization pattern indicates that HURP is actively regulated through its association with microtubules during mitosis and its affinity for chromatin-proximal regions of the spindle. Consistent with its transcriptional induction profile (Whitfield et al., 2002) and its function in mitosis, we found that protein levels of HURP accumulated in G2 and peaked in mitosis at a stage slightly earlier than that of the cyclin B peak (Fig. 5, C and D). HURP was also down-regulated as cells exited from mitosis, which is consistent with a previous study (Hsu et al., 2004).
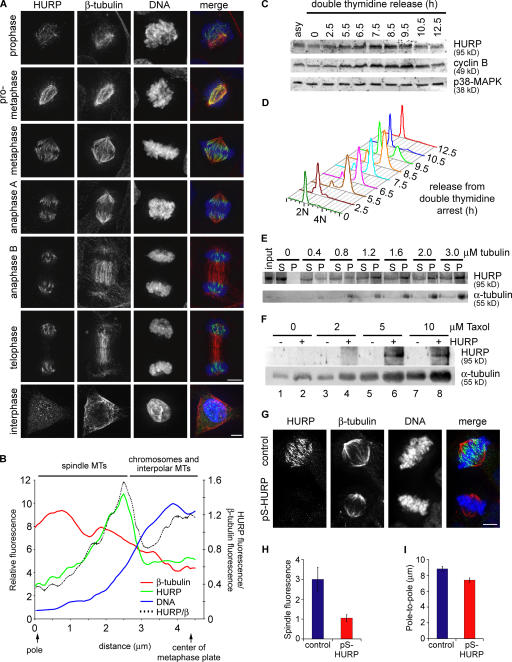
Figure 5.
HURP is a MAP that modulates spindle morphology. (A) Maximum projections from deconvolved z stacks of HeLa cells stained for HURP (green), β-tubulin (red), and DNA (blue). (B) 14 line scans of the relative fluorescence intensity from pole to metaphase plate were measured from the sum projection of the metaphase HeLa cell shown in A, and the averaged fluorescence intensity was plotted. The ratio of HURP to β-tubulin fluorescence intensity is displayed as the dotted black trace. The spindle pole is at 0 μm, and the metaphase plate is centered at 4.3 μm. (C) HeLa S3 cells were synchronized by double-thymidine arrest and harvested at the indicated times after release. Equivalent portions of each sample were analyzed by Western blotting. p38 MAP kinase served as a loading control. (D) Samples from C were stained with propidium iodide and analyzed by flow cytometry to monitor cell cycle progression. 2N, unreplicated diploid content; 4N, replicated diploid content. (E) Purified recombinant GST-HURP was added to taxol-stabilized microtubules in a copelleting assay, and microtubule-associated HURP was analyzed by Western blotting. Equivalent amounts of each sample were loaded. S, supernatant; P, pellet. (F) Purified α/β-tubulin was polymerized in the presence or absence of purified recombinant GST-HURP with a range of taxol concentrations. Polymerized microtubules were pelleted, and equivalent amounts of each sample were analyzed by Western blotting. (G) Maximum projections from deconvolved z stacks of control or HURP-depleted HeLa cells stained as in A. (H) Total β-tubulin immunofluorescence signal intensity on metaphase spindles stained as in A was quantified (n = 10 cells). P = 0.01 (two-tailed t test). (I) Pole-to-pole distances were quantified by γ-tubulin immunofluorescence in control (n = 29) or HURP-depleted (n = 32) HeLa cells at metaphase. P = 9 × 10−4 (two-tailed t test). Error bars represent SEM. Bars, 5 μm.
Given its colocalization with the mitotic spindle, we determined whether HURP is a MAP. Purified recombinant GST-HURP was incubated with polymerized microtubules that were stabilized with taxol, and, upon ultracentrifugation through a glycerol cushion, HURP was found to copellet with microtubules (Fig. 5 E). We also tested the effect of HURP on microtubule polymerization in vitro. The addition of recombinant purified HURP to purified α/β-tubulin near its critical concentration increased the total yield of microtubule polymer pelleted (Fig. 5 F; odd lanes vs. even lanes for microtubule yield), which is an effect similar to the one observed with the addition of taxol. Thus, HURP directly binds to and stabilizes microtubules and promotes their polymerization.
Next, we investigated the in vivo function of HURP on the mitotic spindle by assaying for morphological defects in HURP-depleted metaphase cells. When analyzed for total β-tubulin immunofluorescence, the microtubule mass on the spindle was reduced by threefold in HURP-depleted cells (Fig. 5, G and H). In addition, the interpolar distance, which was marked by γ-tubulin signals, was reduced from 8.84 ± 0.30 to 7.41 ± 0.27 μm in the absence of HURP (Fig. 4 I). Thus, HURP is a mitotic MAP that increases the amount and length of spindle microtubules by enhancing microtubule stability and polymerization.
HURP stabilizes the mitotic spindle and promotes microtubule growth
Given the biochemical activity of HURP as a microtubule-stabilizing factor, we investigated its effects on the dynamics of the mitotic spindle. When treated with 1 μg/ml nocodazole for 10 min, all spindle microtubules were depolymerized in both control and HURP-depleted mitotic cells (Fig. 6 A, left). However, when treated with 100 ng/ml nocodazole, disruption of the mitotic spindle was greater in HURP-depleted cells compared with control cells (Fig. 6 A, right). To corroborate this result, we assayed the stability of spindle microtubules in HeLa cells transiently expressing GFP-HURP approximately five times above the endogenous level (not depicted). At 10 and 50 μg/ml nocodazole, all spindle microtubules were disrupted in control mitotic cells, but cells overexpressing GFP-HURP retained stable spindle microtubules (Fig. 6 B). Thus, increasing the expression of HURP results in a greater stabilization of spindle microtubules in vivo.
Figure 6.
HURP stabilizes the mitotic spindle. (A) Maximum projections from deconvolved z stacks of control or HURP-depleted HeLa cells. Cells were treated with 1 μg/ml or 100 ng/ml nocodazole for 10 min at 37°C, fixed, and stained for β-tubulin (red), HURP (green), and DNA (blue). (B) Maximum projections from deconvolved z stacks of HeLa cells transfected with GFP-HURP. Cells were treated with 10 μg/ml or 50 μg/ml nocodazole for 10 min at 37°C, fixed, and stained for β-tubulin (red), GFP (green), and DNA (blue). (C) Maximum projections from deconvolved z stacks of HeLa cells stained for β-tubulin (red), HURP (green), and DNA (blue). Control or HURP-depleted cells were treated with 1 μg/ml nocodazole for 10 min at 37°C, washed, released into fresh media, and fixed 2 and 30 min after release. Arrowheads indicate centrosomes. (D–F) Still frames from confocal time-lapse microscopy of control or HURP-depleted HeLa cells cotransfected with GFP–α-tubulin (Videos 6–8, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). Cells were treated with 1 μg/ml nocodazole for 5 (D and E) or 15 min (F), washed, and released into fresh media. Time is indicated in minutes/seconds. Bars, 5 μm.
To investigate the role of HURP in microtubule polymerization, control and HURP-depleted cells were treated with 1 μg/ml nocodazole for 10 min and released into fresh media. 2 min after release, control cells formed short microtubules both at the centrosomes as well as at multiple foci within the chromatin (Fig. 6 C). Interestingly, HURP colocalized with β-tubulin foci on both the centrosomes and the chromatin. In contrast, HURP-depleted cells only showed weak β-tubulin staining at the centrosomes, and no microtubule foci were found within the chromatin. 30 min after release, control metaphase cells had reformed a bipolar spindle, whereas HURP-depleted mitotic cells showed only a minimal recovery of microtubules and no bipolar spindles (Fig. 6 C). Thus, HURP enhances the de novo polymerization of microtubules from both centrosomes and chromatin to reform the bipolar spindle.
To further define the role of HURP in microtubule dynamics, the kinetics of microtubule depolymerization and repolymerization were monitored by confocal time- lapse microscopy on HeLa cells transiently expressing GFP– α-tubulin. When the depolymerization phase was monitored in the presence of 1 μg/ml nocodazole, control metaphase cells maintained a bipolar mitotic spindle structure for up to 80 s, whereas the spindle in HURP-depleted metaphase cells was disrupted by 40 s (Fig. 6 D and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). To assay for effects on microtubule polymerization, control and HURP-depleted metaphase cells were treated with 1 μg/ml nocodazole for 5 min to depolymerize all microtubules and were then released into fresh media. Control cells rapidly repolymerized microtubules within 30 s and formed multiple microtubule foci within 1 min (Fig. 6 E and Video 7). However, this recovery was delayed in HURP-depleted cells until 4 min after release, with very few weak microtubule foci even at 5 min after release (Fig. 6 E and Video 7). We also monitored the kinetics of bipolar spindle formation. Control cells reformed a bipolar spindle within 20 min, but this was delayed to 50 min after release in HURP-depleted cells (Fig. 6 F and Video 8). Collectively, these data indicate that HURP increases the stability of the mitotic spindle, enhances the de novo polymerization of microtubules, and promotes timely formation of the bipolar spindle.
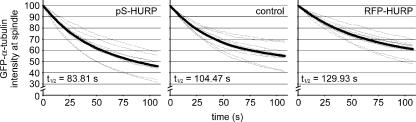
Because HURP stabilizes the mitotic spindle, we measured the turnover rate of α/β-tubulin subunits on the spindles of HeLa cells transiently expressing GFP–α-tubulin. In these fluorescence loss in photobleaching (FLIP) experiments, cytoplasmic GFP–α-tubulin was photobleached continuously while time-lapse images recorded the decrease in fluorescence intensity in the metaphase spindle (Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). In control cells, the half-life of GFP–α-tubulin on the spindle was 104.47 s (Fig. 7, middle). In HURP-depleted cells, the half-life decreased to 83.81 s (Fig. 7, left); however, the half-life increased to 129.93 s in cells overexpressing RFP-HURP (Fig. 7, right). Thus, the presence of HURP decreases the turnover rate of α/β-tubulin subunits on the spindle, thereby stabilizing the microtubules.
Figure 7.
HURP reduces the turnover rate of α/β-tubulin on mitotic spindles. Control, HURP-depleted, or RFP-HURP–transfected HeLa cells were cotransfected with GFP–α-tubulin. For each panel, 12 half-spindles from 10 metaphase cells were quantified for GFP fluorescence intensity every 0.5 s while a photobleaching laser was focused to a diffraction-limited spot in the cytoplasm away from the spindle (Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1). Fluorescence signals for each half-spindle (dotted traces) were normalized to their intensity at 0 s and averaged across the 12 half-spindles at each time point (thick traces). Turnover half-lives for GFP–α-tubulin on the spindle were calculated from the averaged fluorescence signal traces.
Discussion
We have developed a genomic screen for novel mitotic regulators based on a two-filter analysis of gene expression. The first filter selects for novel G2/M up-regulated transcripts that may function in mitosis. The second filter searches for genes that covary with a set of known mitotic regulators that define a core module essential for cell cycle progression. A combination of these two filters selects for candidate mitotic regulators involved in tumorigenesis, which, when coupled to functional assays, allows us to effectively identify novel mitotic regulators. Our two-filter strategy can also be modified to investigate other physiological processes such as the DNA damage response or apoptosis.
Through this screen, we identified HURP as a gene that is up-regulated during G2/M (Whitfield et al., 2002) and that covaried with other mitotic regulators across many tumor tissues (Segal et al., 2004). Compared with controls, spindle microtubules lacking HURP were less stable, polymerized more slowly, and exhibited higher turnover rates. Functionally, this activity is required for the efficient search and capture of kinetochores, timely congression of chromosomes, and generation of proper tension across sister kinetochores. Specifically, HURP-depleted cells had both unattached kinetochores on unaligned chromosomes and kinetochores under partial tension on aligned chromosomes. These defects activated the spindle checkpoint and transiently arrested cells at mitosis. Interestingly, HURP-depleted HeLa cells subsequently bypassed the checkpoint arrest and initiated anaphase without resolution of these deficiencies, leading to the missegregation of their chromosomes.
HURP as a MAP that controls spindle dynamics
HURP is a mitotic MAP that stabilizes the spindle, enhances microtubule polymerization, and promotes the timely formation of a bipolar spindle by reducing the turnover rate of the mitotic spindle (Figs. 5–7 ). Thus, HURP has biochemical activities similar to xMAP215/ch-TOG that control the dynamics of mitotic microtubules (Tournebize et al., 2000; Kinoshita et al., 2002). xMAP215/ch-TOG is localized to the entire mitotic spindle with a high concentration around the centrosomes (Fig. 4 E), where it stabilizes centrosomal microtubules, maintains centrosome integrity, and organizes spindle poles (Lee et al., 2001; Gergely et al., 2003; Cassimeris and Morabito, 2004). Because HURP colocalizes with microtubule foci at centrosomes immediately after release from nocodazole treatment and affects the rate of microtubule repolymerization from centrosomes (Fig. 6 C), HURP also has a direct role in nucleating and stabilizing centrosomal microtubules. However, HURP has a unique localization pattern that forms a gradient with high concentrations on chromatin-proximal regions of the mitotic spindle. Given their complementary localization patterns along the spindle axis, HURP and xMAP215/ch-TOG may stabilize microtubules predominantly near the chromatin and near the poles, respectively.
HURP also shares some characteristics with the microtubule-stabilizing factor TPX2. In particular, the depletion of either HURP or TPX2 resulted in decreased microtubule fluorescence intensity in mitotic spindles (Fig. 5, G and H; Gruss et al., 2002), indicating that both MAPs are required for generating wild-type levels of spindle microtubules. TPX2 is also required for spindle assembly because, in its absence, microtubule asters around the centrosomes do not organize into a bipolar spindle (Gruss et al., 2002). However, HURP-depleted cells remain competent for spindle formation, and the congression of most chromosomes to the metaphase plate is unperturbed. Instead, HURP serves a unique role by controlling the dynamics of spindle microtubules to promote the timely completion of the stochastic mitotic events of kinetochore capture and chromosome congression.
A HURP-mediated biased search mechanism for efficient capture of kinetochores
The lack of HURP activity reveals two phenotypes on chromosome congression and alignment. First, all HURP-depleted cells at metaphase exhibit a reduction in the interkinetochore distance (Fig. 2 C), indicating a reduced tension across sister kinetochores. This apparent decrease in tension results in the poor alignment of chromosomes along the metaphase plate (Fig. 2 F) and in the activation of the spindle checkpoint (Fig. 2, G and H), which delays anaphase onset (Fig. 3 E). Because tension is generated by the poleward pulling forces mediated through active turnover of microtubules at their plus ends and because HURP controls spindle dynamics, a change in the dynamic behavior of microtubules in HURP-depleted cells can directly explain these phenotypes.
Second, HURP depletion leads to a delay in chromosome congression (Fig. 3 E) and the persistence of a few unaligned chromosomes in some metaphase cells (Fig. 1, D–F). Although the cellular basis of this phenotype may be indirect, the microtubule-stabilizing activity of HURP in the vicinity of chromatin is also sufficient to explain the presence of unaligned chromosomes. Mathematical modeling has shown that unbiased microtubule growth from centrosomes cannot capture all kinetochores within the physiological duration of mitosis (Wollman et al., 2005). However, the addition of a hypothetical stabilizing factor that biases microtubule growth toward the chromosomes made kinetochore capture sufficiently rapid. Interestingly, HURP colocalizes with chromatin-proximal microtubules in a concentration gradient that increases toward the chromosomes and is a perfect candidate for such a biased stabilizing factor. In the absence of HURP, a decrease in microtubule stabilization specifically near chromatin may result in an unbiased and inefficient search and capture mechanism, thereby explaining the prolonged duration required for chromosome congression and the persistence of unaligned chromosomes.
Control of HURP activity by a chromatin-associated gradient
In the Xenopus laevis egg system, the Ran-GTP gradient around chromatin activates spindle assembly factors to promote spindle assembly by stabilizing microtubules near the chromatin (Nachury et al., 2001; Wiese et al., 2001; Gorlich et al., 2003; Caudron et al., 2005). The Ran-GTP pathway also functions in spindle assembly in mammalian cells (Gruss et al., 2002; Trieselmann et al., 2003; Tulu et al., 2006), and experimental evidence indicates that cultured cells can maintain a higher chromosomal Ran-GTP concentration relative to the cytoplasm (Li and Zheng, 2004). Thus, the Ran-GTP gradient, if it does exist in somatic cells, is a likely mechanism to control HURP localization and activity because HURP exhibits its own gradient along the mitotic spindle mirroring the proposed Ran-GTP gradient around chromatin (Fig. 5 B). In addition, HURP may also be controlled by chromatin-associated mitotic kinases. We propose that a chromatin-dependent gradient activates HURP to stabilize spindle microtubules near chromosomes during mitosis.
Bypass/adaptation of the spindle checkpoint as a mechanism for genomic instability in tumor cells
Despite activation of the spindle checkpoint in HURP-depleted HeLa cells, the checkpoint arrest was not stably maintained. Instead, these cells initiated anaphase in the presence of kinetochores under partial tension and in the presence of unaligned chromosomes (Fig. 4, A and F). We showed that HeLa cells have an intrinsic ability to bypass the spindle checkpoint because cells partially depleted of ch-TOG or treated with a low concentration of nocodazole also initiated anaphase in the presence of unaligned chromosomes (Fig. 4, B and C). Anaphase onset in HURP-depleted cells was a global event instead of localized sister chromatid disjunction because APC/C was active and Aurora B and Cenp-E redistributed to the central spindle (Fig. 4, G and H). Because HURP-depleted cells at early anaphase still retained Mad2 on kinetochores (Fig. 4 G), this bypass of the spindle checkpoint occurred at a point downstream of kinetochore-associated Mad2, most likely through the modulation of cytosolic APC/C activity.
These observations are in sharp contrast to previous studies in nontransformed cells. For example, a single kinetochore under no tension in meiotic mantid spermatocytes maintains a stable checkpoint arrest (Li and Nicklas, 1995). Similarly, a single unattached kinetochore in PtK1 cells prevents anaphase onset (Rieder et al., 1995). In contrast, HeLa cells have a less sensitive checkpoint response and cannot generate a stable metaphase arrest in the presence of one or a few unaligned chromosomes or in the presence of kinetochores under partial tension.
A persistently active spindle checkpoint can be resolved by apoptosis, by exit from mitosis as a 4N cell, or, as in the case of HURP-depleted cells, by aberrant anaphase initiation (Rieder and Maiato, 2004). One example of improper anaphase onset leading to chromosome missegregation arises from Cenp-I depletion, which also disrupts the localization of Cenp-F, Mad1, and Mad2 to the kinetochores (Liu et al., 2003). Aberrant anaphase initiation also occurs upon the depletion of Cenp-E or Aurora B, which causes a decrease in Mad2 or BubR1 signals on kinetochores (Ditchfield et al., 2003; Weaver et al., 2003). In these cases, the structure of the kinetochore or the checkpoint proteins themselves has been disrupted, making an analysis of the checkpoint bypass mechanism impossible. However, the paradigm for HURP depletion is different for two reasons. First, structural components such as Hec1 remain on the kinetochores, and checkpoint proteins such as Mad2 and BubR1 localize robustly to kinetochores of unaligned chromosomes in HURP-depleted cells. Second, HURP itself is not a checkpoint protein. Thus, the bypass of the spindle checkpoint after HURP depletion unveils a novel mechanism for checkpoint bypass in HeLa cells. Such a bypass mechanism seems unique to tumor cells because we have also observed premature anaphase initiation in the presence of unaligned chromosomes in tumor-derived Hct116 cells but not in the nontransformed hTERT-RPE1 cells (unpublished data).
This less sensitive checkpoint response, likely arising from the altered gene expression patterns in tumor cells, may be a common mechanism to promote genomic instability. In fact, the up-regulation of Mad2 in Hct116 cells has been shown to result in aneuploidy despite the increased checkpoint signal (Hernando et al., 2004). It is possible that a reduction in the sensitivity or response to a persistently active checkpoint signal is permissive for the premature initiation of anaphase, thereby providing a constant mechanism for generating aneuploidy at each cell cycle and for genomic instability in tumor progression.
HURP is up-regulated in hepatomas and other cancers (Tsou et al., 2003), and we have observed that HURP overexpression in HeLa cells results in hyperstabilization of the mitotic spindle (Fig. 6 B) and in reduced tension across sister kinetochores (not depicted). It is possible that the overexpression of HURP can also promote aneuploidy and genomic instability by generating subtle defects in chromosome congression as a result of the misregulation of spindle microtubule dynamics. Alternatively, aneuploidy itself may up-regulate HURP expression, further promoting genomic instability. Elucidating the molecular and cellular function of HURP will explain its function in maintaining genomic stability and tumorigenesis.
Materials and methods
Gene expression analyses
Whitfield et al. (2002) performed microarray analysis on genome-wide gene expression across the cell cycle in HeLa cells and identified 566 genes that were transcriptionally induced in G2 or G2/M. Segal et al. (2004) analyzed gene coexpression profiles among 1,975 published microarrays derived from 22 different types of tumors. Based on statistical analyses of coexpression profiling, they organized human genes into different functional modules, each of which corresponds to a set of genes that act in concert to carry out specific physiological functions. Out of 577 functional modules identified, two modules function in cell cycle regulation and consist of genes that covary with known cell cycle regulators. Our current analyses were based on these two previous studies.
Among 566 G2- or G2/M-induced genes reported by Whitfield et al. (2002), we initially analyzed the following 30 novel genes that have the best induction profile at G2/M: C10orf3, HURP, DKFZp762E1312, FLJ11029, FLJ11252, KIAA0042, ARL6IP, E2-EPF, LRRC17, MELK, MAPK13, PEPP2, HAN11, C9orf100, C9orf140, GAS2L3, FLJ40629, FLJ22624, FLJ20333, FLJ13354, FLJ20364, FLJ20510, FLJ20699, FLJ23293, FLJ21480, KIAA0952, GTSE1, LPP, MDS025, and PMSCL1. We found that the first 12 of the above listed 30 genes were also present in the cell cycle modules reported by Segal et al. (2004). Therefore, we focused our functional analysis on these 12 genes and found that several of them, including C10orf3, DKFZp762E1312, HURP, and LRRC17, were associated with microtubules in vivo during mitosis. At the functional level, we found that C10orf3 is required for cell abscission at the terminal stage of cytokinesis and reported that HURP is required for the efficient capture of kinetochores and congression of chromosomes by the mitotic spindle during mitosis.
Recombinant proteins, antibodies, and cell lines
The full-length HURP cDNA was subcloned into a modified version of the pFastBac vector (Invitrogen) containing an NH2-terminal GST tag, expressed in Sf9 cells, and purified by glutathione–Sepharose 4B (GE Healthcare). GFP-HURP and RFP-HURP were subcloned into a modified version of pCS2+ containing an NH2-terminal GFP or mRFP tag. GFP–α-tubulin was a gift from A. Barth (W. James Nelson Laboratory, Stanford University, Stanford, CA).
HURP fragments (aa 1–280, 81–625, and 626–846) were subcloned into the pGEX vector (GE Healthcare), expressed in Escherichia coli, purified by glutathione–Sepharose 4B, and used to immunize rabbits for the production of antisera. Antibodies were immunopurified using the respective immunogens. The following antibodies were obtained from commercial sources: anti-GFP clone 3E6 (Invitrogen); anti–β-tubulin ascites clone 2–28-33, anti–α-tubulin clone DM1α, and anti–γ-tubulin clone GTU-88 (Sigma-Aldrich); sheep anti–β-tubulin (Cytoskeleton, Inc.); CREST (Antibodies, Inc.); anti-Hec1 (GeneTex, Inc.); anti–Aurora B (BD Biosciences); antiphosphohistone H3 (Upstate Biotechnology); and anti–p38-MAPK, anti-Hps70, anti–cyclin B, and anti–Cenp-E (Santa Cruz Biotechnology, Inc.). Rabbit antibodies against Mad2 and BubR1 were described previously (Fang, 2002). The anti–ch-TOG antibody was provided by L. Cassimeris (Lehigh University, Bethlehem, PA). The HeLa/GFP–Cenp-A cell line was a gift from J.-M. Peters (Research Institute of Molecular Pathology, Vienna, Austria).
RNAi
Two DNA-based pSUPER constructs targeting HURP were transfected into HeLa cells using LipofectAMINE 2000 (Invitrogen). The target sequences for HURP are 5′-GCAATGAGAGAGAGAATTA-3′ and 5′-AGACTAAGATTGATAACGA-3′. The pSUPER empty vector (Brummelkamp et al., 2002) was used as a negative control. siRNAs targeting HURP or ch-TOG were transfected into HeLa or Hct116 cells using Dharmafect 1 (Dharmacon). siRNAs for HURP are 5′-GGTGGCAAGTCAATAATAA-3′, 5′-AGACTAAGATTGATAACGA-3′, and 5′-CGAAATAGACACTTTGGTT-3′. siRNAs for ch-TOG are 5′-GGAAATAGCTGTTCACATA-3′, 5′-GAAGAAACCTCAAGTGGTA-3′, 5′-GGCCAAAGCTCCAGGATTA-3′, and 5′-CAAGAAACCTGGATGGAAA-3′. siCONTROL Non-Targeting siRNA #2 (Dharmacon) was used as a negative control. Partial knockdown of ch-TOG was performed by using 1 nM of specific siRNA with 99 nM of control siRNA.
Immunofluorescence, confocal time-lapse microscopy, and FLIP
HeLa cells on coverglasses were fixed with −20°C methanol for 5 min or 4% PFA for 15 min at room temperature and were permeabilized/blocked with PBS-BT (1× PBS, 0.1% Triton X-100, and 3% BSA) for 30 min at room temperature. Images were acquired and γ adjusted with OpenLab 4.0.3 (Improvision) under a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) using a 1.4 NA plan-Apo 63× or 100× oil immersion objective (Carl Zeiss MicroImaging, Inc.) with a CCD camera (Orca-ER; Hamamatsu). Z stacks were deconvolved and γ adjusted using AutoDeblur 9.1 and AutoVisualize 9.1 (AutoQuant Imaging, Inc.).
For time-lapse microscopy, cells were cultured on coverglasses in Leibovitz's L-15 medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and 1× penicillin-streptomycin-glutamine (Invitrogen). Coverglasses were placed into a sealed or open growth chamber heated to 37°C and observed under an Axiovert 200M microscope with an Orca-ER CCD camera and OpenLab 4.0.3. For confocal time-lapse microscopy, cells were observed using a 1.4 NA plan-Apo 63× oil immersion objective (Carl Zeiss MicroImaging, Inc.) with a confocal scanner unit (UltraView; PerkinElmer). In the spindle checkpoint bypass assays with nocodazole treatment, cells were treated for 1 h with a low concentration of nocodazole (10 ng/ml) that did not grossly disrupt spindle microtubules but did result in a few unaligned chromosomes at metaphase before time-lapse microscopy in the presence of 10 ng/ml nocodazole. In the metaphase spindle depolymerization/repolymerization assays, cells were treated for 10–15 min with 1 μg/ml nocodazole in L-15 prewarmed to 37°C, quickly washed three times with 1× PBS prewarmed to 37°C, and reintroduced to fresh L-15 prewarmed to 37°C. Images were acquired at 0.5-s or 2-min intervals with OpenLab 4.0.3. For epifluorescence time-lapse microscopy, cells were observed using a 0.4 NA Achro-plan 20× air objective, and images were acquired at 2-min intervals.
For FLIP, cells transiently expressing GFP–α-tubulin to <5% of endogenous α-tubulin levels were placed in a sealed growth chamber heated to 37°C. Cytoplasmic GFP–α-tubulin was photobleached with a fiber-optically pumped dye laser, and images were acquired at 0.5-s intervals for 120 s with SlideBook 4.0 (Intelligent Imaging Innovations) under an Axiovert 200M microscope with a 1.4 NA 100× oil immersion objective (Carl Zeiss MicroImaging, Inc.) and a CCD camera (CoolSnap HQ; Photometrics). 12 half-spindles for each transfection were analyzed by measuring the absolute GFP–α-tubulin fluorescence intensity in a defined circular area contained entirely within each half-spindle. Fluorescence intensities were not background corrected because of the minimal contribution to fluorescence signal from cytoplasmic GFP–α-tubulin within the spindle. Fluorescence intensities for each half-spindle were normalized to their maximum intensity at the beginning of the time lapse, and the 12 normalized datasets were averaged to generate a single trace for each condition. Half-lives for GFP–α-tubulin on the spindle were calculated by linear regression of the averaged traces.
Microtubule copelleting and polymerization assay
For the copelleting assay, recombinant HURP protein at a final concentration of 2 μM was added to the reaction mix containing 2 mM GTP, 1× protease inhibitors, 20 μM taxol, and taxol-stabilized microtubules in 1× BRB80 buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA). The reaction was incubated at 30°C for 30 min and pelleted through a 40% glycerol cushion containing 20 μM taxol and 1× protease inhibitors in BRB80 at 100,000 g for 20 min at 30°C. Pellets were washed three times with 1× BRB80 and analyzed by Western blotting.
For the polymerization assay, recombinant HURP protein at a final concentration of 8 μM was added to the reaction mix containing 10 μM of purified α/β-tubulin, 2 mM GTP, and 2 mM DTT. The reaction was warmed to 37°C for 2 min, and taxol was added incrementally during the 25-min polymerization reaction. The reaction was pelleted through a 40% glycerol cushion containing 20 μM taxol in BRB80 at 100,000 g for 20 min at 30°C. Pellets were washed three times with 1× BRB80 and analyzed by Western blotting.
Online supplemental material
Fig. S1 shows that HURP depletion in Hct116 cells generates unaligned chromosomes and an increase in mitotic index compared with control cells. Videos 1 and 3 show control and HURP-depleted HeLa/GFP-H2B cells in mitosis. Video 2 shows control and HURP-depleted HeLa/GFP–Cenp-A cells at metaphase. Video 4 shows HURP-depleted, ch-TOG–depleted, and nocodazole-treated HeLa/GFP-H2B cells initiating anaphase in the presence of unaligned chromosomes. Video 5 shows control and HURP-depleted HeLa/GFP–Cenp-A cells initiating anaphase. Videos 6–8 show control and HURP-depleted HeLa cells transiently expressing GFP–α-tubulin in metaphase that were treated with 1 μg/ml nocodazole (Video 6) and then released (Videos 7 and 8). Video 9 shows FLIP analyses of a control HeLa cell, a HURP-depleted HeLa cell, and a HeLa cell expressing RFP-HURP (all with transient GFP–α-tubulin expression) at metaphase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200511132/DC1.
Supplementary Material
Acknowledgments
We are especially grateful to Jiong Ma for preparing Fig. 1 A. We thank Angela Barth, Elias Spiliotis, and Soichiro Yamada of the W. James Nelson Laboratory for the GFP–α-tubulin construct and for assistance with FLIP experiments, Jan-Michael Peters for the HeLa/GFP–Cenp-A cell line, Lynne Cassimeris for the anti–ch-TOG antibody, and members of the Fang Laboratory for discussions and technical assistance.
J. Wong is a recipient of the Howard Hughes Medical Institute Predoctoral Fellowship. This work was supported by the Burroughs Welcome Career Award in Biomedical Sciences and by a grant from the National Institutes of Health (GM062852) to G. Fang.
Abbreviations used in this paper: APC/C, anaphase-promoting complex/cyclosome; FLIP, fluorescence loss in photobleaching; HURP, hepatoma up-regulated protein; MAP, microtubule-associated protein; NEB, nuclear envelope breakdown.
References
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L., and J. Morabito. 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 15:1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron, M., G. Bunt, P. Bastiaens, and E. Karsenti. 2005. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 309:1373–1376. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., S.A. Jablonski, V. Sudakin, J.C. Hittle, and T.J. Yen. 1999. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., J.C. Waters, E.D. Salmon, and A.W. Murray. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 274:242–246. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- Compton, D.A. 2000. Spindle assembly in animal cells. Annu. Rev. Biochem. 69:95–114. [DOI] [PubMed] [Google Scholar]
- de Lichtenberg, U., L.J. Jensen, S. Brunak, and P. Bork. 2005. Dynamic complex formation during the yeast cell cycle. Science. 307:724–727. [DOI] [PubMed] [Google Scholar]
- Ditchfield, C., V.L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S.S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 13:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadde, S., and R. Heald. 2004. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14:R797–R805. [DOI] [PubMed] [Google Scholar]
- Gergely, F., V.M. Draviam, and J.W. Raff. 2003. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., M.J. Seewald, and K. Ribbeck. 2003. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22:1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss, O.J., M. Wittmann, H. Yokoyama, R. Pepperkok, T. Kufer, H. Sillje, E. Karsenti, I.W. Mattaj, and I. Vernos. 2002. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4:871–879. [DOI] [PubMed] [Google Scholar]
- Hernando, E., Z. Nahle, G. Juan, E. Diaz-Rodriguez, M. Alaminos, M. Hemann, L. Michel, V. Mittal, W. Gerald, R. Benezra, et al. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 430:797–802. [DOI] [PubMed] [Google Scholar]
- Hsu, J.M., Y.C. Lee, C.T. Yu, and C.Y. Huang. 2004. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J. Biol. Chem. 279:32592–32602. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., B. Habermann, and A.A. Hyman. 2002. XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12:267–273. [DOI] [PubMed] [Google Scholar]
- Kops, G.J., B.A. Weaver, and D.W. Cleveland. 2005. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 5:773–785. [DOI] [PubMed] [Google Scholar]
- Lee, M.J., F. Gergely, K. Jeffers, S.Y. Peak-Chew, and J.W. Raff. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643–649. [DOI] [PubMed] [Google Scholar]
- Li, H.Y., and Y. Zheng. 2004. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 18:512–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and R.B. Nicklas. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632. [DOI] [PubMed] [Google Scholar]
- Li, Y., and R. Benezra. 1996. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 274:246–248. [DOI] [PubMed] [Google Scholar]
- Liu, S.T., J.C. Hittle, S.A. Jablonski, M.S. Campbell, K. Yoda, and T.J. Yen. 2003. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5:341–345. [DOI] [PubMed] [Google Scholar]
- Nachury, M.V., T.J. Maresca, W.C. Salmon, C.M. Waterman-Storer, R. Heald, and K. Weis. 2001. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 104:95–106. [DOI] [PubMed] [Google Scholar]
- Rhodes, D.R., J. Yu, K. Shanker, N. Deshpande, R. Varambally, D. Ghosh, T. Barrette, A. Pandey, and A.M. Chinnaiyan. 2004. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl. Acad. Sci. USA. 101:9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., and H. Maiato. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 7:637–651. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., R.W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, E., N. Friedman, D. Koller, and A. Regev. 2004. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 36:1090–1098. [DOI] [PubMed] [Google Scholar]
- Shah, J.V., and D.W. Cleveland. 2000. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 103:997–1000. [DOI] [PubMed] [Google Scholar]
- Skoufias, D.A., P.R. Andreassen, F.B. Lacroix, L. Wilson, and R.L. Margolis. 2001. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA. 98:4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize, R., A. Popov, K. Kinoshita, A.J. Ashford, S. Rybina, A. Pozniakovsky, T.U. Mayer, C.E. Walczak, E. Karsenti, and A.A. Hyman. 2000. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2:13–19. [DOI] [PubMed] [Google Scholar]
- Trieselmann, N., S. Armstrong, J. Rauw, and A. Wilde. 2003. Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 116:4791–4798. [DOI] [PubMed] [Google Scholar]
- Tsou, A.P., C.W. Yang, C.Y. Huang, R.C. Yu, Y.C. Lee, C.W. Chang, B.R. Chen, Y.F. Chung, M.J. Fann, C.W. Chi, et al. 2003. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 22:298–307. [DOI] [PubMed] [Google Scholar]
- Tulu, U.S., C. Fagerstrom, N.P. Ferenz, and P. Wadsworth. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16:536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann, K., and R. Benezra. 2001. Mitotic checkpoints: from yeast to cancer. Curr. Opin. Genet. Dev. 11:83–90. [DOI] [PubMed] [Google Scholar]
- Weaver, B.A., and D.W. Cleveland. 2005. Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell. 8:7–12. [DOI] [PubMed] [Google Scholar]
- Weaver, B.A., Z.Q. Bonday, F.R. Putkey, G.J. Kops, A.D. Silk, and D.W. Cleveland. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M.L., G. Sherlock, A.J. Saldanha, J.I. Murray, C.A. Ball, K.E. Alexander, J.C. Matese, C.M. Perou, M.M. Hurt, P.O. Brown, and D. Botstein. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, C., A. Wilde, M.S. Moore, S.A. Adam, A. Merdes, and Y. Zheng. 2001. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 291:653–656. [DOI] [PubMed] [Google Scholar]
- Wollman, R., E.N. Cytrynbaum, J.T. Jones, T. Meyer, J.M. Scholey, and A. Mogilner. 2005. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol. 15:828–832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.