Abstract
Mitotic disjunction of the repetitive ribosomal DNA (rDNA) involves specialized segregation mechanisms dependent on the conserved phosphatase Cdc14. The reason behind this requirement is unknown. We show that rDNA segregation requires Cdc14 partly because of its physical length but most importantly because a fraction of ribosomal RNA (rRNA) genes are transcribed at very high rates. We show that cells cannot segregate rDNA without Cdc14 unless they undergo genetic rearrangements that reduce rDNA copy number. We then demonstrate that cells with normal length rDNA arrays can segregate rDNA in the absence of Cdc14 as long as rRNA genes are not transcribed. In addition, our study uncovers an unexpected role for the replication barrier protein Fob1 in rDNA segregation that is independent of Cdc14. These findings demonstrate that highly transcribed loci can cause chromosome nondisjunction.
Introduction
Cell survival depends on the accurate transmission of the genetic material to progeny. Coordinating chromosome behavior with the cell cycle machinery guarantees that the products of cell division are two genetically identical cells. Chromosomes are replicated to create two sister chromatids held together by topological and protein-mediated linkages. At the onset of mitosis, chromosomes compact into discrete bodies, converting the chromatids into rod-shaped structures short enough to segregate away from each other. At anaphase, the protein and topological connections between sisters resolve, allowing their segregation from each other to opposite poles of the mitotic spindle. Cohesin is responsible for the protein-mediated linkages. During mitosis, cohesin's cleavage allows separation of sister chromatids (Uhlmann et al., 1999). Although this is the case for most of the genome, the repetitive ribosomal gene cluster also requires the activity of the Cdc14 phosphatase for segregation (Granot and Snyder, 1991; D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004; Machin et al., 2005).
Cdc14 is required for rDNA segregation because it is necessary for the localization of condensin to rDNA (D'Amours et al., 2004; Wang et al., 2004), a protein complex required for chromosome condensation and segregation (Freeman et al., 2000; Bhalla et al., 2002; D'Amours et al., 2004; Lavoie et al., 2004; Sullivan et al., 2004; Machin et al., 2005). However, Cdc14 is better known for its multiple roles during mitotic exit (Stegmeier and Amon, 2004). Cdc14 is itself regulated by an inhibitory protein (Net1) that keeps it bound to nucleolar chromatin for the entire cycle except for anaphase (Visintin et al., 1998; Stegmeier and Amon, 2004), when the Cdc14 early anaphase release (FEAR) network and mitotic exit network (MEN) promote its release, thus allowing Cdc14 to reach its targets (Visintin et al., 1998; Pereira et al., 2002; Stegmeier et al., 2002; Yoshida et al., 2002; Stegmeier and Amon, 2004). Because of these roles, temperature-sensitive mutants of Cdc14 arrest in late anaphase as binucleated cells with unseparated and decondensed rDNA (Granot and Snyder, 1991; Guacci et al., 1994; D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004).
The reason rDNA requires additional segregation mechanisms, dependent on Cdc14, is presently unclear. The locus differs from the majority of the genome in several aspects: (1) it is highly repetitive, which increases chromosome size and the potential to undergo recombination; (2) it replicates unidirectionally as a result of the presence of a replication barrier at the 3′ end of each 35S ribosomal RNA (rRNA) gene (Brewer and Fangman, 1988; Linskens and Huberman, 1988); (3) it is highly transcribed by dedicated polymerases (RNA polymerase I and III), accounting for 60% of all cellular transcription; and (iv) it is repressed for RNA polymerase II transcription (Bryk et al., 1997; Smith and Boeke, 1997). Any or all of these differences could in principle impose segregation constraints in rDNA regions.
We have investigated the reason behind the additional segregation requirements of rDNA. We show the length of the array and the transcriptional hyperactivity of the rRNA genes it contains to be the factors that differentiate its segregation from the rest of the genome. We demonstrate that shortening the array or inactivating RNA polymerase I eliminates the segregation defects of cdc14-1 mutants. In addition to Cdc14, we uncover a second pathway designed to prevent linkages between rDNA on sister chromatids dependent on the replication fork barrier (RFB) gene FOB1.
Results
Mitotic exit in the absence of Cdc14 generates a population “bottleneck”
The function of Cdc14 in rDNA disjunction is probably unrelated to its role in inactivating Cdks, as several mitotic exit mutants can segregate rDNA despite being unable to lower Cdk activity (D'Amours et al., 2004; Machin et al., 2005). However, overexpression of the Cdk inhibitor SIC1 not only forces cdc14-1 mutant cells out of mitosis but also allows their growth on solid media (Jaspersen et al., 1998; Yuste-Rojas and Cross, 2000). To resolve this paradox, we tested whether rDNA segregates correctly when cells are forced out of mitosis without Cdc14. To this aim, we analyzed the segregation of a chromosome tag inserted in the distal flank of rDNA (tetO:487 tags) in cdc14-1 cells expressing SIC1 from the GAL1-10 promoter. Inactivation of Cdc14 through temperature elevation causes arrest at telophase, whereas addition of galactose to these cells induced mitotic exit, as judged by the growth of a new bud. Three different categories were observed, with respect to the segregation of tags, in cdc14-1 cells that had entered a new cycle (Fig. 1 A): (1) unresolved tags (sister chromatids failed to separate), (2) resolved but missegregated tags (separated sisters found in the same nuclear mass), and (3) resolved and segregated tags (sisters found in different nuclear masses). A large proportion of cells showed unresolved tags, indicating rDNA nondisjunction after mitotic exit (Fig. 1 B). Therefore, the function of Cdc14 in rDNA segregation is independent from its role to drive mitotic exit.
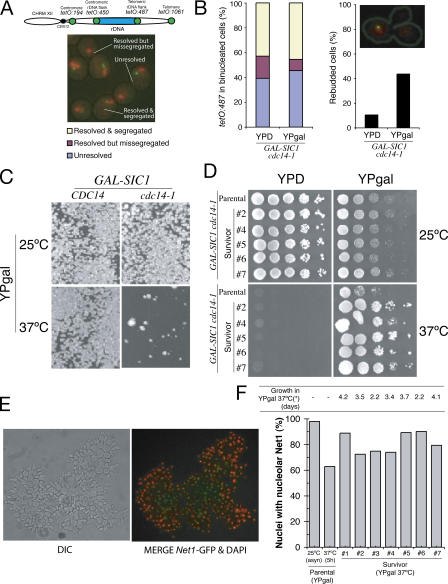
Figure 1.
Sic1-overexpression in cdc14-1 mutants creates a genetic “bottleneck” that corrects rDNA segregation defects. (A) Diagrammatic representation of the location of the chromosome tags along the right arm of chromosome XII used in this study. Representative micrograph of the different categories of tags scored in binucleated cells blocked by Cdc14 inactivation in B. (B) GAL-SIC1 cdc14-1 cells carrying chromosome tags in the centromere-distal flank of rDNA (tetO:487) were arrested in metaphase at 25°C and released to glucose (YPD; SIC1 overexpression off) or galactose (YPgal; SIC1 overexpression on) media at 37°C. The graphs show percentages of cells with segregated tags (left) and rebudding (right) 2 h after the metaphase release. In the top right corner, there is a picture of a cell that has entered a second cell cycle because of SIC1 overexpression but has failed to resolve the distal flank of the rDNA. (C) Yeast strains with the indicated genotypes were sonicated and plated onto YPgal medium (∼20 cells/cm2) and grown at the indicated temperatures for 4 d. A reduced growth of ∼1% of the total amount plated (survivors) is observed for GAL-SIC1 cdc14-1 at 37°C. (D) Different survivor colonies from C were grown in YPD at 25°C for ∼40 generations before they were 10-fold serially diluted, spotted onto different medium, and grown at the indicated temperatures for 3–4 d. (E) Representative micrograph of GAL-SIC1 cdc14-1 survivor cells expressing the nucleolar marker NET1-GFP (green) grown at 37°C in YPgal broth. DAPI is shown in red. Note that survivor cells grow as chains because of defects in cytokinesis. (F) Percentage of nuclei with Net1p-GFP signals in parental (GAL-SIC1 cdc14-1) and survivor (as in E) strains. Note that survivors show better segregation of Net1p-GFP.
The nondisjunction of tags in GAL-SIC1 cdc14-1 cells (Fig. 1 B) is intriguing because these cells have been previously reported to form colonies on solid media containing galactose at 37°C (Jaspersen et al., 1998; Yuste-Rojas and Cross, 2000). To revisit this, we plated GAL-SIC1 cdc14-1 cells on galactose at 37°C (Fig. 1 C). Consistent with previous studies, colonies formed after several days (Jaspersen et al., 1998; Yuste-Rojas and Cross, 2000); however, the amount of colonies corresponded to 1% of the total number of cells (Fig. 1 C). Therefore, the formation of GAL-SIC1 cdc14-1 survivor colonies appears to be a selection process, instead of allelic suppression. Survivor colonies remained able to grow at 37°C in galactose after being passed for 40 generations in glucose-containing media at 23°C (Fig. 1 D). The segregation of rDNA in survivor cells was significantly improved (Fig. 1, E and F); however, these cells were still unable to undergo cytokinesis and consequently grew as chains in culture (Fig. 1 E). These observations show that Cdc14 has at least three independent roles during mitotic exit, namely, Cdk inactivation, nucleolar segregation, and cytokinesis, the former two being the essential functions for cell viability.
Spontaneous gene conversions in rDNA are necessary for survival in the absence of Cdc14
Our results demonstrate that both nucleolar segregation and mitotic exit are the essential functions of Cdc14. We reasoned that the appearance of GAL-SIC1 cdc14-1 survivors might be related to changes that affect the nucleolar segregation function of Cdc14. The frequency of survivors is too high (1%) to be caused by spontaneous gene mutations. Instead, survival is more likely to be associated to changes in rDNA structure that alleviate segregation defects. Compaction of rDNA has been shown to occur during anaphase, and it is required for segregation (Lavoie et al., 2004; Machin et al., 2005). Recently, spontaneous large deletions in the rDNA have been shown to occur in ∼1% of cells (Michel et al., 2005). A large size reduction in rDNA would simulate compaction and could influence segregation. To test this possibility, we compared the size of chromosome XII in GAL-SIC1 cdc14-1 survivors to that of the original strain by pulsed-field gel electrophoresis (PFGE). The chromosome XII size in all survivors was reduced compared with the original strain (Fig. 2 A). No translocations were detected (unpublished data), suggesting that size reduction was associated with rDNA loss in the chromosome. Changes in rDNA array size can also occur through the formation of extrachromosomal ribosomal circles (ERCs; Kobayashi et al., 1998; Defossez et al., 1999). However, we did not detect an increased number of ERCs in the GAL-SIC1 cdc14-1 survivors (Fig. 2 B). Furthermore, the lack of rDNA segregation in cdc14-1 mutants is not affected by the presence of multicopy plasmids carrying rDNA (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200511129/DC1). We conclude that chromosome size reduction in the survivors is caused by a loss in the total rDNA copy number in the cell.
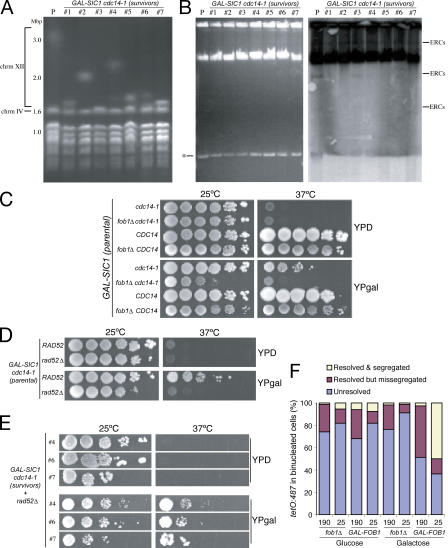
Figure 2.
Recombination-dependent reduction of rDNA size allows growth in the absence of Cdc14 as long as SIC1 is overexpressed. (A) Parental and survivor GAL-SIC1 cdc14-1 strains were grown in YPgal (25°C for parental and 37°C for survivors) and processed for PFGE analysis to visualize all chromosomes. Chromosome sizes are shown in Mbp. Chromosomes XII and IV are indicated. Note that although the parental strain contained ∼200 copies of rDNA (∼3.0 Mbp), the survivor strains contained a mean of 60–70 rDNA copies (∼1.7 Mbp; 1, 3, 5, 6, and 7), with two of them reaching ∼100 copies (∼2.0 Mbp; 2 and 4). Therefore, all survivor strains suffered a significant reduction in chromosome XII size compared with the parental strain. (B) Total DNA was isolated from cultures of parental and survivor GAL-SIC1 cdc14-1 strains grown as in A and electrophoresed (EthBr staining in left panel) to detect ERCs. Transfers (right) were probed with rDNA sequences (RDN25). Identifiable ERC species are indicated. The asterisk denotes an undetermined and unspecific low-weight DNA band. (C–E) Yeast strains with the indicated genotypes were 10-fold serially diluted, spotted onto different medium, and grown at indicated temperatures for 3–4 d. The appearance of SIC1 cdc14-1 survivor colonies requires the FOB1 (C) and RAD52 (D) genes; however, rDNA recombination is not required after the initial selection (E). (F) cdc14-1 cells carrying tetO:487 tags with either 190 or 25 copies of rDNA and with the FOB1 gene deleted (fob1Δ) or under an inducible promoter (GAL-FOB1) were grown initially in YPD, diluted, and transferred to either fresh YPD or YPgal for 12 h before shifting the temperature to 37°C for 4 h to evaluate the resolution and segregation of tags in the cdc14-1 block. Strains with shorter rDNAs segregated better when Fob1 was present, demonstrating that chromosome XII size is an important factor limiting the disjunction of this chromosome in the absence of Cdc14 function.
The reduction of the rDNA array size is therefore a shared phenotype amongst all survivors. However, it is still possible that size reduction is not a requirement for the survival but an indirect effect of the selection that cdc14-1–blocked cells undergo when forced out of mitosis. To distinguish between these two possibilities, we tested whether fixing the size of the rDNA array in the original strain would prevent the appearance of survivors. Changes in rDNA copy number require the FOB1 gene bound to the RFB site on rDNA (Kobayashi et al., 1998). In fob1Δ cells, the rDNA array size is maintained without change in copy number (Kobayashi et al., 1998). Deletion of FOB1 in GAL-SIC1 cdc14-1 cells abolished the appearance of survivors in galactose media at 37°C (Fig. 2 C), suggesting that Fob1 is required for survival. However, Fob1 is an rDNA binding protein with roles that contribute to rDNA segregation (Fig. 2 F and see Fig. 3 A); therefore, it is possible that Fob1 is necessary for survival for reasons other than to mediate array size change. To evaluate this, we investigated whether GAL-SIC1 cdc14-1 survival requires the recombination machinery because the role of Fob1 in rDNA array expansion/contraction also involves mechanisms dependent on recombination (Gangloff et al., 1996; Kobayashi and Horiuchi, 1996; Kobayashi et al., 1998). Like Fob1, deletion of RAD52, an essential protein for recombination, in GAL-SIC1 cdc14-1 cells prevented the appearance of survivors (Fig. 2 D). Interestingly, Rad52 is only required at the time of selection, as deletion of RAD52 in GAL-SIC1 cdc14-1 survivor strains did not affect their ability to grow in galactose media at 37°C (Fig. 2 E). These results demonstrate that a change in rDNA array size is important for the survival of GAL-SIC1 cdc14-1 cells and that such changes are mediated by recombination events.
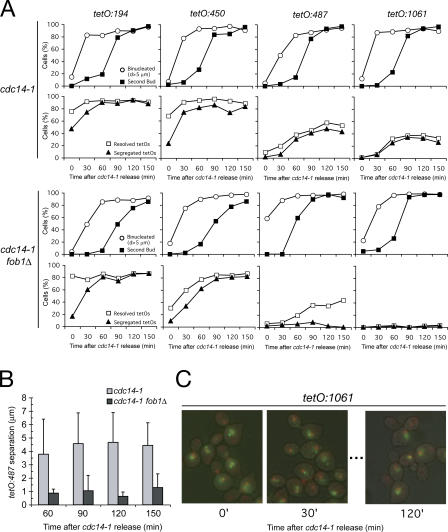
Figure 3.
Deletion of FOB1 worsens cdc14-1 segregation for chromosome XII at its distal regions. (A) cdc14-1 or cdc14-1 fob1Δ strains carrying different tags along the right arm of chromosome XII (tetO:194, tetO:450, tetO:487, or tetO:1061) were arrested in G1 with α-factor, released into 37°C media for 2.5 h to reach cdc14-1 arrest, and shifted back to 25°C for another 2.5 h. Samples were taken every 30 min after the shift to 25°C, and cells were scored for nuclear mass segregation, emergence of a second bud (top), resolution, and segregation of the tetOs (bottom). Note that the shift to 25°C resumes Cdc14 function, although ∼50% of the distal right arms of chromosome XII still undergo incorrect segregation. fob1Δ further impairs the resolution/ segregation defect of distal tags (tetO:487 or tetO:1061). (B) Distances between the resolved tetO:487 were measured for the aforementioned experiment (only time 60 min after the temperature shift onwards). Note that fob1Δ restricts the degree of separation at the rDNA distal flank. (C) Representative micrographs of cells scored in the aforementioned experiment. DAPI is in red, tetO:1061 is in green, and the cell wall is superimposed in black. Note how cdc14-1 blocks cells with diverse nuclear morphology after G1 release. Within 30 min of cells resuming Cdc14 function, nuclear masses are able to completely split apart. The last panel shows an example of the three different fates of the distal chromosome XII regions (full segregation, resolution but missegregation, and lack of resolution).
Long rDNA arrays prevent rDNA disjunction in the absence of Cdc14
Cdc14's role in rDNA segregation is at least in part to target condensin to rDNA regions (D'Amours et al., 2004; Wang et al., 2004), thus promoting compaction of this chromosome, which is an important feature of its segregation (Machin et al., 2005). Reduction of rDNA copy number in GAL-SIC1 cdc14-1 survivor cells shortens chromosome XII, and this might be sufficient to circumvent the need for compaction and, thus, Cdc14's role in the process. To test this model, we investigated whether shortening rDNA arrays would be sufficient to bypass the role of Cdc14 in rDNA segregation. We used two cdc14-1 strains with different rDNA array sizes, a short array of 25 units (RDN1-25) or a long array containing 190 (RDN1-190) copies. Both strains also contained a chromosome tag in the distal flank of rDNA (tetO:487) and carried a FOB1 deletion to prevent any further changes in rDNA size. Surprisingly, we found no differences with respect to segregation between the two strains (Fig. 2 F). However, we noticed a genetic interaction between CDC14 and FOB1 genes at permissive conditions (Fig. 2 C), raising the possibility that Fob1 has additional roles in rDNA segregation that are independent of rDNA size (Fig. 2 F and see Fig. 3 A). To address this, we expressed FOB1 from the GAL1-10 promoter during the last few cell cycles in the RDN1-25 and -190 strains before inactivating Cdc14. Although >50% of cells were able to segregate in the RDN1-25 strain, only 5% segregated in the RDN1-190 strain when Fob1 was present (Fig. 2 F). The results demonstrate that reduction in rDNA length improves rDNA segregation in the absence of Cdc14 function.
Deletion of FOB1 worsens the rDNA segregation defects in cdc14-1 blocks
Our results demonstrate that deletion of Fob1 in a cdc14-1 mutant background impedes rDNA segregation irrespective of array size (Fig. 2 F), suggesting that this protein has a direct role in rDNA segregation. Strains containing the normal number of units (100–200) already show low levels of segregation in the cdc14-1 arrest (Machin et al., 2005), thus making it difficult to quantify the effect of Fob1 in cdc14-1 fob1Δ cells arrested by inactivation of Cdc14. To investigate the contribution of Fob1 to segregation, we used an alternative growth regimen. First, we blocked cdc14-1 fob1Δ cells in anaphase (by temperature) and then returned them to permissive conditions (Machin et al., 2005) to allow mitotic exit. We scored rDNA segregation during mitotic exit (Fig. 3 A). We used different tags along chromosome XII to compare the segregation between cdc14-1 and cdc14-1 fob1Δ cells (Fig. 3 A). Tags in the proximal side of rDNA (tetO:194 and tetO:450) were already resolved in 70–80% of cells arrested in the cdc14-1 block before release (Fig. 3 A) and showed no differences with respect to segregation (with >80% of cells segregated 150 min after release), independent of whether Fob1 was present (Fig. 3 A).
In contrast, the segregation of tags in the distal side of rDNA (tetO:487 and tetO:1061) reached a maximum of ∼50% when Fob1 was present but dropped to <5% in cdc14-1 fob1Δ cells (Fig. 3 A). These results show that Fob1 plays an active role in the segregation of rDNA distal regions in addition to that of Cdc14. These experiments also revealed several interesting observations. It seems that when a culture goes through anaphase without Cdc14, a large proportion of cells show segregation defects for the distal tags even when Cdc14 is added back (Fig. 3 A; tetO:487 and tetO:1061 segregation in cdc14-1). We also noted differences between the tetO:487 and tetO:1061 tags in cdc14-1 fob1Δ cells. Despite the fact that neither tetO:487 nor tetO:1061 tags segregated, tetO:487 resolved in 45% of cells (localized to same nucleus) with a mean distance of 1–2 μm (Fig. 3, B and C), whereas tetO:1061 tags did not resolve from each other (Fig. 3 A).
Deletion of FOB1 induces a delay in rDNA resolution and increases nucleolar topoisomerase II localization
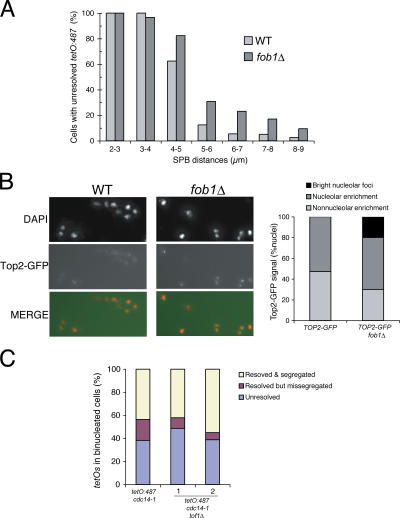
Deletion of Fob1 negatively affects rDNA segregation in a cdc14-1 mutant background (Fig. 3 A), suggesting additive effects for both proteins. However, no segregation phenotypes have been previously described for the single fob1Δ mutant. Next, we tested whether fob1Δ affects rDNA segregation in the presence of Cdc14. We could not detect missegregation of chromosome tags in fob1Δ cells (unpublished data); however, the resolution of tetO:487 tags in fob1Δ cells occurred at longer spindle lengths (Fig. 4 A), suggesting that cells suffered segregation delays. One possibility is that Cdc14 activity is sufficient to mask Fob1's segregation role. To test this, we investigated whether Fob1 interacts with downstream targets of Cdc14. Condensin and Top2 activities in rDNA during anaphase depend on Cdc14 (D'Amours et al., 2004; Sullivan et al., 2004; Wang et al., 2004). Interestingly, fob1Δ shows additive growth defects with temperature-sensitive alleles of the condensin subunit SMC2, smc2-8, as well as TOP2, top2-4 (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200511129/DC1). In addition, we investigated the targeting of condensin and Top2 in fob1Δ cells by chromatin spreads. We did not detect any differences for condensin between wild-type and fob1Δ samples (unpublished data). However, Top2 was present in bright nucleolar foci only in fob1Δ cells (Fig. 4 B). Overexpression of CDC14 in cdc14-1 fob1Δ cells (blocked in a cdc14-1–mediated arrest) induced segregation of rDNA distal tags in >75% of cells (Fig. S3). These results show that the origins of the disjunction defects caused by fob1Δ and Cdc14 inactivation are similar. Our findings suggest that condensin activation and its regulation of Top2 recruitment (Bhalla et al., 2002) in a Cdc14-dependent manner is likely to resolve problems caused by the absence of Fob1, hence masking its contribution to rDNA segregation in fob1Δ strains.
Figure 4.
Deletion of FOB1 gene delays segregation of chromosome XII in anaphase and leads to an accumulation of topoisomerase II at the nucleolus. (A) Wild type (WT) and fob1Δ strains bearing an rDNA distal flank tag (tetO:487) and the SPB marker Tub4-CFP were arrested in G1 and released into 25°C. Samples were taken every 10 min between 80 and 130 min after the release, and resolution of tags was plotted against distances between SPB. Note that there is a small delay in the resolution of the distal flank in anaphase. (B) Wild type and fob1Δ strains carrying a TOP2-GFP allele were processed for chromatin spreads while growing asynchronously in exponential phase. Top2-GFP was detected directly. Note that in a fob1Δ mutant, Top2-GFP appears as bright nucleolar foci (left) in ∼20% of the scored nuclei. (C) cdc14-1 or cdc14-1 tof1Δ strains carrying tetO:487 tags were arrested after 3 h at 37°C, and cells were scored for resolution and segregation of the tetOs. Note that similar levels of missegregation were observed for both strains.
Nucleolar nondisjunction in cdc14-1 mutants is not caused by recombination intermediates or RNA polymerase II silencing
Our results demonstrate that shortening the rDNA array significantly reduces the need for Cdc14 activity to achieve segregation (Fig. 2 F). However, a proportion of cdc14-1 mutant cells with short rDNA arrays still failed to segregate correctly (Fig. 2 F), raising the possibility that additional factors (besides rDNA size) contribute to nondisjunction in cdc14-1 mutants. rDNA differs from the majority of the genome in several aspects, including its potential to undergo recombination (Kobayashi and Horiuchi, 1996; Kobayashi et al., 1998; Johzuka and Horiuchi, 2002), its unidirectional mode of replication (Brewer and Fangman, 1988; Linskens and Huberman, 1988), and the fact that, despite being silenced for RNA polymerase II transcription (Bryk et al., 1997; Smith and Boeke, 1997), it is highly transcribed by RNA polymerase I. Next, we tested whether any of these peculiarities impose the segregation constraints in rDNA that require Cdc14 and Fob1 activities.
First, we considered recombination to be the possible source of nondisjunction because, conceptually, an increased level of recombination between rRNA genes or the inability to remove recombination intermediates could interfere with segregation. However, recombination is unlikely to be the origin of nondisjunction because Fob1 is necessary for rDNA recombination (Kobayashi and Horiuchi, 1996; Kobayashi et al., 1998; Johzuka and Horiuchi, 2002), and we predict that loss of recombination structures would promote segregation and not reduce it as we observed in the cdc14-1 fob1Δ experiment (Fig. 3 A). Nevertheless, we tested the possibility in a more direct way by deleting RAD52 in the cdc14-1 strain and analyzing rDNA segregation in the resulting strain. The resolution and segregation of tetO:487 and tetO:1061 tags in cdc14-1 cells were not affected by rad52Δ (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200511129/DC1), confirming that recombination does not contribute to the rDNA nondisjunction phenotype in the absence of Cdc14. Moreover, the fact that deletion of RAD52 did not worsen segregation as we see in cdc14-1 fob1Δ allowed us to conclude that the phenotype associated to this double mutant is not due to recombination.
Transcriptional silencing in the rDNA gene cluster acts on RNA polymerase II–transcribed genes (Bryk et al., 1997; Smith and Boeke, 1997). Silencing on rDNA requires the silencer protein Sir2 as part of the protein complex called RENT (regulator of nucleolar silencing and telophase exit; Straight et al., 1999). RENT recruitment to rDNA depends on Fob1 (Huang and Moazed, 2003). Deletion of SIR2 does not improve the segregation defect in cdc14-3 mutants released from metaphase (D'Amours et al., 2004). However, it is not known whether sir2Δ worsens segregation as observed for cdc14-1 fob1Δ mutants (Fig. 3 A). To test this possibility, we investigated segregation in cdc14-1 sir2Δ cells at the cdc14-1 block. Segregation of tetO:487 tags in cdc14-1 sir2Δ cells was comparable to that in cdc14-1 (Fig. S4 B). These results confirm that RNA polymerase II–silent chromatin does not interfere with the segregation of nucleolar regions in the cdc14-1 and cdc14-1 fob1Δ mutants.
The role of FOB1 in rDNA disjunction is independent of its FEAR and RFB functions
Our results have revealed a function for Fob1 in nucleolar segregation (Fig. 3 A). Recent work has shown that Fob1 also plays a role regulating the timely activation of Cdc14 (Stegmeier et al., 2004); thus, one possibility is that these two roles are related. Inactivation of FOB1 prematurely releases Cdc14, whereas overexpression causes a delay (Stegmeier et al., 2004). Because the mutant protein Cdc14-1 is rapidly delocalized from the nucleolus at 37°C (Torres-Rosell et al., 2004), it is possible that segregation after cdc14-1–block release (Fig. 3 A) requires passage of the reactivated Cdc14 protein through the nucleolus. If this were the case, fob1Δ could potentially interfere with Cdc14 reactivation and consequently worsen segregation in our experiments. To test this possibility, we analyzed the localization of reactivated Cdc14-1 protein fused to GFP (Torres-Rosell et al., 2004) during the release from a cdc14-1 block (Fig. S4 C). Cdc14 was not observed in the nucleolus until 60–70 min after release (Fig. S4 C), a time when segregation has already reached its maximum levels (Fig. 3 A). Therefore, Cdc14 reactivation does involve passage through the nucleolus before segregation and, hence, Fob1 roles in segregation and Cdc14 activation are independent.
Fob1 is also required for replication fork pausing in the RFB site at the 3′ end of the 35S rRNA gene (Kobayashi and Horiuchi, 1996). This fork barrier is thought to prevent collisions between the replication and transcription machineries (Brewer et al., 1992; Olavarrieta et al., 2002; Takeuchi et al., 2003), thus forcing replication and transcription to occur codirectionally. This function might be important because, at least in plasmids, opposing replication and transcription can generate topological problems (Olavarrieta et al., 2002). Therefore, it is possible that in the absence of Fob1 a high level of collisions between transcription and replication impede mitotic disjunction of rDNA. To test this hypothesis, we investigated whether inactivation of Tof1 in cdc14-1 cells also emulated the rDNA segregation defects of cdc14-1 fob1Δ cells, as Tof1 is also required for fork arrest at the RFB site (Calzada et al., 2005; Tourriere et al., 2005; Mohanty et al., 2006). The levels of tetO:487 tag segregation in cdc14-1 tof1Δ cells are comparable to those in cdc14-1 mutants (Fig. 4 C). We thus conclude that the lack of RFB activity in cdc14-1 fob1Δ cells is not the cause of its segregation defects.
Transcription interferes with rDNA segregation in the absence of Cdc14 function
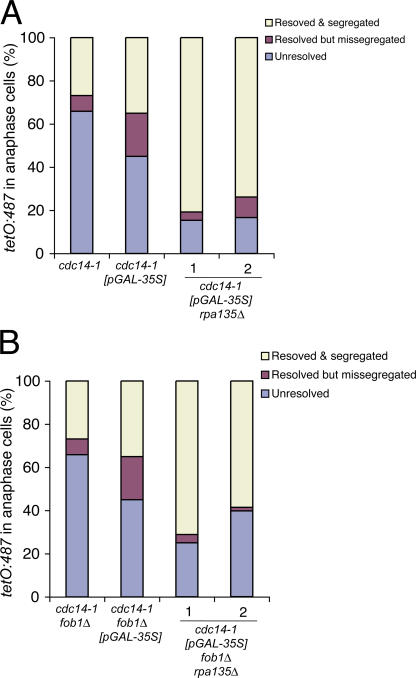
A major difference between rDNA and the rest of the genome is in respect to its transcriptional activity. Despite being silenced for RNA polymerase II transcription (Bryk et al., 1997; Smith and Boeke, 1997), rDNA is also highly transcribed by RNA polymerase I. In higher eukaryotes, a reduction in rRNA transcriptional activity occurs during mitosis, but this is not the case in budding yeast, where rRNA transcription continues through this cell cycle stage (Elliott and McLaughlin, 1979). It is possible that continuous transcription during mitosis requires specialized mechanisms to ensure segregation, perhaps dependent on Cdc14 and Fob1 activities. To test this possibility, we investigated rDNA segregation in cdc14-1 mutants where polymerase I transcription of 35S rRNA was turned off. We deleted RPA135, an essential gene encoding the second largest subunit (A135) of the yeast RNA polymerase I complex in the cdc14-1 strain. The resulting cells are able to grow because they carry a multicopy plasmid with a 35S rRNA gene driven by the RNA polymerase II GAL7 (pGAL-35S) promoter (Nogi et al., 1991). Cells were released from G1 at 37°C to inactivate Cdc14, and the segregation of tetO:487 tags was scored in binucleated cells arrested in the cdc14-1 block. Correct chromosome segregation for both tags was observed in a high proportion (>80%) of cdc14-1 rpa135Δ cells (Fig. 5 A). Next, we asked whether rpa135Δ also suppressed the segregation defects in the cdc14-1 fob1Δ mutant. Segregation of tetO:487 tags was assayed in cdc14-1 fob1Δ rpa135Δ cells (Fig. 5 B). In contrast to the severe missegregation observed in cdc14-1 fob1Δ cells (<5%; Fig. 3 A), >50% of cdc14-1 fob1Δ rpa135Δ cells were able to segregate rDNA regions correctly (Fig. 5 B), suggesting that rpa135Δ bypasses both Cdc14 and Fob1 segregation functions. These results demonstrate that the transcription of rRNA genes imposes segregation constraints in rDNA that require Cdc14 activity for resolution. In addition, the data show that the presence of Fob1 also plays a role in reducing the levels of linkages in the rDNA that need to be resolved by Cdc14. Thus, we identify polymerase I transcription as a novel means of establishing linkages between chromosomes.
Figure 5.
Transcription by RNA polymerase I causes rDNA linkages in cdc14-1 and cdc14-1 fob1Δ. (A) cdc14-1 strains carrying tetO:487 tags and a plasmid that transcribes rRNA 35S precursor from a galactose-inducible promoter (pGAL-35S) with or without the deletion of the second largest subunit of RNA polymerase I (rpa135Δ; two independent clones shown) were grown and arrested as described in Material and methods. Resolution/segregation pattern of rDNA distal flank was quantified in cells arrested by Cdc14 inactivation with an anaphase nucleus (stretched nucleus across the neck or binucleated). (B) cdc14-1 fob1Δ strains carrying a tetO:487 chromosome tag, the pGAL-35S plasmid, and rpa135Δ were grown, arrested, and scored as in A. Note that the lack of RNA polymerase I transcription at the rDNA eliminates missegregation of distal rDNA tags in cdc14-1 and cdc14-1 fob1Δ mutants.
Discussion
To ensure genomic stability through generations, cells need to hold sister chromatids together until metaphase and then remove all the physical connections between them in anaphase. It has long been known that the nucleolus requires Cdc14 to segregate (Granot and Snyder, 1991). However, the reason for this specific requirement was unknown. Here, we have shown that rDNA requires Cdc14 for segregation partly because of its physical length but most importantly because a fraction of rRNA genes are transcribed at very high rates.
We show that the rDNA segregation function of Cdc14 can be bypassed through genetic rearrangements that involve a gross reduction in the number of rDNA copies, thus reducing chromosome size. We also demonstrate that besides rDNA size, the transcription of rRNA genes by RNA polymerase I cells generates linkages between sister chromatids that prevent segregation in the absence of Cdc14. In addition, our study shows that Fob1 has a novel function in rDNA segregation independent from that of Cdc14. Thus, our data not only provide an insight into the mechanisms that give rise to constraints on mitotic rDNA sister chromatid disjunction (i.e., rRNA transcription) but also reveal the presence of two pathways to deal with these problems, one dependent on Fob1 and the second requiring Cdc14. These findings thus explain the reason behind the segregation phenotypes observed in cdc14-1 mutants (Fig. 6).
Figure 6.
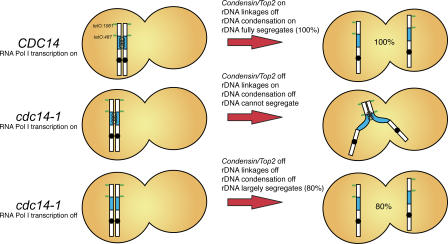
RNA polymerase I transcription of rRNA genes and lack of rDNA condensation constrain chromosome XII disjunction in the absence of Cdc14. Diagrammatic representation of yeast chromosome XII with the rDNA array in blue and the position of various chromosome tags used in this study in green. Cdc14 activity during anaphase recruits condensin (D'Amours et al., 2004; Wang et al., 2004) to rDNA. RNA polymerase I transcription of chromosomal rRNA genes creates linkages between sister chromatids. Condensation of rDNA and removal of transcription-induced linkages are mediated by Cdc14, thus ensuring full segregation of chromosome XII (100% of cells). In the absence of Cdc14, neither rDNA condensation nor transcription-induced linkages are removed (middle); consequently, the mitotic disjunction of the distal regions of chromosome XII is prevented. When RNA polymerase I transcription is inactivated (and growth is supported by 35S RNA polymerase II–mediated transcription from a plasmid copy), transcription-induced linkages between sister chromatids do not arise and, consequently, distal regions of chromosome XII exhibit improved segregation (80% of cells are able to separate tags), even in the absence of Cdc14. Note that full segregation is not achieved (as in top row) because rDNA condensation is not induced. Our findings show that Cdc14-dependent rDNA condensation and resolution mechanisms are required to segregate the long arm of chromosome XII because of its size and the presence of linkages generated by the high transcription rates in rRNA genes.
Cdc14 requirement for nucleolar segregation can be bypassed by reducing rDNA copy number
The main role of Cdc14 during mitotic exit is the dephosphorylation of target proteins that cause the inactivation of Cdks, thereby allowing cells to enter G1 (Stegmeier and Amon, 2004). Consequently, inactivation of Cdc14 causes a telophase arrest where high levels of Cdk activity are retained (Fitzpatrick et al., 1998). Expression of the Cdk inhibitor Sic1 is sufficient to drive cdc14-1–blocked cells out of mitosis (Fitzpatrick et al., 1998; Jaspersen et al., 1998; Yuste-Rojas and Cross, 2000), and it supports growth in solid media (Jaspersen et al., 1998; Yuste-Rojas and Cross, 2000). We previously showed that Cdk inactivation is not required for rDNA to segregate, as cdc15-2 mutants do not inactivate Cdk but are able to segregate rDNA (Machin et al., 2005). Therefore, the genetic suppression of cdc14-1 mutants by Sic1 overexpression (Jaspersen et al., 1998; Yuste-Rojas and Cross, 2000) is not consistent with such a view.
We show that Sic1 overexpression from the galactose promoter does not suppress the cdc14-1 role in segregation but instead forces a genetic bottleneck where a minority of the population is able to bypass Cdc14 requirement by reducing rDNA copy number. We provide an insight into the mechanism used in GAL-SIC1 cdc14-1 cells to contract rDNA. To survive, GAL-SIC1 cdc14-1 cells require recombination dependent on Rad52. In rad52Δ, small rDNA contractions over many generations have been reported and attributed to nonconservative recombination mechanisms, like single-strand annealing (Gangloff et al., 1996). However, we have not detected major changes in rDNA copy number in the single rad52Δ mutant strains grown for >30 generations (unpublished data). Thus, we conclude that GAL-SIC1 cdc14-1 survivor cells contract the rDNA (Fig. 2 A) through spontaneous gene conversion events that significantly reduce the number of rRNA genes in the array. Interestingly, we found that fob1Δ in GAL-SIC1 cdc14-1 caused a heterogeneous phenotype, with half of the colonies requiring Fob1 not only for the establishment of survivors but also for their maintenance (unpublished data). This may indicate that once the spontaneous gene conversion has taken place and the selection for the shorter rDNA array is forced, active maintenance of the reduced size occurs. Alternatively, this unexpected behavior of fob1Δ could also be a consequence of the newly described role in segregation.
How can Cdc14's role in segregation be influenced by a reduction in rDNA copy number? The rDNA repeats make the right arm of chromosome XII the longest in the genome (Machin et al., 2005). We previously showed that, during anaphase, yeast cells hypercondense rDNA to ensure that segregation of this long chromosome takes place before cytokinesis (Lavoie et al., 2004; Machin et al., 2005). Cdc14 is necessary for this step, as it is responsible for the localization of the compaction machinery, the condensin complex, to rDNA (D'Amours et al., 2004; Wang et al., 2004). Thus, the requirement of a reduction in rDNA length to bypass Cdc14's role in segregation in the GAL-SIC1 cdc14-1 survivor strains is consistent with a specific role for Cdc14 in rDNA condensation through condensin targeting. In addition, we have shown that nondisjunction defects in cdc14-1–blocked cells are also alleviated in strains containing fewer copies of rDNA (Fig. 2 F). These findings demonstrate that at least one of the essential roles played by Cdc14 is to mediate rDNA disjunction by ensuring rDNA compaction, thereby shortening the chromosome arms and facilitating segregation.
A novel role for the replication barrier protein Fob1 in rDNA disjunction during anaphase
Our experiments on the segregation of rDNA in strains with short arrays revealed an unexpected role for the replication fork block protein Fob1 in rDNA disjunction (Fig. 2 F). Eliminating Fob1 in cdc14-1 mutant cells causes a dramatic decrease in rDNA resolution (Fig. 3 A). However, fob1Δ cells are able to segregate rDNA efficiently and do not lose cell viability, despite suffering a small delay in segregation and an accumulation of nucleolar Top2 (Fig. 4 B). The fact that Fob1's role in rDNA segregation is only seen in a cdc14-1 mutant background suggests that Cdc14 can compensate for the rDNA segregation defects caused by fob1Δ.
Deletion of FOB1 causes a variety of seemingly unrelated phenotypes, including reduced recombination (Kobayashi and Horiuchi, 1996; Kobayashi et al., 1998; Johzuka and Horiuchi, 2002), loss of silencing in rDNA (Huang and Moazed, 2003), premature release of Cdc14 (Stegmeier et al., 2004), and abrogation of replication fork pausing at the RFB site (Kobayashi and Horiuchi, 1996). We have tested whether Fob1's newly described role in rDNA segregation is caused by an indirect effect from Fob1's function in any of these processes.
Neither rad52Δ nor sir2Δ has an effect on rDNA segregation in cdc14-1 mutants (Fig. S4, A and B), ruling out the possibility that recombination or the presence of silent chromatin interferes with the segregation of rDNA. Recently, Fob1 has been shown to play a role in the nucleolar release of Cdc14 during anaphase (Stegmeier et al., 2004). Two regulatory networks, FEAR and MEN, mediate Cdc14 activation and release (Stegmeier and Amon, 2004). The FEAR network releases nucleolar Cdc14 during early anaphase, whereas MEN promotes and maintains Cdc14 released during the late stages of anaphase (Stegmeier and Amon, 2004). Fob1 is important for Cdc14 activation because it regulates the timing of the FEAR-mediated release. Deletion of Fob1 causes a premature nucleolar release, whereas Fob1 overexpression induces a delay (Stegmeier et al., 2004). Our results show that the segregation role of Fob1 is independent of its role as a regulator of the FEAR network (Fig. S4 C). However, we have also shown that the timely activation of Cdc14 has an effect on the efficiency of rDNA segregation, as illustrated by the fact that a proportion of cells fail to resolve rDNA in our cdc14-1–release experiments (Fig. 3 A) after passage through early anaphase without Cdc14 activity (Fig. 3 A). CDC14 overexpression in cdc14-1 arrests suppressed rDNA segregation defects (Fig. S3), demonstrating that resolution failure in cdc14-1–block release experiments is not irreversible. Therefore, activation during early anaphase by FEAR is important for segregation. These observations are consistent with the reduced viability of FEAR mutants (Stegmeier et al., 2004).
Fob1 is also required for replication fork pausing in the RFB site at the 3′ end of the 35S rRNA gene (Kobayashi and Horiuchi, 1996). The functional significance of these replication blocks is not known. One possibility is that they prevent interference between the transcription and replication machineries, as some reports have demonstrated that head-on collision between these processes can cause both topological entanglements in plasmids (Olavarrieta et al., 2002) and an increase in homologous recombination (Takeuchi et al., 2003; Prado and Aguilera, 2005). However, because eliminating RFB activity in fob1Δ has no deleterious consequences to cells, the function of RFB at the end of rRNA genes has remained mysterious. We have been able to rule out the possibility that Fob1's contribution to rDNA segregation is dependent on its RFB activity because deletion of TOF1, also necessary for fork arrest at the RFB site (Calzada et al., 2005; Tourriere et al., 2005; Mohanty et al., 2006), in cdc14-1 mutants does not cause the segregation defects observed for fob1Δ (Fig. 4 C). Therefore, we conclude that Fob1's effect in rDNA segregation is a novel function unrelated to all its previously described phenotypes. Interestingly, a recent article demonstrated that Fob1 plays a role in the recruitment of condensin to rDNA (Johzuka et al., 2006); therefore, it is possible that Fob1's role in rDNA segregation is related to this function.
Transcription-induced linkages prevent mitotic disjunction in the absence of Cdc14
An important part of the metabolic activity of rDNA is the transcription of rRNA genes. Within the ribosomal gene array, some genes are transcriptionally repressed, whereas others are transcribed at high rates, even during mitosis (Elliott and McLaughlin, 1979). We have demonstrated that the transcriptional hyperactivity in rRNA genes imposes a segregation constraint on rDNA. Inactivation of RNA polymerase I transcription suppressed the nondisjunction defects observed in cdc14-1 and cdc14-1 fob1Δ cells (Fig. 5), demonstrating that transcription causes linkages between sister chromatids that are resolved by Cdc14-mediated processes.
The nature of the transcription-dependent linkages is presently unclear. High transcription rates in some rRNA genes could promote an increase in local catenations that would require specialized pathways for resolution. Cdc14 activity is important for the localization of condensin to rDNA during mitosis (D'Amours et al., 2004; Wang et al., 2004), and condensin has been shown to recruit Top2 to chromatin (Bhalla et al., 2002). Therefore, in cdc14-1 mutants, condensin and Top2 would not be active; thus, neither condensation nor decatenation might be fully achieved. On the other hand, we have shown that Fob1 has a new role in preventing linkages that can be resolved by the action of Cdc14-regulated pathways. The mutant fob1Δ shows Top2 enrichment at the nucleolus (Fig. 4 B), which supports this view. Surprisingly, in our hands, overexpression of Top2 in cdc14-1–arrested cells does not rescue rDNA segregation (unpublished data). This may imply that Cdc14's upstream role in controlling Top2 function is not exclusively linked to targeting through condensin. In agreement with our results, Top2 overexpression does not rescue defects in sister chromatid resolution in condensin mutants, despite Top2 going to chromosomes in such conditions (Bhalla et al., 2002). Another possibility is that rRNA transcripts and protein factors involved in rRNA processing are sufficient to establish linkages between sister chromatids. EM analysis of rDNA in budding yeast has shown that a large number of rRNA molecules are transcribed simultaneously from each gene (Saffer and Miller, 1986). In addition, large protein complexes required for the cleavage and maturation of transcripts assemble onto rRNA molecules cotranscriptionally (Osheim et al., 2004). Because rRNA transcription is maintained during mitosis (Elliott and McLaughlin, 1979), these large protein–RNA complexes might cause entanglements between sister chromatids and thus prevent rDNA segregation unless specialized mechanisms are in place. In higher eukaryotes, mitosis correlates with a reduction in transcriptional activity, thus preventing transcription-induced rDNA linkages during this cell cycle stage.
In summary, nucleolar chromatin differs from the rest of the genome in two main ways: (1) it is present as a large array of tandem repeats and (2) a fraction of rRNA genes are highly transcribed. We have shown that these two features are the reason behind the additional segregation requirements of rDNA. Shortening the array or inactivating RNA polymerase I eliminates the segregation defects of cdc14-1 mutant cells. In addition to Cdc14, we uncover a novel role for the RFB gene FOB1 in promoting rDNA segregation. Our results demonstrate that the high level of transcription in ribosomal genes causes linkages and chromosome nondisjunction in the absence of additional resolution mechanisms dependent on Cdc14 and Fob1. It will be interesting to determine whether highly transcribed loci outside rDNA also generate deleterious effects that interfere with the timely segregation of sister chromatids at anaphase.
Materials and methods
Yeast strains and plasmids
All yeast strains used were S228C background, except for cdc14-1 GAL-SIC1 strains and the strains bearing a fixed number of rDNA units (25 or 190 copies) that were W303 (a gift from F. Cross, The Rockefeller University, New York, NY). Chromosome tags have been described elsewhere (Machin et al., 2005). COOH-terminal epitope tagging with GFP and gene deletions, including rpa135Δ, were performed using PCR allele-replacement methods. The cdc14-1 allele was transferred between strains also using PCR allele-replacement strategy where a 9myc epitope and a selective marker (TRP1) are tagged to the COOH terminus. Western blotting and thermo-sensitivity assays were used to confirm transformations. Plasmid pNOY103 (GAL-35S URA3) was a gift from M. Nomura (University of California, Irvine, Irvine, CA) and K. Kobayashi (National Institute for Basic Biology, Tokyo, Japan). Plasmid for the overexpression of Cdc14 was a gift from A. Amon (Massachusetts Institute of Technology, Cambridge, MA). The top2-4 allele was a gift from M. Sullivan (Cancer Research UK, London, UK). Relevant genotypes of strains used in this study are shown in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200511129/DC1).
Cell cycle and synchronizations
To arrest cells in G1, we used bar1Δ. Cells were treated with 50 ng/ml α-factor for 3 h at 25°C. To release cells from the block, we transferred them to fresh media plus pronase E (0.1 mg/ml). For releases at nonpermissive temperatures, we exposed cells to 37°C for 30 min before their transfer to fresh media (also at 37°C). To release from a cdc14-1 block, G1-released cells were incubated at 37°C for 150 min before shifting them back to 25°C to reactivate Cdc14. G2/M arrest in Fig. 1 B was obtained by adding 15 μg/ml nocodazole to the media and incubating for 3 h. For the experiments in Fig. 5 (lack of RNA polymerase I transcription), parental strains (RPA135 without the pGAL-35S plasmid) and strains bearing rpa135Δ were grown in YPgal at 25°C until log phase (up to 3 d for rpa135Δ strains). Strains RPA135 with the pGAL-35S plasmid were grown in YPgal only for 9 h after a first overnight growth in SC-galactose-ura). RPA135 strains were arrested in α-factor for 3 h and released into 37°C for 3 h (OD600 doubling time in YPgal ∼3 h). RNA polymerase I–deficient strains were arrested in α-factor for 6 h and released into 37°C for 7 h (OD600 doubling time in YPgal ∼7 h). The α-factor block arrests >98% of the cells in G1. About 50% of the RNA polymerase I–deficient cells enter a new cell cycle after the G1 release. Only cells clearly in anaphase (stretched nucleus across the neck or binucleated) were counted.
PFGE and ERC analysis
PFGE to see chromosome XII was performed in a 0.8% agarose gel in 0.5× TBE buffer run for 68 h at 6 V/cm with an initial switching time of 60 s, a final of 120 s, and an angle of 120°. ERC analysis was performed as described by Sinclair and Guarente (1997). The total running time was doubled to 48 h, and a long agarose gel was used.
Microscopy
Yeast cells with GFP-tagged proteins were analyzed by fluorescence microscopy after DAPI staining. Series of z focal plane images were collected on a microscope (IRB; Leica) using a digital camera (C4742-95; Hamamatsu) and OpenLab software (Improvision). A tuneable light source (Polychrome IV) with a Xenon lamp was used. Images in different z axis planes were flattened into a 2D projection and processed in OpenLab. DNA was stained using DAPI (Invitrogen) at a final concentration of 1 μg/ml after short treatment of the cells with 1% Triton X-100. Imaging was done in antifade/DAPI medium (Invitrogen) at room temperature. Micrographs were taken with either 63×/1.4 or 100×/1.35 lenses.
Online supplemental material
Fig. S1 shows that multicopy plasmids bearing the rDNA unit cannot rescue the chromosome XII segregation impairment in cdc14-1 mutants. Fig. S2 shows that condensin and topoisomerase II mutants show synergistic genetic interactions with fob1Δ. Fig. S3 demonstrates overexpression of Cdc14 rescues cdc14-1 and cdc14-1 fob1Δ rDNA segregation defects. Fig. S4 shows that Fob1 function in chromosome XII segregation does not act through its role in rDNA recombination, FEAR network, or rDNA silencing. Table S1 shows relevant genotypes of strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200511129/DC1.
Supplementary Material
Acknowledgments
We are grateful to Angelika Amon, Masayasu Nomura, Takehiko Kobayashi, Fred Cross, and Matt Sullivan for plasmids and strains. We thank Angelika Amon for communicating results before publication. We are grateful to Sarah Farmer for critical reading of the manuscript and other members of the Aragon laboratory for discussions of the work.
The Medical Research Council UK supported this work.
Abbreviations used in this paper: ERC, extrachromosomal ribosomal circle; FEAR, Cdc14 early anaphase release; MEN, mitotic exit network; PFGE, pulsed-field gel electrophoresis; rDNA, ribosomal DNA; RFB, replication fork barrier.
References
- Bhalla, N., S. Biggins, and A.W. Murray. 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 13:632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, B.J., and W.L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 55:637–643. [DOI] [PubMed] [Google Scholar]
- Brewer, B.J., D. Lockshon, and W.L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 71:267–276. [DOI] [PubMed] [Google Scholar]
- Bryk, M., M. Banerjee, M. Murphy, K.E. Knudsen, D.J. Garfinkel, and M.J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255–269. [DOI] [PubMed] [Google Scholar]
- Calzada, A., B. Hodgson, M. Kanemaki, A. Bueno, and K. Labib. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19:1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 117:455–469. [DOI] [PubMed] [Google Scholar]
- Defossez, P.A., R. Prusty, M. Kaeberlein, S.J. Lin, P. Ferrigno, P.A. Silver, R.L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell. 3:447–455. [DOI] [PubMed] [Google Scholar]
- Elliott, S.G., and C.S. McLaughlin. 1979. Regulation of RNA synthesis in yeast. III. Synthesis during the cell cycle. Mol. Gen. Genet. 169:237–243. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, P.J., J.H. Toyn, J.B. Millar, and L.H. Johnston. 1998. DNA replication is completed in Saccharomyces cerevisiae cells that lack functional Cdc14, a dual-specificity protein phosphatase. Mol. Gen. Genet. 258:437–441. [DOI] [PubMed] [Google Scholar]
- Freeman, L., L. Aragon-Alcaide, and A. Strunnikov. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff, S., H. Zou, and R. Rothstein. 1996. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- Granot, D., and M. Snyder. 1991. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil. Cytoskeleton. 20:47–54. [DOI] [PubMed] [Google Scholar]
- Guacci, V., E. Hogan, and D. Koshland. 1994. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 125:517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., and D. Moazed. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17:2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S.L., J.F. Charles, R.L. Tinker-Kulberg, and D.O. Morgan. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 9:2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka, K., and T. Horiuchi. 2002. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 7:99–113. [DOI] [PubMed] [Google Scholar]
- Johzuka, K., M. Terasawa, H. Ogawa, T. Ogawa, and T. Horiuchi. 2006. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent vontraction of a long repetitive array in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., and T. Horiuchi. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1:465–474. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., D.J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12:3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, B.D., E. Hogan, and D. Koshland. 2004. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 18:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens, M.H., and J.A. Huberman. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin, F., J. Torres-Rosell, A. Jarmuz, and L. Aragon. 2005. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J. Cell Biol. 168:209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, A.H., B. Kornmann, K. Dubrana, and D. Shore. 2005. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 19:1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B.K., N.K. Bairwa, and D. Bastia. 2006. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 103:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi, Y., R. Yano, and M. Nomura. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. USA. 88:3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olavarrieta, L., P. Hernandez, D.B. Krimer, and J.B. Schvartzman. 2002. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 322:1–6. [DOI] [PubMed] [Google Scholar]
- Osheim, Y.N., S.L. French, K.M. Keck, E.A. Champion, K. Spasov, F. Dragon, S.J. Baserga, and A.L. Beyer. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 16:943–954. [DOI] [PubMed] [Google Scholar]
- Pereira, G., C. Manson, J. Grindlay, and E. Schiebel. 2002. Regulation of the Bfa1p–Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157:367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado, F., and A. Aguilera. 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 24:1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer, L.D., and O.L. Miller Jr. 1986. Electron microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol. Cell. Biol. 6:1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair, D.A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 91:1033–1042. [DOI] [PubMed] [Google Scholar]
- Smith, J.S., and J.D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241–254. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., and A. Amon. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., J. Huang, R. Rahal, J. Zmolik, D. Moazed, and A. Amon. 2004. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr. Biol. 14:467–480. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., W. Shou, G.J. Dowd, C.W. Turck, R.J. Deshaies, A.D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 97:245–256. [DOI] [PubMed] [Google Scholar]
- Sullivan, M., T. Higuchi, V.L. Katis, and F. Uhlmann. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 117:471–482. [DOI] [PubMed] [Google Scholar]
- Takeuchi, Y., T. Horiuchi, and T. Kobayashi. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17:1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell, J., F. Machin, A. Jarmuz, and L. Aragon. 2004. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle. 3:496–502. [PubMed] [Google Scholar]
- Tourriere, H., G. Versini, V. Cordon-Preciado, C. Alabert, and P. Pasero. 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell. 19:699–706. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 400:37–42. [DOI] [PubMed] [Google Scholar]
- Visintin, R., K. Craig, E.S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718. [DOI] [PubMed] [Google Scholar]
- Wang, B.D., V. Yong-Gonzalez, and A.V. Strunnikov. 2004. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 3:960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., K. Asakawa, and A. Toh-e. 2002. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr. Biol. 12:944–950. [DOI] [PubMed] [Google Scholar]
- Yuste-Rojas, M., and F.R. Cross. 2000. Mutations in CDC14 result in high sensitivity to cyclin gene dosage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:60–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.