Abstract
Tetraspanin uroplakins (UPs) Ia and Ib, together with their single-spanning transmembrane protein partners UP II and IIIa, form a unique crystalline 2D array of 16-nm particles covering almost the entire urothelial surface. A 6 Å–resolution cryo-EM structure of the UP particle revealed that the UP tetraspanins have a rod-shaped structure consisting of four closely packed transmembrane helices that extend into the extracellular loops, capped by a disulfide-stabilized head domain. The UP tetraspanins form the primary complexes with their partners through tight interactions of the transmembrane domains as well as the extracellular domains, so that the head domains of their tall partners can bridge each other at the top of the heterotetramer. The secondary interactions between the primary complexes and the tertiary interaction between the 16-nm particles contribute to the formation of the UP tetraspanin network. The rod-shaped tetraspanin structure allows it to serve as stable pilings in the lipid sea, ideal for docking partner proteins to form structural/signaling networks.
Introduction
The tetraspanins are a family of proteins containing four transmembrane domains (TMs) linked by a small, first extracellular domain (EC1) connecting TM1 and TM2, and a large, second extracellular domain (EC2) connecting TM3 and TM4, as well as the usually short NH2- and COOH-terminal cytoplasmic tails (Hemler, 2003; Stipp et al., 2003; Levy and Shoham, 2005). The tetraspanin proteins are distinguished from other membrane proteins with four TMs (such as sacospan and stargazin) by their highly conserved transmembrane helices and the unique EC2 domain with a Cys-Cys-Gly motif (Stipp et al., 2003). The EC2 domain contains four to eight Cys residues forming disulfide bonds (Kitadokoro et al., 2001; Seigneuret et al., 2001) that can stabilize the structure of EC2 domain. In human, the tetraspanin family consists of ∼30 members; ∼20 of them, including CD9, CD37, CD53, CD63, CD81, CD82, and CD151, are expressed in leukocytes (Tarrant et al., 2003). Tetraspanins can also be found in Drosophila melanogaster, Caenorhabditis elegans, and zebrafish, as well as in plants. Some of the tetraspanins, such as CD9 and CD81, are rather ubiquitous, whereas others, such as uroplakins (UPs) Ia and Ib (in mammalian urothelium) and peripherin/RDS (in retina), are highly tissue restricted.
Tetraspanins have been implicated in many cellular functions, including cell proliferation, fusion, development, motility, and tumor growth and invasion. There is as yet no unified view of how tetraspanins perform these diverse functions. Nevertheless, several pictures have emerged from recent studies of tetraspanins. First, tetraspanins tend to associate with other membrane proteins to laterally organize structural or signaling networks, often referred to as tetraspanin webs, on cell surfaces (Rubinstein et al., 1996). Many integrins, such as α2β1, α3β1, α4β1, α5β1, and α6β1, can associate with one or more tetraspanins (Hemler, 1998; Berditchevski, 2001). Tetraspanins have also been found to associate with the Ig superfamily of proteins, including CD2, CD4, CD8, CD19, MHC I, and MHC II (Hemler, 2003). The ability of tetraspanins to organize membrane networks has earned them the name “molecular facilitators” (Maecker et al., 1997). The tetraspanin networks in cell membranes may actually form a distinct type of tetraspanin-enriched lipid microdomains (Hemler, 2003; Min et al., 2003) that differ from the ordinary raft lipid domains in that they are less resistant to Triton X-100 but more resistant to cholesterol depletion and elevated temperature (Charrin et al., 2003; Hemler, 2003, 2005). The second emerging picture of tetraspanin function is that these proteins may be involved in transmembrane signaling. For example, tetraspanin CD151 can serve as a transmembrane linker between integrin α3β1 and cytoplasmic phosphatidylinositol 4-kinase, playing a role in regulating cell migration (Hemler, 1998; Yauch et al., 1998). In addition, CD151 can stabilize α3β1 integrin in its activated conformation; hence, CD151 may be regarded as a “conformation facilitator” (Nishiuchi et al., 2005). Third, several tetraspanins can serve as pathogen receptors; e.g., tetraspanin UP Ia as the receptor for uropathogenic Escherichia coli (Wu et al., 1996; G. Zhou et al., 2001; Min et al., 2002) and CD81 as the receptor for hepatitis C virus (Cormier et al., 2004). Hence, the tetraspanins have a remarkable capability to interact with diverse partner proteins. However, the structural basis of how the tetraspanins bind to their partner proteins and how they form the tetraspanin web has not yet been elucidated.
The only currently available, high-resolution crystal structure of any tetraspanin is that of the large extracellular loop (EC2) of CD81; this study indicates that EC2 is a mushroom-like structure with a head domain connected to a two-helix stalk that may anchor to the transmembrane helices (Kitadokoro et al., 2001, 2002). Sequence analyses suggest that all tetraspanin EC2 domains share such a mushroom-like structure with a highly variable region embedded in the head domain (Seigneuret et al., 2001). The EC2 structure of CD81 has been used to model the structure of the EC2 domain of other tetraspanins (Bienstock and Barrett, 2001; Seigneuret et al., 2001). Because the crystal structure of the EC2 domain of CD81 contains two EC2 fragments packed against each other to bury a hydrophobic region of the stalk, this dimerization has been assumed to occur naturally and has been used often as a paradigm for predicting tetraspanin–tetraspanin interaction. However, such an EC2–EC2 binary interaction cannot explain the formation of a tetraspanin network. The lack of structural information on the transmembrane helices of tetraspanins and on how these helices are connected to the extracellular loops has severely limited our understanding of the structure-function of tetraspanins and the formation of the tetraspanin network.
UP tetraspanin complex is a crystalline tetraspanin web uniquely suitable for structural studies using EM (Hicks and Ketterer, 1969; Vergara et al., 1969; Brisson and Wade, 1983; Taylor and Robertson, 1984; Walz et al., 1995; Oostergetel et al., 2001; Min et al., 2002, 2003). UP tetraspanin web consists of hexagonal arrays of 16-nm UP protein particles. Each of these particles contains two highly homologous (34% identical) tetraspanins, UP Ia and Ib, which pair specifically and stoichiometrically with their single-spanning transmembrane protein partners, UP II and UP IIIa, respectively (Wu et al., 1995; Liang et al., 2001; Deng et al., 2002; Tu et al., 2002; Min et al., 2003; Hu et al., 2005). The naturally occurring crystalline UP web forms rigid-looking plaques (urothelial plaques; also known as asymmetric unit membrane, or AUM) covering almost the entire apical surface of the mammalian urothelium (Staehelin et al., 1972; Hicks, 1975; Kachar et al., 1999). These 2D membrane plaques can be isolated in milligram quantities from mammalian urothelia (Wu et al., 1990; Liang et al., 2001), and they can provide intermediate resolution structural information of the UP molecules using electron crystallography (Min et al., 2003). Functionally, the urothelial plaques play an important role in the formation of one of the most efficient permeability barriers known to exist in nature, separating the urine from the cellular contents (Negrete et al., 1996). In addition, UP Ia has been shown to be the urothelial receptor for type 1–fimbriated E. coli (Wu et al., 1996; G. Zhou et al., 2001; Min et al., 2002), a major causative agent of urinary tract infection (Foxman and Brown, 2003). The tethering of uropathogenic bacteria to the urothelial surface UP Ia receptor, via the lectin adhesin FimH located at the tip of bacterial fimbria, can trigger a transmembrane signaling transduction cascade leading to the urothelial engulfment of the bacteria (Mulvey et al., 1998). Relatively little is known, however, about the mechanism of this transmembrane signal transduction (Min et al., 2003).
We present here a 3D structure of the 16-nm UP tetraspanin complexes at 6-Å resolution. Our data revealed the secondary structural elements of the UP molecules and enabled us to construct a model of UP tetraspanins, showing that UP tetraspanins interact intimately with their single-spanning transmembrane protein partners through their transmembrane as well as extracellular domains. Our results have implications on the structural basis for the formation of the tetraspanin network in general (Hemler, 2005).
Results
Structure determination and the overall feature of the 16-nm particle at 6-Å resolution
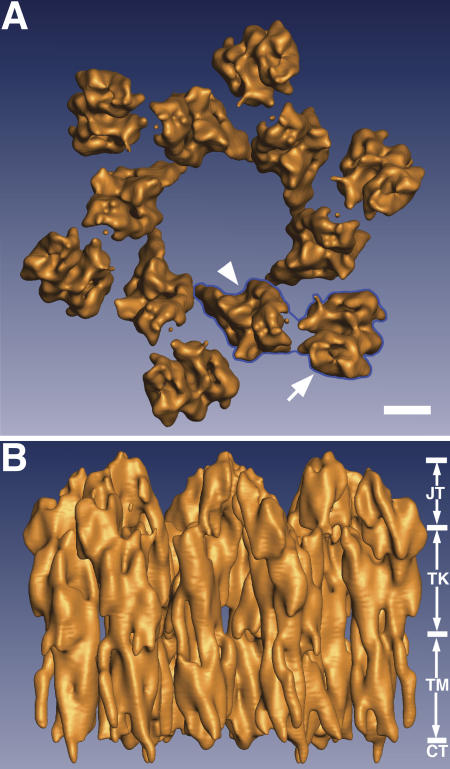
To obtain the 3D structure of the 16-nm UP particle, we recorded, under low-dose conditions, tilted series of images of frozen-hydrated, purified mouse urothelial plaques that have a diameter of up to ∼1 μm (Min et al., 2003). This enabled us to reconstruct a 3D structure of the mouse UP particle at 6-Å resolution in the membrane plane and 12.5 Å in the vertical direction (Fig. 1, Fig. S1, and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200602086/DC1). The overall shape of the 16-nm particle is very similar to the structure visualized at 10-Å resolution (Min et al., 2003 [compare with Oostergetel et al., 2001]). A top view of the 3D map showed that the 16-nm particle has a hexagonal stellate shape with six subunits (Fig. 1 A); each subunit (Fig. 1 A, blue outline) in turn consists of an inner (Fig. 1 A, arrowhead) and an outer subdomain (Fig. 1 A, arrow). The largest diameter of the particle is ∼17.5 nm. The six inner subdomains encircle an area of ∼6 nm in diameter in the center of the particle. This central region is likely to be occupied by lipids, as it cannot be penetrated by negative stains (Walz et al., 1995; Min et al., 2002). The electron density map–enclosed volume, when contoured at the 1-σ level, of one 16-nm particle is ∼6.13 × 106 Å3, which can accommodate ∼650 kD of protein, consistent with the estimated total molecular mass (702 kD) based on the sequences of the four major UPs (Walz et al., 1995; Min et al., 2003). A side view of the 3D map showed that the 16-nm UP particle has a cylindrical shape of ∼12.5 nm in height (Fig. 1 B). This cylindrical shape of the particle has about the same diameter throughout its length. The density constriction at the region of the exoplasmic surface of the lipid bilayer, previously seen in a 10 Å–resolution 3D reconstruction, is less evident in the current 3D model (Min et al., 2003); whether this density constriction reflects a flexibility in this region remains to be investigated. Consistent with our previous results (Min et al., 2003), the 16-nm particle can be divided vertically into four zones (Fig. 1 B): from the top down, the joint, the trunk, the transmembrane, and the cytoplasmic.
Figure 1.
The 3D structure of the mouse UP tetraspanin complexes at 6-Å resolution. (A) The top view of the hexagonal 16-nm particle shows that it consists of six subunits (one of them is outlined in blue) that encircle a central area of ∼6 nm in diameter filled with lipids. Each subunit in turn consists of an inner subdomain (arrowhead) and an outer subdomain (arrow) that are connected at the top of the particle (Fig. 3 C). Bar, 2 nm. (B) The UP particle has a cylindrical shape and a height of ∼12.5 nm. The vertical dimension of the particle can be approximately divided into four zones: from the top, the joint (JT; ∼3 nm in height), the trunk (TK; ∼5 nm), the TM (∼4 nm), and cytoplasmic domain (CT).
The transmembrane zone of the 16-nm particle consists of five-helix bundles
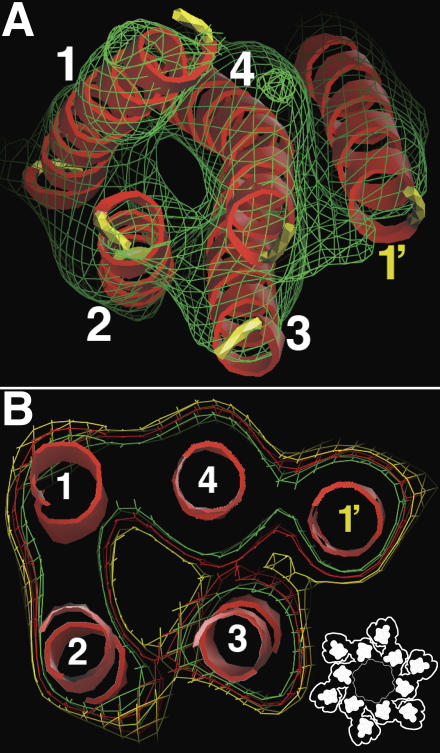
Our 3D reconstruction clearly revealed, for the first time, secondary structural elements of the 16-nm particle. This is particularly obvious for the transmembrane zone of the particle (Figs. 2 and 3). The transmembrane zone (Fig. 1 B, TM; and Fig. 3, TM; ∼4 nm in height) of the 16-nm particle in the 3D reconstruction consists of only rod-shaped electron densities, which likely represent the transmembrane helices of the UP molecules. The TMs of the inner and outer subdomains, with five helices each, have almost the same shape (Fig. 3, A and B), consistent with our previous prediction that each subdomain is formed by a heterodimer of UPs consisting of a tetraspanin UP (Ia or Ib) and its single-spanning transmembrane UP partner (II or IIIa; Min et al., 2002, 2003). The distances between the centers of the nearest neighbor transmembrane helices in the middle of the transmembrane region range from ∼9.3 to 11 Å. For both the inner and outer subdomains, the electron densities of four of the transmembrane helices (Fig. 2, helices 1–4; see below for the assignment of helix number) are rather closely connected, whereas an additional single transmembrane helix (helix 1') is clearly less connected to the other helices (Fig. 2 and Fig. 3, A and B). Together with our previous localization of UP Ia to the inner subdomain (Min et al., 2002), this suggests that the four closely packed transmembrane helices belong to the tetraspanin UP Ia (for the inner subdomain) or Ib (outer subdomain), whereas the somewhat detached, single transmembrane helix belongs to the single-spanning transmembrane UP II (inner subdomain) or IIIa (outer subdomain). The four transmembrane helix bundles of the UP tetraspanins are left-handed, although the cross-angles, as defined according to Bowie (1997a), between these TMs are somewhat variable (Fig. 2 A and Fig. 3, A and B, top). In this regard, the four transmembrane helices of the UP tetraspanins can be grouped into two pairs, the TM1–TM2 pair and the TM3–TM4 pair (Fig. 2 A). The cross-angle between the two helices within each pair is ∼10°, which is relatively small, whereas the cross-angle between the two pairs is ∼25°, which is slightly larger than the 20° common cross-angle in transmembrane proteins in general (Bowie, 1997b). The small packing cross-angles between the helices in the two pairs allow the helices within a pair to be closely associated with each other along the entire length (Eilers et al., 2002).
Figure 2.
The TM of the primary UP tetraspanin pair as a five-TM helix bundle. (A) The transmembrane helices of the UP Ia/II tetraspanin pair viewed from the cytoplasmic side. The electron density is represented by a green mesh contoured at the 2.0-σ level. The four helices of the tetraspanin UP Ia are labeled 1–4, whereas the single helix of UP II is labeled 1' (see text for details). (B) The section of the electron density in the middle of the transmembrane region of the UP Ia/II pair. Models of the five transmembrane helices are shown as red ribbons. The yellow, red, and green meshes represent the electron density contoured at the 1.0-, 1.5-, and 2.0-σ levels, respectively. The inset shows the relative positions of the TM regions in the 16-nm particle.
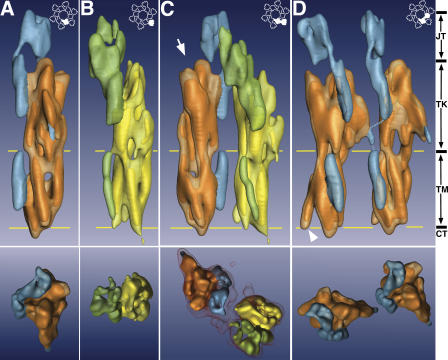
Figure 3.
The subdomains of the 16-nm particle and the relation between the UP tetraspanins and their partners. The top and bottom panels represent the side and top views, respectively, of the electron densities contoured at the 2-σ level. (A and B) The two primary UP pairs, i.e., the inner (A; UP Ia/II) and the outer (B; UP Ib/IIIa) subdomains, viewed separately. The tetraspanin UPs (inner and outer subdomains are shown in brown and yellow, respectively) are segmented from that of their partners (inner and outer subdomains are shown in blue and green, respectively). (C) A subunit of the 16-nm UP particle is made of an inner and an outer subdomain linked through the top “joint.” A 1.0-σ level contour of the electron density is shown in the top view (bottom) to illustrate the connection between the inner and outer domains. A possible FimH binding site on UP Ia is indicated by an arrow (top; see Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200602086/DC1). (D) Two neighboring inner domains are connected through a weak link at the exoplasmic boundary of the lipid bilayer. This link joins the tetraspanin UP Ia with the UP II of the neighboring primary UP Ia/II pair. The arrowhead indicates the NH2-terminal end of TM1. The two parallel lines (yellow) in the top panels approximately mark the boundaries of the TM, and the four vertical zones are indicated on the right as the joint (JT), trunk (TK), transmembrane (TM), and cytoplasmic (CT) domains. The inset in the top right corner of each top panel illustrates the position of the shown subdomains within the 16-nm particle.
The single-spanning transmembrane UPs interact with tetraspanin UPs through both transmembrane and extracellular domains
The electron densities of inner and outer subdomains of the 16-nm particle can be clearly segmented according to the connectivity of the densities; i.e., closely connected densities are considered to be parts of the same molecule, into the tetraspanin region (Fig. 3, UP Ia and Ib [brown and yellow, respectively]) and the single-spanning UP region (Fig. 3, UP II and IIIa [blue and green, respectively]). The single-spanning transmembrane protein region of the inner and outer domains have roughly an inverted L-shape, with the vertical arm of the L interrupted by a region of discontinuity, most likely reflecting a flexible local structure. This inverted L is anchored by its transmembrane helix (Fig. 3, A and B, blue and green densities), which is packed against the four transmembrane helices of the partner tetraspanins, forming the five-helix bundle of the transmembrane zone of each subdomain. The long arm of the inverted L continues up against the cylindrical UP tetraspanins and makes a bend at the top to form the short arm of the inverted L. This short arm joins the other short arm of the inverted L from the paired subdomain within the same subunit (Fig. 3 C, blue and green densities; and Fig. 1 A), hence the name “joint” (Fig. 1 B and Fig. 3; Min et al., 2003). Interestingly, the joint is the only contact between the two subdomains within a subunit; the two tetraspanins, UP Ia and Ib, do not appear to have direct contacts (Fig. 3 C). This kind of loose connection between the two subdomains within a subunit suggests a flexible interaction between the inner and outer subdomains, thus providing a basis for possible structural changes of UPs upon binding by bacterial fimbria (Min et al., 2003; see Discussion). Consistent with this, the volume of the electron density of the joint goes down much faster than the rest of the particle, when contoured with increased contour levels (Fig. 3 C, bottom), suggesting that the joint is flexible.
The TM helices of the tetraspanin UPs extend into the extracellular domains, allowing the construction of a molecular model of the UP tetraspanins
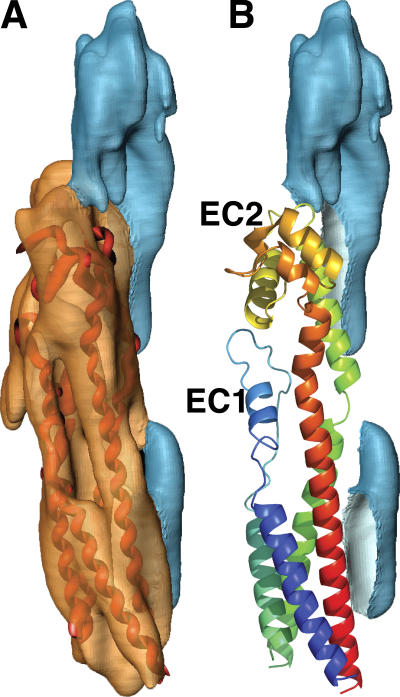
Visualization of the secondary structural elements of the 16-nm particle enabled us to construct a molecular model of the UP tetraspanins (Fig. 4) by following the electron densities, as well as several additional considerations. First, the rod-shaped electron densities of the transmembrane helices of the two UP tetraspanins extend continuously into the extracellular region of the particle (Fig. 3, top; densities of UP Ia and Ib are colored brown and yellow, respectively). Second, The x-ray structure of the EC2 of the homologous tetraspanin CD81 shows that the stem of the EC2 domain of tetraspanins, which are contiguous with TM3 and TM4, are made up of two closely packed helices (Kitadokoro et al., 2001). Thus, TM3 and TM4 are likely packed against each other. Third, because TM2 and TM3 are connected by a short, five-amino-acid cytoplasmic loop (Yu et al., 1994; Stipp et al., 2003), they must be packed closely against each other. Fourth, the shape of the EC2 from that of the x-ray structure of CD81 allows only a unique placement of EC2 of the UP tetraspanins into the electron density of the extracellular portion (TK and JT) of a subdomain. These constraints allowed us to assign the transmembrane helices of the UP tetraspanins (Fig. 2). They also allowed us to construct a poly-alanine atomic model of the tetraspanin UP Ia and Ib by building α-helices into the electron densities of the TMs and by placing the models of the extracellular loops based on homologous x-ray structure of the EC2 domain of CD81 (Fig. 4). Our model of tetraspanin UPs has an overall cylindrical shape (Fig. 4) with a largely four-helix bundle structure capped at the top by a head domain, which consists of the disulfide-stabilized region of EC2 (Fig. 4 B). The small extracellular loop EC1 extended from TM1 and TM2 is packed against the hydrophobic part of the large loop (Fig. 4 B). This model differs from the dimeric crystal structure of the CD81 EC2, where this hydrophobic region of EC2 is packed against that of the other EC2 to form a homodimer (Kitadokoro et al., 2001). Our cylindrically shaped model of UP tetraspanins is similar to a just-published theoretical 3D model of CD81 (Seigneuret, 2006) but very different from a model of CD82 (KAI1) that has skewed (side displaced) extracellular domains (Bienstock and Barrett, 2001). Interestingly, our electron density shows that the TM1 and TM2 of the UP tetraspanins are packed more tightly at the exoplasmic end than the cytoplasmic end (most visible in the orientation shown by the arrowhead in Fig. 3 D). This is consistent with a recent modeling study of another tetraspanin, CD9, by Kovalenko et al. (2005). These authors showed that TM1 and TM2 of CD9, and of tetraspanins in general, have conserved complementary heptad repeats, and their model suggested that TM1 and TM2 pack against each other left-handedly, with the extracellular ends packed more tightly (Kovalenko et al., 2005). Our assignment of the transmembrane helices of the UP tetraspanins is also consistent with the general observation that neighboring transmembrane helices often contact each other and that the antiparallel packing is preferred (Bowie, 1997b).
Figure 4.
Molecular model of the UP tetraspanins. (A) A molecular model of UP Ia tetraspanin is fitted into the electron density map. (B) The density of the UP Ia tetraspanin is removed to show the model's relation to the density of the single-spanning transmembrane protein. The model is colored so that the color spectra start with blue at the NH2 terminus and end with red at the COOH terminus.
Discussion
Tetraspanin structure: a stable, bundled piling in the lipid sea
Our 3D reconstruction shows that the UP tetraspanins Ia and Ib have an overall cylindrical structure, formed by the bundled TM helices that extended into the extracellular loops and capped at the top by the disulfide-stabilized regions of EC2 (Figs. 2–4 ). This cylindrical bundle likely represents a stable protein structure. First, the four-helix bundles in the TM of UP Ia and Ib are closely packed. They have axial distances of ∼9.3 to 11 Å, comparable with the average 9.6-Å axial distance in the closely packed transmembrane helices in other membrane proteins (Bowie, 1997b). They have left-handed cross-angles, which are more closely packed in general than the right-handed ones (Eilers et al., 2002), also suggesting stability. The first three transmembrane helices of tetraspanin family of proteins exhibit a heptad repeat of Gly residues (Kovalenko et al., 2005), which may allow the helices to interact closely by favoring van de Waals contacts with surrounding residues and forming backbone CαH-amide carbonyl hydrogen bonds (Javadpour et al., 1999; Senes et al., 2001; Eilers et al., 2002). The very short, five-residue cytoplasmic loop connecting TM2 and TM3 ensures that these two TMs are closely packed together. Second, tetraspanin proteins, including UP Ia and Ib, have a highly conserved Asn in TM1 and a Glu/Gln in TM3 that may be able to form interhelical hydrogen bonds. These conserved polar/charged residues are located on the faces of the helices containing many conserved residues (unpublished data). It has been proposed that residues mediating interhelical contacts would be more evolutionarily conserved than those that face the lipids because mutations of the former are more likely to destabilize the protein (Donnelly et al., 1993; Stevens and Arkin, 2001; Beuming and Weinstein, 2004). Hence, these conserved polar/charged residues are likely involved in the interhelical interactions in tetraspanin TMs (Choma et al., 2000; F.X. Zhou et al., 2000, 2001). In fact, mutation of the conserved Glu residue in TM3 of UP Ib disturbs its transmembrane structure, leading to its ER retention (Kreibich, G., personal communication). Third, the extracellular domains of UP Ia and Ib also form a closely packed structure. In our model, the small extracellular loop covers up the hydrophobic part of the large extracellular loop (Fig. 4 B), forming an integral part of the cylindrical tetraspanin structure. Although this proposed structure is different from what was suggested by the x-ray structure of the EC2 domain of CD81 (Kitadokoro et al., 2001), it is consistent with the finding that the small loop is necessary for optimal surface expression of the large loop and that it may contribute to the stability of the tetraspanin structure (Masciopinto et al., 2001; Drummer et al., 2005). Collectively, these data suggest that the four-helix bundle of the UP tetraspanins, and likely that of the tetraspanins in general, is a stable rod-shaped structure. With the lower half of the bundle embedded in the lipid bilayer, the tetraspanins may serve as protein pilings in the lipid sea, ideal for docking other transmembrane proteins.
Structural roles of tetraspanins: transmembrane signal transduction
We have previously identified tetraspanin UP Ia as the urothelial receptor for the type 1–fimbriated bacteria by protein and bacterial overlay assays (Wu et al., 1996; G. Zhou et al., 2001) and by EM localization (Min et al., 2002). We have proposed a mechanism by which the urothelial cell senses the bacterial attachment, i.e., through the bacterium binding–induced conformational changes of the TMs of the 16-nm particle (Min et al., 2003). Our current model of the 16-nm particle, with secondary structural elements, provides some insights into the possible mechanism of this transmembrane signal transduction. Because UP Ia tetraspanin is shorter than UP II and IIIa, for the bacterial adhesin FimH molecule to bind to UP Ia (Fig. 3 C, arrow; and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200602086/DC1), the FimH has to reach into a crevice created by the two joints (made up of UP II and IIIa) of the neighboring subunits of the 16-nm particle. This crevice is ∼3 nm wide, just large enough to accommodate the lectin domain (∼2.5 nm in diameter and 5 nm long) of the FimH molecule (Choudhury et al., 1999). Therefore, the FimH binding to UP Ia most likely involves additional protein–protein contacts to joints of the 16-nm particle. These broad contacts would be ideal for exerting a force at the joint region of the particle and for inducing a conformational change that can propagate to the TMs. Interestingly, the binding between FimH and the mannose moiety of UP Ia can exert a mechanical force in the range of pico-Newtons (Liang et al., 2000). A mechanical force of this magnitude should be sufficiently strong to be sensed by cytoskeletal elements that interact with the cytoplasmic tails of the UPs (Staehelin et al., 1972; Balaban et al., 2001). Because the tetraspanin UP Ia is basically a closely packed four-helix bundle, likely to be relatively rigid, it is possible that conformation changes induced by the bacterial binding will lead to changes in the relative orientation (twisting), or sliding, between the tetraspanin UP Ia and its single-spanning transmembrane UP II partner. Interestingly, a “twisted ribbon” model of the 16-nm particle was proposed based only on the negative-stained structure of the extracellular portion of the 16-nm particle (Walz et al., 1995). Some other members of the tetraspanin family are also known to serve as receptors, e.g., CD81 as the receptor for hepatitis C virus and, as has recently been proposed, CD9 as an alternative receptor for interleukin 16 (Cormier et al., 2004; Qi et al., 2006). Additional studies are needed to elucidate the mechanistic details of how these receptors transmit signals.
The extracellular part of the tetraspanin UPs extends to ∼5 nm above the lipid bilayer (the height of the trunk domain in Fig. 1 B; Fig. 3). Because this height is about the same as that of an Ig or Ig-like domain, the extracellular domain of tetraspanins is ideally suited for binding to the tetraspanin-associated proteins, many of which contain an Ig or Ig-like (such as Calf-1 and -2 in integrins) extracellular domain immediately above the lipid bilayer (Hemler, 2001; Le Naour et al., 2004). The ability of a tetraspanin to bind its partner through both its extracellular domain and TM helices may play an important role in supporting its tall partners for signaling purpose. For example, CD151 can stabilize its partner, α3β1 integrin, in its activated configuration, as has recently been reported by Nishiuchi et al. (2005).
Tetraspanin network formation
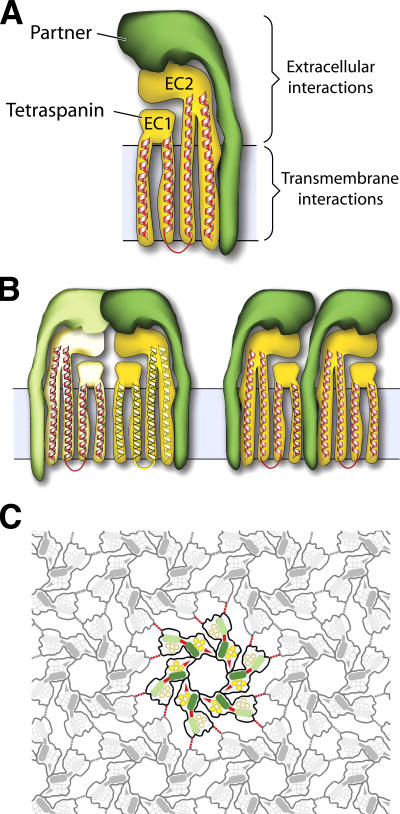
One of the key concepts in tetraspanin functions is the formation of a tetraspanin network (or web) by tetraspanins and their partner proteins (Rubinstein et al., 1996; Maecker et al., 1997; Hemler, 2005). Studies have been performed to delineate the levels and strengths of interactions in these networks (Claas et al., 2001; Charrin et al., 2003; Yang et al., 2004). The existing data indicate that there are three levels of interactions: the primary interaction between a tetraspanin and its partner, the secondary interaction between the primary complexes, and the tertiary interaction between these secondary complexes (Hemler, 2003; Levy and Shoham, 2005; Martin et al., 2005). However, most of the existing studies relied on examinations of the tetraspanin complexes based on their detergent resistance, which are relatively nonspecific and insensitive. Our current structural data and molecular model of the UP tetraspanin complexes have revealed in molecular details several levels of interactions in the UP tetraspanin network (Fig. 5). The first level of interaction is clearly the primary interaction between UP tetraspanins Ia and Ib and their single-spanning transmembrane partners UP II and IIIa, respectively (Fig. 5 A). This interaction is very extensive, involving both TM helices and extracellular domains, and forms the primary complexes—the subdomains in the 16-nm particle (Fig. 1 A and Fig. 5 A). Supporting this concept are the findings that UP Ia and Ib can be readily cross-linked with UP II and IIIa, respectively, to form heterodimers UP Ia/II and Ib/IIIa (Wu et al., 1995) and that the formation of the UP Ia/II and Ib/IIIa heterodimers is a prerequisite for their ER exit (Deng et al., 2002; Tu et al., 2002; Hu et al., 2005). There are two types of secondary interactions between the primary complexes in the UP tetraspanin network (Fig. 5 B). One is between the UP Ia/II and Ib/IIIa primary complexes (i.e., the inner and outer subdomains), via the contact of UP II and IIIa at the joint (Fig. 5, B [left] and C [red bars]), to form a subunit (six of which form a 16-nm particle; Fig. 1 A and Fig. 5 C). The other secondary interaction is between the UP Ia/II complexes (the inner subdomains; Fig. 5, B [right] and C [red arrowheads]), via the contact between UP Ia of one primary complex and UP II of a neighboring primary complex; this secondary interaction is responsible for linking the six inner subdomains to form the inner ring of the 16-nm particle (Fig. 1 A). Strong detergents such as octyl glucoside can break up these secondary complexes but not the primary complexes (Liang et al., 2001), indicating that the secondary interactions are weaker than the primary ones. Although the first type of secondary interaction (Fig. 5 B, left) within a subunit can bridge only two primary complexes, the second type of interaction (Fig. 5 B, right) can potentially bridge more than six primary complexes when involving non-UP tetraspanins. It is conceivable that these two types of secondary interactions also exist in other tetraspanin networks so that multiple primary complexes can link together to form an extensive tetraspanin web. One of the widely reported secondary interactions in tetraspanin network is the formation of homo- or heterodimer of tetraspanins; some of them can be cross-linked using short, or even “zero-length” cross-linking reagents (Kovalenko et al., 2004, 2005). Although we do not find direct contacts between UP tetraspanins in the structure of the 16-nm particle, the distances between UP Ia and Ib within a subunit or between the neighboring subunits are quite small. Moreover, our data do not exclude the possibility of forming homo- or heterodimer of tetraspanins in other tetraspanin networks because the cylindrical shape of the tetraspanin structure can clearly allow direct contacts between tetraspanins in both transmembrane and extracellular domains. In fact, isolated UP Ia and Ib can readily form SDS-resistant multimers (Wu et al., 1990, 1995). The third level of interaction is between the outer subdomains of neighboring 16-nm particles (Fig. 5 C, dotted red lines). This interaction is quite weak and can be visualized only in the electron density map at very low contour levels. This is consistent with the observation that the 16-nm particle as a single unit can be separated from the urothelial plaque with a mild detergent wash (unpublished data). We have therefore observed three levels of interactions in the UP tetraspanin network, and our data provide the structural basis, at the molecular level, of these interactions. Although the details may vary in other tetraspanin networks, our model provides a framework for a better understanding of the formation of tetraspanin networks in general.
Figure 5.
Interactions in the UP complexes and the formation of the tetraspanin networks. (A) Primary interaction. A UP tetraspanin (UP Ia or Ib; yellow) interacts with its partner (UP II or IIIa; green) via both their transmembrane and head domains to form primary pair complexes Ia/II or Ib/IIIa (Fig. 3, A and B; and Fig. 4), which occupy the inner or outer subdomains of the 16-nm particle, respectively. (B) Two types of secondary interactions. (left) Two primary tetraspanin pairs interact mainly via the extracellular head domains of the single-spanning transmembrane partners to form a heterotetramer complex (Fig. 3 C), one of the six subunits of the 16-nm particle. (right) The tetraspanin of one primary pair interacts with the single-spanning transmembrane protein of another primary pair, as occurring in the inner ring of the 16-nm particle (Fig. 3 D). Note that only the complexes formed by the second type of interaction (right) can extend to incorporate more than two primary pairs. (C) Formation of the urothelial tetraspanin network. The formation of the 16-nm particle involves the secondary interactions between two primary heterodimeric complexes (red bars) and between the neighboring UP Ia/II heterodimers of the inner subdomains (red arrowheads). Tertiary interactions between the particles (dotted red lines) further extend the UP tetraspanin network. For clarity, only one particle is colored.
Materials and methods
Preparation and EM examination of mouse urothelial plaques
Mouse urothelial plaques were isolated by sucrose density gradient and differential detergent wash as described previously (Wu et al., 1990; Liang et al., 1999); the quality of the purified urothelial plaques was assessed by negative staining and EM. For cryo-EM, 5 μl of the purified plaques, adjusted to ∼0.1 mg/ml, was applied to a molybdenum grid (300 meshes) with a layer of newly prepared carbon film and transferred to a tannic acid (0.75%) solution. After the excess liquid was blotted with filter paper, the sample was quickly immersed into liquid nitrogen. The frozen sample was loaded onto a Gatan cryo-holder and transferred to an electron microscope (CM200 FEG; Philips) operated at a voltage of 200 kV.
Image recording and processing
Electron micrographs were taken at a magnification of 50,000 in low-dose mode and with 0.5–1.7 μm defocus, at up to 50° tilt angles. The micrographs were screened using an optical diffractometer to select the regions with the best diffraction spots. Because of the heterogeneity of the sample, <10% of the images provided high-resolution information. Selected micrographs were digitized using a scanner (Carl Zeiss MicroImaging, Inc.) at a step size of 14 μm, which corresponds to 2.8 Å in the crystal. A total of 55 images were selected from >1,000 micrographs, and the unbending of the images was performed using MRC software suites (Crowther et al., 1996). The 3D density map was visualized using O (Jones et al., 1991) or AMIRA (Mercury Computer System, Inc.) software packages.
Density segmentation and model building
Segmentation of the density map of the 16-nm particle was performed using AMIRA. For model building of the UP tetraspanins, transmembrane poly-alanine helices were first built into the electron density using O and then the poly-alanine model of EC1 (based on the homology region of casein kinase-1; Protein Data Bank accession no. 2CSN) and EC2 (based on the structure of CD81 large extracellular loop; Protein Data Bank accession no. 1G8Q) were manually docked into the electron density map. The modeled extracellular loops were connected to the TMs following the density map and were locally adjusted using O.
Online supplemental material
Fig. S1 shows calculated diffractions and several lattice lines of 2D crystals of mouse UPs. Fig. S2 is a hypothetical model of the FimH–uroplakin interaction, illustrating that FimH has to reach into the crevice formed by two neighboring joints. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200602086/DC1.
Supplementary Material
Acknowledgments
We thank Matthieu Schapira for his help in the early stage of the tetraspanin model building, Gert Kreibich for useful discussions, and Gordon Cook for illustrations.
This work is supported by National Institutes of Health grants DK52206 (to X.-P. Kong and T.-T. Sun) and DK39753 (to T.-T. Sun).
Abbreviations used in this paper: TM, transmembrane domain; UP, uroplakin.
References
- Balaban, N.Q., U.S. Schwarz, D. Riveline, P. Goichberg, G. Tzur, I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi, and B. Geiger. 2001. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3:466–472. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114:4143–4151. [DOI] [PubMed] [Google Scholar]
- Beuming, T., and H. Weinstein. 2004. A knowledge-based scale for the analysis and prediction of buried and exposed faces of transmembrane domain proteins. Bioinformatics. 20:1822–1835. [DOI] [PubMed] [Google Scholar]
- Bienstock, R.J., and J.C. Barrett. 2001. KAI1, a prostate metastasis suppressor: prediction of solvated structure and interactions with binding partners; integrins, cadherins, and cell-surface receptor proteins. Mol. Carcinog. 32:139–153. [DOI] [PubMed] [Google Scholar]
- Bowie, J.U. 1997. a. Helix packing angle preferences. Nat. Struct. Biol. 4:915–917. [DOI] [PubMed] [Google Scholar]
- Bowie, J.U. 1997. b. Helix packing in membrane proteins. J. Mol. Biol. 272:780–789. [DOI] [PubMed] [Google Scholar]
- Brisson, A., and R.H. Wade. 1983. Three-dimensional structure of luminal plasma membrane protein from urinary bladder. J. Mol. Biol. 166:21–36. [DOI] [PubMed] [Google Scholar]
- Charrin, S., S. Manie, M. Billard, L. Ashman, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304:107–112. [DOI] [PubMed] [Google Scholar]
- Choma, C., H. Gratkowski, J.D. Lear, and W.F. DeGrado. 2000. Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Biol. 7:161–166. [DOI] [PubMed] [Google Scholar]
- Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S.J. Hultgren, and S.D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 285:1061–1066. [DOI] [PubMed] [Google Scholar]
- Claas, C., C.S. Stipp, and M.E. Hemler. 2001. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 276:7974–7984. [DOI] [PubMed] [Google Scholar]
- Cormier, E.G., F. Tsamis, F. Kajumo, R.J. Durso, J.P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA. 101:7270–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther, R.A., R. Henderson, and J.M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9–16. [DOI] [PubMed] [Google Scholar]
- Deng, F.M., F.X. Liang, L. Tu, K.A. Resing, P. Hu, M. Supino, C.C. Hu, G. Zhou, M. Ding, G. Kreibich, and T.T. Sun. 2002. Uroplakin IIIb, a urothelial differentiation marker, dimerizes with uroplakin Ib as an early step of urothelial plaque assembly. J. Cell Biol. 159:685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, D., J.P. Overington, S.V. Ruffle, J.H. Nugent, and T.L. Blundell. 1993. Modeling alpha-helical transmembrane domains: the calculation and use of substitution tables for lipid-facing residues. Protein Sci. 2:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummer, H.E., K.A. Wilson, and P. Poumbourios. 2005. Determinants of CD81 dimerization and interaction with hepatitis C virus glycoprotein E2. Biochem. Biophys. Res. Commun. 328:251–257. [DOI] [PubMed] [Google Scholar]
- Eilers, M., A.B. Patel, W. Liu, and S.O. Smith. 2002. Comparison of helix interactions in membrane and soluble alpha-bundle proteins. Biophys. J. 82:2720–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, B., and P. Brown. 2003. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect. Dis. Clin. North Am. 17:227–241. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 1998. Integrin associated proteins. Curr. Opin. Cell Biol. 10:578–585. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 2001. Specific tetraspanin functions. J. Cell Biol. 155:1103–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler, M.E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397–422. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6:801–811. [DOI] [PubMed] [Google Scholar]
- Hicks, R.M. 1975. The mammalian urinary bladder: an accommodating organ. Biol. Rev. Camb. Philos. Soc. 50:215–246. [DOI] [PubMed] [Google Scholar]
- Hicks, R.M., and B. Ketterer. 1969. Hexagonal lattice of subunits in the thick luminal membrane of the rat urinary bladder. Nature. 224:1304–1305. [DOI] [PubMed] [Google Scholar]
- Hu, C.C., F.X. Liang, G. Zhou, L. Tu, C.H. Tang, J. Zhou, G. Kreibich, and T.T. Sun. 2005. Assembly of urothelial plaques: tetraspanin function in membrane protein trafficking. Mol. Biol. Cell. 16:3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javadpour, M.M., M. Eilers, M. Groesbeek, and S.O. Smith. 1999. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys. J. 77:1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., J.Y. Zou, S.W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 47:110–119. [DOI] [PubMed] [Google Scholar]
- Kachar, B., F. Liang, U. Lins, M. Ding, X.R. Wu, D. Stoffler, U. Aebi, and T.T. Sun. 1999. Three-dimensional analysis of the 16 nm urothelial plaque particle: luminal surface exposure, preferential head-to-head interaction, and hinge formation. J. Mol. Biol. 285:595–608. [DOI] [PubMed] [Google Scholar]
- Kitadokoro, K., D. Bordo, G. Galli, R. Petracca, F. Falugi, S. Abrignani, G. Grandi, and M. Bolognesi. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitadokoro, K., M. Ponassi, G. Galli, R. Petracca, F. Falugi, G. Grandi, and M. Bolognesi. 2002. Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop. Biol. Chem. 383:1447–1452. [DOI] [PubMed] [Google Scholar]
- Kovalenko, O.V., X. Yang, T.V. Kolesnikova, and M.E. Hemler. 2004. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 377:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko, O.V., D.G. Metcalf, W.F. Degrado, and M.E. Hemler. 2005. Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct. Biol. 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour, F., S. Charrin, V. Labas, J.P. Le Caer, C. Boucheix, and E. Rubinstein. 2004. Tetraspanins connect several types of Ig proteins: IgM is a novel component of the tetraspanin web on B-lymphoid cells. Cancer Immunol. Immunother. 53:148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5:136–148. [DOI] [PubMed] [Google Scholar]
- Liang, F., B. Kachar, M. Ding, Z. Zhai, X.R. Wu, and T.T. Sun. 1999. Urothelial hinge as a highly specialized membrane: detergent-insolubility, urohingin association, and in vitro formation. Differentiation. 65:59–69. [DOI] [PubMed] [Google Scholar]
- Liang, F.X., I. Riedel, F.M. Deng, G. Zhou, C. Xu, X.R. Wu, X.P. Kong, R. Moll, and T.T. Sun. 2001. Organization of uroplakin subunits: transmembrane topology, pair formation and plaque composition. Biochem. J. 355:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, M.N., S.P. Smith, S.J. Metallo, I.S. Choi, M. Prentiss, and G.M. Whitesides. 2000. Measuring the forces involved in polyvalent adhesion of uropathogenic Escherichia coli to mannose-presenting surfaces. Proc. Natl. Acad. Sci. USA. 97:13092–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker, H.T., S.C. Todd, and S. Levy. 1997. The tetraspanin superfamily: molecular facilitators. FASEB J. 11:428–442. [PubMed] [Google Scholar]
- Martin, F., D.M. Roth, D.A. Jans, C.W. Pouton, L.J. Partridge, P.N. Monk, and G.W. Moseley. 2005. Tetraspanins in viral infections: a fundamental role in viral biology? J. Virol. 79:10839–10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciopinto, F., S. Campagnoli, S. Abrignani, Y. Uematsu, and P. Pileri. 2001. The small extracellular loop of CD81 is necessary for optimal surface expression of the large loop, a putative HCV receptor. Virus Res. 80:1–10. [DOI] [PubMed] [Google Scholar]
- Min, G., M. Stolz, G. Zhou, F. Liang, P. Sebbel, D. Stoffler, R. Glockshuber, T.T. Sun, U. Aebi, and X.P. Kong. 2002. Localization of uroplakin Ia, the urothelial receptor for bacterial adhesin FimH, on the six inner domains of the 16 nm urothelial plaque particle. J. Mol. Biol. 317:697–706. [DOI] [PubMed] [Google Scholar]
- Min, G., G. Zhou, M. Schapira, T.T. Sun, and X.P. Kong. 2003. Structural basis of urothelial permeability barrier function as revealed by Cryo-EM studies of the 16 nm uroplakin particle. J. Cell Sci. 116:4087–4094. [DOI] [PubMed] [Google Scholar]
- Mulvey, M.A., Y.S. Lopez-Boado, C.L. Wilson, R. Roth, W.C. Parks, J. Heuser, and S.J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 282:1494–1497. [DOI] [PubMed] [Google Scholar]
- Negrete, H.O., J.P. Lavelle, J. Berg, S.A. Lewis, and M.L. Zeidel. 1996. Permeability properties of the intact mammalian bladder epithelium. Am. J. Physiol. 271:F886–F894. [DOI] [PubMed] [Google Scholar]
- Nishiuchi, R., N. Sanzen, S. Nada, Y. Sumida, Y. Wada, M. Okada, J. Takagi, H. Hasegawa, and K. Sekiguchi. 2005. Potentiation of the ligand-binding activity of integrin α3β1 via association with tetraspanin CD151. Proc. Natl. Acad. Sci. USA. 102:1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostergetel, G.T., W. Keegstra, and A. Brisson. 2001. Structure of the major membrane protein complex from urinary bladder epithelial cells by cryo-electron crystallography. J. Mol. Biol. 314:245–252. [DOI] [PubMed] [Google Scholar]
- Qi, J.C., J. Wang, S. Mandadi, K. Tanaka, B.D. Roufogalis, M.C. Madigan, K. Lai, F. Yan, B.H. Chong, R.L. Stevens, and S.A. Krilis. 2006. Human and mouse mast cells use the tetraspanin CD9 as an alternate interleukin-16 receptor. Blood. 107:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, E., F. Le Naour, C. Lagaudriere-Gesbert, M. Billard, H. Conjeaud, and C. Boucheix. 1996. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur. J. Immunol. 26:2657–2665. [DOI] [PubMed] [Google Scholar]
- Seigneuret, M. 2006. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 90:212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneuret, M., A. Delaguillaumie, C. Lagaudriere-Gesbert, and H. Conjeaud. 2001. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 276:40055–40064. [DOI] [PubMed] [Google Scholar]
- Senes, A., I. Ubarretxena-Belandia, and D.M. Engelman. 2001. The Calpha—H⋅⋅⋅O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. USA. 98:9056–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin, L.A., F.J. Chlapowski, and M.A. Bonneville. 1972. Lumenal plasma membrane of the urinary bladder. I. Three-dimensional reconstruction from freeze-etch images. J. Cell Biol. 53:73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, T.J., and I.T. Arkin. 2001. Substitution rates in alpha-helical transmembrane proteins. Protein Sci. 10:2507–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp, C.S., T.V. Kolesnikova, and M.E. Hemler. 2003. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28:106–112. [DOI] [PubMed] [Google Scholar]
- Tarrant, J.M., L. Robb, A.B. van Spriel, and M.D. Wright. 2003. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 24:610–617. [DOI] [PubMed] [Google Scholar]
- Taylor, K.A., and J.D. Robertson. 1984. Analysis of the three-dimensional structure of the urinary bladder epithelial cell membranes. J. Ultrastruct. Res. 87:23–30. [DOI] [PubMed] [Google Scholar]
- Tu, L., T.T. Sun, and G. Kreibich. 2002. Specific heterodimer formation is a prerequisite for uroplakins to exit from the endoplasmic reticulum. Mol. Biol. Cell. 13:4221–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara, J., W. Longley, and J.D. Robertson. 1969. A hexagonal arrangement of subunits in membrane of mouse urinary bladder. J. Mol. Biol. 46:593–596. [DOI] [PubMed] [Google Scholar]
- Walz, T., M. Haner, X.R. Wu, C. Henn, A. Engel, T.T. Sun, and U. Aebi. 1995. Towards the molecular architecture of the asymmetric unit membrane of the mammalian urinary bladder epithelium: a closed “twisted ribbon” structure. J. Mol. Biol. 248:887–900. [DOI] [PubMed] [Google Scholar]
- Wu, X.R., M. Manabe, J. Yu, and T.T. Sun. 1990. Large scale purification and immunolocalization of bovine uroplakins I, II, and III. Molecular markers of urothelial differentiation. J. Biol. Chem. 265:19170–19179. [PubMed] [Google Scholar]
- Wu, X.R., J.J. Medina, and T.T. Sun. 1995. Selective interactions of UPIa and UPIb, two members of the transmembrane 4 superfamily, with distinct single transmembrane-domained proteins in differentiated urothelial cells. J. Biol. Chem. 270:29752–29759. [DOI] [PubMed] [Google Scholar]
- Wu, X.R., T.T. Sun, and J.J. Medina. 1996. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl. Acad. Sci. USA. 93:9630–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., O.V. Kovalenko, W. Tang, C. Claas, C.S. Stipp, and M.E. Hemler. 2004. Palmitoylation supports assembly and function of integrin- tetraspanin complexes. J. Cell Biol. 167:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch, R.L., F. Berditchevski, M.B. Harler, J. Reichner, and M.E. Hemler. 1998. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell. 9:2751–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., J.H. Lin, X.R. Wu, and T.T. Sun. 1994. Uroplakins Ia and Ib, two major differentiation products of bladder epithelium, belong to a family of four transmembrane domain (4TM) proteins. J. Cell Biol. 125:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F.X., M.J. Cocco, W.P. Russ, A.T. Brunger, and D.M. Engelman. 2000. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 7:154–160. [DOI] [PubMed] [Google Scholar]
- Zhou, F.X., H.J. Merianos, A.T. Brunger, and D.M. Engelman. 2001. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. USA. 98:2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G., W.J. Mo, P. Sebbel, G. Min, T.A. Neubert, R. Glockshuber, X.R. Wu, T.T. Sun, and X.P. Kong. 2001. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J. Cell Sci. 114:4095–4103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.