Abstract
Many copies of mammalian mitochondrial DNA contain a short triple-stranded region, or displacement loop (D-loop), in the major noncoding region. In the 35 years since their discovery, no function has been assigned to mitochondrial D-loops. We purified mitochondrial nucleoprotein complexes from rat liver and identified a previously uncharacterized protein, ATAD3p. Localization studies suggested that human ATAD3 is a component of many, but not all, mitochondrial nucleoids. Gene silencing of ATAD3 by RNA interference altered the structure of mitochondrial nucleoids and led to the dissociation of mitochondrial DNA fragments held together by protein, specifically, ones containing the D-loop region. In vitro, a recombinant fragment of ATAD3p bound to supercoiled DNA molecules that contained a synthetic D-loop, with a marked preference over partially relaxed molecules with a D-loop or supercoiled DNA circles. These results suggest that mitochondrial D-loops serve to recruit ATAD3p for the purpose of forming or segregating mitochondrial nucleoids.
Introduction
Mitochondrial DNA (mtDNA) forms nucleoprotein complexes (Chen and Butow, 2005). In yeast, several candidate mitochondrial nucleoid proteins have been identified by in organello formaldehyde cross-linking experiments (Kaufman et al., 2000). Several of them associate closely with mtDNA and contribute to its stability (MacAlpine et al., 2000; Kaufman et al., 2003; Chen et al., 2005). Less is known about mammalian mitochondrial nucleoids; they contain Tfam, which is believed to be the major mtDNA packaging protein (Alam et al., 2003), and Twinkle, an mtDNA helicase (Spelbrink et al., 2001), mitochondrial single-strand binding protein, and DNA polymerase γ (Garrido et al., 2003). Additional proteins copurify with frog oocyte mtDNA (Bogenhagen et al., 2003), although their roles in mtDNA maintenance are uncertain.
In mammals, many molecules of mtDNA contain a short triple-stranded region, or displacement loop (D-loop; Arnberg et al., 1971; Kasamatsu et al., 1971), located in the major noncoding region (NCR). The third strand of the D-loop, 7S DNA, is ∼0.65 kb long in humans, spanning from approximately nt 16,111 to nt 191 (Andrews et al., 1999). D-loops are synthesized via transcription initiating at the light strand promoter and transition to DNA synthesis at the origin of heavy strand replication (Clayton, 1982). They have been proposed to represent stalled or aborted replication intermediates (Clayton, 1982). Hitherto, there has been no evidence that mitochondrial D-loops are functional entities.
Tfam/Abf2 are members of the HMG family of DNA binding proteins, which bend DNA. Subunit α of bacterial HU is a histone-like protein, which is capable of binding to a variety of nucleic acid substrates (Balandina et al., 2002) and of complementing Abf2-deficient yeast (Megraw and Chae, 1993). Because HU is simpler than its eukaryotic counterparts and more readily expressed in Escherichia coli, we used it as a bait to affinity purify mammalian mtDNA with its associated proteins; this strategy led to the isolation of the protein TOB3 (ATAD3p), found but not characterized previously in a proteomic screen of rat liver mitochondria (Mootha et al., 2003) and, more recently, in enriched mitochondrial nucleoprotein preparations (Wang and Bogenhagen, 2006). Here, we show that ATAD3p is a DNA binding protein, which is present in mitochondrial nucleoids; siRNA of ATAD3 decreased the number of mtDNA multimers, and in vitro part of the protein binds preferentially to D-loop–containing molecules, suggesting that mitochondrial D-loops play a role in mtDNA organization.
Results and discussion
Preparations of HU affinity-purified rat liver mtDNA contained six identifiable proteins (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200609158/DC1): only Tfam was a known mitochondrial nucleoid protein. One of the six, ATAD3p, has an AAA+ domain (Finn et al., 2006), located toward its C terminus, and a conserved hydrophobic region of 20 amino acids (residues 246–265 of human ATAD3B); however, it lacks a canonical N-terminal mitochondrial targeting signal. The sequence of ATAD3 is 85% identical between rat and humans, and homologues are spread throughout animals, plants, and protists (Fig. S1). The hydrophobic region is the appropriate length to span a lipid bilayer, but DAS-TMFilter, Phobius, and Minnou programs do not predict it will form a transmembrane helix; thus, in silico analysis suggested that ATAD3p was not an integral membrane protein. Nevertheless, the protein is tightly associated with mitochondrial membranes based on alkaline carbonate treatment of mitochondria (Fig. S2 A).
To determine the cellular location and properties of ATAD3, recombinant human protein was produced. Human ATAD3B consists of 648 amino acids with a calculated molecular mass of 72 kD. The protein expressed poorly in E. coli (unpublished data), but two fragments, ATAD3-f1 (residues 44–247) and ATAD3-f2 (residues 264–617), were expressed readily as soluble GST fusion proteins (unpublished data). Antibodies raised against ATAD3-f1 recognized two proteins in human 143B osteosarcoma cells but only one protein (ATAD3A) in A549 adenocarcinoma cells (Fig. S2 B). Immunocytochemistry with antibody to ATAD3-f1 revealed a punctate staining pattern within mitochondria (Fig. S2 C), which frequently coincided with mtDNA (Fig. 1 A). Notwithstanding, many mitochondrial nucleoids appeared to lack ATAD3p (Fig. 1 A). Thus, the amount of ATAD3p associating with mtDNA appears to vary from nucleoid to nucleoid.
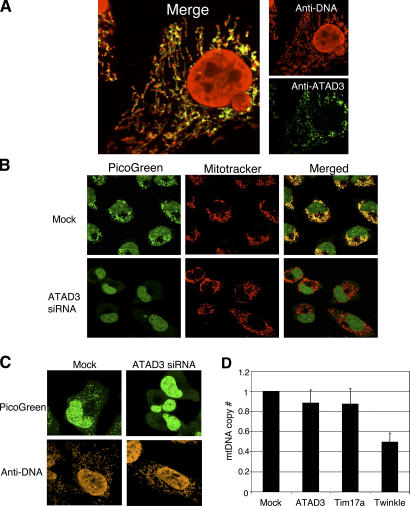
Figure 1.
ATAD3 colocalizes with mtDNA, and gene silencing alters PicoGreen staining of mitochondrial nucleoids. (A) 143B osteosarcoma cells were incubated with primary antibodies to ATAD3 and DNA and secondary antibodies emitting green and red light, respectively, on confocal microscopy. (B) siRNA targeted to ATAD3 led to a marked decrease in PicoGreen staining of mtDNA (green), whereas mitochondrial morphology was not visibly affected based on MitoTracker staining (red). Human 143B osteosarcoma cells were twice mock transfected (mock) or transfected with dsRNA-452 targeting ATAD3 (ATAD3 siRNA) at 72-h intervals. Cells were stained with MitoTracker and PicoGreen 144 h after the first transfection and examined by confocal microscopy. MitoTracker orange signal was pseudocolored red to improve contrast. (C) Comparison of PicoGreen-stained mtDNA in living cells (top) with fixed cells labeled with anti-DNA antibody (bottom). Images were acquired with a Nikon 60×/1.40 oil-immersion objective, set at zoom 2 (B) and zoom 4 (A and C) at 22–24°C in Immersol (Carl Zeiss MicroImaging, Inc.). (D) qPCR estimation of mtDNA copy number in control, ATAD3, Twinkle, and Tim17A siRNA–treated cells.
Two rounds of transfection of 143B osteosarcoma cells with double-stranded RNA (dsRNA)–452 targeted to ATAD3 decreased PicoGreen staining of mitochondrial nucleoids markedly (Fig. 1 B). However, the copy number of mtDNA after ATAD3 siRNA was ∼88% of control values (Fig. 1 D), suggesting that PicoGreen staining does not provide a direct measure of mtDNA mass, an inference confirmed by immunofluorescent detection of DNA (Fig. 1 C). Therefore, PicoGreen staining of DNA must depend on the DNA's topological state. Relaxation of supercoiled plasmid DNA in vitro was accompanied by a substantial increase in PicoGreen signal (unpublished data), substantiating this view. Therefore, we conclude that ATAD3p depletion leads to an increase in negative supercoiling and that the more condensed form of mtDNA largely excludes PicoGreen. Mitochondria had an essentially normal morphology in ATAD3 siRNA–treated 143B cells (Fig. 1 B and not depicted), implying that ATAD3p has no role in mitochondrial fission or fusion; hence, the observed change in mtDNA associated with gene silencing is not an indirect consequence of mitochondrial disorganization.
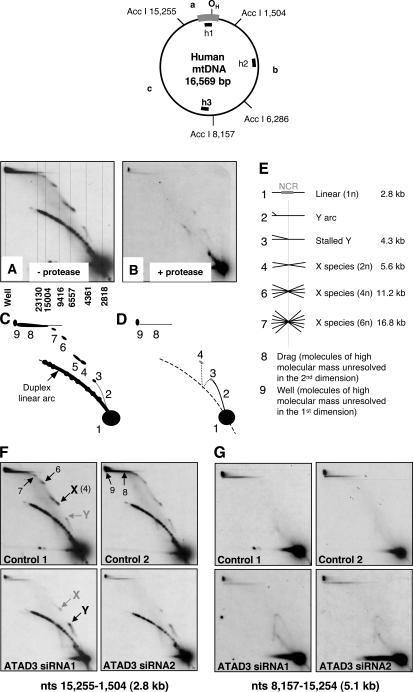
2D agarose gel electrophoresis (AGE; Brewer and Fangman, 1987) has been used to characterize mitochondrial replication intermediates (Yang et al., 2002; Yasukawa et al., 2005). Here, the bulk of the protein was removed by treatment with detergent and phenol without the usual proteinase K digestion. Under these conditions, an AccI restriction fragment (nt 15,255–1,504) of human mtDNA did not enter the gel readily compared with protease-treated samples, and there were numerous prominent spots on the linear duplex arc, indicating that protein remained associated with mtDNA (Fig. 2, A and B). In addition, residual protein gave rise to a series of spots resolving well above the linear duplex arc. The position of the first of these (Fig. 2 A, species 4) was coincident with the apex of an X arc (Friedman and Brewer, 1995; Yasukawa et al., 2005); that is, its mobility implied it comprised two fragments of DNA joined near the center (Fig. 2 A [interpreted in Fig. 2, C–E]). The mass of the other spots (species 6, 7, and 8) is consistent with higher order multimers, and the increasing distance from the linear duplex arc implies that these are also joined near their center (as interpreted in Fig. 2 C). If ATAD3p is a bona fide component of mitochondrial nucleoids, its depletion via RNAi might alter the mobility of protein-bound mtDNA fragments. Therefore, total cellular DNA was extracted, without a protease step, from control cells and ATAD3 siRNA–transfected cells, and restriction digested, and the fragments were separated by 2D-AGE. After Southern blotting, nylon membranes were hybridized sequentially to probes for three regions of mtDNA covering most of the human mitochondrial genome: nt 15,255–1,504 (a); nt 1,505–6,286 (b); and nt 8,157–15,254 (c). In ATAD3 siRNA samples, there was a significant decrease (P = 0.00005) in signal from mtDNA trapped near the well, and at the boundary of the 1D and 2D gels, specifically for the NCR-containing fragment (a; Fig. 2 F versus Fig. 2 G and Fig. S2 D). The iterated spots corresponding to the apex of a simple X arc, and higher multimers thereof, were also significantly decreased (P < 0.0065), whereas the protein-bound fragments of mtDNA resolving on or close to the linear duplex arc were substantially the same as controls (Fig. 2 F), and no significant alteration was observed in other regions of mtDNA in controls and ATAD3 siRNA–treated samples (Fig. 2 G and not depicted). Therefore, ATAD3p binds preferentially within the region defined by the AccI fragment spanning nt 15,255–1,504 of human mtDNA, and ATAD3 is implicated in the maintenance or formation of mtDNA multimers. Although the 2D-AGE data (Fig. 2) suggested that ATAD3p binds to multiple molecules of mtDNA, other factors presumably contribute to mitochondrial nucleoid stability in vivo, as there was no apparent nucleoid fragmentation in ATAD3 siRNA–treated cells stained with anti-DNA antibody (Fig. 1 C).
Figure 2.
Multiple NCR-containing fragments of mtDNA are held together by detergent-resistant protein in control cells but not in ATAD3 siRNA–treated cells. DNA was extracted from 143B cells with (B) or without the use of proteinase K (A, F, and G) and digested with AccI. The restriction fragments were separated by 2D-AGE and hybridized with probe h1, detecting fragment a, nt 15,255–1,504 (A, B, and F), or probe h2 to fragment c, nt 8,157–15,254 (G). AccI restriction sites and probes are shown on a schematic map of human mtDNA at the head of the figure. Sketches of A and B appear in C and D, respectively, and are interpreted in E, except for molecular species 5, which probably represents forms of species 4 with greater amounts of associated protein. (F and G) Controls 1 and 2 are mock-transfected 143B cells, and ATAD3 siRNA1 and -2 are 143B cells subjected to two rounds of transfection with dsRNA-452. There were many spots aligned along the linear duplex arc in the case of fragment a (nt 15,255–1,504; F), but not fragment c (nt 8,157–15,254; G) or fragment b (1,505–6,286; not depicted), irrespective of ATAD3 RNAi. These spots comprise fragment a and proteins of unknown identity. ATAD3 siRNA led to a marked decrease in X-like species resolving well above the linear duplex arc (species 4, 6, and 7), suggesting that ATAD3p is specifically associated with X-like forms of fragment a.
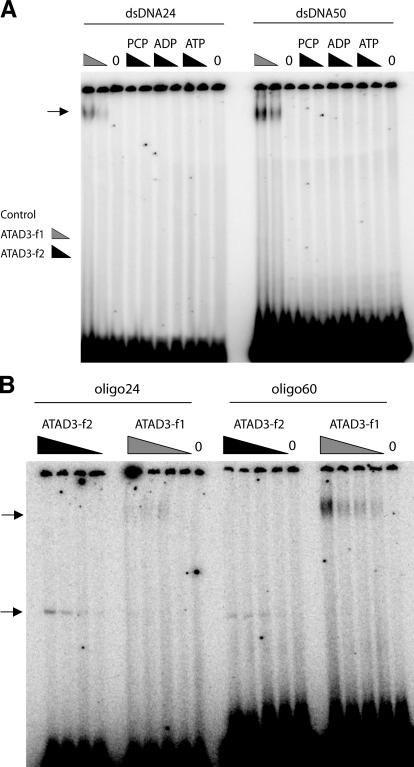
Based on the analysis of AccI fragments of human mtDNA described above, ATAD3p might bind at any number of sites from nt 15,255 to nt 1,504. However, the fact that the depleted species were X-like structures led us to focus on the central portion of the fragment, the NCR. The NCR encompasses the mitochondrial D-loop, a triple-stranded region of ∼600 nucleotides. Therefore, the human NCR was cloned in Bluescript and synthetic D-loops were produced to investigate whether the two recombinant fragments of ATAD3 had specific DNA binding properties. First, however, the f1 and f2 fragments of human ATAD3 were incubated with random pieces of single-stranded and duplex DNA and analyzed by electrophoretic mobility shift assay (EMSA) to determine their nonspecific DNA binding properties. Only ATAD3-f1 was capable of binding to duplex DNA, whereas both fragments bound to single-stranded oligodeoxynucleotides (Fig. 3 and Fig. S2 E). In all cases, the vast majority of linear DNA remained unbound; therefore, neither duplex nor single-stranded DNA is a good substrate for ATAD3. The first synthetic D-loop (C) tested used an oligonucleotide (oligo C) spanning 120 nucleotides, from nt 16,081 to nt 16,200, near the 3′ end of the native D-loop. D-loop C was incubated with RecA protein, or ATAD3-f1 and -f2, and subjected to 1D- AGE. ATAD3-f1 and RecA bound to D-loop C to a much greater extent than ATAD3-f2 (Fig. 4 A); the pronounced mobility shift effected by hATAD3-f1 compared with RecA suggests either that the former accrues multiple DNA molecules or that each D-loop is bound by a large number of ATAD3p molecules.
Figure 3.
ATAD3-f1 binds both single-stranded and duplex DNA weakly, whereas ATAD3-f2 binds only single-stranded DNA. (A) ATAD3-f2 contains a putative ATPase domain, so DNA binding reactions involving this protein fragment were performed in the presence of 3 mM ATP, 3 mM ADP, or PCP (2,3-Methyleneadenosine 5′-triphosphate disodium salt), a nonhydrolysable analogue of ATP (2 mM). Radiolabeled double-stranded oligonucleotides of 24 (dsDNA24) or 50 (dsDNA50) base pairs were incubated with 0, 264, or 528 ng of ATAD3-f2 protein. In parallel, dsDNA24 or -50 were incubated with 0, 145, and 290 ng of ATAD3-f1 for 20 min at 37°C with (not depicted) or without 3 mM ATP. The reaction mixtures were fractioned by nondenaturing PAGE (3.5% gel) and exposed to phosphorimager screens. A small percentage of dsDNA24 and -50 was retarded after incubation with ATAD3-f1, forming a band close to the well (arrows). (B) Oligo24 and -60 were incubated separately with increasing concentrations of ATAD3-f1 (72.5, 145, 290, and 580 ng) or ATAD3-f2 (68, 132, 264, and 528 ng), and the products were resolved on a 3.5% TBE polyacrylamide gel.
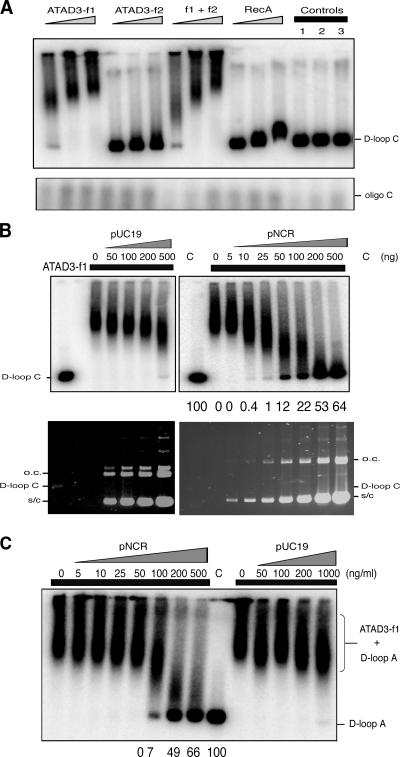
Figure 4.
ATAD3-f1 binds D-loop structures. (A) A fixed amount of D-loop C, comprising pNCR and oligo C (120 mer corresponding to nucleotides 16,081–16,200 of the human mtDNA H-strand), was incubated with increasing concentrations of ATAD3-f1 (145, 290, and 580 ng), ATAD3-f2 (132, 264, and 528 ng), ATAD3-f1 and ATAD3-f2 (145 + 132, 290 + 264, and 580 + 528 ng), or RecA (500, 1,000, and 2,000 ng). DNA–protein mixtures were fractioned on a 1% TAE agarose gel. Controls 1–3 show synthetic D-loop C without protein (1) and with 5 μg/ml ethidium bromide (2) or heated to 42°C (3). None of these treatments, nor incubation with protein, had an appreciable effect on the amount of free oligo C at the bottom of the gel (see inset). (B) ATAD3-F1 preferentially binds to synthetic D-loop C. A fixed amount of D-loop C was mixed with increasing amounts of competitor pUC19 (0, 50, 100, 200, and 500 ng) or plasmid containing the NCR of human mtDNA, pNCR (0, 5, 10, 25, 50, 100, 200, and 500 ng), and incubated with 290 ng of ATAD3-f1. Protein–DNA complexes were separated in a 1% TAE agarose gel. In the case of pNCR, the series of numbers at the base of the phosphorimage refers to the amount of D-loop C (B) or D-loop A (C) not bound by ATAD3-f1, taking the control (C), which was incubated without protein, as 100%. Thus, there was no appreciable (0) unbound D-loop C when the incubation reaction contained a 30-fold excess of pNCR (10 ng), whereas 64% of D-loops failed to associate with ATAD3-f1 in the presence of 500 ng of pNCR (>1,000-fold excess). A second synthetic D-loop (A), formed by the action of RecA on pNCR and oligoA (nucleotides 72–191), behaved similarly (C).
In competition experiments, all molecules of D-loop C associated with ATAD3-f1 when mixed with a >1,000-fold excess of pUC19, plasmid DNA (Fig. 4 B). Plasmid including the NCR sequence (pNCR) was a more effective competitor, yet there remained a preference for D-loop structures over supercoiled plasmid without a D-loop, as ∼80% of D-loops bound to protein in the presence of a 100-fold excess of pNCR (Fig. 4 B). The increased competition from pNCR, relative to pUC19, suggested that there might be an element of sequence- specific binding to the action of ATAD3-f1. However, a second synthetic D-loop (A) corresponding to the other end of the D-loop (nt 72–191) displayed the same competitive advantage in binding ATAD3-f1 as D-loop C (Fig. 4 C). The sequences of oligonucleotides A and C used to generate the two D-loops are not alike (Andrews et al., 1999); therefore, ATAD3-f1 does not display sequence-specific binding.
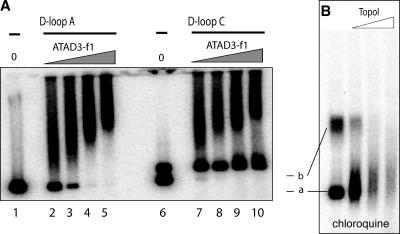
D-loop C occasionally formed two bands in the absence of protein, particularly when lower amounts of RecA were used to generate the synthetic D-loop (Fig. 5 A, lane 6); both species had greater mobility than open circular DNA (Fig. 2 B). Differences in supercoiling are more readily apparent when DNA molecules are separated on chloroquine gels (Richardson et al., 1984); therefore, the same products were separated on a chloroquine gel. As expected, this amplified the difference in mobility between forms a and b, consistent with band a being a less tightly supercoiled version of synthetic D-loop C than band b (Fig. 5 B). ATAD3p showed a marked preference for the most highly supercoiled D-loop–containing molecules (Fig. 5 A, lanes 7 and 8); this feature indicates that D-loop context is crucial to ATAD3p recruitment or retention, which may be necessary to avoid the protein interfering with the processes of replication and transcription. The highest concentrations of protein produced only a modest shift in some of the more relaxed molecules (Fig. 5 A, lanes 9 and 10). Therefore, tightly supercoiled DNA with a D-loop is the strongly preferred substrate for ATAD3p, and such DNA molecules permit oligomerization of either the protein or DNA, or both.
Figure 5.
ATAD3 binds to highly supercoiled DNA with a D-loop in preference to relaxed molecules with a D-loop. (A) 0, 72.5, 145, 290, or 580 ng of ATAD3-f1 was incubated with a fixed amount of D-loop A or C. The DNA–protein complexes were separated from unbound DNA molecules in a 1% TAE agarose gel. D-loop A formed one discrete band without protein (lane 0), and almost all D-loop A was bound by 290 ng protein. In contrast, D-loop C alone yielded two bands, ATAD3-f1 displayed a marked preference for the lower band (a): 72.5 ng protein bound >95% of band a, whereas 580 ng produced only a modest shift in a fraction band b. The difference in mobility between band a and b was accentuated by separating the two species on chloroquine gels (B). Treatment of D-loop C with E. coli topoisomerase I (1, 2, or 3 units) at 37°C for 30 min resulted in smearing and an overall reduction in signal, suggesting that relaxation of the plasmid destabilizes the D-loop. Topoisomers were separated in a 1% TAE agarose gel containing 5 μg/ml chloroquine (B).
In summary, ATAD3p frequently colocalizes with mtDNA, and supercoiled DNA with a D-loop is the preferred substrate of a recombinant fragment of ATAD3p. The pronounced preference of ATAD3p for triple-stranded DNA predicts it will associate with a specific class of mtDNA molecules, those containing a D-loop. Hence, the key prediction of this report is that D-loop (7S DNA) synthesis occurs to recruit ATAD3p to mtDNA via the f1 portion of the protein.
Much remains to be elucidated about the function of ATAD3. Several AAA family members are involved in DNA transactions, including the bacterial nucleoid protein FtsK, clamp loaders, Cdc6, and components of the origin recognition complex. The AAA domain of ATAD3 might well confer on the protein the ability to translocate DNA as elsewhere (Singleton and Wigley, 2003). The ATPase of ATAD3 is functional (Fig. S2 F), and ATP but not ADP enables ATAD3-f2 to bind single-stranded DNA, although ATP hydrolysis is not required for binding, as the protein also binds to single-stranded DNA in the presence of PCP (Fig. S2 E). Presumably, the binding of ATP to ATAD3-f2 induces a conformational change that enables the protein to bind to single-stranded DNA.
ATAD3p is tightly associated with mitochondrial membranes (Fig. S2 A) and, therefore, is likely to contribute to the association of mtDNA with membranes; yet, the in silico prediction is that ATAD3p lacks transmembrane helices, so other proteins are probably required to tether mtDNA to the inner membrane. The amount of ATAD3p associated with mitochondrial nucleoids is highly variable, and some nucleoids appear to lack the protein entirely (Fig. 1 A), which suggests that ATAD3p associates only transiently with mtDNA or with a distinct subpopulation of mtDNAs. The limited effect of ATAD3 gene silencing on mtDNA copy number implies it is not required for replication. Therefore, we favor a role for ATAD3p in nucleoid formation or segregation.
Materials and methods
Isolation of mitochondrial nucleoprotein complexes
Rat liver mitochondria prepared as described previously (Yang et al., 2002) were treated with 0.2 mg/ml RNase A and DNase I for 2 h at 4°C, washed, and sedimented by centrifugation at 8,000 g max for 10 min. 40 mg of mitochondria were disrupted by suspension in 4 ml lysis buffer (10 mM Hepes-NaOH, pH 7.6, 0.2 mM PMSF, 1 mM EDTA, 1 mM DTT, and 0.8% n-Dodecyl-β-d-maltopyranoside) and centrifuged at 1,000 g max for 10 min, and the supernatant was incubated with 4 ml recombinant-HU–coated beads in lysis buffer supplemented with 200 mM NaCl. After washing the beads with lysis buffer containing 100 mM NaCl and elution with 80 mM glutathione, the eluate was separated on a 20–45% iodixanol gradient, the fractions enriched in mtDNA were pooled, and the affiliated proteins were analyzed by mass spectrometry.
Constructs
Fragments of ATAD3(B) corresponding to amino acids 44–247 (ATAD3-f1) and 264–617 (ATAD3-f2) were amplified from a full-length cDNA (IMAGE Clone ID 3138578; Mammalian Genome Collection) and fused with a GST gene. Expressed protein was purified by sequential chromotography (glutathione Sepharose, HiTrap SP FF/HiTrap Q FF, and Superose 12 gel-filtration [GE Healthcare]). GST-HU protein was expressed similarly, except that induction was at 37°C, and purification was via Ni-Sepharose HP followed by SP Sepharose FF (GE Healthcare).
Cell culture and RNAi
Human cells were grown in DME with 10% fetal bovine serum. For RNAi, 143B human osteosarcoma cells growing on 6-well plates at 25–30% confluency were transfected with 10 nM dsRNA and 3 μl of Lipofectamine 2000 (Invitrogen). Cells were transfected a second time, at 72 h, and examined by confocal microscopy at 144 h or lysed for total RNA or DNA extraction. In preliminary tests, dsRNA-452 (5′-UCAAUGAGGAGAAUUUACGGAAGCAAG-3′; 5′-UAAGUUACUCCUCUUAAAUGCCUUCGU-3′) reduced mRNA levels by 65% after 72 h (based on qPCR analysis). Twinkle and Tim17A siRNA and qPCR were performed as described previously (Tyynismaa et al., 2004).
Confocal microscopy
143B cells were washed and live stained with 3 μl PicoGreen reagent (Invitrogen) and 100 nM of MitoTracker orange (Invitrogen) as described previously (Tyynismaa et al., 2004; Ashley et al., 2005) or fixed and stained with anti-DNA antibody (PROGEN Biotechnik) and 1:5,000 anti–ATAD3-f1. A confocal microscopy system (Radiance 2000; Bio-Rad Laboratories) was used for cell imaging, and images were edited using Photoshop Element (Adobe).
DNA analysis and EMSA
2D-AGE of AccI-digested human DNA was done as described previously (Yasukawa et al., 2005). The mtDNA copy number was estimated by qPCR, as described previously (Tyynismaa et al., 2004). Synthetic D-loops were generated by incubating end-labeled oligo C nt 191–72 or A nt 16,081–16,200 with pNCR as described previously (Shibata et al., 1980). pNCR comprised Bluescript plasmid (Stratagene) and a 1.2-kb fragment of human mtDNA encompassing the NCR (nt 16,024–576) of human mtDNA. Purified proteins were incubated with labeled DNA substrates, the products were separated on 1% AGE or 3.5% native PAGE, and phosphorimages were produced using a Typhoon detector (GE Healthcare). The sequences of the oligonucleotides used in the EMSAs are listed below, except for oligos A and C, which were based on the revised human mtDNA sequence (Andrews et al., 1999). Oligo24 top, 5′-GATCTTGTACACGGCCGACTAGTG-3′; Oligo24 bottom, 5′-CATGTGCCGGCTGATCACCTAG-3′; Oligo50 top, 5′-ATCCGGAATCTCCACGCAAACGGCGCCTCATTCTTCTTCATCTGTATCTTC-3′; and Oligo50 bottom, 5′-GAAGATACAGATGAAGAAGAATGAGGCGCCGTTTGCGTGGAGATTCCGGAT-3′.
Online supplemental material
Table S1 lists the six proteins identified by mass spectrometry analysis that consistently copurified with mtDNA. Fig. S1 shows that ATAD3 is an evolutionary conserved member of the AAA family. Fig. S2 shows that ATAD3p is a mitochondrial membrane–bound ATPase with two single-stranded DNA binding domains, which stabilizes extracted mtDNA multimers. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200609158/DC1.
Supplementary Material
Acknowledgments
We are indebted to numerous members of the Dunn Human Nutrition Unit for their help and encouragement, in particular, Drs. Edmund Kunji, Joe Carroll, and Takehiro Yasukawa.
The study was funded by the UK Medical Research Council, the European Union MitoCombat Integrated Programme (FP6), and a grant from Chang Gung Memorial Hospital, Taiwan (CMRP 1331), to Chih-Chieh (Chris) Mao. Johannes Spelbrink is a member of the FinMIT centre of excellence and is supported by the Academy of Finland, Sigfrid Juselius Foundation, and Tampere University Hospital Medical Research Fund.
J. He and C.-C. Mao contributed equally to this paper.
Abbreviations used in this paper: AGE, agarose gel electrophoresis; D-loop, displacement loop; dsRNA, double-stranded RNA; EMSA, electrophoretic mobility shift assay; mtDNA, mitochondrial DNA; NCR, noncoding region.
References
- Alam, T.I., T. Kanki, T. Muta, K. Ukaji, Y. Abe, H. Nakayama, K. Takio, N. Hamasaki, and D. Kang. 2003. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 31:1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, R.M., I. Kubacka, P.F. Chinnery, R.N. Lightowlers, D.M. Turnbull, and N. Howell. 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23:147. [DOI] [PubMed] [Google Scholar]
- Arnberg, A., E.F. van Bruggen, and P. Borst. 1971. The presence of DNA molecules with a displacement loop in standard mitochondrial DNA preparations. Biochim. Biophys. Acta. 246:353–357. [DOI] [PubMed] [Google Scholar]
- Ashley, N., D. Harris, and J. Poulton. 2005. Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp. Cell Res. 303:432–446. [DOI] [PubMed] [Google Scholar]
- Balandina, A., D. Kamashev, and J. Rouviere-Yaniv. 2002. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 277:27622–27628. [DOI] [PubMed] [Google Scholar]
- Bogenhagen, D.F., Y. Wang, E.L. Shen, and R. Kobayashi. 2003. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell. Proteomics. 2:1205–1216. [DOI] [PubMed] [Google Scholar]
- Brewer, B.J., and W.L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 51:463–471. [DOI] [PubMed] [Google Scholar]
- Chen, X.J., and R.A. Butow. 2005. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6:815–825. [DOI] [PubMed] [Google Scholar]
- Chen, X.J., X. Wang, B.A. Kaufman, and R.A. Butow. 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 307:714–717. [DOI] [PubMed] [Google Scholar]
- Clayton, D.A. 1982. Replication of animal mitochondrial DNA. Cell. 28:693–705. [DOI] [PubMed] [Google Scholar]
- Finn, R.D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, et al. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, K.L., and B.J. Brewer. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262:613–627. [DOI] [PubMed] [Google Scholar]
- Garrido, N., L. Griparic, E. Jokitalo, J. Wartiovaara, A.M. van der Bliek, and J.N. Spelbrink. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 14:1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu, H., D.L. Robberson, and J. Vinograd. 1971. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc. Natl. Acad. Sci. USA. 68:2252–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, B.A., S.M. Newman, R.L. Hallberg, C.A. Slaughter, P.S. Perlman, and R.A. Butow. 2000. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl. Acad. Sci. USA. 97:7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, B.A., J.E. Kolesar, P.S. Perlman, and R.A. Butow. 2003. A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. J. Cell Biol. 163:457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine, D.M., P.S. Perlman, and R.A. Butow. 2000. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 19:767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T.L., and C.B. Chae. 1993. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem. 268:12758–12763. [PubMed] [Google Scholar]
- Mootha, V.K., J. Bunkenborg, J.V. Olsen, M. Hjerrild, J.R. Wisniewski, E. Stahl, M.S. Bolouri, H.N. Ray, S. Sihag, M. Kamal, et al. 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 115:629–640. [DOI] [PubMed] [Google Scholar]
- Richardson, S.M., C.F. Higgins, and D.M. Lilley. 1984. The genetic control of DNA supercoiling in Salmonella typhimurium. EMBO J. 3:1745–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, T., C. DasGupta, R.P. Cunningham, and C.M. Radding. 1980. Homologous pairing in genetic recombination: formation of D loops by combined action of recA protein and a helix-destabilizing protein. Proc. Natl. Acad. Sci. USA. 77:2606–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, M.R., and D.B. Wigley. 2003. Multiple roles for ATP hydrolysis in nucleic acid modifying enzymes. EMBO J. 22:4579–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink, J.N., F.Y. Li, V. Tiranti, K. Nikali, Q.P. Yuan, M. Tariq, S. Wanrooij, N. Garrido, G. Comi, L. Morandi, et al. 2001. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 28:223–231. [DOI] [PubMed] [Google Scholar]
- Tyynismaa, H., H. Sembongi, M. Bokori-Brown, C. Granycome, N. Ashley, J. Poulton, A. Jalanko, J.N. Spelbrink, I.J. Holt, and A. Suomalainen. 2004. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 13:3219–3227. [DOI] [PubMed] [Google Scholar]
- Wang, Y., and D.F. Bogenhagen. 2006. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 281:25791–25802. [DOI] [PubMed] [Google Scholar]
- Yang, M.Y., M. Bowmaker, A. Reyes, L. Vergani, P. Angeli, E. Gringeri, H.T. Jacobs, and I.J. Holt. 2002. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 111:495–505. [DOI] [PubMed] [Google Scholar]
- Yasukawa, T., M.Y. Yang, H.T. Jacobs, and I.J. Holt. 2005. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 18:651–662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.