Abstract
PR-Set7 is a histone methyltransferase that specifically monomethylates histone H4 lysine 20 (K20) and is essential for cell proliferation. Our results show that in PR-Set7 mutants, the DNA damage checkpoint is activated. This phenotype is manifested by reduction in both the mitotic and the S phase indexes, a delay in the progression through early mitosis, and strong reduction of cyclin B. Furthermore, in a double mutant of PR-Set7 and mei-41 (the fly ATR orthologue), the abnormalities of mitotic progression and the cyclin B protein level were rescued. PR-Set7 also showed a defect in chromosome condensation that was enhanced in the double mutant. We therefore propose that monomethylated H4K20 is involved in the maintenance of proper higher order structure of DNA and is consequently essential for chromosome condensation.
Introduction
Dynamic changes in chromatin structure are directly influenced by the posttranslational modifications of the N-terminal histone tails (Luger and Richmond, 1998). Specific amino acids within the tails are modified by phosphorylation, ubiquitination, ADP ribosylation, acetylation, and methylation (Strahl and Allis, 2000; Zhang and Reinberg, 2001). Methylation of different lysine and arginine residues in histone H3 and H4 tails is associated with actively transcribed or repressed chromatin (Fischle et al., 2003).
Histone H4 lysine 20 (K20) can be mono-, di-, or trimethylated. PR-Set7 is a histone methyltransferase that specifically monomethylates histone H4K20 (Fang et al., 2002; Nishioka et al., 2002; Couture et al., 2005; Xiao et al., 2005). Trimethylation of the same lysine is controlled by other histone methyltransferases, Suv4-20h1, and Suv4-20h2 (Schotta et al., 2004). Coincident with the conservation of the H4K20 methyl modifications in higher eukaryotes, both enzymes show substantial homology in species ranging from flies to humans. A null mutation in Drosophila melanogaster PR-Set7 suppresses position effect variegation, indicating that H4K20 methylation plays a role in silencing of gene expression (Karachentsev et al., 2005). Several observations suggest that PR-Set7–dependent methylation of H4K20 also plays an important role in cell proliferation. In HeLa cells, expression of PR-Set7 increases during S phase and peaks at mitosis (Rice et al., 2002). In D. melanogaster larvae, tissues with higher rates of cell divisions, such as imaginal discs, are severely affected by the depletion of PR-Set7. Homozygous PR-Set7 mutant discs are smaller than wild type because they contain only ∼25% as many cells as wild type (Karachentsev et al., 2005).
Here, we investigated the function of PR-Set7–dependent methylation in more detail by studying the cell cycle in mutant neuroblasts. Neuroblasts are diploid, and their cell cycle progression has been well documented (Gatti and Baker, 1989). In the PR-Set720–null allele, used in the experiments described here, the PR-Set7 protein is missing from the first-instar larval stage onward, homozygous or hemizygous (PR-Set720/Df[3R]red31) animals survive until the larval/pupal transition, and the reduction of methylated H4K20 is only observed in late-stage larvae (Karachentsev et al., 2005).
In PR-Set7 mutant third-instar larval brains, monomethylated H4K20 was strongly reduced. We found that in the mutant brains, the mitotic index was reduced, progression through early mitosis was delayed, and cyclin B was reduced. The abnormalities in mitotic progression and in the level of cyclin B were rescued when the DNA damage checkpoint was abolished, indicating that the DNA damage checkpoint is activated in PR-Set7.
Results
Monomethylated H4K20 is strongly reduced in PR-Set7 mutant brains
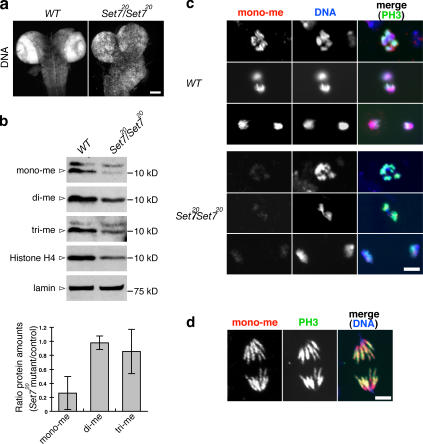
The organization of the third-instar larval brains used in all the experiments here is affected in the mutant (Fig. 1 a). The wild-type brain hemispheres contain two rings with high rates of cell divisions, called the optic lobes. These regions are clearly disorganized in both homozygous PR-Set7 and hemizygous PR-Set7/Df(3R)red31 larval brains (Fig. 1 a and not depicted).
Figure 1.
Monomethylated H4K20 is strongly reduced in PR-Set7 brains. (a) Wild-type (WT) and PR-Set7/PR-Set7 third-instar larval brains were stained with Hoechst. The two strongly staining rings (dense nuclei) observed in wild type are the optic lobes. (b) Western blots of extracts from wild-type and PR-Set7/PR-Set7 third-instar larval brains probed with anti–mono-, anti–di-, and anti–trimethylated H4K20 (mono-, di-, and tri-me), anti–histone H4, and anti-lamin antibodies. The intensity of the bands was quantified by ImageJ, and the value of mono-, di-, or trimethylated H4K20 was normalized to the values of both histone H4 and lamin (see Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1). The graph shows the ratio of PR-Set7/PR-Set7 mutant to wild-type values. Error bars show two SDs (n = 3). (c) Neuroblasts were costained with anti–monomethylated H4K20 (mono-me; red) and anti-PH3 antibodies (green). DNA was stained with Hoechst (blue). (d) Monomethylated H4K20 (red) is distributed all along the chromosomes. Bars: (a) 100 μm; (c and d) 5 μm.
We initiated our studies by determining whether histone H4K20 methylation is reduced in homozygous PR-Set7 brains. Western blots of mutant third-instar larval brain lysates showed that mono-, di-, and trimethylated H4K20 and total histone H4 are reduced compared with wild type, when each value is normalized to the value of lamin (Fig. 1 b and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1). However, when each value is normalized to the values of both histone H4 and lamin, monomethylated H4K20 is reduced to ∼26% of wild-type levels, whereas di- and trimethylation is only down <15% (Fig. 1 b, bottom; and Table S1). This result indicates that monomethylated H4K20 is strongly reduced in the mutant brains. The reason the histone H4 level was lower in the mutant extracts is not clear. Staining of neuroblasts with anti–monomethylated H4K20 antibody (anti-mono) gave similar results (Fig. 1 c).
Monomethylated H4K20 is uniformly distributed along mitotic chromosomes
We next observed the distribution of monomethylated H4K20 during the cell cycle by staining wild-type neuroblasts. Anti-mono staining was detected only on condensed DNA or chromosomes (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1). Monomethylated H4K20 is first detected in cells in late G2-prophase at the very onset of DNA condensation and peaks at metaphase, similar to the detection of phosphorylated histone H3 Ser 10 (PH3), known to be associated with condensed DNA (Fig. 2, c–g; Hendzel et al., 1997). Interestingly, the monomethylation mark thought to be associated with repressed chromatin is not increased at the centromere region but distributed evenly along the chromosomes (Fig. 1 d).
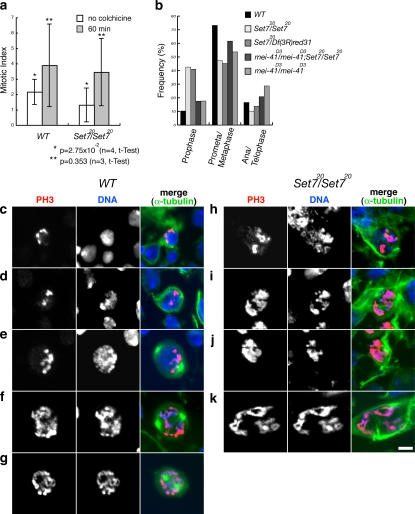
Figure 2.
PR-Set7/PR-Set7 neuroblasts show a delay in early mitotic stages. (a) Mitotic indexes of wild type (WT) and PR-Set7/PR-Set7 before and after incubation with colchicine for 1 h. The mitotic index was determined as the number of PH3-positive cells over the total number of cells (see Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1). (b) Quantification of mitotic parameters in wild-type (n = 272), PR-Set720/PR-Set720 (n = 275), PR-Set720/Df(3R)red31 (n = 363), mei-41D3/mei-41D3;PR-Set720/PR-Set720 (n = 365), and mei-41D3/mei-41D3 (n = 404) neuroblasts. We considered cells in prophase if they were positive for PH3 staining and showed interphase-like organization of microtubules without visible asters. Percentage was defined as the number of mitotic cells at a specific stage over total number of mitotic cells (Table S3). (c–k) Cells were costained with anti-PH3 antibody (red), Hoechst (blue), and anti–α-tubulin antibody (green). Wild-type prophase (c and d), wild-type prometaphase (e–g), and typical prophase figures observed in PR-Set7 (h–k) are shown. Bar, 5 μm.
Monomethylated H4K20 is present throughout the cell cycle in HeLa cells (Karachentsev et al., 2007) and is detected on polytene salivary gland chromosomes (Karachentsev et al., 2005). Salivary gland cells undergo a modified cell cycle, lacking G2 and M (Makunin et al., 2002). This staining therefore suggests that monomethylated H4K20 is also present in nonmitotic cells. It is likely that monomethylated H4K20 is present throughout the cell cycle and that it is not immunocytologically detectable until chromosome condensation concentrates the signal. Monomethyl mark may be uniformly distributed throughout the cell cycle and throughout the genome as detected on mitotic chromosomes (Fig. 1 d).
PR-Set7 neuroblasts show a delay in early mitotic stages
We studied the progression through mitosis of PR-Set7 neuroblasts to determine when the mutant cells have a defect. In wild type, the mitotic index (percentage of cells in mitosis) was 2.16%, and it was significantly reduced in PR-Set7 to 1.30% (P = 2.75 × 10−2; Fig. 2 a and Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1). To identify cells in the different mitotic stages, neuroblasts were stained with both anti-PH3 and anti–α-tubulin antibodies. We considered cells in prophase if they were positive for PH3 staining and showed interphase-like organization of microtubules without visible asters. As shown in Fig. 2 b (Table S3), both PR-Set7 and PR-Set7/Df(3R)red31 had a fourfold higher frequency of cells in prophase (PR-Set7, 42.5%; PR-Set7/Df(3R)red31, 41.0%) than wild type (10.3%) and a correspondingly lower frequency of cells in the other mitotic phases. After a 1-h treatment with colchicine the mitotic index in the PR-Set7 mutant increased 2.7-fold (Fig. 2 a and Table S2), indicating that the spindle assembly checkpoint is not disrupted in PR-Set7. However, with cholchicine, the ratio of prophase and prometaphase cells in the mutant was still high (wild type, 2.6%; PR-Set7, 43.6%; Fig. S2 a). These results show that the mutant cells are delayed in early mitotic stages.
In wild type, the PH3 signal appears at prophase (Fig. 2 c), increases with chromosome condensation (Fig. 2, d–f), and covers the entire chromosomes at late prometaphase (Fig. 2 g). The formation of mitotic spindles always correlates with both PH3 staining and chromosome condensation, and two asters are observed before the PH3 signal covers the entire chromosome (Fig. 2 e). However, in PR-Set7, the PH3 signal covered most of the chromatin before the asters appeared (Fig. 2, h–k). In wild type, 79% of prophase cells showed no chromosome condensation, as shown in Fig. 2 c (see also Fig. S2 b), whereas in PR-Set7, most of the prophase cells (65%) had condensed DNA (or chromosomes), as shown in Fig. 2 (h–k). These results indicate that nuclear and cytoplasmic events, namely, chromosome condensation and spindle formation, are uncoupled in PR-Set7. These events may keep mutant cells from entering metaphase or from chromosome separation.
Cyclin B is down-regulated in PR-Set7 by anaphase-promoting complex/cyclosome (APC/C)–dependent proteolysis
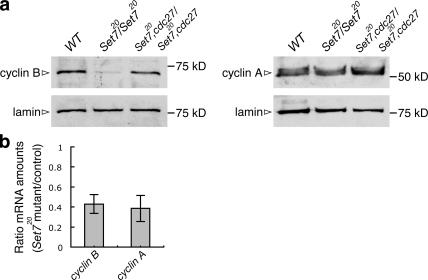
The PR-Set7 phenotypes described so far could be caused by the failure to accumulate mitotic cyclins. It has been well established that accumulation of mitotic cyclins and the activation of the Cdk–cyclin complexes are essential for entry into mitosis and formation of mitotic spindles (Zachariae and Nasmyth, 1999; Lopes et al., 2005). We therefore examined cyclin A and B protein levels by Western blotting and found that cyclin B was reduced in PR-Set7, whereas cyclin A was present at normal levels (Fig. 3 a). In D. melanogaster, the expression pattern of cyclin A is similar to cyclin B, and both accumulate during G2 (Knoblich and Lehner, 1993). Because the cyclin A protein level was not reduced in the mutant, the lower mitotic index observed in the mutant does not explain the reduction of cyclin B. We also found that the cyclin B protein level was still down-regulated in the mutant when the mitotic index was increased after a 1-h colchicine treatment (Fig. 2 a and not depicted). To investigate whether the low level of cyclin B was controlled at the transcriptional level, we measured mRNA levels of cyclin A and B by quantitative real-time PCR. Both mRNAs showed similar reductions in the mutant compared with wild type (Fig. 3 b), probably because of the lower mitotic index, indicating that the reduction of the mRNA level does not fully explain the reduction of cyclin B protein. The APC/C subunit cdc27 is required for the degradation of cyclin B in D. melanogaster (Deak et al., 2003). Hence, we made double-homozygous PR-Set7 and cdc27 mutants and found that the cyclin B level recovered in the double mutant (Fig. 3 a). These results indicate that the reduction of cyclin B in PR-Set7 is mediated by the APC/C proteolysis and is not regulated at the transcriptional level.
Figure 3.
Cyclin B is down-regulated in PR-Set7 by APC/C-dependent proteolysis. (a) Western blots of extracts from wild-type (WT), homozygous PR-Set7, and double-homozygous PR-Set7,cdc27 third-instar larval brains were probed with anti–cyclin B, anti–cyclin A, and anti-lamin antibodies. (b) Cyclin A and cyclin B mRNA levels in wild type and PR-Set7/PR-Set7 were measured by quantitative real-time PCR and normalized to the level of rp-49 mRNA. Total RNA was isolated twice independently, and each RNA was measured twice at different dilutions. The graph shows the ratio of PR-Set7 mutant values to wild type. Error bars show two SDs (n = 4).
Monomethylation of histone H4K20 is essential for proper chromosome condensation
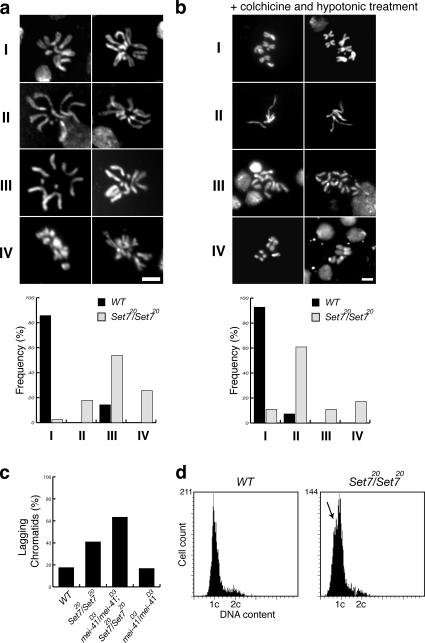
Mitotic stages other than prophase also showed abnormalities in PR-Set7 neuroblasts. Many prometaphase figures had irregular chromosomes (Fig. S2, c and g). Although most metaphase figures looked surprisingly normal in the mutant (Fig. S2, d and h), a large proportion of PR-Set7 anaphase and telophase figures contain lagging chromatids (40.9%; Fig. 4 c and Fig. S2, e, f, i, and j), raising the possibility that PR-Set7 has a defect in chromosome condensation.
Figure 4.
Most of the PR-Set7/PR-Set7 chromosomes show a defect in chromosome condensation. (a) Representative figures of metaphase chromosomes were assigned to distinct categories, with I showing chromosomes with normal morphology and II–IV representing categories of abnormal chromosomes. The histogram shows the percentage of figures in each category (wild type [WT], n = 28; PR-Set7/PR-Set7, n = 39). At least four brains were quantified for each genotype. Bar, 5 μm. (b) Chromosome spreads with hypotonic treatment after a 1-h colchicine treatment. Mitotic chromosomes were defined by staining of PH3. Representative chromosome figures are shown in categories I (normal) and II–IV (abnormal). The histogram shows the percentage of figures in each category (wild type, n = 55; PR-Set7/PR-Set7, n = 64). At least three brains were quantified for each genotype. Bar, 5 μm. (c) Frequency of anaphase and telophase cells containing lagging chromatids (wild type, n = 63; PR-Set7/PR-Set7, n = 44; mei-41D3/mei-41D3;PR-Set7/PR-Set7; n = 49; mei-41D3/mei-41D3, n = 48). At least four brains were quantified for each genotype. (d) The DNA contents of wild-type and PR-Set7/PR-Set7 nuclei were measured by a laser-scanning cytometer, iCys (CompuCyte Corp.). In the mutant, some cells with less DNA than 2n (arrow) were detected.
We next looked at the morphology of metaphase chromosomes in wild-type and PR-Set7 brains (Fig. 4 a). We focused on the length and width of chromosomes and boundaries between sister chromatids to assess whether chromosome condensation is affected in the mutant. Chromosome figures were subdivided into categories I–IV, depending on phenotypes. Most of the wild-type chromosomes (85.7%) had clearly defined sister chromatids (normal chromosomes, category I). 97.3% of mutant chromosomes displayed aberrant morphology. 17.9% of chromosomes were thinner and longer than category I chromosomes and showed vaguely defined sister chromatids (category II). In 53.8%, the sister chromatids are not well defined enough to be apparent (category III). Other chromosomes (25.6%) were entangled (category IV). All these observations point to a defect in chromosome condensation in the mutant.
We also prepared chromosome spreads after a 1-h colchicine treatment and hypotonic shock to better define the degree of chromosome condensation (Fig. 4 b). Most of the wild-type chromosomes (92.7%) showed clearly defined sister chromatids as expected (category I). 60.9% of the mutant chromosomes were strikingly abnormal, considerably longer and thinner than wild type, and showed no defined sister chromatids (category II). 10.9% of mutant chromosomes were longer and thicker and lost the sister chromatid borders, similar to what is observed in category III in Fig. 4 a. Because the length of mutant chromosomes in category IV (17.2%) is similar to normal chromosomes (category I), some mutant chromosomes seem to complete chromosome axis shortening but still have an obvious defect in defining sister chromatids. The simplest explanation of these results is that the PR-Set7–dependent monomethylation of H4K20 is required for proper chromosome condensation. We found that despite the abnormal chromosome organization, the mutant chromosomes contain the condensin component Barren (the fly orthologue of XCAP-H) and DNA topoisomerase II, both important for chromosome architecture (Sakaguchi and Kikuchi, 2004; Yu et al., 2004; Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1).
To determine whether the abnormal chromosome condensation and separation in the mutant results in polyploidy, we measured the DNA content of wild-type and mutant brain cells using a laser-scanning cytometer (Darzynkiewicz et al., 1999), and found that the number of polyploid cells was not increased in the mutant (Fig. 4 d). In the cytometer analysis, we noticed some cells with less DNA than 2n in PR-Set7 (Fig. 4 d, arrow). We investigated whether apoptotic cells are increased in the mutant by TUNEL and found that positive cells were not significantly increased in the mutant (wild type, 40/5,012, and PR-Set7/PR-Set7, 56/4,056; P = 0.108).
DNA damage checkpoint is activated in PR-Set7
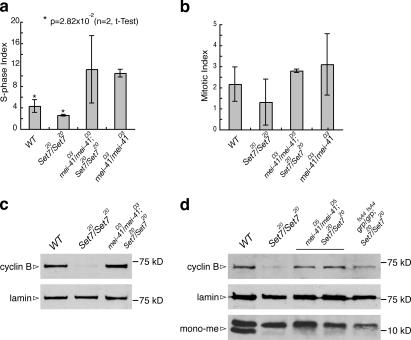
The observed abnormalities in cell cycle progression and the reduction of cyclin B protein levels mediated by APC/C-dependent proteolysis in PR-Set7 led us to hypothesize that the DNA damage checkpoint is activated. We also checked the S phase index (percentage of cells in S phase) in PR-Set7 and found that it was significantly reduced (P = 2.82 × 10−2; Fig. 5 a and Table S4, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1).
Figure 5.
The DNA damage checkpoint is activated in PR-Set7. (a) S phase indexes of wild type (WT), PR-Set7/PR-Set7, mei-41D3/mei-41D3; PR-Set7/PR-Set7, and mei-41D3/mei-41D3 (see Table S4, available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1). (b) Mitotic indexes of wild type, PR-Set7/PR-Set7, mei-41D3/mei-41D3;PR-Set7/PR-Set7, and mei-41D3/mei-41D3 (Table S2). (c) Western blots of extracts from wild-type, PR-Set7/PR-Set7, and mei-41D3/mei-41D3;PR-Set7/PR-Set7 third-instar larval brains were probed with anti–cyclin B and anti-lamin antibodies. (d) Western blots of extracts from wild-type, PR-Set7/PR-Set7, mei-41D5/mei-41D5;PR-Set7/PR-Set7, and grpfsA4/grpfsA4;PR-Set7/PR-Set7 third-instar larval brains were probed with anti–cyclin B, anti-lamin and anti–monomethylated H4K20 (mono-me) antibodies. Two different extracts from mei-41D5/mei-41D5;PR-Set7/PR-Set7 were used.
To investigate whether these phenotypes are caused by the activation of the DNA damage checkpoint, we made a homozygous double mutant of PR-Set7 and the D. melanogaster ATR orthologue mei-41 (mei-41D3/mei-41D3;PR-Set720/PR-Set720), essential for the DNA damage checkpoint (Hari et al., 1995; O'Connell et al., 2000; Song, 2005). The mei-41D3 allele used here has a defect in the checkpoint, allowing cells with damaged DNA to enter mitosis (Hari et al., 1995). In the mei-41D3;PR-Set720 double mutant, the mitotic index was rescued, similar to that observed in wild type and the homozygous mei-41D3 mutant (Fig. 5 b and Table S2). The number of prophase cells was reduced compared with the number observed in PR-Set7 homozygotes and became similar to wild type and mei-41D3/mei-41D3 (Fig. 2 b and Table S3), and the “uncoupled cells” observed in PR-Set7 (Fig. 2, h–k) disappeared (not depicted). These results indicate that the abnormalities of mitotic progression in PR-Set7 are rescued in the double mutant. The S phase index in the mei-41D3/mei-41D3 is increased 2.4-fold compared with wild type, suggesting that the mei-41D3 mutant has a defect in DNA replication (Fig. 5 a and Table S4). Therefore, we cannot determine whether the decrease of the S phase index in PR-Set7 is rescued in the double-homozygous mei-41D3;PR-Set720 mutant.
The protein levels of cyclin B also recovered in the double-homozygous mei-41D3;PR-Set720 mutant (Fig. 5 c). To confirm this result, we also made a homozygous double mutant of PR-Set7 and another allele of mei-41, also affecting the G2/M checkpoint, mei-41D5 (mei-41D5/mei-41D5;PR-Set720/PR-Set720; Laurencon et al., 2003). Further, we constructed a double mutant with PR-Set7 and the D. melanogaster Chk1 orthologue grp (grpfsA4/grpfsA4;PR-Set720/PR-Set720). The grp gene functions downstream of mei-41, and mutants in both genes have similar phenotypes (Sibon et al., 1999; Song, 2005). On Western blots of brain extracts of both double mutants, monomethylated H4K20 is not detected, similar to the PR-Set7 extract, but the cyclin B protein levels are rescued (Fig. 5 d). These results show that in PR-Set7, the DNA damage checkpoint, especially the ATR pathway, is activated and that this activation is responsible for the down-regulation of cyclin B and the abnormal mitotic progression. These results also suggest that a specific mechanism exists in D. melanogaster that down-regulates cyclin B through APC/C proteolysis in response to DNA damage.
The ratio of anaphase/telophase cells with lagging chromatids was increased when the checkpoint was abolished in the double-homozygous mei-41;PR-Set7 mutant (Fig. 4 c), indicating that the defect in chromosome condensation is independent of checkpoint activation and that, in the absence of the checkpoint, the severity of the chromosome condensation defect is enhanced.
DNA double-strand breaks (DSBs) are not increased in PR-Set7
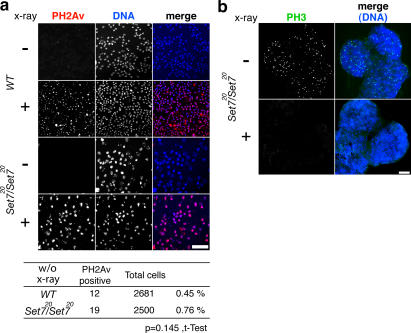
Because we found that the DNA damage checkpoint is activated in PR-Set7, we investigated whether DNA DSBs are increased in the mutant. We stained cells with anti–phosphorylated histone H2Av (PH2Av) antibody. Like H2A.X, H2Av, an essential fly histone variant, becomes phosphorylated at sites of DSBs by DNA damage recognizing factors (Madigan et al., 2002). Thus, PH2Av is well established as a marker for DSBs. We found that the PH2Av-positive cells were not significantly increased in the mutant (P = 0.145), indicating that endogenous DSBs do not occur in PR-Set7 (Fig. 6 a). To confirm that the PR-Set7 cells react normally to DNA damage, we irradiated both wild-type and mutant brains with x-ray and found that the PH2AV staining increased in both (Fig. 6 a). Further, after γ-irradiation PH3-positive mitotic cells were drastically decreased in the mutant (Fig. 6 b), similar to what was shown in wild type (Hari et al., 1995), indicating that the PR-Set7 cells react in similar ways as wild-type cells to γ-irradiation. We conclude that DSBs are not increased in PR-Set7.
Figure 6.
Endogenous DNA DSBs are not increased in PR-Set7 mutant. (a) Cells before and 15 min after γ-irradiation treatment (2000 rad) were stained with anti-PH2Av (red) as a maker for DNA DSBs and Hoechst (blue). Bar, 50 μm. (b) The PR-Set7 mutant brains before and 15 min after γ-irradiation treatment (2000 rad) were stained with anti-PH3 (green) and Hoechst (blue). Bar, 100 μm.
Discussion
What activates the DNA damage checkpoint in PR-Set7? Histone methylation has been thought to control gene expression by packaging DNA into open and closed chromatin. PR-Set7 indeed suppresses position effect variegation, indicating that it functions as a transcriptional suppressor (Karachentsev et al., 2005). However, abnormal regulation of gene expression in PR-Set7 does not explain the activation of the checkpoint. Because the mutation in mei-41 (ATR) abolished the phenotypes caused by the activated checkpoint in PR-Set7, the expression of genes downstream of mei-41 is apparently unchanged in PR-Set7. Furthermore, ATR is one of the proteins that initiate checkpoint signaling and localize to sites of DNA damage, suggesting that the ATR protein is directly activated by DNA damage (Vidanes et al., 2005). Also, we showed that the DNA repair pathway after γ-irradiation was activated in PR-Set7; PH2Av staining is increased and the number of mitotic cells is decreased, similar to what is observed in wild type (Fig. 6; Hari et al., 1995). Collectively, our results indicate that the expression of genes involved in the DNA damage checkpoint is normal in PR-Set7 and that control of gene expression is not involved in activation of the checkpoint.
We observed reduction of both S phase and mitotic indexes in the mutant (Fig. 5, a and b), suggesting that both G1 and G2 checkpoints are activated. Because the mutant shows many defects in chromosome condensation and separation (Fig. 4, a, b, and c), these defects could be one of the reasons the G1 checkpoint is activated after mitosis. What activates the G2 checkpoint? Because DSBs are not increased in the mutant (Fig. 6 a), they cannot be cause for G2 checkpoint activation. Several checkpoints appear to exist in mammalian cells that monitor chromatin structure as well as DSBs. A decatenation checkpoint that monitors chromatid decatenation has been demonstrated in human cells, with progression from G2 to mitosis being inhibited when chromatids are insufficiently decatenated (Deming et al., 2001). Furthermore, in mammalian cells, ATM (DNA damage checkpoint kinase) activation is not necessarily dependent on direct binding to DSBs but may result from changes in the structure of chromatin (Bakkenist and Kastan, 2003). As in mammalian cells, abnormal higher order structure of DNA or chromatin may activate the DNA damage checkpoint, as observed in PR-Set7. We therefore hypothesize that monomethylated H4K20 is involved in the maintenance of proper higher order structure of DNA or proper chromatin structure.
Crystal structural analysis showed that K20 is not part of the N-terminal tail of histone H4, like other sites of methylation (e.g., histone H3 lysine 9; Luger et al., 1997). K20 lies close to the histone-fold domain and is covered by DNA. The results in Schizosaccharomyces pombe agree with the crystal structure. It has been proposed that methylated H4K20 lies normally inside the DNA and that it is exposed only when DSBs occur, creating a binding site for Crb2 (Sanders et al., 2004). The structural analysis further showed that H4K20 makes an interparticle contact with an H2A-H2B dimer (Davey et al., 2002), suggesting that H4K20 could be involved in the maintenance of proper histone structure. In Saccharomyces cerevisiae, the loss of acetylation of H3 lysine 56 affects nucleosome structure (Masumoto et al., 2005). H3 lysine 56 is also close to the histone-fold domain and weakens histone–DNA interactions (Luger et al., 1997). Like acetylation of H3 lysine 56, monomethylation of H4K20 may affect nucleosome structure.
We showed that the loss of monomethylated H4K20 activates the DNA damage checkpoint, which may be induced by abnormal higher order structure of DNA or abnormal chromatin structure in the absence of DSBs. The abnormal DNA or chromatin structure probably causes the abnormal mitotic chromosomes observed in PR-Set7. Our results suggest that monomethylation of H4K20 has a more global effect on chromatin structure than described so far.
Materials and methods
Fly strains
The w1118 stock was used as the wild-type control. Homozygous PR-Set720 larvae were recognized by the absence of the TM3 Sb GFP balancer (Karachentsev et al., 2005). Df(3R)red31, Cdc27l(3)L7123, and mei-41D5 were obtained from the Bloomington Stock Center. mei-41D3 (Hari et al., 1995) and grpfsA4 (Sibon et al., 1999) were obtained from K. McKim (Rutgers University, Piscataway, NJ) and W.E. Theurkauf (University of Massachusetts Medical School, Worcester, MA), respectively.
Immunoblotting
Brains were dissected, and immunoblotting was performed as previously described (Karachentsev et al., 2005). Rabbit polyclonal anti-monomethylated, anti-dimethylated, anti-trimethylated, and histone H4 antibodies (Upstate Biotechnology) were used at 1:1,000 dilution (Schotta et al., 2004). Rabbit polyclonal anti-lamin obtained from P.A. Fisher (State University of New York, Stony Brook, Stony Brook, NY) was used at 1:1,000. Mouse monoclonal anti–cyclin B antibody, F2F4 (Developmental Studies Hybridoma Bank), and anti–cyclin A antibody, A12 (Developmental Studies Hybridoma Bank), were used at 1:200.
Cytology and microscopy
Third-instar larval brains were dissected, fixed, and stained as previously described (Bonaccorsi et al., 2000). Whenever required, brains were dissected in PBS and incubated in 10 μM colchicine (Sigma-Aldrich) for 1 h before fixation. Rabbit polyclonal anti-monomethylated (Upstate Biotechnology) and mouse monoclonal anti-PH3 antibody (Upstate Biotechnology) were used at 1:200 dilution. Rabbit polyclonal anti-PH3 antibody (Upstate Biotechnology) was used at 1:1,000. Anti–α-tubulin conjugated with FITC (Sigma-Aldrich) was used at 1:50. Rabbit polyclonal anti–PH2Av (Ser139) antibody (Upstate Biotechnology) was used at 1:250. Rabbit polyclonal anti-Barren antibody was obtained from H. Bellen (Baylor College of Medicine, Houston, TX) and used at 1:50, and mouse monoclonal anti-Top2 antibody was obtained from A. Kikuchi (Nagoya University, Nagoya, Japan) and used at 1:5. Secondary antibodies were Cy3-conjugated goat anti–rabbit or anti–mouse (Jackson ImmunoResearch Laboratories) used at 1:500 and Alexa Fluor 489 goat anti–mouse or Alexa Fluor 488 donkey anti–rabbit (Invitrogen) used at 1:400. For TUNEL assay, fixed cells were incubated with TUNEL enzyme (Roche) for 1 h, and the Alexa Fluor 488 Signal-Amplification kit (Invitrogen) was used for detection of the signals. The samples were mounted in Vectashield (Vector Laboratories), and Immersol 518F (Carl Zeiss MicroImaging, Inc.) was used as the imaging medium. Immunostained preparations were studied using a microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) with a 63× Plan-Apochromat NA 1.4 and 100× and a 40× Plan-Neofluar NA 1.3 objective lenses (room temperature) equipped with a digital camera (SensiCam; Cooke Corp.). Digital images were collected using the Image Pro Plus imaging software (MediaCybernetics). To measure the DNA content, we stained brain cells with DAPI and used a laser-scanning cytometer, iCys (CompuCyte Corp.; Darzynkiewicz et al., 1999).
BrdU incorporation
Brains were dissected and incubated for 45 min in serum-free Schneider's insect medium containing 100 μg/ml BrdU. They were rinsed three times in PBS, fixed, and stained with the mouse monoclonal anti-BrdU antibody (1:100 dilution; Becton Dickinson) as previously described (Savvidou et al., 2005).
Chromosome preparation with hypotonic treatment
After a 1-h treatment with colchicine, brains were incubated for 3 min in 0.5% trisodium citrate for hypotonic shock. They were then fixed and stained with rabbit polyclonal anti-PH3 antibody to identify mitotic chromosomes.
Irradiation experiments
Third-instar larvae were transferred to yeast paste-supplemented agar plates and irradiated with 2000 rad of x-rays. After 15 min, the brains were dissected, squashed, and stained.
Online supplemental material
Fig. S1 shows that monomethylated H4K20 is detected only on condensed DNA or chromosomes. Fig. S2 shows typical mitotic figures in PR-Set7. Fig. S3 shows that PR-Set7 chromosomes contain the condensin component Barren and Top2. Table S1 shows the numbers behind the graph shown in Fig. 1 b. Table S1 shows the ratio of intensity of the bands from PR-Set7/ PR-Set7 relative to wild type in Fig. 1 b. Tables S2 and S3 show mitotic index and parameters in squashed brains of wild type, PR-Set7/ PR-Set7, mei-41/mei-41;PR-Set7/PR-Set7, and mei-41/mei-41 and PR-Set7/ Df(3R)red31, respectively. Table S4 shows S phase index in squashed brains of wild type, PR-Set7/PR-Set7, mei-41/mei-41;PR-Set7/PR-Set7, and mei-41/mei-41. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200607178/DC1.
Acknowledgments
We thank M.S. Heck, K. McKim, S. Minakhina, and N. Walworth for helpful advice; Z. Darzynkiewicz, X. Huang, and T. Tanaka for use and help with the iCys; and M. Druzhinina and L. Nguyen for technical help and fly food. We thank G. Deshpande, P. Schedl, and D. Reinberg for comments on the manuscript; W.E. Theurkauf for the fly stock; and H. Bellen, P.A. Fisher, and A. Kikuchi for the antibodies.
This work was supported by a grant from the National Institutes of Health (PHS HD 18055) and the Horace W. Goldsmith Foundation. A. Sakaguchi thanks the Charles and Johanna Busch Fund for support.
Abbreviations used in this paper: APC/C, anaphase-promoting complex/cyclosome; DSB, double-strand break.
References
- Bakkenist, C.J., and M.B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi, S., M.G. Giansanti, and M. Gatti. 2000. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2:54–56. [DOI] [PubMed] [Google Scholar]
- Couture, J.F., E. Collazo, J.S. Brunzelle, and R.C. Trievel. 2005. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 19:1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz, Z., E. Bedner, X. Li, W. Gorczyca, and M.R. Melamed. 1999. Laser-scanning cytometry: a new instrumentation with many applications. Exp. Cell Res. 249:1–12. [DOI] [PubMed] [Google Scholar]
- Davey, C.A., D.F. Sargent, K. Luger, A.W. Maeder, and T.J. Richmond. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319:1097–1113. [DOI] [PubMed] [Google Scholar]
- Deak, P., M. Donaldson, and D.M. Glover. 2003. Mutations in makos, a Drosophila gene encoding the Cdc27 subunit of the anaphase promoting complex, enhance centrosomal defects in polo and are suppressed by mutations in twins/aar, which encodes a regulatory subunit of PP2A. J. Cell Sci. 116:4147–4158. [DOI] [PubMed] [Google Scholar]
- Deming, P.B., C.A. Cistulli, H. Zhao, P.R. Graves, H. Piwnica-Worms, R.S. Paules, C.S. Downes, and W.K. Kaufmann. 2001. The human decatenation checkpoint. Proc. Natl. Acad. Sci. USA. 98:12044–12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, J., Q. Feng, C.S. Ketel, H. Wang, R. Cao, L. Xia, H. Erdjument-Bromage, P. Tempst, J.A. Simon, and Y. Zhang. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12:1086–1099. [DOI] [PubMed] [Google Scholar]
- Fischle, W., Y. Wang, and C.D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172–183. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and B.S. Baker. 1989. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 3:438–453. [DOI] [PubMed] [Google Scholar]
- Hari, K.L., A. Santerre, J.J. Sekelsky, K.S. McKim, J.B. Boyd, and R.S. Hawley. 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 82:815–821. [DOI] [PubMed] [Google Scholar]
- Hendzel, M.J., Y. Wei, M.A. Mancini, A. Van Hooser, T. Ranalli, B.R. Brinkley, D.P. Bazett-Jones, and C.D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360. [DOI] [PubMed] [Google Scholar]
- Karachentsev, D., K. Sarma, D. Reinberg, and R. Steward. 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 19:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev, D., M. Druzhinina, and R. Steward. 2007. Free and chromatin-associated mono-, di-, and trimethylation of histone H4-lysine 20 during development and cell cycle progression. Dev. Biol. In press. [DOI] [PMC free article] [PubMed]
- Knoblich, J.A., and C.F. Lehner. 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J. 12:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon, A., A. Purdy, J. Sekelsky, R.S. Hawley, and T.T. Su. 2003. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics. 164:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, C.S., P. Sampaio, B. Williams, M. Goldberg, and C.E. Sunkel. 2005. The Drosophila Bub3 protein is required for the mitotic checkpoint and for normal accumulation of cyclins during G2 and early stages of mitosis. J. Cell Sci. 118:187–198. [DOI] [PubMed] [Google Scholar]
- Luger, K., and T.J. Richmond. 1998. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8:140–146. [DOI] [PubMed] [Google Scholar]
- Luger, K., A.W. Mader, R.K. Richmond, D.F. Sargent, and T.J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 389:251–260. [DOI] [PubMed] [Google Scholar]
- Madigan, J.P., H.L. Chotkowski, and R.L. Glaser. 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30:3698–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunin, I.V., E.I. Volkova, E.S. Belyaeva, E.N. Nabirochkina, V. Pirrotta, and I.F. Zhimulev. 2002. The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics. 160:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 436:294–298. [DOI] [PubMed] [Google Scholar]
- Nishioka, K., J.C. Rice, K. Sarma, H. Erdjument-Bromage, J. Werner, Y. Wang, S. Chuikov, P. Valenzuela, P. Tempst, R. Steward, et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 9:1201–1213. [DOI] [PubMed] [Google Scholar]
- O'Connell, M.J., N.C. Walworth, and A.M. Carr. 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10:296–303. [DOI] [PubMed] [Google Scholar]
- Rice, J.C., K. Nishioka, K. Sarma, R. Steward, D. Reinberg, and C.D. Allis. 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 16:2225–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, A., and A. Kikuchi. 2004. Functional compatibility between isoform α and β of type II DNA topoisomerase. J. Cell Sci. 117:1047–1054. [DOI] [PubMed] [Google Scholar]
- Sanders, S.L., M. Portoso, J. Mata, J. Bahler, R.C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 119:603–614. [DOI] [PubMed] [Google Scholar]
- Savvidou, E., N. Cobbe, S. Steffensen, S. Cotterill, and M.M. Heck. 2005. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J. Cell Sci. 118:2529–2543. [DOI] [PubMed] [Google Scholar]
- Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon, O.C., A. Laurencon, R. Hawley, and W.E. Theurkauf. 1999. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9:302–312. [DOI] [PubMed] [Google Scholar]
- Song, Y.H. 2005. Drosophila melanogaster: a model for the study of DNA damage checkpoint response. Mol. Cells. 19:167–179. [PubMed] [Google Scholar]
- Strahl, B.D., and C.D. Allis. 2000. The language of covalent histone modifications. Nature. 403:41–45. [DOI] [PubMed] [Google Scholar]
- Vidanes, G.M., C.Y. Bonilla, and D.P. Toczyski. 2005. Complicated tails: histone modifications and the DNA damage response. Cell. 121:973–976. [DOI] [PubMed] [Google Scholar]
- Xiao, B., C. Jing, G. Kelly, P.A. Walker, F.W. Muskett, T.A. Frenkiel, S.R. Martin, K. Sarma, D. Reinberg, S.J. Gamblin, and J.R. Wilson. 2005. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 19:1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., S.L. Fleming, B. Williams, E.V. Williams, Z. Li, P. Somma, C.L. Rieder, and M.L. Goldberg. 2004. Greatwall kinase: a nuclear protein required for proper chromosome condensation and mitotic progression in Drosophila. J. Cell Biol. 164:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039–2058. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343–2360. [DOI] [PubMed] [Google Scholar]