Abstract
How centrosome removal or perturbations of centrosomal proteins leads to G1 arrest in untransformed mammalian cells has been a mystery. We use microsurgery and laser ablation to remove the centrosome from two types of normal human cells. First, we find that the cells assemble centrioles de novo after centrosome removal; thus, this phenomenon is not restricted to transformed cells. Second, normal cells can progress through G1 in its entirety without centrioles. Therefore, the centrosome is not a necessary, integral part of the mechanisms that drive the cell cycle through G1 into S phase. Third, we provide evidence that centrosome loss is, functionally, a stress that can act additively with other stresses to arrest cells in G1 in a p38-dependent fashion.
Introduction
In mammalian cells, the interrelationship between centrosomes and cell cycle is multifaceted. Because the activities of the centrosome are temporally linked to, and dependent on, cell-cycle progression, the centrosome was traditionally thought to be controlled by the cytoplasmic changes accompanying progress through the cell cycle. For example, the number of microtubules that are nucleated by centrosomes start out low in early interphase and increase markedly as the cell approaches mitosis. The amount of γ-tubulin at the centrosome, and the number of microtubules that grow from it in vitro in lysed cell models, increases as the cells approach mitosis (Snyder and McIntosh, 1975; Kuriyama and Borisy, 1981; Khodjakov and Rieder, 1999; Young et al., 2000). Also, the precise duplication of the centrosome is initiated at the onset of S phase by the rise in the activities of cyclin-dependent kinase 2 coupled to cyclin E and/or A, the kinase complexes that drive the cell into S phase (Sluder, 2004).
However, the centrosome is more than just a follower of the cell cycle. Evidence has been accumulating that the centrosome has an activity that is essential for the cell to progress through G1 and enter S phase. The first indication came from the finding that microsurgical removal of the interphase centrosome from BSC-1 cells did not prevent the acentrosomal cells from entering mitosis, but almost all of them arrested in G1 thereafter (Maniotis and Schliwa, 1991; Hinchcliffe et al., 2001). Similarly, after laser ablation of one centrosome at metaphase, CV-1 cells divided but the daughter cells that inherited no centrosome arrested in G1 (Khodjakov and Rieder, 2001). Subsequent work indicated that acentrosomal BSC-1 cells after mitosis arrest with elevated levels of p21, an absence of the Ki-67 proliferation antigen, and hypophosphorylated retinoblas toma protein, which imply an early G1 arrest involving p53 (Srsen et al., 2006; unpublished data). In contrast, removal of centrosomes from HeLa cells does not block G1 progression (La Terra et al., 2005). However, these are transformed cells with dysfunctional G1 controls caused by the expression of human papillomavirus proteins E6 and E7 (for review see zur Hausen, 2002). Importantly, several recent studies report that the knockdown or displacement from the centrosome of a variety of proteins associated with the centrosome leads to a p53-dependent G1 arrest of a large proportion of the cell population (for reviews see Sluder, 2005; Doxsey et al., 2005a,b; Srsen et al., 2006). Together, these studies point to a role for the centrosome in the mechanisms that control the untransformed cell's progress through G1 into S phase.
The way in which the centrosome influences G1 progression in untransformed cells is a mystery because such a wide variety of seemingly disparate experimental perturbations all lead to a G1 arrest. Possibilities include a novel checkpoint that monitors centrosome absence or damage, disorganization/dysfunction of the interphase cytoskeleton, and disabling of the centrosome's possible role in promoting the efficiency of signaling reactions that may be necessary for G1 progression (Hinchcliffe et al., 2001; Murray, 2001; Doxsey et al., 2005a,b; Sluder 2005). These possibilities, however, are presently ideas awaiting experimental investigation. Given our previous observation that HeLa cells can progress through G1 without a centrosome (La Terra et al., 2005), we investigate the consequences of centrosome removal in normal human cells. We were particularly interested in determining if untransformed human cells without a centrosome can progress through G1.
Results
Centrosome removal in G1: de novo centriole assembly
We initially worked with hTERT RPE1 and human mammary epithelial cells (HMECs) stably expressing human centrin-1/GFP to tag the centrioles. These normal human cells expressing centrin-1/GFP progress through the cell cycle at the same rate as the native cells, with a doubling time of 15–18 h. Centriole duplication and mitosis are normal. These cells have an intact p53 pathway, as indicated by cell cycle arrest with elevated levels of p21 in response to DNA damage (unpublished data).
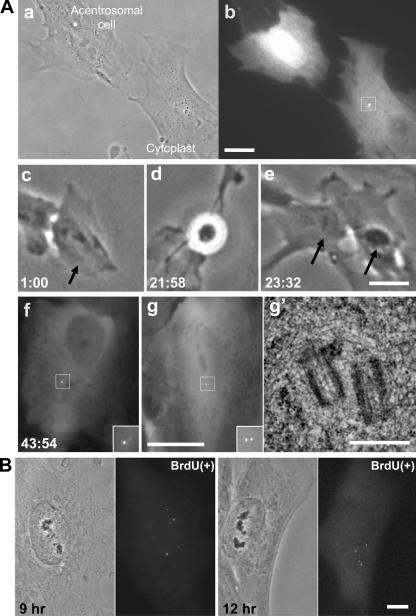
We identified RPE1 cells that were in G1 by the presence of two bright focal centrin-1/GFP spots (centrioles); in phase contrast, we cut between the nucleus and the centrioles with a glass needle, as previously described (Hinchcliffe et al., 2001; La Terra et al., 2005), to form an acentrosomal cell and a centriole-containing cytoplasmic fragment called a cytoplast (Fig. 1 A, a). 15–30 min after the operation, cytoplasts were examined for ∼1 s in fluorescence to confirm the presence of the centrioles (Fig. 1 A, b). Individual acentrosomal cells were followed with phase-contrast time-lapse video recordings after the coverslips were transferred from micromanipulation preparations to closed chambers, which allow cells to proliferate normally for at least 100 h, or until confluency is reached (Sluder et al., 2005).
Figure 1.
De novo centriole assembly in an acentrosomal RPE1 cell. (A, a) Acentrosomal cell and cytoplast 30 min after G1 microsurgery. (b) Fluorescence image of the GFP-tagged centriole pair (box) in the cytoplast. (c) The same acentrosomal cell at 1 h after microsurgery (arrow). (d and e) This acentrosomal cell enters mitosis and divides into two daughters (arrows). (f) Fluorescence images of one daughter showing bright focal centrin-1/GFP spots (inset). The inset shows the centrin foci at higher magnification. (g) The other daughter cell that was serial-sectioned for EM analysis. (g′) Electron micrograph of a section showing two centrioles in the location of the fluorescent dots. Hours:minutes after the microsurgery are shown in the lower corner of each frame taken from the time-lapse recording. (B) Paired phase and fluorescence images of two cells 9 and 12 h after G1 centrosome removal. These cells are in S phase, as determined by BrdU incorporation. The fluorescence images show the several centrin-1/GFP foci (precentrioles) that have assembled. Bars: (A, a–g) 20 μm; (A, g′) 0.5 μm; (B) 10 μm.
Before we fully describe the cell cycle progression of acentrosomal RPE1 cells (see next section), we first describe our observation that untransformed cells assemble centrioles de novo. Surprisingly, we found that acentrosomal RPE1 cells progressed through interphase and divided one or more times. During the first interphase and after the first or second mitosis, the cells contained 0–6 puncta of centrin-1 (Fig. 1 A, f and g, and Fig. 2, H–K). The bright focal appearance and variable number of centrin foci is characteristic of de novo centriole assembly (Khodjakov et al., 2002; La Terra et al., 2005). Serial section electron microscopy of three acentrosomal cells previously followed in vivo revealed that the bright centrin foci assembled de novo corresponded to morphologically normal centrioles (Fig. 1 A, g and g′). HMEC cells also formed bright centrin foci in the first interphase after G1 ablation of both centrioles. Serial section electron microscopy of two of these cells confirmed the de novo formation of centrioles (unpublished data).
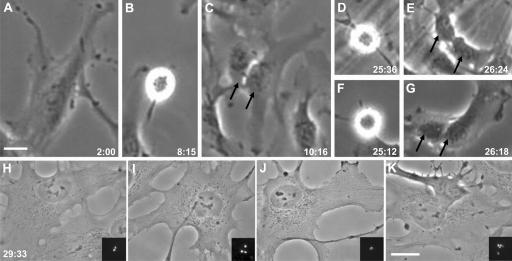
Figure 2.
Acentrosomal RPE1 cell progressing from G1 through two mitoses. (A) Acentrosomal cell 2 h after the microsurgery. (B) First mitosis. (C) Progeny of the first mitosis. The two daughters are indicated by arrows. (D and E and F and G) Second mitosis of both daughters. (H–K) The four granddaughters are shown at higher magnification, with insets showing the variable number of centrin-1/GFP foci that are indicative of centrioles assembled de novo. Phase-contrast images with GFP fluorescence (z series, maximum intensity point projections) shown in the insets. Hours:minutes after the microsurgery are shown in the bottom corner of each frame taken from the time-lapse recording. Bars, 20 μm.
To determine when acentrosomal cells start to assemble centrioles de novo, we cut RPE1 cells in G1, added BrdU to the medium, and examined them at various times thereafter. We observed the formation of 2–7 centrin foci (precentrioles) starting ∼9 h after the microsurgery (Fig. 1 B), which was temporally coincident with S phase as determined by BrdU incorporation. To test if the early, and perhaps invisible, formation of precentrioles occurs during G1 in RPE1 cells, we microsurgically removed the centrosome from 15 cells that were pretreated with 1 mM mimosine to arrest them in G1 (Krude, 1999; Wang et al., 2000). Because precentriole maturation into morphological centrioles is a time-dependent process in HeLa cells (La Terra et al., 2005), there would be sufficient time for nascent precentrioles to mature and become readily visible in acentrosomal cells arrested in G1. 11 acentrosomal cells remained arrested in G1 for at least 24 h and, with one exception, none contained any visible centrin foci. The other four progressed into S phase, and all contained two or four centrin foci; these serve as internal controls, demonstrating that mimosine does not have an activity that shuts down the de novo centriole assembly pathway. Separately, we laser ablated the centrosome in five G1 RPE1 cells, and then hit the nucleus with the laser to induce DNA damage to hold the cells in G1. All arrested in interphase for at least 72 h, and none formed centrin foci. Together, these observations indicate that precentriole formation occurs in S phase, and thus, the G1 progression of acentrosomal cells reported in the next section is not supported by the presence of precentrioles.
Centrosome removal in G1: no cell cycle arrest
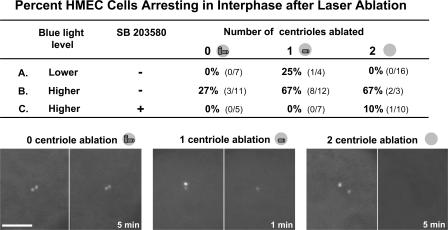
In 38 trials, all acentrosomal cells, even those produced 1 h after the completion of mitosis, progressed through interphase, entered mitosis (mean 15 h after cut), and completed cleavage into two daughters in a normal fashion (Fig. 1 A). However, instead of arresting in G1 after mitosis, as we would have expected from previous studies (Hinchcliffe et al., 2001), out of 72 daughters that stayed in view, 2 arrested in G1, 5 progressed into S phase, and 65 divided at least one more time within 48 h (Fig. 2). To determine if G1 progression after centrosome removal is peculiar to microsurgery, we used laser ablation of the centrosome, which destroys only a small volume of the cell. Using a spinning disk confocal at low laser power setting (107 μW output at the objective lens) to visualize the GFP-tagged centrioles, we found that 7/8 RPE1 cells progressed through interphase to mitosis after G1 ablation of both centrioles. We also conducted G1 centrosome ablations on a p53-positive clone of HMECs expressing centrin-1/GFP to tag the centrioles. Using the same confocal power setting to visualize the GFP-tagged centrioles, we found that after ablation of both centrioles during G1, all 16 cells progressed through interphase into mitosis (Fig. 3, top, line A). In these experiments, we used pairs of sister G1 cells in which one received a cytoplasmic control ablation and the other a directed centrosome ablation; we found that the time from ablation to mitosis was the same (control irradiated cell mean = 26.9 h, n = 15; experimental cell mean = 26.8 h, n = 16). Together, these results demonstrate that G1 progression without a centrosome is not specific to the type of untransformed cell or the means used to remove the centrosome. Furthermore, when we induced physical damage to the centrosome by ablating one G1 centriole, three fourths of the cells progressed through interphase to mitosis (Fig. 3, top, line A).
Figure 3.
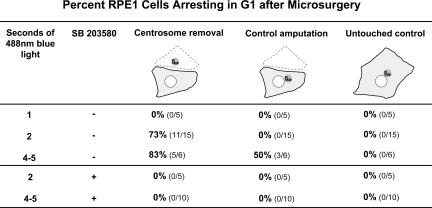
Laser ablation of one or two centrioles during G1 predisposes HMEC cells to a p38-dependent interphase arrest. (top) The blue light level column shows the intensity of the confocal blue light power used to position cells at the coordinates of the laser beam and, later, follow the cells for 72 h. Lower, 107 μW output at the objective; higher, 450 μW output at the objective. The SB 203580 column shows the presence (+) or absence (−) of the p38 inhibitor. In the number of centrioles ablated columns, a near miss did not ablate any centrioles (0) and these serve as controls; other ablations eliminated one centriole (1), and others eliminated both centrioles (2). Line A summarizes the results for cells positioned in the coordinates of the laser beam and later followed for 72 h at the lower 488-nm blue light level in the absence of the p38 inhibitor SB 203580. Line B shows the results for cells positioned in the coordinates of the laser beam and, later, followed at the higher blue light level without p38 inhibitor. Line C shows the results for cells exposed to the higher blue light level in the presence of SB 203580 starting 30 min before laser ablation and followed thereafter in the presence of the inhibitor. (bottom) Before and after fluorescence images of 0, 1, and 2 centriole laser ablations. Times in the right hand panels indicate minutes after ablation. Bar, 5 μm.
Cells “born” without centrioles progress through G1
It is formally possible that we removed the centrosome from G1 cells after the point at which the centrosome becomes dispensable for cell cycle progression. To directly test whether or not RPE1 acentrosomal cells can progress through G1 in its entirety without a centrosome, we removed one of the two centrosomes from cells during late S–G2 (after centriole replication). The de novo pathway is inhibited as long as cells contain even a single centriole (La Terra et al., 2005), and cells that enter mitosis with a single centrosome will divide into two daughters, one inheriting a centrosome and the other entering G1 without a centrosome (Khodjakov et al., 2000; Khodjakov and Rieder, 2001).
The 20 cells that entered mitosis within 8 h after the microsurgery all divided in a bipolar fashion. Shortly after mitosis, we added BrdU to the medium and later used an ∼1-s fluorescence examination to identify which daughter did not contain centrioles. We followed the acentrosomal daughter cells for at least 36 h after mitosis to determine whether they progressed through interphase to mitosis. Those that did not were fixed at 36 h to assay for BrdU incorporation to determine if they progressed into S phase. All centrosome-containing daughters progressed to the next mitosis.
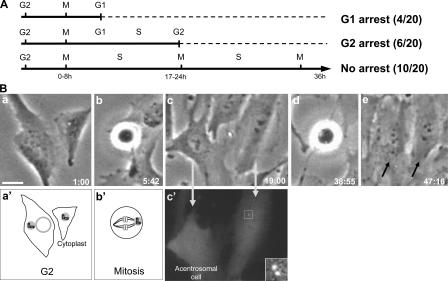
4 out of 20 acentrosomal daughter cells arrested in G1 after the first mitosis, as determined by lack of BrdU incorporation. Six acentrosomal daughters progressed into S phase, but did not enter mitosis within 36 h. The remaining 10 progressed through interphase and through the next mitosis (Fig. 4 A). An example of such an acentrosomal cell progressing from one mitosis to the next is shown in Fig. 4 B (an additional example is shown in Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200607073/DC1). These observations reveal that 80% of the acentrosomal daughter cells progress through the entirety of G1 without a centrosome. Because precentrioles do not form during G1 (La Terra et al., 2005; this study), the G1 progression we observed in this study is not supported by the assembly of precentrioles. This notion is further supported by one case of a G2 microsurgery in which we observed that the daughter cell “born” without centrioles progressed through the next mitosis, and no centrin foci were observed in the granddaughter cells.
Figure 4.
RPE1 cells “born” without centrosomes progress through G1 in its entirety. (A) Summary of data. G2 cells were identified by the presence of four centrin dots (two centrosomes) and were cut to remove one centrosome. They were then followed by phase-contrast time-lapse video microscopy for at least 36 h. After first mitosis, BrdU was added to the medium and acentriolar daughters were identified. Those acentrosomal daughters that failed to enter second mitosis were fixed to assay for BrdU incorporation. First and second mitoses occurred between 0–8 h and 17–24 h, respectively, after the microsurgery. (B) Cell cycle progression of a cell from which one centrosome was removed in G2. (a and a′) Image and diagram of cell/cytoplast 1 h after microsurgery. (b and b′) Image and diagram of cell with one centrosome in mitosis. (c and c′) Phase and centrin-1/GFP fluorescence images of the daughter cells. The cell on the left has inherited no centrioles, and the one on the right has inherited the centriole pair (box and inset at higher magnification; z series maximum intensity projection). The acentrosomal cell enters mitosis (d) and produces two daughters, indicated by arrows (e). Hours:minutes after microsurgery are shown in the lower corners of each frame taken from time-lapse recording. Bar, 20 μm.
These results are not peculiar to microsurgery or the cell type. We ablated one of the two centrosomes in G2 HMEC cells. In three experiments, all cells divided in a bipolar fashion between 1.5 and 7 h after the ablations, and the daughters born without centrioles progressed through interphase into the second mitosis. In two separate experiments on HMEC cells, we laser ablated one centrosome in metaphase cells, and then followed the daughters not inheriting centrioles. We found that both acentriolar daughters proceeded through interphase into mitosis within 24 h, as did their centriole-containing sisters (unpublished data).
Stress influences G1 progression
These observations are in clear contrast to previous reports that centrosome removal or the knockdown/displacement of a wide variety of centrosomal proteins lead to a G1 arrest in untransformed cells (for reviews see Sluder, 2005; Doxsey et al., 2005a,b). Insight into a possible reason for these fundamentally different observations was first suggested by our anecdotal observations that prolonged exposure of acentrosomal cells to 488-nm blue light, which is used to excite GFP, correlated with a G1 arrest, whereas control cells in the same microscope field continued through multiple cell cycles. This observation led us to ask if loss of the centrosome, by itself, is a stress for the cell, and if any additional stress (in this study, blue light) causes it to arrest in G1.
To test this notion, we used microsurgery of G1 cells to produce acentrosomal cells, and, in the same microscope field, performed control amputations of equivalent cell areas on other cells. The untouched cells in the same fields served as controls. 30 min after the cutting operations, we exposed the field of cells to various durations of 488-nm blue light (18 nW/μm2 at the field plane; 580 μW output at the objective lens) to controllably stress all the cells. The field was then followed by time-lapse microscopy to determine which cells progressed through interphase into mitosis and which did not. We used blue light as an exogenous stress, because it is deleterious to cells and dosages can be precisely controlled by computer control of the shutter on the epifluorescence pathway. Our results do not depend on knowing the details of how blue light stresses a cell; we use blue light only as an experimental tool. In this regard, other stressors, such as pH and composition of the media, could, in principle, be used in our application.
Our results, which are summarized in Fig. 5, reveal that the acentrosomal cells are most sensitive to blue light–induced stress, the control-amputated cells are sensitive, but less so, and the untouched control cells are not affected by blue light exposures within the range we used. The brevity of the blue light exposures that lead to a G1 arrest of acentrosomal cells and control-amputated cells reveals how sensitive they are to blue light, relative to the untouched controls. All untouched control cells exposed in G1 to 20–40 s exposures of blue light progress through interphase to mitosis (n = 34).
Figure 5.
Microsurgery and centrosome removal predispose RPE1 cells to p38-dependent G1 arrest. For each experiment, a G1 cell was microsurgically cut to remove the centrosome and, in the same field, a cell was cut to amputate an equivalent portion of the cytoplasm without removing the centrosome. Untouched cells in the same field served as controls. BrdU was added to allow for later analysis for entry into S phase. 30 min after the microsurgery, the field was exposed for defined durations to the 488-nm light. Each field was followed by phase-contrast time-lapse microscopy for at least 48 h to determine if the acentrosomal cell, control amputation, or untouched controls progressed to mitosis in the presence of BrdU. The acentrosomal cells and control cut cells that arrested in interphase were fixed and assayed for BrdU incorporation; none showed any BrdU incorporation, demonstrating that the cells never progressed out of G1. The last two lines of this table show the results of experiments in which the preparations continuously contained the p38 stress kinase inhibitor SB 203580 starting 30 min before the microsurgery.
Stresses such as UV light, heat, and osmotic shock result in activation of the MAP kinase p38, which in turn leads to G1 arrest by influencing cyclin D1 stability, as well as the phosphorylation of p53 and pRb (for reviews see Ambrosino and Nebreda, 2001; Zarubin and Han, 2005; Harris and Levine, 2005). To assess the involvement of the p38 MAPK pathway in a G1 arrest of acentrosomal cells, we added SB 203580, which is an inhibitor of the p38 stress kinase (Kumar et al., 1999), to the medium 30 min before the G1 cuts were made. 30 min after the microsurgery, the fields were exposed to blue light for either 2 or 4–5 s. We found that none of the acentrosomal cells or control amputees arrested in G1, even after 4–5 s exposures (Fig. 5, bottom).
These results are not peculiar to centrosome removal by microsurgery. When widefield excitation with blue light was used to position the centrosomes of RPE1 cells at the coordinates of the laser beam, and to later observe the cells, we found that laser ablation of centrosomes in G1 produced an interphase arrest in 14/16 cells, whereas the adjacent control cells progressed through interphase to mitosis. More recently, system upgrades, including the use of a spinning disk confocal (used at 107 μW output at the objective lens), allowed us to significantly reduce the intensity of blue light used to image the centrosomes. Under such conditions, we found that 7/8 RPE1 cells progressed to mitosis after complete centrosome ablation during early G1.
We also investigated how blue light exposure influences the G1 cell cycle progression of acentrosomal HMEC cells when two intensities of blue light were used to image centrosomes and monitor their ablation (the percentage of cells arresting in interphase under various conditions is summarized in Fig. 3). At the lower blue light level (107 μW output at the objective lens), G1 laser irradiation of the cytoplasm adjacent to the centrosome (no damage to centrosome), damage to the centrosome in the form of ablation of one centriole, or the ablation of both centrioles did not give a substantial incidence of cell cycle arrest. Only one cell arrested (Fig. 3, top, line A). In contrast, at an approximately fourfold higher blue light level used to observe the cells (450 μW output at the objective lens for the same total amount of time), laser irradiation during G1 of the cytoplasm adjacent to the centrosome (control irradiation) led to an interphase arrest in approximately one third of the cells (Fig. 3, top, line B). Ablation of one or both centrioles during G1 arrested two thirds of the cells in interphase.
To determine if activation of the p38 stress-activated kinase plays a role in these observed interphase arrests at the higher blue light level, we repeated these ablations with cells continuously exposed to the p38 inhibitor SB 203580. Cytoplasmic laser irradiations, ablation of one centriole, or ablation of both centrioles did not lead to a G1 arrest in any of the cells (Fig. 3, top, line C). Together, these results indicate that physical damage to the centrosome, or its complete ablation, promotes a p38 stress-activated kinase–mediated interphase arrest when the HMEC cells are additionally stressed by blue light.
G1 progression of acentrosomal BSC-1 cells
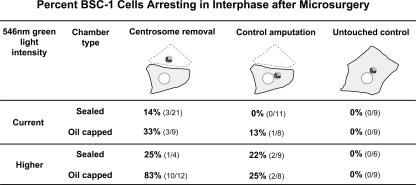
We previously reported that after microsurgical removal of the centrosome during interphase, BSC-1 cells progressed through mitosis and 88% arrested in G1 after that mitosis (Hinchcliffe et al., 2001). To test how our previous results fit with our current findings, we reinvestigated the consequences of microsurgical removal of the interphase centrosome from BSC-1 cells using our current methodology. Our current methods involve several system upgrades, such as the use of a mechanically more stable micromanipulator and more sensitive video cameras that allow ∼64-fold lower green light (546 nm) intensities for time-lapse imaging (4.7 nW output from the condenser vs. 302 nW condenser output previously used). Also, after microsurgery, we now remount the cell bearing coverslips into sealed observation chambers (Sluder et al., 2005) for time-lapse observations, rather than leaving them in oil-capped micromanipulation preparations. The sealed chambers contain an approximately threefold higher volume of medium (600 μl).
We cut a BSC-1 cell to remove the centrosome, and we performed a control amputation of cytoplasm from another in the same field of view. The untouched cells served as controls. For some experiments, the coverslips were transferred after the microsurgery to sealed observation chambers, as we have done after the microsurgery of RPE1 cells. For other experiments, we left the cells in the oil-capped micromanipulation preparations for time-lapse observations. Using our current observation conditions, we found that 14% of the acentrosomal cells arrested in interphase after mitosis, whereas none of the control amputation or untouched controls arrested in interphase (Fig. 6). When the cells were left in the micromanipulation chambers for time-lapse filming, 33% of the acentrosomal cells and 13% of the control-amputated cells arrested in interphase after mitosis; none of the untouched controls arrested.
Figure 6.
G1 progression of acentrosomal and control cut BSC-1 cells under various experimental conditions. For each experiment, an interphase cell was microsurgically cut to remove the centrosome and, in the same field, a cell was cut to amputate an equivalent portion of the cytoplasm without removing the centrosome. Untouched cells in the same field served as controls. After microsurgery in oil-capped micromanipulation chambers, cells were continuously cultured in the same chamber or transferred into a sealed filming chamber, and observed for ∼90 h at the indicated intensity of 546-nm green light. After microsurgery, all cells went through mitosis, but thereafter some arrested in interphase. The Green light level column shows green light intensity used for time-lapse imaging. Current, 4.75 nW condenser output; Higher, 1170 nW condenser output. In the Chamber type column shows the chamber types used. Sealed, the sealed chamber currently used for time-lapse observation; Oil capped, the oil-capped chamber used for micromanipulation.
To test if acentrosomal BSC-1 cells are sensitive to the level of continuous green light used for time-lapse observations, we performed the same experiments, but raised the illumination intensity to 1,170-nW condenser output (3.8-fold higher than the Hinchcliffe et al. (2001) study). Our results (Fig. 6) show that for both the sealed and oil-capped micromanipulation chambers used for filming, a higher percentage of the acentrosomal cells and control cut cells arrest in interphase after mitosis under these higher green light conditions. Notably, none of the untouched control cells arrested under any of these conditions.
Discussion
De novo centriole assembly
De novo centriole assembly after centrosome removal has been, as of yet, observed only in transformed cells (Khodjakov et al., 2002; La Terra et al., 2005). The fact that de novo centriole assembly was not found in untransformed cells suggested that transformation abrogates the normal limits on spontaneous centrosome assembly (Maniotis and Schliwa, 1991; Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001). We demonstrate that de novo centriole assembly is a general phenomenon for mammalian somatic cells, not a peculiarity of cell transformation. In RPE1 and HMEC cells, this process appears to have the same characteristics as in HeLa cells. For centrosome removal during G1, a variable number of GFP fluorescent centrin foci (called precentrioles) appear at the time of first S phase, become brighter with time, and eventually develop into morphologically recognizable centrioles. Although the number of centrioles assembled de novo in RPE1 cells is variable, we note that fewer (up to 6) form in these cells than in HeLa cells (up to 14). For RPE1 cells, we observed 40 cases in which the daughters of acentrosomal cells contained two centrioles after the second mitosis. This could point to the existence of a mechanism that controls centriole copy number or reflects the de novo assembly of a single centriole that later duplicates. Lastly, we observed four acentrosomal RPE1 cells and seven acentrosomal HMEC cells that did not form bright centrin foci in the first cell cycle, something not seen in the HeLa experimental system (La Terra et al., 2005).
G1 progression without a centrosome in untransformed human cells
A surprising aspect of our study was the finding that normal human cells progress through G1 in its entirety without a centrosome, as long as they are not subjected to exogenous stress, such as 488-nm light. Cells progress through G1 to mitosis whether the centrosome is removed early in G1 or the cells are “born” at the end of mitosis without a centrosome. The identical behaviors of RPE1 and HMEC cells after centrosome removal indicate that our results are not specific to cell type or the means used to remove the centrosome. Although these cells assemble centrioles de novo during S phase after centriole removal, we tested for the early stages of centriole formation during G1 by grossly prolonging this cell cycle phase, and we could not find the formation of any centrin foci indicative of precentrioles. Thus, it does not appear that progression through G1 in acentrosomal cells was supported by the formation of precentrioles soon after mitosis. Our finding that 1 RPE1 and 5 HMEC acentrosomal cells went through more than a complete cell cycle without any centrin foci and no centriole structures were found by serial section electron microscopy (HMEC; n = 3; unpublished data) provides additional evidence that G1 progression was not supported by precentriole assembly.
To gain insight into the apparent difference between our present results and those of Hinchcliffe et al. (2001), we characterized the behavior of microsurgically produced BSC-1 acentrosomal cells and control-amputated cells using our current experimental conditions. Our observations reveal that acentrosomal BSC-1 cells behave in a qualitatively similar fashion to acentrosomal RPE1 and HMEC cells. BSC-1 cells can progress through G1 without a centrosome under our current conditions, but not at as high a frequency as RPE1 cells. Together, our observations indicate that G1 progression in BSC-1 cells may be more sensitive to the loss of the centrosome than in RPE1 cells, and micromanipulation chambers provide a less favorable environment than sealed chambers for the G1 progression of BSC-1 cells that have been stressed by microsurgery and loss of the centrosome. Also, the previous use of higher green light intensities for time-lapse observations than we currently use may have contributed to the previously observed G1 arrest of acentrosomal cells.
Together, our results with human and BSC-1 cells reveal that the centrosome is not required for G1 progression in normal cells that have a functional p53 pathway. This means that the centrosome and its activities are not an integral part of the mechanisms that drive the cell cycle through G1 into S phase. Our finding that interphase progression after centrosome removal occurred with normal kinetics indicates that the normal human cell does not have a traditional checkpoint mechanism that monitors the presence or function of the intact centrosome.
Centrosome damage, stress, and G1 arrest
Our finding that the centrosome is not needed for G1 cell cycle progression raises the question of why knockdown or displacement of a variety of centrosomal proteins in untransformed cells leads to a G1 arrest in a substantial proportion of the population of cells (Sluder 2005; Doxsey et al., 2005a,b; Srsen et al., 2006). We envision two possibilities to initially consider separately. The first is that the cell has a mechanism that can detect damage to the centrosome and/or sense-compromised centrosome function (Murray, 2001; Doxsey et al., 2005a,b). If some portion of the centrosome must remain present to act as a signaling platform to trigger the p53 pathway, complete centrosome removal would not be sensed by the cell as centrosome damage or dysfunction. Although this reasoning can explain why experimental centrosomal protein knockdowns could arrest cells in G1, one wonders if cells in an organism would face the loss or disabling mutation of centrosomal proteins often enough to drive the evolution of a distinct checkpoint. Thus, we raise the speculative possibility that a centrosome-damage–sensing mechanism could have evolved to deal with other cellular defects, and that it uses the centrosome as a device to stop the cell cycle until the problem is resolved. For example, when cells are exposed to heat, which is a common environmental hazard, the proteotoxic stress causes centrosomes to lose some of their proteins, which can lead to abnormal mitosis (Hut et al., 2005). Also, irreversibly damaged proteins can accumulate at the centrosome as an “aggresome” to be degraded by proteosomes concentrated there (Johnston et al., 1998). There is evidence that excess protein accumulation at the centrosomes leads to the fragmentation of the centrosomal microtubule-organizing center and the consequent generation of multipolar spindles if the cell enters mitosis (Ehrhardt and Sluder, 2005). In this scenario, possible disruption of the centrosome by denatured protein accumulation could lead to a G1 arrest until the damaged proteins are degraded and the centrosome is restored to its intact state.
The second possibility is that damage to the centrosome or its removal leads to centrosome dysfunction, which is a stress for the cell that can act additively with other stresses to trigger a p38-p53 response. Normal mammalian cells in G1 monitor a variety of intracellular and extracellular conditions that, together, lead to a commitment, or not, to enter the cell cycle (Giaccia and Kastan, 1998; Hulleman and Boonstra, 2001; Sherr and McCormick, 2002; Ingber, 2003; Massagué, 2004). For example, the cell is sensitive to serum growth factors, cell–cell contacts, and substrate adhesion. In addition, even slight perturbations of the actin cytoskeleton can cause a durable G1 arrest (Hansen et al., 1994; Ingber et al., 1995; Bohmer et al., 1996; Lohez et al., 2003). Restrictions on cell spreading and disassembly of the interphase microtubule array have also been reported to arrest untransformed cells in G1 in a p53-p21–dependent fashion (Jimenez et al., 1999; Sablina et al., 2001; Ingber, 2003).
We tested this stress hypothesis and obtained functional evidence that centrosome removal significantly sensitizes both RPE and HMEC cells to exogenous stress, leading to a p38-dependent arrest. Acentrosomal RPE1 cells are more sensitive to blue light–induced stress than control-amputated cells, and both are substantially more sensitive than the untouched controls in the same fields. Neither the loss of the centrosome nor short exposures to blue light acting singly is sufficient to cause a G1 arrest, but they can work additively to tip the balance toward such an arrest. Furthermore, the use of two intensities of blue light to observe HMEC cells after laser ablation of the centrosome provides additional evidence that partial or complete centrosome removal makes G1 progression more sensitive to exogenous stress, in this case blue light. Together, our results provide assurance that the phenomena we observe are not dependent on the means used to remove the centrosome or the specific cell type. We used blue light as the exogenous stressor, but recognize that a wide variety of other suboptimal conditions could also act in concert with centrosome loss or damage to cause a G1 arrest. Indeed, the recent finding that siRNA depletions of the centrosome-associated proteins PCM1 or pericentrin lead to a p38-dependent G1 arrest of only ∼50% of the cells (Srsen et al., 2006) led these authors to propose that the arrest is not a specific centrosome-dependent cell cycle control, but rather, a stress-driven response to centrosome defects.
The two possible ways in which centrosome damage or loss could be sensed by the cell are clearly not mutually exclusive, and, mechanistically, they could share common pathways leading to the activation of p38. Conceptually, the essential difference between these two proposals is that, in the first case, a centrosomal remnant must be present to signal centrosome damage/dysfunction, and in the second case, compromising centrosomal function through centrosome damage or removal is, by itself, a stress that can act additively with other stresses to trigger a p38-mediated G1 arrest. It would not be surprising if both possibilities are factors for the cell. In any case, although the centrosome is not needed for G1 progression in its entirety, the status of the centrosome is not irrelevant for G1 cell cycle control either.
Materials and methods
Cell culture and drug treatment
Centrin-1/GFP in pEGFP-N1 vector (Piel et al., 2000) was transfected into telomerase-immortalized normal human cells (hTERT-RPE1; CLONTECH Laboratories, Inc.), and stable clones were isolated via G418 selection and limited-dilution cloning. Cells were cultured as described in Uetake and Sluder (2004). HMEC clone 184A1 (wild-type for p53 and pRb) were obtained from M. Stampfer (Lawrence Berkeley National Laboratory, Berkeley, CA). These cells were transfected with centrin-1/GFP in LentiLox3.7 vector as directed (Stampfer and Bartley, 1984). HMEC cells were grown according to protocols available at http://www.lbl.gov/∼mrgs/other/types.html. BSC-1 cells were obtained from the American Type Culture Collection. The cells were cultured in MEM containing 12.5 μM Hepes, 10% fetal calf serum (Invitrogen), 100 U penicillin (Invitrogen), and 100 μg streptomycin (Invitrogen). BrdU (Sigma-Aldrich) was added to a final concentration of 5 μg/ml immediately after centrosome removal from G1 cells or just after first mitosis after centrosome removal from G2 cells. The incorporation of BrdU was determined as previously described (Uetake and Sluder, 2004). 1 mM mimosine (Calbiochem) was prepared from a 10-mM stock solution in culture media (Krude, 1999; Wang et al., 2000). SB 203580 (10 μM [Sigma-Aldrich] for RPE1 and 20 μM [Calbiochem] for HMEC) was prepared by dilution of a 10-mM DMSO stock into culture media.
Glass-needle microsurgery and long-term imaging
Glass-needle microsurgery and phase-contrast time-lapse recording were conducted as described in Hinchcliffe et al. (2001) and La Terra et al. (2005). In brief, coverslips bearing cells were assembled into open-faced micromanipulation chambers filled with culturing media kept in the CO2 incubator for several hours to equilibrate the pH, and capped with mineral oil (Sigma-Aldrich). Microsurgery was performed at 37°C with a custom-built piezoelectric micromanipulator on an ACM microscope (Carl Zeiss MicroImaging, Inc.) equipped with phase-contrast optics and epifluorescence. First, the position of centrin-1/GFP dots was confirmed by a few exposures to 488-nm blue light. The cell was cut between the nucleus and the centrosome with a glass microneedle under phase-contrast optics. 15–30 min after the operation, the removal of centrosome was confirmed by, at most, a 1-s exposure to 488-nm blue light. After the microsurgery, the acentrosomal cell was circled with a diamond scribe, and the coverslip was removed from the manipulation chamber and assembled into a closed chamber (Sluder et al., 2005) with buffered culture media containing BrdU, and then followed at 37°C with Universal (16×, NA 0.32 objective; Carl Zeiss MicroImaging, Inc.) or BH-2 (10×, NA 0.3 objective; Olympus) microscopes equipped with phase-contrast optics. Images were recorded with Orca ER (Hamamatsu), Orca 100 (Hamamatsu), 1300 (Retiga), 2000R (Retiga), or EXi (QImaging Corp.) cameras. Image sequences were written to the hard drives of PC computers using C-imaging software (Compix, Inc.) and were exported as .avi movies. Centrin-1/GFP foci in the acentrosomal cells were characterized by z series (20 optical planes separated by 0.2 μm or 8 optical planes separated by 0.5 μm) fluorescent imaging with a microscope (DMR; Leica; 63×, NA 1.32 objective or 100×, NA 1.30). Maximal intensity projections were compiled with SlideBook software (Intelligent Imaging Innovations, Inc.). The light intensities at each wavelength were measured as total output at the stage level with a LaserMate-Q laser power meter (Coherent, Inc.).
Laser microsurgery and long-term imaging
Laser microsurgery was conducted on a custom-assembled microscopy workstation centered on a microscope (TE2000-E2; Nikon). 532-nm, 8-ns laser pulses were generated by a Q-switched Nd:YAG laser (Diva II; Thales Lasers, Paris, France) run at 20-Hz repetition rate. Collimated laser beam was expanded to ∼8 mm to fill the aperture of a 100× 1.4 NA PlanApo lens and delivered through a dedicated epi-port. It takes ∼10 laser pulses (1 s) to destroy the centrosome. Fluorescence images were recorded with a Cascade512B back-illuminated EM-CCD camera (Photometrics) attached to the left microscope port (100% transmission) in confocal mode (spinning disk confocal; Perkin-Elmer). 3D datasets were taken at 0.25-μm z steps. All light sources were shuttered by either fast mechanical shutters (Vincent Associates) or AOTF (Solamere Technology Group) so that cells were exposed to blue light only during laser operations and/or image acquisition. The system was driven by IP Lab software (BD Biosciences). After laser ablation of the centrosome, the position of the experimental cell was marked with a diamond scribe and filmed as previously described (La Terra et al., 2005).
Electron microscopy
Fixation, embedding, and serial sectioning were performed according to established procedures (Rieder and Cassels, 1999). 100-nm sections were examined in a microscope (910;Carl Zeiss MicroImaging, Inc.) at 100-KV and photographed on film. Film negatives were subsequently scanned and contrast-adjusted in PhotoShop CS (Adobe).
Online supplemental material
Fig. S1 shows the progression of a RPE1 cell born without a centrosome through G1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200607073/DC1.
Acknowledgments
We thank Drs. Dannel McCollum, Anna Krzywicka-Racka, and Conly Rieder for useful discussions. We thank Ms. Polla Hergert for invaluable help with EM. We acknowledge the use of the Wadsworth Center EM Core Facility.
This work was supported by National Institutes of Health GM030758 to G. Sluder and GM59363 to A. Khodjakov.
Abbreviation used in this paper: HMEC, human mammary epithelial cell.
References
- Ambrosino, C., and A.R. Nebreda. 2001. Cell cycle regulation by p38 MAP kinases. Biol. Cell. 93:47–51. [DOI] [PubMed] [Google Scholar]
- Bohmer, R.M., E. Scharf, and R.K. Assoian. 1996. Cytoskeletal integrity is required throughout the mitogen stimulation phase of the cell cycle and mediates the anchorage-dependent expression of cyclin D1. Mol. Biol. Cell. 7:101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey, S., D. McCollum, and W. Theurkauf. 2005. a. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21:411–434. [DOI] [PubMed] [Google Scholar]
- Doxsey, S., W. Zimmerman, and K. Mikule. 2005. b. Centrosome control of the cell cycle. Trends Cell Biol. 15:303–311. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, A.G., and G. Sluder. 2005. Spindle pole fragmentation due to proteasome inhibition. J. Cell. Physiol. 204:808–818. [DOI] [PubMed] [Google Scholar]
- Giaccia, A.J., and M.B. Kastan. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973–2983. [DOI] [PubMed] [Google Scholar]
- Hansen, L.K., D.J. Mooney, J.P. Vacanti, and D.E. Ingber. 1994. Integrin binding and cell spreading on extracellular matrix act at different points in the cell cycle to promote hepatocyte growth. Mol. Biol. Cell. 5:967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.L., and A.J. Levine. 2005. The p53 pathway: positive and negative feedback loops. Oncogene. 24:2899–2908. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., F.J. Miller, M. Cham, A. Khodjakov, and G. Sluder. 2001. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 291:1547–1550. [DOI] [PubMed] [Google Scholar]
- Hulleman, E., and J. Boonstra. 2001. Regulation of G1 phase progression of growth factors and the extracellular matrix. CMLS. 58:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hut, H.M., H.H. Kampinga, and O.C. Sibon. 2005. Hsp70 protects mitotic cells against heat-induced centrosome damage and division abnormalities. Mol. Biol. Cell. 16:3776–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber, D.E. 2003. Tensegrity II: How structural networks influence cellular information processing networks. J. Cell Sci. 116:1397–1408. [DOI] [PubMed] [Google Scholar]
- Ingber, D.E., D. Prusty, Z. Sun, H. Betensky, and N. Wang. 1995. Cell shape, cytoskeletal mechanics, and cell cycle control in angiogenesis. J. Biomech. 28:1471–1484. [DOI] [PubMed] [Google Scholar]
- Jimenez, G.S., S.H. Khan, J.M. Stommel, and G.M. Wahl. 1999. p53 regulation by post-translational modification and nuclear retention in response to diverse stresses. Oncogene. 18:7656–7665. [DOI] [PubMed] [Google Scholar]
- Johnston, J.A., C.L. Ward, and R.R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and C.L. Rieder. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and C.L. Rieder. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., R.W. Cole, B.R. Oakley, and C.L. Rieder. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., C.L. Rieder, G. Sluder, G. Cassels, O. Sibon, and C.L. Wang. 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 158:1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude, T. 1999. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 247:148–159. [DOI] [PubMed] [Google Scholar]
- Kumar, S., M.S. Jiang, J.L. Adams, and J.C. Lee. 1999. Pyridinylimidazole compound SB 203580 inhibits the activity but not the activation of p38 mitogen-activated protein kinase. Biochem. Biophys. Res. Commun. 263:825–831. [DOI] [PubMed] [Google Scholar]
- Kuriyama, R., and G.G. Borisy. 1981. Microtubule-nucleating activity of centrosomes in Chinese hamster ovary cells is independant of the centriole cycle but coupled to the mitotic cycle. J. Cell Biol. 91:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra, S., C.N. English, P. Hergert, B.F. McEwen, G. Sluder, and A. Khodjakov. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohez, O.D., C. Reynaud, F. Borel, P.R. Andreassen, and R.L. Margolis. 2003. Arrest of mammalian fibroblasts in G1 in response to actin inhibition is dependent on retinoblastoma pocket proteins but not on p53. J. Cell Biol. 161:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis, A., and M. Schliwa. 1991. Microsugical removal of centrosomes blocks cell reproduction and centriole generation in BSC-1 cells. Cell. 67:495–504. [DOI] [PubMed] [Google Scholar]
- Massagué, J. 2004. G1 cell-cycle control and cancer. Nature. 432:298–306. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. 2001. Cell cycle. Centrioles at the checkpoint. Science. 291:1499–1502. [DOI] [PubMed] [Google Scholar]
- Piel, M., P. Meyer, A. Khodjakov, C.L. Rieder, and M. Bornens. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., and G. Cassels. 1999. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods Cell Biol. 61:297–315. [DOI] [PubMed] [Google Scholar]
- Sablina, A.A., P.M. Chumakov, A.J. Levine, and B.P. Kopnin. 2001. p53 activation in response to microtubule disruption is mediated by integrin-Erk signaling. Oncogene. 20:899–909. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell. 2:103–112. [DOI] [PubMed] [Google Scholar]
- Sluder, G. 2004. Centrosome duplication and its regulation in the higher animal cell. In Centrosomes in Development and Disease. E.A. Nigg, editor. Wiley-VCH, Weinheim. 167–189.
- Sluder, G. 2005. Two-way traffic: centrosomes and the cell cycle. Nat. Rev. Mol. Cell Biol. 6:743–748. [DOI] [PubMed] [Google Scholar]
- Sluder, G., J. Nordberg, F. Miller, and E. Hinchcliffe. 2005. A sealed preparation for long-term observations of cultured cells. In Live Cell Imaging: A Laboratory Manual. R.D. Goldman and D.L. Spector, editors. Cold Spring Harbor Laboratory Press, Cold Spring Habor. 345–349 pp.
- Stampfer, M.R., and J.C. Bartley. 1984. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc. Natl. Acad. Sci. USA. 82:2394–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, J.A., and J.R. McIntosh. 1975. Initiation and growth of microtubules from mitotic centers in lysed mammalian cells. J. Cell Biol. 67:744–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srsen, V., N. Gnadt, A. Dammermann, and A. Merdes. 2006. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 174:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake, Y., and G. Sluder. 2004. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint.” J. Cell Biol. 165:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., R. Miskimins, and W.K. Miskimins. 2000. Mimosine arrests cells in G1 by enhancing the levels of p27Kip1. Exp. Cell Res. 254:64–71. [DOI] [PubMed] [Google Scholar]
- Young, A., J.B. Dictenberg, A. Purohit, R. Tuft, and S.J. Doxsey. 2000. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol. Biol. Cell. 11:2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin, T., and J. Han. 2005. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 15:11–18. [DOI] [PubMed] [Google Scholar]
- zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2:342–350. [DOI] [PubMed] [Google Scholar]