Abstract
During male germ cell postmeiotic maturation, dramatic chromatin reorganization occurs, which is driven by completely unknown mechanisms. For the first time, we describe a specific reprogramming of mouse pericentric heterochromatin. Initiated when histones undergo global acetylation in early elongating spermatids, this process leads to the establishment of new DNA packaging structures organizing the pericentric regions in condensing spermatids. Five new histone variants were discovered, which are expressed in late spermiogenic cells. Two of them, which we named H2AL1 and H2AL2, specifically mark the pericentric regions in condensing spermatids and participate in the formation of new nucleoprotein structures. Moreover, our investigations also suggest that TH2B, an already identified testis-specific H2B variant of unknown function, could provide a platform for the structural transitions accompanying the incorporation of these new histone variants.
Introduction
During the postmeiotic maturation of male haploid germ cells, or spermiogenesis, the DNA is repackaged in a process involving a dramatic chromatin reorganization (Caron et al., 2005). At the onset of spermiogenesis, round spermatids inherit a nucleosome-based chromatin organization, which is progressively restructured while the genome undergoes condensation. A wave of global histone acetylation marks the initial steps of this process in elongating spermatids and precedes their replacement in condensing spermatids, first by transition proteins, TP1 and TP2, and then by protamines. The latter ensure tight DNA packaging by the establishment of multiple intraprotein cross-links (Oliva and Dixon, 1990; Lewis et al., 2003b).
This textbook vision of mammalian spermiogenesis is now challenged by findings suggesting that the DNA is actually not homogeneously packed within the spermatozoa (Rousseaux et al., 2005). Approximately 10–15% of histones are retained in the human sperm nucleus, heterogeneously distributed within the genome, with an enrichment in specific loci such as imprinted genes or genes expressed during early embryogenesis (Gardiner-Garden et al., 1998; Wykes and Krawetz, 2003). Moreover, a large diversity of somatic-type or testis-specific histone variants become associated with the DNA in germ cells (Lewis et al., 2003a; Govin et al., 2004; Kimmins and Sassone-Corsi, 2005), some of them presenting a heterogeneous distribution within the spermatids or mature sperm nucleus (Zalensky et al., 2002; Martianov et al., 2005). In addition, heterochromatin regions seem to maintain a distinct organization during spermiogenesis, as telomeres were shown to be enriched in somatic-type core histones and H2B variants (Gineitis et al., 2000; Zalensky et al., 2002; Wykes and Krawetz, 2003; Churikov et al., 2004), and centromeres maintain some of their somatic-specific marks, such as an enrichment in the histone H3 variant CENP-A in spermatozoa (Palmer et al., 1990).
These observations suggest that the genome undergoes a regional differentiation during mammalian spermiogenesis. The exact nature of this differential reorganization of the genome and the molecular mechanisms driving it are unknown. One possibility is that this process could initially be built from differential marks inherited from early germ cells. In fact, pericentric heterochromatin is characterized by a specific histone code including a K9 trimethylation of histone H3 (H3K9me3) and its association with nonhistone proteins including the HP1 family members (Maison and Almouzni, 2004). These large regions surrounding the centromeres are mainly composed of satellite repeats, named the major satellites in the mouse, generally assembled in clusters known as “chromocenters” (Guenatri et al., 2004). After the completion of meiosis, the pericentric heterochromatin still harbors somatic features (O'Carroll et al., 2000; Peters et al., 2001). However, in mouse round spermatids, it undergoes a very unique reorganization characterized by the assembly of all pericentric regions into a single large chromocenter. A very interesting and unanswered question is whether this distinct feature could be the first step of a specific reprogramming of pericentric heterochromatin in male haploid germ cells.
To investigate this issue, we undertook a step-by-step exploration of the chromocenter organization during mouse spermiogenesis. An increase in histone acetylation had previously been observed in early elongating spermatids (Hazzouri et al., 2000). A close inspection of histone modifications during spermatid elongation reveals that pericentric heterochromatin exhibits very unusual characteristics combining active and repressive histone marks. Moreover, at later stages of spermiogenesis, nucleosomal structures containing acetylated histones are retained on the major satellites when most histones have been removed elsewhere. Finally, the investigation of nucleoprotein structures organizing the genome in condensing spermatids has led to the identification of several new H2A and H2B histone variants. Two of them, named H2AL1 and H2AL2, were found in new DNA packaging structures, which specifically reorganize the major satellite DNA in condensed spermatids. Altogether, these data highlight specific processes activated after meiosis and establish a differential organization of pericentric heterochromatin during mouse spermiogenesis.
Results
Pericentric heterochromatin acetylation during postmeiotic reorganization of the male genome
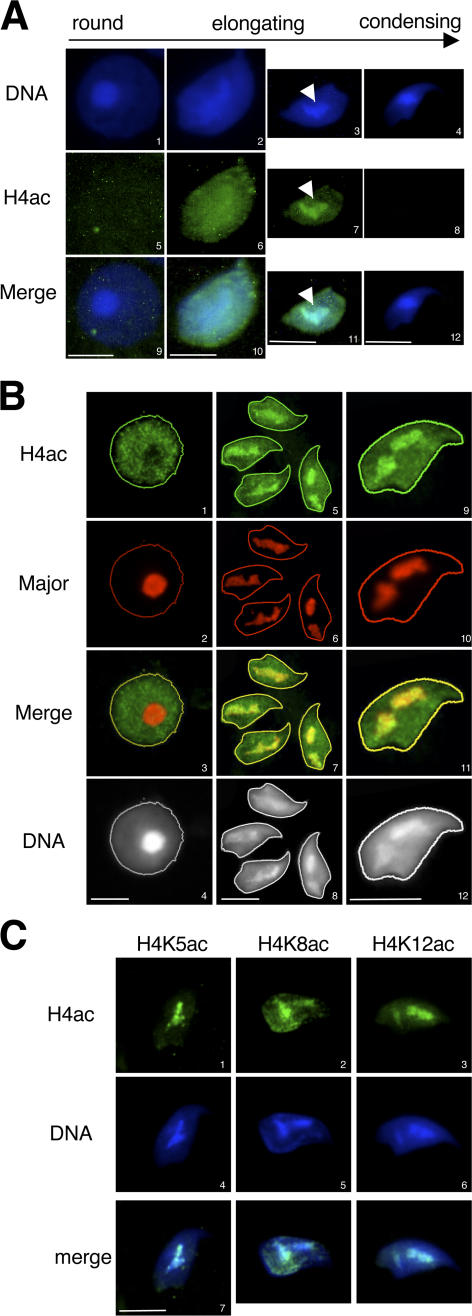
During postmeiotic chromatin reorganization in male germ cells, a global hyperacetylation of histones occurs, which precedes their removal (Hazzouri et al., 2000). To better characterize these events, the kinetics of the core histones' hyperacetylation and disappearance has been followed by immunofluorescence (IF) on microdissected squash preparations of seminiferous tubules, using an antibody recognizing the tetra-acetylated H4 N-terminal tail (H4ac). Round spermatids (steps 2–6) are weakly stained by the antibody (Fig. 1 A). In early elongating spermatids (step 8), H4 acetylation clearly increases and is homogeneously distributed throughout the whole nucleus. At later stages (steps 9–10), the signal for acetylated H4 globally decreases, except in a central domain, which is also intensely stained with DAPI (Fig. 1 A, arrows). Finally, in step 11 condensed spermatids, acetylated H4 has completely disappeared.
Figure 1.
Pericentric chromatin becomes acetylated in elongating spermatids. (A) H4 acetylation pattern in germ cells was analyzed by IF on spermatids at the indicated stages of maturation, using an anti–tetra-acetylated H4 antibody (H4ac). DNA was counterstained with DAPI. Arrows indicate the redistribution of the acetylated signal in an intensely DAPI-stained region in elongating spermatids. (B) Immuno-FISH assays, detecting H4 acetylation (H4ac) by IF and major satellites by FISH, were performed on round (1–4) and elongating (5–12) spermatids. Acquisition of the H4 acetylation staining has been enhanced to increase the signal intensity, compared with A, to show the hypoacetylation state of pericentric heterochromatin in round spermatids. (C) The acetylation of lysines 5, 8, or 12 of histone H4 (H4K5ac, H4K8ac, and H4K12ac, respectively) were analyzed in elongating spermatids by IF as in A using the corresponding antibodies. Bars, 5 μm.
In elongating step 9–10 spermatids, this restricted central area of the nucleus, where acetylated H4 remains, could correspond to a region where the genome is differentially reorganized. Because of its intense DAPI staining, we hypothesized that this domain could correspond to the A/T-rich pericentric constitutive heterochromatin, composed of the mouse major satellites repeats. To investigate this point, immuno-FISH assays were performed, where acetylated H4 was first detected by IF and the major satellites were then localized by FISH on the same germ cells. As expected, the major satellites were detected in the identifiable chromocenter of round spermatids (Fig. 1 B, 2). The DAPI-dense regions of step 9–10 elongating spermatids were also completely stained by the major satellites probes (Fig. 1 B, 6 and 10), confirming that they indeed correspond to pericentric heterochromatin. A close analysis of pericentric heterochromatin shows that it undergoes important changes during spermatid maturation. In round spermatids, it is underacetylated and colocalizes with the round chromocenter. In elongating spermatids, it becomes enriched in acetylated histone H4 while undergoing decompaction and spreading within the nucleus (Fig. 1 B, compare 1 to 5 and 9; note that the acquisition of the H4 acetylation signal in round spermatids was enhanced compared with that in Fig. 1 A to give better evidence of the unacetylated state of pericentric heterochromatin in round spermatids compared with the hypoacetylated state of the rest of the genome).
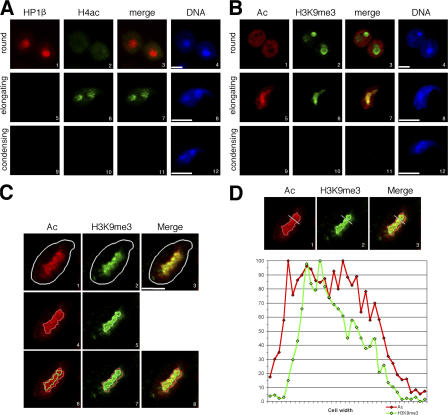
Figure 2.
A restricted domain of pericentric chromatin harbors both euchromatic and heterochromatic marks in elongating spermatids. (A) HP1β (red) and tetra-acetylated H4 (H4ac; green) were analyzed by IF in round (1–4), elongating (5–8), and condensing (9–12) spermatids. Merge corresponds to HP1β and H4ac codetection. Bars, 5 μm. (B) Acetylation of histones (Ac; red) and trimethylation of H3K9 (H3K9me3; green) were analyzed by IF in round (1–4), elongating (5–8), and condensing (9–12) spermatids. Merge corresponds to H4ac and H3K9me3 codetection. Bars, 5 μm. (C and D) Intensity of histone Ac (red; Alexa 546) and H3K9me3 (green; Alexa 488) fluorescence signals were quantified on step 9 spermatids using an analysis software (MetaMorph). For each detection, a region containing values >50% of maximal fluorescence was delimited (C, 4–8; red and green borders for Ac and H3K9me3 signals, respectively), and quantification performed along the gray axis (D, 1–3) was reported along a diagram. A representative picture of the analysis of five different cells shows that the acetylated domain and the H3K9me3 do not perfectly overlap. Bar, 2.5 μm.
We further aimed to identify the H4 lysines targeted by acetylation within pericentric heterochromatin. We had previously observed that the global acetylation increase in elongating spermatids mainly affects K5, K8, and K12 residues of histone H4, but not K16 (unpublished data). Here, IF with specific antibodies shows that all three acetylated lysines—acK5, acK8, and acK12—are associated with the major satellite region in elongating spermatids (Fig. 1 C).
A new combination of histone marks in pericentric heterochromatin of elongating spermatids
This unusual accumulation of acetylated histones in spermatids' pericentric heterochromatin led us to investigate the fate of known heterochromatin marks, such as HP1 binding and H3K9 trimethylation in these cells. HP1β was present in the chromocenter of round spermatids but disappeared at later stages, when H4 acetylation accumulated (Fig. 2 A), showing a tight relationship between the presence of acetylated H4 in pericentric heterochromatin and the removal of HP1β at the beginning of the elongation process.
In contrast, trimethylation of H3K9, easily detectable in the round spermatids' chromocenter, does not disappear in elongating spermatids when H4 acetylation takes place (Fig. 2 B). Interestingly, during a short period of their developmental stage, corresponding to step 9 spermatids, both marks were localized in pericentric heterochromatin of all cells. A detailed analysis of both modifications in these cells was performed by confocal microscopy. The intensity of H4Ac and H3K9me3 signals, revealed by Alexa 546 and Alexa 488 fluorochromes, respectively, were quantified: for each detection, a region containing values >50% of maximal fluorescence was delimited (Fig. 2 C, red and green borders, panels 4–8), and a quantification of fluorescence was shown along an axis and reported on a diagram (Fig. 2 D). Similar experiments were performed by reversing the fluorochromes (H4Ac signal in green and H3K9me3 signal in red), so that artifacts due to differential bleaching or sensitivity could be ruled out (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200604141/DC1). It shows that the signals for H4 acetylation and H3K9 trimethylation actually do not strictly colocalize (Fig. 2 D and Fig. S1 B, diagrams). This observation suggests the existence within the analyzed domain of subregions where nucleosomes harbor either one or the other modification, but probably not both simultaneously. This partial colocalization of the two histone marks might reflect a dynamic process reorganizing pericentric heterochromatin at this specific stage of maturation (step 9 spermatids), with a progressive invasion of the major satellite region by H4 acetylation.
Altogether, these data show that during early spermiogenesis, pericentric heterochromatin undergoes a transition from a somatic-like epigenetic state (which includes histone hypoacetylation, H3K9 trimethylation, and HP1 binding) into a completely new state, with persistent H3K9me3 but an absence of HP1 and an unusual accumulation of H4 acetylation. These observations clearly indicate a distinct behavior of pericentric heterochromatin during postmeiotic chromatin reorganization in male germ cells.
Nucleosomes and new DNA packaging structures organize pericentric heterochromatin during late spermiogenesis
We further aimed to determine whether pericentric regions would also harbor specific chromatin features at later stages of spermiogenesis, after disappearance of acetylated H4 and trimethylated H3K9 in condensing spermatids (Fig. 2, A and B, 10). Indeed, at this stage, either the modifications could be removed, leaving the histones in place, or the modified histones themselves could be displaced and degraded. To investigate the possibility of a nucleosomal structure remaining in the pericentric heterochromatin of condensing spermatids, we chose a biochemical approach. Step 12–16 condensing spermatids were purified from mouse testes. Their extreme compaction renders their nuclei completely resistant to MNase digestion by the classical assays developed for somatic cells. Therefore, we first had to partially decondense them in a detergent-containing buffer, to obtain a soluble nuclear extract, and a chromatin-containing pellet, which was then submitted to MNase fractionation (Fig. S2 A, protocol 1, available at http://www.jcb.org/cgi/content/full/jcb.200604141/DC1). The MNase-solubilized fraction was then separated by centrifugation from an insoluble pellet.
The analysis of the DNA present in the MNase-solubilized fraction shows that a prolonged digestion by MNase produces a single MNase-resistant DNA fragment of the expected nucleosomal size, ∼150 bp, indicating that at least part of the nucleosomal chromatin of condensing spermatids was indeed obtained with this procedure (Fig. 3 B). Accordingly, core histones were abundantly released by MNase digestion (Fig. S2 B, H2A, H2B, and H4).
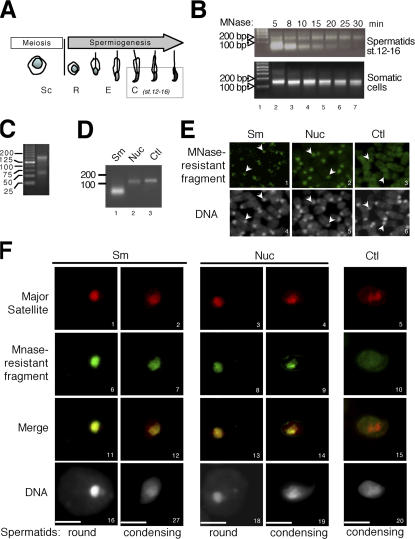
Figure 3.
Two MNase-resistant structures are present in condensing spermatids and mainly correspond to pericentric heterochromatin. (A) Schematic representation of germ cells at different stages of spermatogenesis (Sc, spermatocytes; R, round spermatids; E, elongating spematids; C, condensing and condensed spermatids [steps 12–16]) used in MNase-digestion assays. (B) Chromatin of purified step 12–16 spermatids (top) or somatic cells (bottom) were submitted to MNase digestion during the indicated times. The DNA fragments present in MNase-resistant nucleoprotein structures were purified and analyzed by electrophoresis on a 2% agarose gel. Lane 1 corresponds to the size markers. (C) MNase-resistant DNA fragments obtained as in B, after 15 min of MNase digestion, were analyzed on a 4% agarose gel, with small size markers. (D) The small DNA fragment (Sm) and the nucleosomal DNA fragment (Nuc) obtained after 8 and 30 min, respectively, of MNase digestion of condensing spermatids chromatin were purified to be used as probes for FISH. The control probe (Ctl) corresponds to the genome of whole testis cells entirely digested into mononucleosomes by a prolonged action of MNase. (E and F) FISH using probes described in C (green) were performed on mouse metaphase chromosomes (E) and on male germ cells in codetection with a major satellite probe (F, red). DNA was counterstained with DAPI. Arrows in E indicate centric and pericentric chromatin domains. Bars, 5 μm.
Surprisingly, this procedure also generated an additional DNA band that migrated faster than the nucleosomal DNA fragment and that was more sensitive to MNase digestion (Fig. 3 B). An electrophoretic analysis on a concentrated agarose gel with small size DNA markers confirmed that this band essentially corresponds to a discrete DNA fragment of ∼60 bp (Fig. 3 C). It is of note that this smaller DNA fragment was not produced from somatic cells (Fig. 3 B, bottom) or from germ cells at earlier stages of spermiogenesis (not depicted), after MNase treatment in the same conditions. We concluded, therefore, that this fragment could correspond to a condensing spermatid-specific DNA packaging structure.
We then further investigated the nature of the DNA associated with the nucleosomal and the new spermatid-specific DNA packaging structures. For this purpose, the two types of MNase-resistant DNA fragments were purified and used as probes in FISH assays, on mouse metaphase chromosomes, and on mouse spermatogenic cells. Interestingly, both fragments detected the pericentric regions and perfectly colocalized with the major satellite regions both on metaphase chromosomes and in spermatogenic cells (Fig. 3, E and F; and Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200604141/DC1). As controls, probes obtained either by sonication of spermatid total DNA or after MNase digestion of chromatin of mouse somatic cells using the stringent conditions of protocol 1 or of a whole testis cell suspension did not label any specific domain (Fig. 3, E and F; and Fig. S4 A). The quantification of the fluorescence signals given by each probe show that ∼72% of the nucleosomal fragment and 51% of the small fragment correspond to major satellite sequences (Fig. S4 B). These results indicate that in condensing spermatids, nucleosomes remain in pericentric heterochromatin regions, where they coexist with another MNase-resistant structure of an unknown nature. We therefore hypothesized that these new DNA packaging structures may contain condensing spermatid-specific proteins, which remained to be identified.
Identification of three new histone variants accumulating during late spermiogenesis
To find new proteins involved in the organization of the genome in late spermatids, a proteomic approach was undertaken. Basic proteins were extracted with acid from nuclei of purified step 12–16 spermatids and compared with basic proteins of germ cells at earlier maturation stages on SDS-PAGE. Coomassie staining revealed global changes in the basic protein constitution of condensing spermatids compared with that of germ cells at earlier stages, with a decrease in histone content, and the appearance of two bands ∼8 and 18 kD, corresponding to TP1 and TP2, respectively (Fig. 4 A). The proteins present in this particular cell fraction were identified by mass spectrometry (MS). As expected, transition proteins TP1 and TP2 and four canonical core histones were found, as well as known somatic and testis-specific histone variants. This proteomic analysis also revealed for the first time the presence of the mouse testis histone variant H3t (available from GenBank/EMBL/DDBJ under accession no. XP_356549). The mRNA encoding the human orthologue was previously shown to be expressed in spermatocytes and spermatids (Govin et al., 2005).
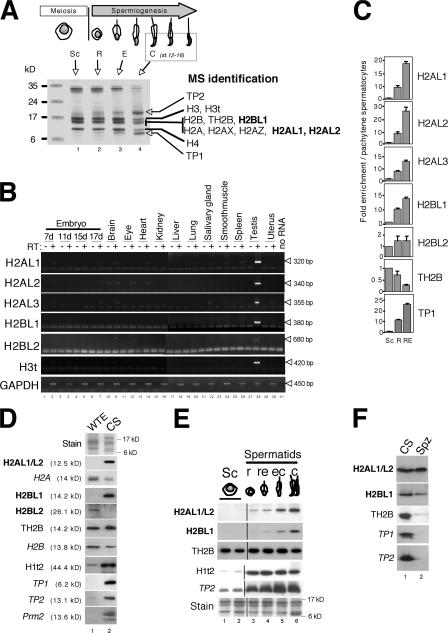
Figure 4.
Identification of three new testis-specific histone variants specifically expressed during late spermiogenesis. (A) Acidosoluble extracts were prepared from male germ cells purified according to their maturation stages (Sc, spermatocytes; R, or round; E, elongating and condensing; C, condensing and condensed spermatids) and separated on 15% SDS-PAGE stained with Coomassie blue. Proteins present in step 12–16 spermatid extracts (lane 4) have been excised from the gel and identified by MS. (B) PCR using specific primers of the newly identified H2AL1, H2AL2, H2AL3, H2BL1, and H2BL2, as well as H3t and GAPDH as controls, were performed on reverse-transcription products (+) obtained from total RNA of the indicated mouse tissues, in comparison to negative controls without (−) the reverse transcriptase (RT). (C) Reverse transcription was performed on total RNA extracted from either whole testis or fractions enriched in germ cells at different maturation stages. Enrichment of the indicated cDNA in each fraction compared with the spermatocytes was evaluated by qPCR using the appropriate primers. The mean value of at least two independent assays is presented. Error bars indicate mean ± SD. (D, E, and F) Western blots using the indicated antibodies were performed on various testis or germ cell extracts. Coomassie staining of the extracts is also shown (Stain panels). (D) Whole testis extract (WTE) and step 12–16 spermatid nuclei. CS, condensing/condensed spermatids. Note that after a longer exposure, the anti-H2AL1/L2, -H2BL1, -Prm2, -TP1, and -TP2 blots all showed a weak signal in the whole testis extract (not depicted). (E) Germ cells fractionated according to their maturation stages by sedimentation on BSA gradient. (F) Step 12–16 spermatid nuclei (CS) and mature epididymal sperm heads (Spz).
More important, three new proteins containing substantial sequence similarity with the H2A or H2B histone folds were also identified. These new histone variants were named H2Alike1 (H2AL1; available from GenBank/EMBL/DDBJ under accession no. AAH87913), H2Alike2 (H2AL2; accession no. NP_080903), and H2Blike1 (H2BL1; accession no. XP_127485), respectively. H2AL1 and H2AL2 are related proteins and show a better homology to each other than to H2A (Table I). H2BL1 is very similar to the bovine testis–specific H2B variant, known as SubH2Bv (Aul and Oko, 2001). In addition to the MS analysis, an in silico search in testis ESTs was performed and led to the identification of two supplementary H2A and H2B variants named H2AL3 (accession no. NP_080372) and H2BL2 (accession no. BAB24353), respectively (Table I).
Table I.
The new identified histone variants
| Name | Accession no. | Length (aa) | % Similarities (% Identities) | Sequence |
|---|---|---|---|---|
| H2AL1 | AAH87913 | 105 | 65 (30) with H2A; 83 (75) with H2AL2 | MAKKMQRRRRQKRTRSQRGELPFSLVDRFLREEFHSSRLSSSALSFLTSVLEYLTSNIL ELAGEVAQTTGRKRIAPEDVRLVVQNNEQLRQLFKPGGTSVNEDDN |
| H2AL2 | NP_080903 | 111 | 75 (41) with H2A; 83 (75) with H2AL1 | MARKRQRRRRRKVTRSQRAELQFPVSRVDRFLREGNYSRRLSSSAPVFLAGVLEYLT SNILELAGEVAHTTGRKRIAPEHVCRVVQNNEQLHQLFKQGGTSVFEPPEPDDN |
| H2AL3 | NP_080372 | 117 | 60 (24) with H2A | MSEKKSQEKPCSDNNQIEDPSSRPEVQVPVNYVYRILQEEQYTPCIGSTTSDFLL AMLDYLTDYILEVVGSEANINNQQNISQDRERQRDNDREPSRGFKNAPFSLFD EMPGPRRNG |
| H2BL1 | XP_127485 | 123 | 72 (38) with H2B; 82 (58) with SubH2Bv | MAKPTFKRQCYIKRHLRPLYRKHSRCSSINLGHGNYSLYINRVLKEVVPNRGIS SYSVDIMNILINDIFERIATEACQQMFLRKRCTLTPGDIQQAVHLLLPKKLATL AVTFGSKAVHRFIHS |
| H2BL2 | BAB24353 | 224 | 64 (31) with H2B | MASTTAMDVLEELSSDSSEKQVQPRKPEKAKREKDKPKKGGPEKKAKKEKQEK AKPEKKPKKKPEKEKPEGEKLEKKPKKDKREKAKPKKKPEQENREQETPEQEKPE VQRRRSLHQSIREDERRARLIRRRKNSFAIYFPKVLKNIHVGLSLSQRSVNILDSFVK DMFERIASEASFLARQARNSTINSREIQTAIRLLLPGELCRRAVAEGTMAMVRYISNK |
MS peptides are indicated by underlining.
The expression pattern of the genes encoding the five new histone variants was analyzed by RT-PCR on RNAs from various mice tissues. Fig. 4 B shows that, like the H3t variant, all the new histone variants are mainly expressed in the testis. To test whether all these new variants could be postmeiotically expressed, RT-qPCR (quantitative PCR) was performed on male germ cells fractionated according to their maturation stages on a BSA gradient. Fig. 4 C shows that H2AL1, H2AL2, H2AL3, and H2BL1 mRNA were strongly enriched in round and elongating spermatids (compare to pachytene spermatocytes, used as a reference), in contrast to TH2B used as a control, when mRNA was most abundant in meiotic cells. H2BL2 mRNA was detected at a very low level in meiotic, as well as postmeiotic, germ cells.
To study the accumulation of the encoded proteins during spermatogenesis, antibodies against each of the new histone variants were generated, and their specificity was checked on cells transfected with the corresponding expression vectors (unpublished data). All were highly specific in Western blots, except H2AL1 and H2AL2, which could not be distinguished from one another by their respective antibodies. They will, therefore, be referred as H2AL1/L2 hereafter.
H2BL2, although present in whole testis extracts, was not detected in germ cells (Fig. 4 D and not depicted), and H2AL3 was not detected either in whole testis extracts or in germ cells (not depicted). These data suggest that these in silico identified histones probably do not play a role in the postmeiotic chromatin reorganization of male germ cells. In contrast, H2AL1/L2 and H2BL1 are strongly enriched in step 12–16 spermatids compared with the whole testis (Fig. 4 D) and accumulate during late spermiogenesis, in condensing spermatids (Fig. 4 E). Their expression pattern is therefore different from that of TH2B, detected in meiotic cells and afterward, but similar to that of TP1, TP2, Protamine2, and H1t2, suggesting that H2AL1/L2 and H2BL1 could, like these proteins, be involved in chromatin organization during spermatid condensation. Finally, and very interestingly, H2AL1/L2 and H2BL1 remain present in mature spermatozoa isolated from epididymis, whereas all of the other known spermatid proteins, such as TP1, TP2, and TH2B, disappear (Fig. 4 F).
H2AL1/L2 and H2BL1 therefore behave differently from the other histones and histone variants present in germ cells, most of which are expressed earlier during spermatogenesis and removed in condensing spermatids, just as these new variants are produced. This pattern of expression pointed to H2AL1/L2 and H2BL1 as good candidates for specific genome reorganizers in condensing spermatids.
H2AL1/L2 are present in the new nucleoprotein structures specifically organizing pericentric heterochromatin
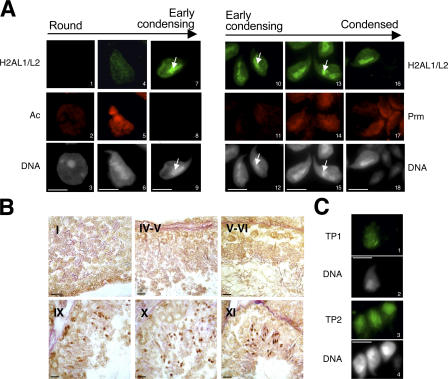
The subnuclear distribution of H2AL1/L2 in germ cells was further analyzed by IF on testis imprints (Fig. 5). These proteins first appear in step 9 elongating spermatids and strongly accumulate in early condensing spermatids (step 11) after the disappearance of the acetylation signal and before and during protamine incorporation (Fig. 5 A). These observations are in good agreement with the Western blot data (Fig. 4 E) and with immunohistochemistry (IH) data showing H2AL1/L2 accumulation in step 9–11 spermatids (Fig. 5 B).
Figure 5.
H2AL1/L2 are localized on pericentric heterochromatin in early condensing spermatids. (A) H2AL1/L2 were analyzed by IF on round (1–3), elongating (4–6), early condensing (7–12), and condensing and condensed (13–18) spermatids in codetection with acetylated histones (Ac; 1–9) or protamine (Prm; 10–18). The DNA panels correspond to the DAPI counterstaining. Arrows indicate the distribution of H2AL1/L2 on densely DAPI-stained regions. Bars, 5 μm. (B) H2AL1/L2 were analyzed by IH on testis sections. The tubule sections were staged according to the association of the corresponding germinal cells (Russell et al., 1990). The stage of each section is indicated with Roman numbers. The antibodies stained the spermatids found in stages IX, X, and XI tubules, respectively (corresponding to step 9, 10, and 11 spermatids). Bars, 20 μm. (C) TP1 or TP2 were detected by IF on early condensing spermatids. DNA panels correspond to the DAPI counterstaining. Bars, 5 μm.
Interestingly, H2AL1/L2, in contrast to TP1 and TP2, show a heterogeneous distribution in early condensing spermatids, with a preferential localization in the intensely DAPI-stained region, previously identified as pericentric heterochromatin (Fig. 5, A and C). These data highly suggest that during late spermiogenesis, H2AL1/L2 could differentially organize pericentric heterochromatin after the disappearance of acetylated nucleosomes. However, although clearly detectable in Western blots, H2AL1/L2 could not be observed by IF or IH in both step 12–16 spermatids and mature spermatozoa, probably because of the high genome compaction of these cells. Moreover, H2BL1 could not be detected in germ cells by these in situ approaches, probably also because of limited antibody accessibility.
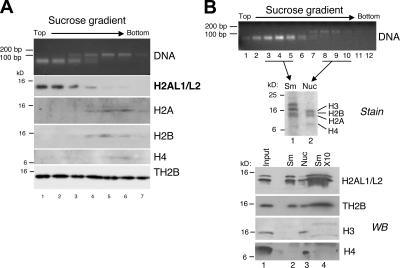
Because H2AL1/L2 were localized to pericentric heterochromatin, we checked for their presence in the major satellite-organizing nucleoprotein structures previously isolated from step 12–16 spermatids. In preliminary analyses, we observed that H2AL1/L2, as well as somatic-type core histones and TH2B, were released in the MNase-digested fractions of condensing spermatids prepared according to protocol 1 (Fig. S2 B), suggesting an association of these histones with at least one of the two MNase-resistant nucleoprotein structures shown in Fig. 3 B. However, in contrast to the somatic core histones, a substantial amount of H2AL1/L2—and, to a lesser extent, TH2B—was also present in the soluble nuclear extract, suggesting that the decompacting buffer (Fig. S2 A, protocol 1) had partially disrupted the corresponding nucleoprotein structures (Fig. S2 B). We therefore set up another procedure using a less stringent decondensing buffer (Fig. S2 A, protocol 2), which prevented the partial disruption of histone variants (Fig. S2 C) but gave the same MNase-digestion pattern (Fig. S3 B and not depicted). To investigate the association of H2AL1/L2 within the nucleosomal or the smaller structure, the two structures (obtained using the less stringent protocol 2) were separated on a sucrose gradient, and the associated histones or histone variants were analyzed by Western blot (Fig. 6). As expected, the somatic-type histones H2A, H2B, H3, and H4 cofractionated with the larger fragment, confirming its nucleosomal nature (Fig. 6 A, lanes 4–7; and Fig. 6 B, stain [lane 2] and H3 and H4 [lane 3]). In contrast, the H2AL1/L2 variants specifically cofractionated with the small MNase-resistant DNA fragment, indicating that they could be major constituents of the new DNA packaging structure identified in condensing spermatids (Fig. 6 A, H2AL1/L2 [lanes 1–4]; and Fig. 6 B, V [lane 2]). Remarkably, neither H3 nor H4 could be detected in this structure, even when higher concentrations of proteins from these fractions were analyzed (Fig. 6 B, stain [lane 1] and H3 and H4 [lanes 2 and 4]). Interestingly, TH2B cofractionated with both DNA fragments, suggesting that, unlike H2B, it associates not only with nucleosomes but also with the new structures.
Figure 6.
In condensing spermatids, H2AL1/L2 are associated with the new nucleoprotein structure identified by MNase digestion. Nucleoprotein structures released from step 12–16 spermatids after MNase digestion were fractionated by ultracentrifugation on a sucrose gradient. DNA fragments and proteins of each collected fraction were analyzed, respectively, by electrophoresis on an agarose gel (DNA) and by Western blot using the appropriate antibodies. A and B correspond to two independent experiments. In B, fractions 3–5 (containing the small fragment [Sm]) and fractions 8–10 (containing the nucleosomal fragment [Nuc]) of the gradient were pooled for Western blots (bottom, lanes 2 and 3) and Coomassie staining (top, lanes 1 and 2) analyses, respectively. To confirm the absence of H3 and H4, the proteins from the small fragment fraction were also loaded in tenfold excess (Western blot [WB], lane 4).
To confirm the association between the DNA fragments and histones, further purifications of the nucleoprotein structures were performed using hydroxyapatite. This ion-exchange medium presents a high affinity for DNA, which allows the capture and purification by phosphate elution of nucleosomes or other nucleoprotein structures (Rickwood and MacGillivray, 1975). In these experiments, H2AL1/L2 not only were pulled down on hydroxyapatite but also perfectly coeluted with the small DNA fragment (Fig. S3, B and C), showing that the small DNA fragment and H2AL1/L2 are part of the same structure. Moreover, it was confirmed that TH2B, present within the nucleosomes, also associates with the new structure.
H2AL1/L2 specifically dimerize with TH2B and can form regular but unstable nucleosomes when expressed in somatic cells
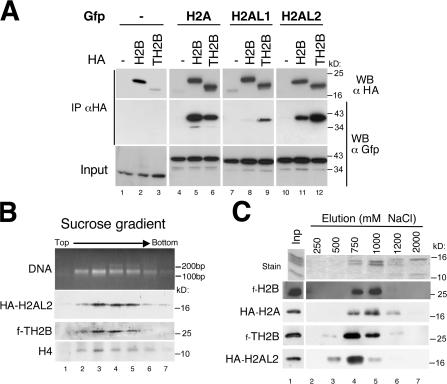
The association of TH2B and the novel histone variants within an unknown DNA packaging structure prompted us to investigate their ability to reorganize the genome when ectopically expressed in somatic cells. We first monitored the ability of H2AL1 and H2AL2 to dimerize with either H2B or TH2B. HA-tagged H2B and TH2B were coexpressed with H2A, H2AL1, and H2AL2 fused to GFP. H2B and TH2B were then immunoprecipitated using an anti-HA antibody, and the presence of H2A, H2AL1, and H2AL2 was detected by Western blot (Fig. 7). The data show that H2A is able to form dimers with TH2B but less efficiently than with H2B. In contrast, H2AL2 displayed a strong preference for dimerization with TH2B compared with H2B (Fig. 7 A). H2AL1 also dimerized more efficiently with TH2B than with H2B, albeit with a lower affinity. These findings nicely correlate with the results obtained after the analysis of the chromatin of condensing spermatids (Fig. 6), showing the presence of TH2B and the absence of H2B in the histone H2AL1/L2–containing structures.
Figure 7.
H2AL2–TH2B dimers can be assembled into nucleosomes when ectopically expressed in somatic cells. (A) Cos7 cells were cotransfected with vectors expressing GFP-fused H2A, H2AL1, H2AL2, or GFP alone as a control, and HA-tagged H2B or TH2B, or an empty vector (−). After immunoprecipitation (IP) performed on whole cell extracts using an anti-HA antibody, the immunoprecipitated proteins were denatured in Laemmli loading buffer and analyzed by Western blot (WB). A Western blot probed with an anti-HA antibody showed the efficiency of the immunoprecipitations (top). The coimmunoprecipitation of histones H2A, H2AL1, or H2AL2 was detected by Western blot with an anti-GFP antibody and compared with the input. (B) Oligonucleosomes were prepared by MNase digestion from Cos7 cells expressing HA-H2AL2 and Flag-TH2B and fractionated by ultracentrifugation on a sucrose gradient. DNA fragments and proteins of each collected fraction were analyzed, respectively, by electrophoresis on an agarose gel (DNA) and by Western blots using the indicated antibodies. (C) Oligonucleosomes prepared from Cos7 cells expressing HA-H2A/Flag-H2B or HA-H2AL2/Flag-TH2B were captured by hydroxyapatite. After elution by increasing NaCl concentrations, endogenous somatic histones were detected on SDS-PAGE by Coomassie staining (top) and the ectopically expressed H2A/H2B or H2AL2/TH2B by Western blots.
An important question is whether the H2AL1/L2–TH2B dimers are capable of forming regular nucleosomes and/or could induce the formation of a different type of structure in somatic cells. To investigate this issue, the chromatin from cells expressing H2AL2/TH2B was digested by MNase, and the resulting oligonucleosomes were fractionated on a sucrose gradient. Fig. 7 B shows that all the H2AL2 and TH2B were efficiently incorporated into regular nucleosomes, and no smaller DNA packaging structure was found. We then analyzed the stability of the H2AL2–TH2B–containing nucleosomes compared with nucleosomes containing only somatic histones. Oligonucleosomes from cells expressing tagged H2A–H2B or tagged H2AL2–TH2B were immobilized on hydroxyapatite and eluted with increasing concentrations of salt. The results show that H2AL2–TH2B dimers are released at lower salt concentrations than H2A–H2B dimers, suggesting that H2AL2–TH2B–containing nucleosomes are less stable than nucleosomes containing somatic histones (Fig. 7 C). Altogether, these data show that TH2B and H2AL2 preferentially associate with each other, are able to form nucleosomes when expressed in somatic cells, and are found in new nucleoprotein structures specifically organizing pericentric heterochromatin in condensing spermatids.
Discussion
This study shows for the first time that pericentric heterochromatin exhibits very specific features at all stages of mouse spermiogenesis. Upon completion of meiosis, round spermatid chromatin retains somatic-type epigenetic marks, which then undergo reprogramming at the beginning of the elongation phase. A wave of histone acetylation affecting the whole genome marks the initiation of the chromocenter decompaction and spreading. In elongating spermatids, pericentric heterochromatin acquires a novel and unique organization where it is both marked by H4ac and H3K9me3 and loss of HP1. The observed general histone hyperacetylation and the removal of HP1 from pericentric regions are reminiscent of the reported disappearance of HP1 from chromocenter after prolonged trichostatin A treatment in somatic cells (Taddei et al., 2001). It is therefore very probable that at the beginning of spermatid elongation, the global histone acetylation triggers the disruption of HP1 binding as well as the induction of pericentric heterochromatin reprogramming.
Interestingly, although HP1 is lost, the H3K9me3 mark remains when H4 acetylation invades the pericentric regions and could be involved in delaying the completion of histone acetylation within the major satellite region. Indeed, although both marks colocalize in pericentric heterochromatin, a more detailed analysis suggests that they are not present on the same subregions and, consequently, on the same nucleosomes. This implies the occurrence of an active removal of the H3K9me3 mark before or simultaneously with H4 acetylation. It could be either demethylated or widely exchanged with unmethylated histone H3 or H3 variants. Interestingly, the proteomic identification of the surviving histones isolated from condensing spermatids (steps 12–16) presented here showed the presence of the H3 variants H3.3 and H3t. Hence, the exchange of trimethylated H3K9 with H3.3 and H3t in the pericentric regions could also account for the progressive removal of the H3K9me3 mark.
Later in spermiogenesis, the surviving nucleosomal organization of the pericentric regions could provide a basis for the preferential recruitment of new H2A variants identified here in condensing spermatids. Interestingly, our data point to TH2B as a key player in the incorporation of H2AL1/L2. First, TH2B is expressed much earlier than H2AL1/L2 during spermatogenesis but is still present at late spermiogenesis stages, when H2AL1/L2 accumulate. Second, using an ectopic expression approach, we show that H2AL1 and H2AL2 largely prefer TH2B to H2B, as a dimerization partner. Moreover, unlike H2B, TH2B was found with H2AL1/L2 associated to the small MNase-resistant DNA fragments, strongly suggesting that H2AL1 or H2AL2 also dimerize with TH2B in spermatogenic cells. Third, TH2B possesses the ability to induce nucleosome instability when incorporated in vitro into nucleosomes containing somatic-type histones (Li et al., 2005), and H2AL2–TH2B–containing nucleosomes were found here less stable than those containing H2A–H2B. Altogether, these observations support the hypothesis that TH2B-containing nucleosomes would be preferential sites for H2AL1/L2 incorporation through direct dimerization. These unstable nucleosomes would then become targets for important structural reorganization in condensing spermatids, leading to the formation of the new structures evidenced here by MNase digestion. Several testis-specific factors could potentially play a role in such a process. For example, the bromodomain-containing factor, TIF1-∂, is expressed in elongating spermatids (steps 9–11) concomitantly with the apparition of H2AL1/L2 and preferentially localizes on pericentric heterochromatin (Khetchoumian et al., 2004).
Although H2AL1/L2–TH2B dimers were incorporated into the nucleosomes in somatic cells, neither H3 nor H4 could be found in the H2AL1/L2–TH2B–containing structure in condensed spermatids. These observations are reminiscent of previous studies showing that a MNase digestion of chromatin from somatic cells could also produce subnucleosomal particles, some of which are composed of H2A–H2B dimers associated to a DNA fragment of ∼50 bp (Nelson et al., 1977; Rill and Nelson, 1978; Nelson et al., 1982). Whether the H2AL1/L2–TH2B are the testis-specific counterparts of these somatic subnucleosomal particles or whether they associate with other sperm basic proteins to form completely new DNA packaging structures is to be addressed in the future.
In contrast to H2AL1/L2, the presence of TH2B is strongly reduced in epididymal spermatozoa (Fig. 4 F). However, longer Western blot exposures and MS analyses reveal that a small amount of TH2B remains (unpublished data), and an IF approach also suggested that hTSH2B (the human homologue of TH2B) could be detected in a subset of spermatozoa (Zalensky et al., 2002). The exact nature of the H2AL1/L2-containing structures in sperm thus remains to be established. The possibility that H2BL1, which is synthesized later, partly replaces TH2B in the very late stages of spermiogenesis is an attractive hypothesis requiring further investigation.
In the human genome, no H2A and H2B variants were found with substantial sequence similarity to the mouse H2AL1, H2AL2, and H2BL1. However, other testis-specific variants could act as functional homologues in human germ cells. For example, H2BFWT, a human testis–specific H2B variant present in mature sperm, is found uniquely in primates and, like H2BL1, shows a relatively low similarity with H2B and TH2B (Churikov et al., 2004). Interestingly, several human sperm H2B variants preferentially accumulate on telomeres (Gineitis et al., 2000; Zalensky et al., 2002; Churikov et al., 2004). Thus, although not conserved, specific histone variants could organize particular regions of the genome, like telomeres, centromeres, or pericentric heterochromatin, within the globally protamine-packaged genome.
This sperm-specific packaging of pericentric heterochromatin could be important for postfertilization chromatin reorganization events. Indeed, after fertilization, a genome-wide epigenetic reprogramming occurs and establishes the totipotency of the zygote from the differentially organized paternal and maternal genomes, inherited from the highly specialized male and female gametes. Interestingly, recent data report important differences between the onset of paternal and maternal pericentric heterochromatin epigenetic marks in the first steps of preimplantation embryogenesis in the mouse (Santos et al., 2005; Martin et al., 2006). Therefore, it is tempting to speculate that the presence of H2AL1/L2 in a novel organizational unit of major satellites in sperm would, after fertilization, act as a guide for epigenetic reprogramming of paternal pericentric heterochromatin. It is also likely that histone variants play a role in the establishment of other epigenetically inherited structures in the male genome. Indeed, in addition to a clear enrichment in pericentric heterochromatin, H2AL1/L2-containing structures are also dispersed elsewhere in the genome (Fig. S4 B). Characterization of these regions will constitute an additional, exciting challenge for the future, which should shed further light on both the nature and transmission of paternal epigenetic information.
Materials and methods
Antibodies
Antibodies against H2AL1, H2AL2, H3AL3, H2BL1, and H2BL2 were generated in rabbits by three injections of 200 μg of purified His-tagged recombinant proteins. Sera were diluted at 1:1,000 for Western blot and 1:250 for IF. Anti-H1t2 is described by Martianov et al. (2005). Anti-H3 was provided by Abcam. Anti-TP1 and anti-TP2, anti-protamine, and anti-acetylated antibodies were provided by S. Kistler (University of South Carolina, Columbia, SC), R. Balhorn (Lawrence Livermore National Laboratory, Livermore, CA), and M. Yoshida (RIKEN, Wako, Japan), respectively. Other anti-histones antibodies were provided by Upstate Biotechnology and used as advised by the supplier.
Purification of germ cells
Three different protocols were used, depending on the required quantity, purity degree, and maturation stage of male germ cells. Fractions enriched in spermatogenic cells at different stages of maturation were obtained by sedimentation on a BSA gradient as previously described (Pivot-Pajot et al., 2003). Pure fractions of nuclei from step 12–16 spermatids were obtained by sonication of mice testis as described by Marushige and Marushige (1983). Pure fractions of epididymal sperm heads were obtained as follows. Epididymes were opened with a razor blade to free spermatozoa in a DME drop. The spermatozoa were then pelleted by centrifugation (4°C, 1,300 g, 10 min), resuspended in 1.5 ml DME containing 1 mg/ml salmon sperm DNA, sonicated at 250 J to break flagella, and centrifuged (1,300 g, 10 min, 4°C) on a discontinuous Percoll gradient (100%/70%/40%). The pellet, containing pure sperm heads, was washed once in DME/1 M NaCl and then in DME. The quality of each fraction or preparation was controlled by observation under a phase-contrast microscope.
IF and IH on germ cells
Microdissected tubules were prepared as described by Kotaja et al. (2004). Testis imprint preparations were performed by gently pressing the testis (previously cut in two and frozen in liquid nitrogen) onto glass slides, air-drying, incubating in 90% ethanol for 3 min, and air-drying again. Permeabilization of cells was allowed in 0.5% saponine, 0.25% Triton X-100, and 1× PBS for 15 min at room temperature. Nonspecific binding was blocked with 5% dried milk, 0.2% Tween 20, and 1× PBS for 30 min at room temperature. Primary antibodies were diluted in 1% dried milk, 0.2% Tween 20, and 1× PBS (dilutions are indicated in Antibodies section). Incubations were performed overnight at 4°C in a humidified chamber. Slides were then washed three times for 5 min in the antibodies dilution buffer. Secondary antibodies (anti–rabbit cross-linked to Alexa 488 and/or anti–mouse cross-linked to Alexa 546 [Invitrogen]) were diluted at 1:500 in the same buffer as the primary antibodies and incubated for 30 min at 37°C. Washes were performed as for primary antibodies. DNA was counterstained by DAPI, and slides were mounted in MOWIOL. The protocol of IH experiments is described in detail by Faure et al. (2003).
FISH on germ cells
Slide preparation.
The germinal cells from one mouse testis were obtained by dilacerating the seminiferous tubules in ∼500 μl DME/HamF12 medium (1:1), washed by centrifugation at 1,000 rpm for 10 min, resuspended, incubated for 10 min in 1% sodium citrate at room temperature, and centrifuged again. The germinal cells of the pellet were carefully dissociated and fixed twice in methanol/acetic acid (3:1) for 10 min at room temperature and then spread onto dry, clean slides. The slides were air-dried and kept at room temperature for up to 1 wk, until FISH was done. For the immuno-FISH experiments, IF was performed as usual, the positions of the acquired IF images were recorded, and the slides were washed in 2× SSC at 37°C for 30 min, dehydrated by immersing in a series of ethanol (70%/90%/100%), and air-dried.
FISH procedure.
The slides were denatured in 70% formamide/2× SSC for 12 min at 82°C (germinal cell preparation) or 1 min at 70°C (metaphase), dehydrated by passage through a cold ethanol series and air-dried (20× SSC: 175.3 g/l NaCl and 88.2 g/l sodium citrate in water, pH adjusted to 7 with NaOH). The DNA probes were labeled with either Biotin 11 dUTP or Digoxigenin dUTP by Nick Translation kit (Roche). A 10-μl sample of hybridization mix (50% formamide, 20% dextran sulfate, 1× SSC, and 1× SSPE; 20× SSPE: 174 g/l NaCl, 27.6 g/l NaH2PO4H2O, and 7.2 g/l EDTA, pH adjusted to 7.4), containing 50–100 ng of each of the labeled probes, 10 μg sonicated salmon sperm DNA, and, when needed, 5–10 μg mouse cot DNA, was heated at 72°C for 10 min, preincubated at 37°C for 30 min, and applied to each slide. The preparations were then placed in the dark, under sealed coverslips, for hybridization during 24–48 h in a moist chamber at 37°C. The coverslips were then carefully removed, and the slides were washed in 2× SSC for 2 min at 70°C and preincubated for 15 min in phosphate-buffered detergent (PBD; Qbiogene) at room temperature. The digoxigenin-labeled and biotin-labeled probes were detected, respectively, by a 15-min incubation at 37°C with anti–digoxigenin-rhodamine (1:100; Boehringer) or streptavidin–Alexa 488 (1:200; Invitrogen), diluted in PBD, and washed three times for 5 min in PBD at room temperature. The preparations were finally counterstained with 250 ng/ml DAPI in Vectashield (Vector Laboratories).
The quantification of the enrichment of each probe in major satellite sequences was performed on FISH slides as follows. The quantification was performed on mouse metaphases hybridized with the small fragment, the nucleosomal fragment, or control probes in codetection with major satellites. For each probe, the fluorescence signals on each whole chromosome and on its corresponding pericentric region (i.e., the area defined by major satellite hybridization) were quantified using MetaMorph software (Molecular Devices). The “pericentric/whole chromosome” signal ratio for the major satellite probe was considered 100% enrichment. For each of the other probes, this ratio (pericentric/whole chromosome) was also determined and normalized to the value obtained for a major satellite probe.
Microscope image acquisitions
A microscope (Axiophot; Carl Zeiss MicroImaging, Inc.) coupled to a −40°C chilled charge-coupled device camera (Hamamatsu) was used for 2D image acquisitions. Images were acquired at room temperature with a 63× objective (1.4 NA). Confocal images were taken using a confocal laser scanning microscope (Carl Zeiss MicroImaging, Inc.) and quantified using MetaMorph. The images presented in the figures were processed using Photoshop (Adobe).
MNase digestion of chromatin from step 12–16 condensing spermatids
Step 12–16 spermatids obtained from 10 testes, or 107 NIH 3T3 cells used as control, were lysed (15 min, in ice) in 150 μl buffer 1 (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% DOC, and 0.1% SDS) or buffer 2 (50 mM Tris, pH 7.4, 300 mM NaCl, 0.1% NP-40, 0.1% DOC, 1 mM DTT, and antiprotease cocktail [Complete EDTA Free; Roche]). After centrifugation (20,000 g, 4°C, 10 min), the pellet was resuspended in 150 μl of the initial buffer. In the case of buffer 2, a short sonication (80 J) was performed to allow the suspension of chromatin fragments of ∼5,000 bp, and unlysed spermatids were eliminated by an additional centrifugation. MNase digestion was performed on the buffer 1 or 2 lysates by the addition of 150 μl MNase buffer (10 mM Tris, pH 7.5, 10 mM KCl, and 1 mM CaCl2) and 15 U of micrococcal nuclease S7 (Roche), and incubation at 37°C for the indicated times. Reaction was stopped by 5 mM EDTA (final concentration). MNase-digested fractions were separated by ultracentrifugation (80,000 g, 20°C, 7 h) on a 10–30% linear sucrose gradient (in 1 mM phosphate buffer, pH 7.4, 80 mM NaCl, 0.2 mM EDTA, and antiprotease cocktail). DNA analyses were performed on 10 μl of MNase-digested fractions by treatment with proteinase K followed by electrophoresis on a 2% agarose gel. Proteins of the collected fractions were analyzed Western blots.
MS and protein identification
After separation by SDS-PAGE, discrete bands were excised from the Coomassie blue–stained gel. The in-gel digestion was performed as previously described (Ferro et al., 2000). Gel pieces were then sequentially extracted with 5% (vol/vol) formic acid solution, 50% acetonitrile, 5% (vol/vol) formic acid, and acetonitrile. After drying, the tryptic peptides were resuspended in 0.5% aqueous trifluoroacetic acid. The samples were injected into a CapLC nanoLC system (Waters) and first preconcentrated on a 300 μm × 5 mm precolumn (PepMap C18; Dionex). The peptides were then eluted onto a C18 column (75 μm × 150 mm; Dionex). The chromatographic separation used a gradient from solution A (2% acetonitrile, 98% water, and 0.1% formic acid) to solution B (80% acetonitrile, 20% water, and 0.08% formic acid) for >35 or 60 min at a flow rate of 200 nl/min. The LC system was directly coupled to a mass spectrometer (QTOF Ultima; Waters). MS and MS/MS data were acquired and processed automatically using MassLynx 4.0 software. Database searching was performed using the MASCOT 2.1 program. Two databases were used: a homemade list of well-known contaminants (keratines and trypsin) and an updated compilation of SwissProt and Trembl databases with specifying Mus as the species. For searching the Mus database, the variable modifications allowed were as follows: acetyl-lysine, N-ter acetylation, dimethyl-lysine, methyl-lysine, protein N-acetylation, methionine oxidation, serine and threonine phosphorylation, methionine sulphone, and cysteic acid. Because of the potential high frequency of basic amino acid clusters (e.g., for histone proteins), four missed cleavages were also allowed. Proteins, which were identified with at least two peptides, both showing a score >40, were validated without any manual validation. For proteins identified by only one peptide having a score >40, the peptide sequence was checked manually. Peptides with scores >20 and <40 were systematically checked and/or interpreted manually to confirm or cancel the MASCOT suggestion.
Reverse transcription, PCR, and qPCR
Reverse transcription reactions were performed with the StrataScript First- Strand Synthesis system (Stratagene) using random hexamer primers. qPCR reactions were performed using Brilliant SYBR Green qPCR MasterMix on an Mx3005p cycler (Stratagene). cDNA from total testis was used for the standard curve, and data were normalized using Brdt cDNA abundance, as Northern blot attests the constant level of this mRNA in meiotic and postmeiotic cells (Pivot-Pajot et al., 2003).
Ex vivo studies of histone variants
The coding sequences of H2AL1, H2AL2, H2A, H2B, and TH2B were inserted into pCDNA3.1 (Invitrogen) modified by insertion of HA or Flag tags or into peGFP-C (BD Biosciences). Plasmids were cotransfected in Cos7 cells by the Fugene transfection system (Roche), and cells were collected 24 h after transfection. Coimmunoprecipitations were performed as described by Caron et al. (2003). Preparation of oligonucleosomes and capture on hydroxyapatite were performed as described by Sun et al. (2003).
Online supplemental material
Fig. S1 shows codetection and quantification of H4Ac and H3K9me3 in elongating spermatids by immunofluorescence. Fig. S2 shows protocols used for the MNase digestion of condensed spermatids and analysis of the resulting release of chromatin-associated proteins. Fig. S3 shows fractionation and capture on hydroxyapatite of the MNase-resistant structures of condensed spermatids. Fig. S4 demonstrates quantification of FISH experiments, showing an enrichment of the two MNase-resistant structures of condensed spermatids in major satellite sequences. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200604141/DC1.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. C. David Allis for the gift of the anti-TH2B antibody, generated and provided ahead of publication in collaboration with Upstate/Serologicals. We are also grateful to Drs. Steve Kistler, Rod Balhorn, and Minoru Yoshida for the generous gifts of anti-TP1, anti-TP2, anti-protamine, and anti-acetylated lysines antibodies, respectively. We wish to thank Sandrine Curtet-Benitski, Christine De Robertis, Roberte Pelletier, and Alexei Grichine for expert technical assistance.
The S. Khochbin Laboratory is supported by grants from the Regulome consortium (ANR-05-BLAN-0396-04), the Action Concertée Incitative, and Canceropole Lyon Auvergne Rhone-Alpes (EpiMed and EpiPro programs).
J. Govin and E. Escoffier contributed equally to this paper.
Abbreviations used in this paper: IF, immunofluorescence; IH, immunohistochemistry; MS, mass spectrometry; qPCR, quantitative PCR.
References
- Aul, R.B., and R.J. Oko. 2001. The major subacrosomal occupant of bull spermatozoa is a novel histone H2B variant associated with the forming acrosome during spermiogenesis. Dev. Biol. 239:376–387. [DOI] [PubMed] [Google Scholar]
- Caron, C., C. Pivot-Pajot, L.A. Van Grunsven, E. Col, C. Lestrat, S. Rousseaux, and S. Khochbin. 2003. Cdyl: a new transcriptional co-repressor. EMBO Rep. 4:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, C., J. Govin, S. Rousseaux, and S. Khochbin. 2005. How to pack the genome for a safe trip. Prog. Mol. Subcell. Biol. 38:65–89. [DOI] [PubMed] [Google Scholar]
- Churikov, D., J. Siino, M. Svetlova, K. Zhang, A. Gineitis, E. Morton Bradbury, and A. Zalensky. 2004. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics. 84:745–756. [DOI] [PubMed] [Google Scholar]
- Faure, A.K., C. Pivot-Pajot, A. Kerjean, M. Hazzouri, R. Pelletier, M. Peoc'h, B. Sele, S. Khochbin, and S. Rousseaux. 2003. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol. Hum. Reprod. 9:757–763. [DOI] [PubMed] [Google Scholar]
- Ferro, M., D. Seigneurin-Berny, N. Rolland, A. Chapel, D. Salvi, J. Garin, and J. Joyard. 2000. Organic solvent extraction as a versatile procedure to identify hydrophobic chloroplast membrane proteins. Electrophoresis. 21:3517–3526. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden, M., M. Ballesteros, M. Gordon, and P.P. Tam. 1998. Histone- and protamine-DNA association: conservation of different patterns within the beta-globin domain in human sperm. Mol. Cell. Biol. 18:3350–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gineitis, A.A., I.A. Zalenskaya, P.M. Yau, E.M. Bradbury, and A.O. Zalensky. 2000. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J. Cell Biol. 151:1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin, J., C. Caron, C. Lestrat, S. Rousseaux, and S. Khochbin. 2004. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 271:3459–3469. [DOI] [PubMed] [Google Scholar]
- Govin, J., C. Caron, S. Rousseaux, and S. Khochbin. 2005. Testis-specific histone H3 expression in somatic cells. Trends Biochem. Sci. 30:357–359. [DOI] [PubMed] [Google Scholar]
- Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri, M., C. Pivot-Pajot, A.K. Faure, Y. Usson, R. Pelletier, B. Sele, S. Khochbin, and S. Rousseaux. 2000. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur. J. Cell Biol. 79:950–960. [DOI] [PubMed] [Google Scholar]
- Khetchoumian, K., M. Teletin, M. Mark, T. Lerouge, M. Cervino, M. Oulad-Abdelghani, P. Chambon, and R. Losson. 2004. TIF1delta, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J. Biol. Chem. 279:48329–48341. [DOI] [PubMed] [Google Scholar]
- Kimmins, S., and P. Sassone-Corsi. 2005. Chromatin remodelling and epigenetic features of germ cells. Nature. 434:583–589. [DOI] [PubMed] [Google Scholar]
- Kotaja, N., S. Kimmins, S. Brancorsini, D. Hentsch, J.L. Vonesch, I. Davidson, M. Parvinen, and P. Sassone-Corsi. 2004. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat. Methods. 1:249–254. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., D.W. Abbott, and J. Ausio. 2003. a. A haploid affair: core histone transitions during spermatogenesis. Biochem. Cell Biol. 81:131–140. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., Y. Song, M.E. de Jong, S.M. Bagha, and J. Ausio. 2003. b. A walk though vertebrate and invertebrate protamines. Chromosoma. 111:473–482. [DOI] [PubMed] [Google Scholar]
- Li, A., A.H. Maffey, W.D. Abbott, N. Conde e Silva, A. Prunell, J. Siino, D. Churikov, A.O. Zalensky, and J. Ausio. 2005. Characterization of nucleosomes consisting of the human testis/sperm-specific histone H2B variant (hTSH2B). Biochemistry. 44:2529–2535. [DOI] [PubMed] [Google Scholar]
- Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5:296–304. [DOI] [PubMed] [Google Scholar]
- Martianov, I., S. Brancorsini, R. Catena, A. Gansmuller, N. Kotaja, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2005. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl. Acad. Sci. USA. 102:2808–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., N. Beaujean, V. Brochard, C. Audouard, D. Zink, and P. Debey. 2006. Genome restructuring in mouse during reprogramming and early development. Dev. Biol. 292:317–332. [DOI] [PubMed] [Google Scholar]
- Marushige, Y., and K. Marushige. 1983. Proteolysis of somatic type histones in transforming rat spermatid chromatin. Biochim. Biophys. Acta. 761:48–57. [DOI] [PubMed] [Google Scholar]
- Nelson, D.A., D.K. Oosterhof, and R.L. Rill. 1977. Histones H2A and H2B are neighbors along DNA in chromatin: characterization of subnucleosomal particles containing H2A+H2B. Nucleic Acids Res. 4:4223–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.A., A.J. Mencke, S.A. Chambers, D.K. Oosterhof, and R.L. Rill. 1982. Subnucleosomes and their relationships to the arrangement of histone binding sites along nucleosome deoxyribonucleic acid. Biochemistry. 21:4350–4362. [DOI] [PubMed] [Google Scholar]
- O'Carroll, D., H. Scherthan, A.H. Peters, S. Opravil, A.R. Haynes, G. Laible, S. Rea, M. Schmid, A. Lebersorger, M. Jerratsch, et al. 2000. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 20:9423–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva, R., and G.H. Dixon. 1990. Vertebrate protamine gene evolution I. Sequence alignments and gene structure. J. Mol. Evol. 30:333–346. [DOI] [PubMed] [Google Scholar]
- Palmer, D.K., K. O'Day, and R.L. Margolis. 1990. The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma. 100:32–36. [DOI] [PubMed] [Google Scholar]
- Peters, A.H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, et al. 2001. Loss of the suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 107:323–337. [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot, C., C. Caron, J. Govin, A. Vion, S. Rousseaux, and S. Khochbin. 2003. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol. Cell. Biol. 23:5354–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickwood, D., and A.J. MacGillivray. 1975. Improved techniques for the fractionation of non-histone proteins of chromatin on hydroxyapatite. Eur. J. Biochem. 51:593–601. [DOI] [PubMed] [Google Scholar]
- Rill, R.L., and D.A. Nelson. 1978. Histone organization in chromatin: comparison of nucleosomes and subnucleosomal particles from erythrocyte, myeloma, and yeast chromatin. Cold Spring Harb. Symp. Quant. Biol. 42:475–482. [DOI] [PubMed] [Google Scholar]
- Rousseaux, S., C. Caron, J. Govin, C. Lestrat, A.K. Faure, and S. Khochbin. 2005. Establishment of male-specific epigenetic information. Gene. 345:139–153. [DOI] [PubMed] [Google Scholar]
- Russell, L., R. Ettlin, A. Sinha-Hikim, and E. Clegg. 1990. Histological and histopathological evaluation of the testis. Cache River Press, Clearwater, FL. 286 pp.
- Santos, F., A.H. Peters, A.P. Otte, W. Reik, and W. Dean. 2005. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 280:225–236. [DOI] [PubMed] [Google Scholar]
- Sun, J.M., V.A. Spencer, H.Y. Chen, L. Li, and J.R. Davie. 2003. Measurement of histone acetyltransferase and histone deacetylase activities and kinetics of histone acetylation. Methods. 31:12–23. [DOI] [PubMed] [Google Scholar]
- Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114–120. [DOI] [PubMed] [Google Scholar]
- Wykes, S.M., and S.A. Krawetz. 2003. The structural organization of sperm chromatin. J. Biol. Chem. 278:29471–29477. [DOI] [PubMed] [Google Scholar]
- Zalensky, A.O., J.S. Siino, A.A. Gineitis, I.A. Zalenskaya, N.V. Tomilin, P. Yau, and E.M. Bradbury. 2002. Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J. Biol. Chem. 277:43474–43480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.