Abstract
Mutations in Hydin cause hydrocephalus in mice, and HYDIN is a strong candidate for causing hydrocephalus in humans. The gene is conserved in ciliated species, including Chlamydomonas reinhardtii. An antibody raised against C. reinhardtii hydin was specific for an ∼540-kD flagellar protein that is missing from axonemes of strains that lack the central pair (CP). The antibody specifically decorated the C2 microtubule of the CP apparatus. An 80% knock down of hydin resulted in short flagella lacking the C2b projection of the C2 microtubule; the flagella were arrested at the switch points between the effective and recovery strokes. Biochemical analyses revealed that hydin interacts with the CP proteins CPC1 and kinesin-like protein 1 (KLP1). In conclusion, C. reinhardtii hydin is a CP protein required for flagellar motility and probably involved in the CP–radial spoke control pathway that regulates dynein arm activity. Hydrocephalus caused by mutations in hydin likely involves the malfunctioning of cilia because of a defect in the CP.
Introduction
Hydrocephalus describes the lethal accumulation of cerebrospinal fluid (CSF) in the brain. The incidence is 0.12–2.5 per 1,000 live and stillbirths, and the majority of human hydrocephalus is of genetic origin (Schrander-Stumpel and Fryns, 1998). A number of genetic mouse models of congenital hydrocephalus have been described. These include hy3 mice; recently, the affected gene, Hydin, was identified using an insertional mutant (Davy and Robinson, 2003). The corresponding chromosomal region in the human genome has also been implicated in hydrocephalus (Callen et al., 1990). Hydin is a conserved gene present in the genomes of various protists and metazoans and encoding a large protein of >500 kD.
In the neonatal brain of the mouse, Hydin is expressed in the ciliated epithelial cells lining the lateral, third, and fourth ventricles (Davy and Robinson, 2003). Hydin is also expressed in the ciliated epithelia of bronchi and oviduct and in developing spermatocytes. This expression pattern suggests a ciliary/flagellar function for hydin. Indeed, mutations in several other genes encoding ciliary proteins cause hydrocephalus in mice. These include Mdnah5, which encodes an axonemal dynein heavy chain; Spag6, a homologue of Chlamydomonas reinhardtii central pair (CP) component PF16; and Tg737, which encodes the intraflagellar transport protein IFT88/Polaris (Sapiro et al., 2002; Ibanez-Tallon et al., 2004; Banizs et al., 2005). Comparative genomics revealed that homologues of Hydin are present in species with cilia/flagella and absent in species lacking these organelles, such as Saccharomyces cerevisiae and Arabidopsis thaliana (Li et al., 2004; Pazour et al., 2005; Broadhead et al., 2006). Hydin was identified in the flagellar proteomes of C. reinhardtii (Pazour et al., 2005) and Trypanosoma brucei (Broadhead et al., 2006). In the proteomic analysis of fractionated C. reinhardtii flagella (Pazour et al., 2005), hydin was identified by 69 different peptides, primarily from axonemal fractions, suggesting that it is abundant and tightly associated with the axoneme. Finally, in T. brucei, hydin ablation by RNAi resulted in reduced motility (Broadhead et al., 2006). However, despite the evidence indicating a function in flagella, neither hydin's location within the organelle nor its role in flagellar assembly and motility are known. In this study, we use a combination of biochemical, reverse genetic, and structural approaches to examine the function and location of hydin in the C. reinhardtii flagellum.
Results
Hydin is located in the CP apparatus
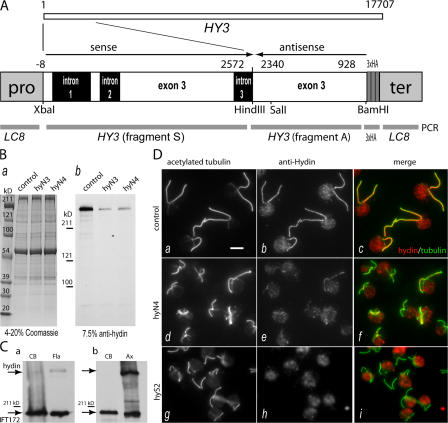
C. reinhardtii hydin is a polypeptide of ∼540 kD encoded by a single copy gene, HY3, spanning ∼17,700 bp on linkage group I. To generate an antibody to C. reinhardtii hydin, a HY3 fragment (Fig. 1 A, fragment A), representing exon 3, was amplified from genomic DNA by PCR and cloned into the bacterial expression vector pMAL-cRI v.2 encoding maltose-binding protein (MBP). The MBP-hydin peptide was expressed in Escherichia coli, purified, and used for antibody production. A BamHI–SalI piece of fragment A was cloned into pGEX-6P-1, and the purified GST-hydin peptide was used for affinity purification of the antibody. The purified anti- hydin antibody stained a single band of ∼540 kD in Western blots of axonemes of control cells (Fig. 1 B, control lane).
Figure 1.
HY3 gene-silencing vector and anti-hydin antibody. (A) C. reinhardtii HY3, which encodes hydin, is a gene of 17.7 kb. Fragment A, corresponding to exon 3 of HY3, and a BamHI–SalI piece of fragment A were cloned into bacterial expression vectors, and the fusion proteins were used for antibody production and purification. A gene-silencing vector was constructed from fragment A, fragment S (another PCR product of HY3), a triple HA tag, and the promoter and terminator region of the LC8 gene. (B) Coomassie-stained gel (a; 4–20% SDS-PAGE) and Western blot (b; 7.5% SDS-PAGE) of isolated axonemes of CC3395 (control) and the HY3 RNAi strains hyN3 and hyN4. Anti-hydin specifically stained a band of ∼540 kD that was strongly reduced in the HY3 RNAi strains. (C) Western blots probed with anti-hydin and anti-IFT172 (Cole et al., 1998) comparing the amount of hydin present in deflagellated cells (CB) and isolated flagella (Fla) or axonemes (Ax). (a) Equivalent numbers of cell bodies and flagellar pairs from ∼106 cells were loaded. (b) Equal amounts (∼25 μg) of cell body and axonemal protein were loaded. IFT172, an intraflagellar transport protein used as a control, is present in the cell body and flagella; a considerable amount remains with the axonemes (Hou et al., 2004). (D) Immunofluorescence images of methanol-fixed cells of strains CC3395 (control), hyN4, and hyS2 labeled with anti-acetylated tubulin (a, d, and g) and anti-hydin (b, e, and h). Merged images (c, f, and i) reveal the localization of hydin to the flagella of wild-type cells and the reduction of hydin in the hydin RNAi cells. Note the shorter flagella in the latter. At least part of the fluorescence in the cell bodies stained with anti-hydin is background caused by chlorophyll autofluorescence. Bar, 5 μm.
Similar to other flagellar proteins, expression of HY3 is strongly induced after deflagellation by pH shock (Li et al., 2004; Pazour et al., 2005; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200611115/DC1). Western blot analysis of cell bodies and isolated flagella revealed that hydin was highly enriched in the flagella (Fig. 1 C) and attached to the axonemes (Fig. 1 C b and see Fig. 7 B). Immunofluorescence microscopy combining anti-hydin with anti-acetylated tubulin confirmed that hydin is present in the flagella of wild-type cells (Fig. 1 D, a–c). When isolated flagella were extracted with detergent, anti-hydin staining remained associated with the axonemes and overlapped with the anti-tubulin signal over the entire length of the axonemes (Fig. 2 A, a–c). Therefore, hydin is an axonemal protein and one of the largest proteins so far identified in the flagellum.
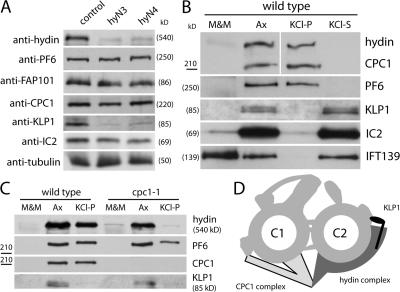
Figure 7.
Hydin interacts with KLP1 and the CPC1 complex. (A) Western blots of isolated axonemes from wild-type, hyN3, and hyN4 probed with antibodies to the CP proteins hydin, PF6, FAP101, CPC1, and KLP1. Antibodies to α-tubulin and IC2 were used as loading controls. Both hydin and KLP1 were significantly reduced in the axonemes of hydin RNAi cells. (B) Western blot comparing the amounts of hydin, PF6, CPC1, KLP1, IC2, and IFT139 in the detergent-soluble flagellar membrane-plus-matrix fraction (M&M), axonemes (Ax), residual axonemes after 0.6 M KCl extraction (KCl-P), and the KCl extract of axonemes (KCl-S). Protein from equivalent numbers of flagella were loaded for each sample. (C) Western blot comparing the solubility of hydin, PF6, CPC1, and KLP1 in wild-type and cpc1 flagella. Protein from equal numbers of flagella was loaded for each sample. (D) Schematic representation of the CP apparatus. In this model, hydin is an essential component of the C2b projection. It extends to the C2a projection, where it stabilizes the association of KLP1 with the C2 microtubule, and extends in the other direction to interact with the CPC1 complex of the C1 microtubule.
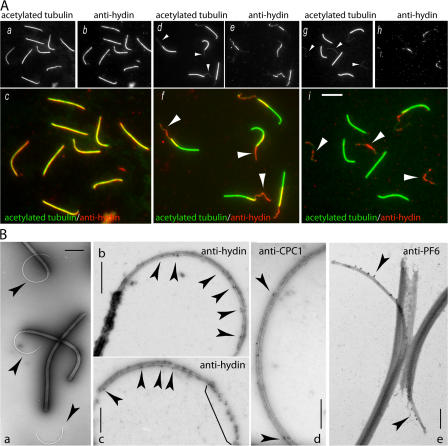
Figure 2.
Hydin localizes to the CP. (A) Immunofluorescence microscopy of isolated axonemes (a–c) and axonemes treated with ATP and trypsin to induce partial (d–f) or complete (g–i) extrusion of the CP. Staining with anti-acetylated tubulin (a, d, and g) reveals the CP as a thin microtubular bundle projecting (arrowheads in d and f) or completely extruded (arrowheads in g and i) from the axonemes. Anti-hydin staining (b, e, and h) colocalized with the thin microtubular structures (arrowheads in f and i) and was absent from regions of axonemes now vacated by the CP (f) as well as entire axonemes from which the CP was completely extruded (i). Because of the prolonged incubation with protease, signal strength had declined in panels h and i. Bar, 5 μm. (B) Whole mount immunogold EM of extruded CPs. (a) Negatively stained axonemes with protruding CPs (arrowheads) showing the preparation used for immunogold labeling experiments. (b and c) 15-nm gold complexes (arrowheads) labeling hydin were observed mostly along the C2 microtubule on the concave side of the arc formed by the CP. The bracket shows increased label density on disintegrated CP. (d and e) In contrast, gold particles labeling the C1 proteins CPC1 (d) and PF6 (e) were predominately located on the convex side of the CP. Immunostaining with anti-PF6 was performed as described by Bernstein et al. (1994). Bars: (a) 2 μm; (b–d) 500 nm; (e) 350 nm.
To determine whether hydin is associated with the outer doublets or the CP of microtubules, we used a modified CP extrusion assay. When isolated flagella were treated with 1 mM ATP, trypsin, and 1% NP-40 for 2 min, the CP was partially expelled from the axonemes (Fig. 2 A d). Double labeling with anti-hydin and anti-acetylated tubulin revealed that hydin was concentrated on the CP, which was visible as a thin microtubular structure projecting from the distal end of the axoneme (Fig. 2 A, e and f); accordingly, anti-hydin staining was absent from the proximal region of the axoneme that had been vacated by the CP. Treatment with an additional 1 mM of ATP after 2 and 4 min often resulted in the complete extrusion of the CP from the axonemes (Fig. 2 A g). Hydin colocalized with the extruded CP but was absent from the axonemes that were now devoid of the CP (Fig. 2 A, h and i). These data show that C. reinhardtii hydin is located exclusively in the CP apparatus.
Hydin is associated with the C2 microtubule
In whole mount immunogold EM of extruded CPs (Fig. 2 B a), gold complexes representing hydin decorated the projecting CP microtubules (Fig. 2 B, b and c). Labeling was present but sparse on well-preserved CPs and increased as the CPs disintegrated, suggesting that parts of hydin targeted by the antibody are not readily accessible in the native CP. The majority of the gold particles (76%; n = 275) was present on the microtubule located on the concave side of the arc formed by the CP; for this analysis, only gold complexes on CPs with well-preserved, intact microtubules were scored. Previous studies have shown that this is the C2 microtubule (see Fig. 6 K for a schematic drawing of the CP; Bernstein et al., 1994; Bernstein and Rosenbaum, 1994). As a control, we carried out a similar immunolocalization using antibodies to CPC1 (Zhang and Mitchell, 2004) and PF6 (Wargo et al., 2005), both components of the C1 microtubule (Mitchell and Sale, 1999; Rupp et al., 2001; Fig. 2, d and e). As expected, these antibodies decorated the CP microtubule on the convex side of the arc (PF6, 94% and n = 56; CPC1, 91% and n = 23). Thus, that portion of hydin encoded by exon 3 is closely associated with the C2 microtubule.
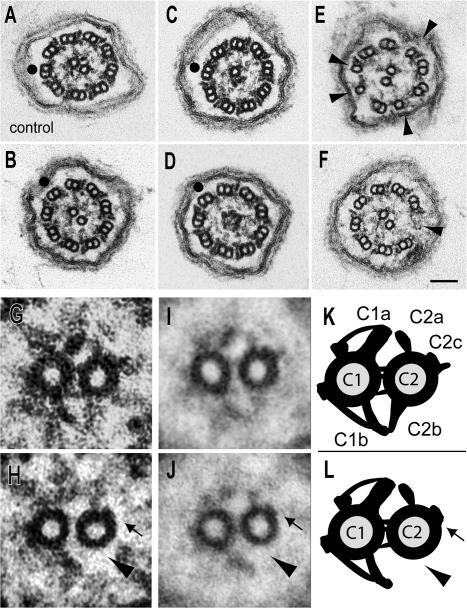
Figure 6.
Flagella of HY3 RNAi cells lack the C2b projection and part of the C2c projection. (A–H) Axonemal cross sections of wild type (A and G; CC3395) and the HY3 RNAi strains hyN4 (B, D, and H), hyN3 (C), and hyD2 (E and F). (G and H) Detail from A and B, respectively. (I and J) Image averages based on six wild-type (I) and six hyN4 images (J). (K and L) Schematic representations of the CP apparatus of wild type (K) and HY3 RNAi cells (L). Similar defects were observed in all four hydin RNAi strains analyzed by EM. In H, J, and L, the defects in the CP of the hydin RNAi cells are marked with arrowheads (absence of projection C2b) and arrows (absence of density of C2c). Arrowheads in E point out defective and missing doublets; note the presence of dynein arms on some of the singlet microtubules in E. Black dots indicate doublet No. 1 lacking the outer dynein arm. Bar, 0.1 μm.
Hydin is greatly reduced in mutants lacking the CP
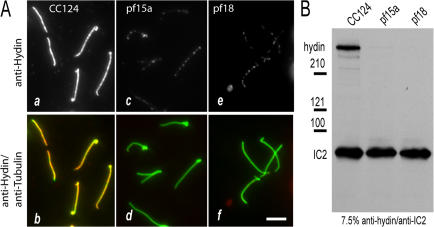
In C. reinhardtii, the mutants pf15a and pf18 fail to assemble the CP (Witman et al., 1972, 1978; Adams et al., 1981). We examined the flagella of these mutants to confirm that hydin is a component of the CP. In immunofluorescence microscopy, the hydin signal was strong in detached flagella of wild type (CC124; Fig. 3 A, a and b) but greatly reduced in detached flagella of pf15a and pf18 (Fig. 3 A, c–f). In the flagella of these mutants, the CP is replaced by an amorphous core (Witman et al., 1972, 1978; Adams et al., 1981), which may account for the residual hydin signal. This amorphous core is lost from most (80–90%) of the isolated axonemes of the mutants (Witman et al., 1978; Adams et al., 1981), and indeed, Western blot analysis of isolated axonemes from pf15a and pf18 revealed only traces of hydin in both mutants (Fig. 3 B). These results provide strong independent evidence that hydin is a component of the CP apparatus.
Figure 3.
Hydin is greatly reduced in flagella of CP mutants. (A) Immunofluorescence microscopy of detached flagella. Wild-type (CC124; a and b), pf15a (c and d), and pf18 (e and f) cells were allowed to settle onto polyethylenimine-treated slides that were then submerged into −20°C methanol. This procedure frequently caused the flagella to detach, and the cell bodies were subsequently washed off. (top) Anti-hydin signal; (bottom) merged images of anti-hydin and anti-acetylated tubulin. The hydin signal was nearly undetectable in flagella of pf15a and pf18. Bar, 5 μm. (B) Western blot of isolated axonemes of CC124 (wild type), pf15a, and pf18. The blot was probed with anti-hydin and an antibody to the outer dynein arm intermediate chain IC2 (King and Witman, 1990) as a loading control.
HY3 RNAi effectively reduced hydin mRNA and protein
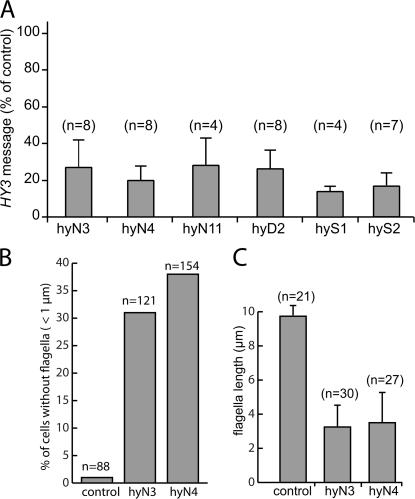
Mutations of HY3 in C. reinhardtii have not yet been identified. To investigate the function of hydin, an RNAi vector targeting C. reinhardtii HY3 was constructed by cloning HY3 fragment S in sense and HY3 fragment A in antisense orientation downstream of the LC8 promoter and upstream of a triple HA tag and the LC8 terminator (Fig. 1 A). The resulting plasmid, pKL3-hyAS, was cotransformed into C. reinhardtii cells of strain CC3395. 39 of 204 independently derived transformants scored from three experiments showed a severe motility phenotype, with the majority of the cells resting at the bottom of the culture vessels. HY3 message levels were tested by quantitative RT-PCR in 11 of the nonmotile transformants. In 10 of the strains, HY3 was down-regulated by up to 87% (Fig. 4 A). The remaining strain had normal HY3 message levels and, in contrast to the others (see the following section), completely lacked flagella, suggesting that its phenotype was due to an insertion of the RNAi vector into a gene necessary for flagellar assembly.
Figure 4.
HY3 knockdown affects flagellar assembly. (A) HY3 mRNA is reduced in HY3 RNAi cells. Quantitative PCR was used to determine the amount of HY3 message in several transformants. The data are shown as a percentage of wild type for strains hyN3, hyN4, hyN11, hyD2, hyS1, and hyS2 and are based on one (hyS1), two (hyD2 and hyS2), or three independent isolates of total RNA. (B and C) HY3 RNAi cells have short flagella or lack flagella. Data are based on immunofluorescence images obtained with anti-acetylated tubulin. (B) Frequency of stumpy flagella (<1 μm) and bald cells in control (CC3395) and the HY3 RNAi strains hyN3 and hyN4. (C) Flagellated cells of strains hyN3 and hyN4 have shorter flagella than those of the control strain CC3395. Only flagella >1 μm were measured.
In Western blots of isolated axonemes of selected hydin RNAi strains, hydin was strongly reduced (Fig. 1 B b). In immunofluorescence microscopy using the anti-hydin antibody, labeling of the flagella in the RNAi strains was much weaker than in wild type, although the strength of the residual staining varied somewhat between RNAi strains and individual cells (Fig. 1 D, d–i). Thus, hydin depletion was confirmed by both Western blotting and immunofluorescence microscopy. Similar to earlier observations for RNAi of other genes in C. reinhardtii (Koblenz et al., 2003), hydin knockdown was not stable over an extended period of time. All strains returned to wild-type amounts of hydin and wild-type phenotype within 2–6 mo after transformation (unpublished data).
Hydin knockdown causes an unusual form of flagellar paralysis
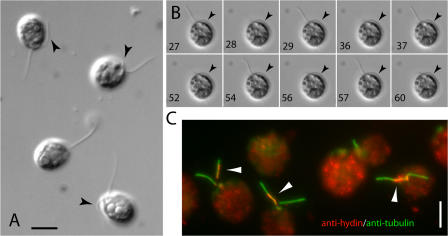
The CP apparatus is thought to regulate dynein arm activity, allowing coordinated flagellar movement (Smith and Yang, 2004). Light microscopic examination of living cells from 10 independent hydin RNAi strains revealed that the flagella were mostly paralyzed and only occasionally formed bends. Flagella also were shorter than those of wild-type cells, with some cells lacking flagella completely (Fig. 1 D and Fig. 4, B and C). Strikingly, the flagella of approximately half of the cells were arrested with one flagellum in the “hands-up” position, i.e., pointing away from the cell body, and one flagellum in the “hands-down” position, i.e., lying alongside the cell body (Fig. 5 A). The remaining cells either had both flagella arrested in the hands-down position (hyN3, 30%; hyS1, 26%) or both flagella in the hands-up position (hyN3, 14%; hyS1, 27%). This is distinctly different from the phenotype of other CP mutants: the CP mutants pf15a, pf16, pf18, and pf19 were paralyzed with both flagella in the hands-up position (95–100%; see Table I; pf6, cpc1, and pf20 were too motile to analyze). The relative position of the eyespot was analyzed in 222 hydin RNAi cells showing asymmetric arrest of the two flagella (living or rapidly fixed as described by Mitchell [2003]); 45% of these had the trans flagellum and 55% had the cis flagellum, which is closer to the eyespot, in the hands-up position. Therefore, the tendency to arrest in the hands-up or hands-down position does not correlate with the cis or trans flagellum.
Figure 5.
The flagella of HY3 RNAi strains arrest in the hands-up or hands-down position. (A) DIC image of living cells of hyN3 showing the resting positions of the two flagella of each cell. Arrowheads indicate the hands-down flagellum positioned along the cell body. (B) Residual flagellar movement of a hyS1 cell documented by DIC microscopy. The hands-down flagellum (arrowheads) is relatively inactive. Frame numbers are indicated and approximately equal the time in seconds. The complete sequence is shown in Video 3 (available at http://www.jcb.org/cgi/content/full/jcb.200611115/DC1). (C) Methanol-fixed hyN3 cells were double labeled with anti-hydin (red) and anti-acetylated tubulin (green). Frequently, residual hydin accumulated in one of the two flagella (arrowheads). Bars, 5 μm.
Table I.
Flagellar position in paralyzed cells of hydin RNAi and other CP mutant strains
| Strain | Both up | One up and one down |
Both down |
|---|---|---|---|
| % | % | % | |
| hyN3 (n = 103) | 14 | 56 | 30 |
| hyS1 (n = 199) | 27 | 47 | 26 |
| pf15a (n = 108) | 97 | 2 | 1 |
| pf18 (n = 131) | 95 | 5 | — |
| pf16 (n = 143) | 98 | 2 | — |
| pf19 (n = 76) | 100 | — | — |
The data are based on living cells observed by DIC or phase contrast. Only cells in which both flagella were visible were scored.
Although the majority (90–99%) of the hydin RNAi cells were paralyzed, some cells showed residual flagellar movements resulting in twitching or spinning. We noticed that the flagellum in the hands-up position was the more active one, which typically would undergo a quick power stroke, a resting period of variable length, and a recovery stroke, after which it stopped again in the hands-up position (Fig. 5 B and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200611115/DC1). The flagellum in the hands-down position was either completely paralyzed or beat at a lower frequency than the hands-up flagellum (Videos 1 and 2). We further observed that residual hydin in the cells examined by immunofluorescence microscopy tended to be located in just one of the two flagella (Fig. 5 C). Because hydin is required for flagellar motility, it is possible that the asymmetrical motility of flagella observed in some HY3 RNAi strains is related to the unequal distribution of residual hydin in the two flagella.
Ultrastructural and biochemical defects of hydin-depleted flagella
To determine whether flagellar paralysis was accompanied by ultrastructural defects of the axoneme, four strains (hyD2, hyN3, hyN4, and hyS2) were processed for transmission EM, and flagellar cross sections were analyzed. In all four strains, similar ultrastructural defects were observed (Fig. 6). Most notably, the C2b projection and parts of the C2c projection of the C2 microtubule were missing from almost all 9 + 2 axonemes (∼90%; n = 34 for strain hyS2 and 14 for hyN4) of the hydin RNAi strains (Fig. 6, B and H). Image averages generated from digitized images clearly revealed the differences in the CP apparatus of 9 + 2 axonemes of wild-type and HY3 RNAi cells (Fig. 6, I–L). In 5–12% of the cross sections, one CP microtubule was missing; an additional 3–5% of cross sections lacked both CP microtubules, which were replaced by a core of amorphous material similar to that described in other mutants lacking the CP (Fig. 6, C and D; Witman et al., 1972, 1978; Adams et al., 1981). Finally, ∼2% of the cross-sectioned axonemes lacked one to four B-tubules or up to three entire doublet microtubules (Fig. 6, E and F).
Previous ultrastructural analysis of KLP1-depleted axonemes suggested that KLP1 is a component of the C2c projection (Yokoyama et al., 2004). A part of this projection was largely missing in the axonemes of hydin RNAi cells, raising the question of whether hydin depletion affected the presence of KLP1 and other known CP proteins. Western blotting showed no difference in the amounts of the C1 proteins PF6, FAP101 (Wargo et al., 2005), or CPC1 in axonemes isolated from wild-type or hydin RNAi cells (Fig. 7 A). In contrast, probing with anti-KLP1 (Bernstein et al., 1994) showed that KLP1 was strongly reduced in the hydin RNAi axonemes. These data indicate that localization of KLP1 to the C2 tubule depends on hydin.
Hydin interacts with the CPC1 complex
To obtain biochemical evidence for the location of hydin within the CP, we took advantage of the fact that the C2 microtubule can be selectively depolymerized by extraction of axonemes with high salt, e.g., 0.6 M KCl (Mitchell and Sale, 1999). These conditions solubilized most of the KLP1 and the outer dynein arm intermediate chain IC2 (Fig. 7 B, lane KCl-S), whereas most of the hydin and the C1 proteins PF6 and CPC1 stayed in the insoluble fraction (lane KCl-P), in good agreement with data from the proteomic study of C. reinhardtii flagella (Pazour et al., 2005).
This result, indicating that hydin is associated with the C1 microtubule when the C2 microtubule is solubilized, was surprising given the effect of hydin depletion on the C2b and C2c projections, as well as on the C2 protein KLP1, and the immunogold localization of hydin to the C2 microtubule. However, ultrastructural analysis has shown that the C1b and C2b projections physically interact and has revealed that axonemes isolated from the mutant cpc1, which lack the C1b projection, also frequently lack the C2b projection (Mitchell and Sale, 1999). Therefore, if hydin is a component of the C2b projection, one would predict that hydin would be missing or destabilized in the cpc1 mutant. To test this, flagella were isolated from cpc1 and wild-type cells, and the detergent-soluble membrane-plus-matrix fraction, axonemes, and KCl-extracted axonemes were compared by Western blotting (Fig. 7 C). As shown above, hydin fractionated with the axonemes in wild-type cells and still remained with the axonemes after high-salt extraction. In contrast, in cpc1, some of the hydin was released into the detergent-soluble membrane-plus-matrix fraction, and little remained in the axonemes after high-salt extraction. The majority of PF6, a component of the C1a projection, remained attached to the wild-type and cpc1 axonemes after high-salt extraction. These data show that hydin interacts with the CPC1 complex of the C1 microtubule. Fig. 7 D shows a model for the location of hydin within the CP that is consistent with the immunolocalization, ultrastructural, and biochemical data.
Discussion
In this study, we analyzed the location and function of hydin in the flagella of C. reinhardtii. An antibody raised against a polypeptide encoded by exon 3 of C. reinhardtii HY3 decorated the CP in immunofluorescence microscopy and immunogold EM. Western blotting and immunofluorescence microscopy using anti-hydin revealed that hydin was strongly reduced in axonemes of pf15a and pf18, two mutants that lack the CP apparatus. In strains depleted of hydin by RNAi, EM revealed defects in the CP ranging from the absence of individual CP projections to loss of the entire CP apparatus. Hydin knockdown resulted in paralyzed flagella, a phenotype characteristic of radial spoke and CP mutants of C. reinhardtii. Biochemical studies showed that KLP1, another CP protein, was dependent on hydin for assembly into the axoneme. In summary, evidence from several independent experimental approaches all indicate that hydin is a CP protein required for flagellar motility.
Hydin is a component of the C2b projection
In whole mounts labeled with anti-hydin, the majority of the gold particles was located on the C2 microtubule. Ultrastructural analysis of C. reinhardtii HY3 RNAi cells revealed that almost all flagella lacked the C2b projection on the C2 microtubule, strongly suggesting that hydin is a component of this projection. Previously, it was shown that the C2b and C1b projections physically interact, and that the stability of the C2b projection is partly dependent on the C1b projection (Mitchell and Sale, 1999). Based on these observations, our model (Fig. 7 D) would predict that the stability of hydin, in the C2b projection, would depend in part on the C1b projection. Indeed, we found that much of the hydin could be eluted from cpc1 axonemes, which lack the C1b projection, but not from wild-type axonemes, by dissolving the C2 microtubule using 0.6 M KCl. We propose that hydin is a core component of the C2b projection but also is anchored to the C1 microtubule via the C1b projection and the CPC1 complex.
Hydin knockdown cells also lack an element of the C2c projection. The latter is probably constituted at least in part by KLP1, because KLP1 has been localized to the C2 microtubule (Bernstein et al., 1994) and the ablation of KLP1 by RNAi results in the loss of parts of projection C2c (Yokoyama et al., 2004). In many KLP1 RNAi axonemes, the position of projection C2b was shifted, suggesting that its attachment to the C2 microtubule was destabilized. We propose that hydin also interacts with the C2c protein KLP1 and that this interaction is important for the stability of both the C2b and C2c projections. Consistent with this, we found that the amount of KLP1 is strikingly reduced in isolated axonemes of hydin-depleted cells, indicating that hydin anchors KLP1 to the C2 microtubule. Anchorage of KLP1 to hydin on C2 would explain the observation that isolated KLP1 binds tightly to microtubules in the presence of AMPPNP and is released by ATP, but cannot be extracted from axonemes by ATP (Yokoyama et al., 2004).
It should be noted that our model, in which hydin partially circumscribes the CP apparatus by stretching from the C1b projection to the C2c projection, would allow “cross talk” between projections in different halves of the CP apparatus, thereby potentially coordinating regulation of dynein arms in both halves of the axoneme (see below).
Hydin shares a proposed microtubule-binding domain with other axonemal and CP proteins
Functional domains of hydin are unknown, but a recent in silico analysis identified four ASH (ASPM, SPD-2, Hydin) domains (Ponting, 2006). ASH domains were found in several centrosomal and flagellar proteins, including abnormal spindle-like microcephaly-associated protein (ASPM), C. reinhardtii PF6, and several human orthologues of proteins identified in the C. reinhardtii flagellar proteome. The ASH domain consists of eight β-strands and is proposed to be involved in microtubule binding. Consistent with this, four of five ASH family members identified in the proteomic analysis of C. reinhardtii flagella (Pazour et al., 2005) are tightly bound to the axoneme. Thus, the ASH domain of hydin may mediate its attachment to the C2 microtubule. Interestingly, the two characterized ASH family members (hydin and PF6) in the flagellar proteome are CP proteins, raising the possibility that within the flagellum the ASH domain targets proteins to the CP.
Hydin RNAi cells reveal a possible role for hydin in switching activity
C. reinhardtii mutants completely lacking the CP complex or the radial spokes are generally paralyzed, although their dynein arms are capable of generating interdoublet sliding after treatment of the isolated axonemes with trypsin (Witman et al., 1978). Moreover, axonemes of these mutants can be reactivated at low ATP concentrations in vitro (Omoto et al., 1999), and extragenic suppressors, some of which involve dynein arm components, can restore some motility in the absence of the missing structures. These observations indicate that the CP and radial spokes are involved in the regulation of the dynein arms (Smith and Yang, 2004). The phenotype of mutants lacking one of the CP projections is more variable: pf6 mutants (lacking the C1a projection) “twitch in place as a result of flagella that beat slowly with a slightly abnormal waveform” (Rupp et al., 2001), cpc1 mutants (lacking projection C1b) swim slowly and their flagella beat at a reduced frequency with normal waveform (Mitchell and Sale, 1999), and KLP1 RNAi cells (lacking parts of C2c) have either paralyzed flagella or a reduced flagellar beat frequency (Yokoyama et al., 2004). Paralysis and low-beat frequency residual movements also were characteristic of flagella of our HY3 RNAi cells. However, the HY3 RNAi cells were unique among known CP mutants in that the two flagella rested in different positions (one “hands up” and one “hands down”) in ∼50% of the cells analyzed. We observed almost equal numbers of cis and trans flagella in the hands-up position, and ∼25% of cells with both flagella in either the hands-up or hands-down position, indicating that the position of arrest (hands up or hands down) was random and did not correlate with whether the flagellum was cis or trans relative to the eyespot (Kamiya and Witman, 1984). The flagella arrested in the hands-up position are at the end of their recovery stroke/beginning of their effective stroke, whereas those arrested in the hands-down position are at the end of their effective stroke/beginning of their recovery stroke. These positions correspond to the two switch points in the C. reinhardtii flagellar beat cycle, when the dynein arms in one half of the axoneme are turned off and the arms in the opposite half of the axoneme are turned on (Satir and Matsuoka, 1989). Based on the association of the CP with groups of doublet microtubules from split axonemes, where the activity state of the arms could be deduced, it has been proposed that a radial spoke–CP attachment cycle is involved in turning the arms on and off at these switch points (Satir and Matsuoka, 1989). The unique behavior of the hydin knockdown flagella indicates that hydin may have an important role in this switching function. It has been proposed that KLP1 acts “as a conformational switch to signal spoke-dependent control of dynein activity” (Yokoyama et al., 2004). Inasmuch as KLP1 appears to interact with hydin, it is tempting to speculate that KLP1 motor activity generates a conformational change in hydin, resulting in altered contact with the radial spokes and thereby transmitting a signal to the dynein arms.
Hydin RNAi results in short flagella
C. reinhardtii cells partially depleted of hydin by RNAi often had short or stumpy flagella, which has been not reported for other CP mutants of C. reinhardtii. However, sperm of mice deficient in Spag6, which has been localized to the CP apparatus and is an orthologue of the C. reinhardtii CP component PF16, frequently had an altered morphology, including truncation of the tail, loss of CP microtubules, and abnormal outer dense fibers and fibrous sheath (Sapiro et al., 2002). Mutations of Kpl2 protein, a homologue of the C. reinhardtii CP protein CPC1, cause the immotile short-tail sperm (ISTS) defect of Yorkshire boars (Sironen et al., 2006). ISTS is characterized by shorter sperm flagella, often lacking one or two CP microtubules, and displaying ultrastructural defects of the outer doublets, such as defective B-tubules or the absence of entire doublets (Andersson et al., 2000). It is currently unknown why mutation of certain CP proteins can result in truncation of flagella. About 2% of the flagella from hydin RNAi cells had defects in the outer doublet microtubules in addition to the much more frequent loss of CP projections. Examination of both cross sections and longitudinal sections of the flagella indicate that the outer doublet abnormalities are localized to the flagellar tip. Therefore, certain defects in the CP may affect some aspect of axonemal assembly or turnover, which occur at the tip.
Defects in Hydin likely cause hydrocephalus through a mechanism involving cilia
Mice homozygous for mutations in Hydin develop hydrocephalus (Davy and Robinson, 2003). Hydrocephalus is thought to be caused either by overproduction or insufficient absorption of CSF or by insufficient transport of fluid between the ventricles of the brain. The latter could be caused by defects in motile cilia present on the surfaces of the ventricles and aqueducts. The movement of these cilia causes an ependymal flow necessary to maintain open cerebral aqueducts (Ibanez-Tallon et al., 2004; Banizs et al., 2005). Indeed, mice with mutations in several other genes encoding flagellar proteins develop hydrocephalus. These include Tg737orpk mutant mice, which exhibit multiple cilia-related defects, including cystic kidney disease (Moyer et al., 1994). Tg737 encodes Polaris/IFT88, a conserved protein required for the assembly of primary and motile cilia (Pazour et al., 2000). In Tg737orpk mutant mice, ependymal cilia are severely malformed, leading to disorganized beating and impaired movement of CSF (Banizs et al., 2005). Mice with primary ciliary dyskinesia caused by dysfunctional Mdnah5, an axonemal outer arm dynein heavy chain, develop hydrocephalus at early postnatal ages (Ibanez-Tallon et al., 2002). 50% of mice lacking the CP protein Spag6 develop lethal hydrocephalus within 8 wk of birth (Sapiro et al., 2002). Finally, loss of cilia because of mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in abnormalities in organ situs and, in some cases, hydrocephalus (Chen et al., 1998). In summary, cilia function is important for the CSF ventricular system, and at least certain forms of hydrocephalus belong to the ciliopathies, an emerging class of human genetic disorder (Zariwala et al., 2006).
In an earlier study, the role of cilia in the pathogenesis of hydrocephalus in the hy3 mouse was questioned. Scanning EM studies of hy3 mice found that the cilia were lost, but only from the ependymal surfaces that were most affected by the raised intraventricular pressure. Moreover, the development of hydrocephalus seemed to precede the loss of cilia. It was concluded that these ciliary defects are “most probably the result of the hydrocephalus, and not its cause” (Bannister and Chapman, 1980). However, based on our finding that hydin is a CP protein necessary for flagellar motility in C. reinhardtii, we postulate that hydrocephalus caused by mutations in Hydin involves defects in the CP apparatus, resulting in impaired ciliary motility and probably subsequent ciliary degeneration.
The literature also strongly suggests a connection between ciliary defects and hydrocephalus in humans (Kosaki et al., 2004; for reviews see Ibanez-Tallon et al., 2003; Afzelius, 2004). For example, in humans with mutations in the axonemal dynein heavy chain DNAH5, the incidence of hydrocephalus is 83 times higher than that in the general population (Ibanez-Tallon et al., 2004), although not all patients with ciliary defects develop hydrocephalus. At least one case of human hydrocephalus has been mapped to within 1.2 Mb of HYDIN in human chromosome band 16q22.2 (Callen et al., 1990). This, together with the clear connection between mutations in Hydin and hydrocephalus in the mouse, make HYDIN a strong candidate for causing hydrocephalus in humans. Our findings indicate that, if hydrocephalus is indeed caused by mutations in HYDIN, then these patients probably develop the disease as a result of defects in the CP of the ependymal cilia.
Materials and methods
Strains and culture conditions
C. reinhardtii strains used in the work include CC3395 (arg7-8, cwd, mt−), CC124 (agg1, nit1, nit2, mt−), and cpc1-1 (CC-3706; arg7, mt+), all from the Chlamydomonas Genetics Center. pf15a was obtained from R.P. Levine (Harvard University, Cambridge, MA), whereas pf16, pf18, pf19, and pf20 were R. Lewin isolates originally obtained from the Culture Collection of Algae and Protozoa (Cambridge, UK); all have been maintained in this laboratory since 1974. Cells were grown in M medium I with 2.2 mM KH2PO4 and 1.71 mM K2HPO4 (Sager and Granick, 1953) or TAP (Gorman and Levine, 1965) at 23°C with aeration and a light/dark-cycle of 14/10 h (Witman, 1986), or shaken in constant light.
Antibody production
A 1.3-kb fragment (fragment A) corresponding to exon 3 of HY3 in the U.S. Department of Energy's Joint Genome Institute's C. reinhardtii genome v. 2 was amplified by PCR from genomic DNA of CC3395 using primer pair hyf2a and hyr2a (see Tables S1 and S2, available at http://www.jcb.org/cgi/content/full/jcb.200611115/DC1, for primers and PCR conditions). The PCR product was digested with HindIII and BamHI and ligated into pMAL-cR1 v. 2 digested with the same enzymes (New England Biolabs, Inc.). The construct was transformed into E. coli XL1 blue, and, for expression of the maltose-binding∷hydin fusion protein, into BL21. The fusion protein was purified by amylose affinity chromatography, and antibodies were produced in rabbits (CRP, Inc.) using the company's standard protocol. Fragment A was digested with BamHI and SalI and ligated into pGEX-6P-1 (GE Healthcare) restricted with the same enzymes. After transformation into E. coli XL1blue, the GST∷hydin-fusion protein was purified by affinity chromatography, subjected to SDS-PAGE (7.5%), transferred onto polyvinylidene difluoride membrane, stained with Ponceau S, and excised from the membrane using a razor blade. The immobilized fusion protein was used for affinity purification of anti-hydin from bleeds 1, 2, and 4.
RNAi construct and transformation
We constructed a novel expression vector based on the C. reinhardtii LC8 gene; this is an intronless gene that encodes the dynein light chain LC8 and is inducible by deflagellation. The upstream region of LC8, including the first and second codon, was amplified from genomic DNA by PCR using primers LC8f1-3 and LC8r1-2 and GoTaq Flexi DNA polymerase (Promega). The downstream region of LC8 was amplified using primers LC8r2_2 and LC8f1-3, and the products were restricted with HindIII–BamHI and BamHI–EcoRI, respectively. Both products were then ligated into pUC119 digested with HindIII and EcoRI. The resulting plasmid (pLC8S) was digested with BamHI, and a triple HA tag, amplified from p3xHA (Silflow et al., 2001) using primers HAf2tm68 and Har2 and restricted with BamHI and BglII, was inserted, resulting in plasmid pKL3. Fragment S covering the first three exons and introns of HY3 was amplified by PCR using primers hyf1s and hyr1s. Fragments A (see above) and S were restricted with BamHI–HindIII and HindIII–XbaI, respectively, and ligated into pKL3 digested with NheI and BamHI. The resulting plasmid, pKL3-hyAS, was linearized with EcoRI or DraI. Cotransformation was performed using the glass bead method (Kindle, 1990) and ARG7 as a selectable marker.
Preparation of axonemes and CP extrusion
Flagella were isolated from C. reinhardtii as described previously by Witman (1986). For Western blot analysis, flagella were extracted with 1% NP-40 for 5 min on ice, axonemes were collected by centrifugation (30,000 g, 20 min, 4°C), and the detergent-soluble membrane-plus-matrix and axoneme fractions were prepared for SDS-PAGE. For further fractionation, axonemes were extracted with 0.6 M KCl for 30 min on ice, pelleted, and extracted with 0.5 M KI for 20 min on ice. Extrusion of the CP was induced as previously described (Kamiya, 1982; Hosokawa and Miki-Noumura, 1987) with the following modifications: ATP (1 mM final concentration) and trypsin (5 μg/ml final concentration; Invitrogen) were added to isolated flagella in HMDEK-PEG (30 mM Hepes, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM KCl, and 0.5% PEG 20,000) at room temperature. Flagella were then demembranated by addition of 1% Nonidet NP-40 (Calbiochem). After 2 min, aliquots were fixed with formaldehyde (2% final concentration), allowed to settle onto poly-l-lysine–coated multiwell slides for 2–8 min, and submerged into −20°C methanol for 6–10 min. For complete extrusion of CP microtubules, more ATP (1 mM) was added 2 and 4 min after flagellar demembranation, and samples were processed as described after a total of 6 min of incubation. Methanol-fixed specimens were processed for immunofluorescence microscopy.
RNA and DNA isolation
DNA was isolated from C. reinhardtii using PlantDNAzol reagent (Invitrogen) following the instructions of the manufacturer. TRIzol LS reagent (Invitrogen) was used for RNA isolation, and cDNA synthesis was performed using PowerScript Reverse Transcriptase (CLONTECH Laboratories, Inc.) using the manufacturer's protocol, except that cDNA synthesis was performed for 50 min at 42°C, 20 min at 48°C, and 20 min at 55°C. The relative amount of cDNA representing HY3 message in samples was determined by real-time PCR using primers C_410060F and C_410060R that spanned intron 4; QuantiTect SYBR green PCR master mix (QIAGEN) was used to monitor amplification. The relative amount of the G protein β subunit (Schloss, 1990), which is constantly expressed under various conditions, was measured in each trial and used to correct for slight differences in amount of cDNA in each sample. Up to three independent sets of RNA were isolated and analyzed.
Electron and immunofluorescence microscopy
Cells were fixed in glutaraldehyde for EM (Hoops and Witman, 1983) and processed as described previously (Wilkerson et al., 1995). For whole mount immunogold EM, CPs were extruded as described above but omitting trypsin. After 22–40 min, the suspension was applied to carbon/formvar-coated grids and fixed with 2.5–3% formaldehyde. Immunostaining was performed as described by Bernstein et al. (1994), and specimens were analyzed using electron microscopes (CM10 or -12; Philips). Cells were fixed and stained for immunofluorescence microscopy as described by Lechtreck and Geimer (2000) with the following modifications: 1% polyethylenimine was used to immobilize strains with a cell wall, and primary antibodies were applied overnight at 4°C. See Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200611115/DC1, for antibodies and dilutions used in this study. After washing, specimens were mounted with ProLong Gold (Invitrogen). Images were acquired at room temperature using AxioVision software and a camera (AxioCam MRm) on a microscope (Axioskop 2 Plus) equipped with a 100×/1.4 oil differential interference contrast (DIC) Plan-Apochromat objective (Carl Zeiss Microimaging, Inc.) and epifluorescence. Image brightness and contrast were adjusted using Photoshop 5.0 and 6.0 (Adobe). Figures for publication were assembled using Illustrator 8.0 (Adobe). Capture times and adjustments were similar for images mounted together. Averaged images were prepared in Photoshop by setting the original images to 16% opacity and aligning them manually.
Online supplemental material
Fig. S1 shows the induction of hydin transcripts after deflagellation, providing additional evidence that hydin is a flagellar protein. Video 1 shows a C. reinhardtii hydin RNAi cell of strain hyN3 in which the flagellum in the hands-up position beats more frequently than the other flagellum. Video 2 shows a hydin RNAi cell of strain hyN4 with the more active flagellum in the hands-up position and the other in the hands-down position. Video 3 shows a hydin RNAi cell of strain hyS1, in which the more active flagellum rests mostly in the hands-up position and the other flagellum in the hands-down position. Tables S1, S2, and S3 show PCR primers, PCR conditions, and antibodies, respectively, used in this work. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200611115/DC1.
Acknowledgments
We thank David Mitchell (State University of New York), Joel Rosenbaum (Yale University), and Elizabeth Smith (Dartmouth College) for antibody gifts, and Gregory Hendricks (University of Massachusetts Medical School) for help with the electron microscopy.
This work was supported by National Institutes of Health grant G30626 and by the Robert W. Booth Fund at the Greater Worcester Community Foundation. The University of Massachusetts Medical School EM Facility used in this research was partially supported by grant P30 DK32520.
Abbreviations used in this paper: CP, central pair; CSF, cerebrospinal fluid; DIC, differential interference contrast.
References
- Adams, G.M., B. Huang, G. Piperno, and D.J. Luck. 1981. Central-pair microtubular complex of Chlamydomonas flagella: polypeptide composition as revealed by analysis of mutants. J. Cell Biol. 91:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzelius, B.A. 2004. Cilia-related diseases. J. Pathol. 204:470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, M., O.A.T. Peltoniemi, A. Maekinen, A. Sukura, and H. Rodriguez-Martinez. 2000. The hereditary ‘short tail’ sperm defect—a new reproductive problem in Yorkshire boars. Reprod. Domest. Anim. 35:59–63. [Google Scholar]
- Banizs, B., M.M. Pike, C.L. Millican, W.B. Ferguson, P. Komlosi, J. Sheetz, P.D. Bell, E.M. Schwiebert, and B.K. Yoder. 2005. Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development. 132:5329–5339. [DOI] [PubMed] [Google Scholar]
- Bannister, C.M., and S.A. Chapman. 1980. Ventricular ependyma of normal and hydrocephalic subjects: a scanning electronmicroscopic study. Dev. Med. Child Neurol. 22:725–735. [DOI] [PubMed] [Google Scholar]
- Bernstein, M., and J.L. Rosenbaum. 1994. Kinesin-like proteins in the flagella of Chlamydomonas. Trends Cell Biol. 4:236–240. [DOI] [PubMed] [Google Scholar]
- Bernstein, M., P.L. Beech, S.G. Katz, and J.L. Rosenbaum. 1994. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhead, R., H.R. Dawe, H. Farr, S. Griffiths, S.R. Hart, N. Portman, M.K. Shaw, M.L. Ginger, S.J. Gaskell, P.G. McKean, and K. Gull. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 440:224–227. [DOI] [PubMed] [Google Scholar]
- Callen, D.F., E.G. Baker, and S.A. Lane. 1990. Re-evaluation of GM2346 from a del(16)(q22) to t(4;16)(q35;q22.1). Clin. Genet. 38:466–468. [DOI] [PubMed] [Google Scholar]
- Chen, J., H.J. Knowles, J.L. Hebert, and B.P. Hackett. 1998. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J. Clin. Invest. 102:1077–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D.G., D.R. Diener, A.L. Himelblau, P.L. Beech, J.C. Fuster, and J.L. Rosenbaum. 1998. Chlamydomonas kinesin-II–dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy, B.E., and M.L. Robinson. 2003. Congenital hydrocephalus in hy3 mice is caused by a frameshift mutation in Hydin, a large novel gene. Hum. Mol. Genet. 12:1163–1170. [DOI] [PubMed] [Google Scholar]
- Gorman, D.S., and R.P. Levine. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA. 54:1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops, H.J., and G.B. Witman. 1983. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol. 97:902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, Y., and T. Miki-Noumura. 1987. Bending motion of Chlamydomonas axonemes after extrusion of central-pair microtubules. J. Cell Biol. 105:1297–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., G.J. Pazour, and G.B. Witman. 2004. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 15:4382–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Tallon, I., S. Gorokhova, and N. Heintz. 2002. Loss of function of axonemal dynein Mdnah5 causes primary ciliary dyskinesia and hydrocephalus. Hum. Mol. Genet. 11:715–721. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon, I., N. Heintz, and H. Omran. 2003. To beat or not to beat: roles of cilia in development and disease. Hum. Mol. Genet. 12:R27–R35. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon, I., A. Pagenstecher, M. Fliegauf, H. Olbrich, A. Kispert, U.P. Ketelsen, A. North, N. Heintz, and H. Omran. 2004. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Hum. Mol. Genet. 13:2133–2141. [DOI] [PubMed] [Google Scholar]
- Kamiya, R. 1982. Extrusion and Rotation of the central-pair microtubules in detergent-treated Chlamydomonas flagella. Prog. Clin. Biol. Res. 80:169–173. [DOI] [PubMed] [Google Scholar]
- Kamiya, R., and G.B. Witman. 1984. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J. Cell Biol. 98:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle, K.L. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 87:1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M., and G.B. Witman. 1990. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J. Biol. Chem. 265:19807–19811. [PubMed] [Google Scholar]
- Koblenz, B., J. Schoppmeier, A. Grunow, and K.F. Lechtreck. 2003. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 116:2635–2646. [DOI] [PubMed] [Google Scholar]
- Kosaki, K., K. Ikeda, K. Miyakoshi, M. Ueno, R. Kosaki, D. Takahashi, M. Tanaka, C. Torikata, Y. Yoshimura, and T. Takahashi. 2004. Absent inner dynein arms in a fetus with familial hydrocephalus-situs abnormality. Am. J. Med. Genet. A. 129:308–311. [DOI] [PubMed] [Google Scholar]
- Lechtreck, K.F., and S. Geimer. 2000. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil. Cytoskeleton. 47:219–235. [DOI] [PubMed] [Google Scholar]
- Li, J.B., J.M. Gerdes, C.J. Haycraft, Y. Fan, T.M. Teslovich, H. May-Simera, H. Li, O.E. Blacque, L. Li, C.C. Leitch, et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 117:541–552. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R. 2003. Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil. Cytoskeleton. 56:120–129. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R., and W.S. Sale. 1999. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144:293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, J.H., M.J. Lee-Tischler, H.Y. Kwon, J.J. Schrick, E.D. Avner, W.E. Sweeney, V.L. Godfrey, N.L. Cacheiro, J.E. Wilkinson, and R.P. Woychik. 1994. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 264:1329–1333. [DOI] [PubMed] [Google Scholar]
- Omoto, C.K., I.R. Gibbons, R. Kamiya, C. Shingyoji, K. Takahashi, and G.B. Witman. 1999. Rotation of the central pair microtubules in eukaryotic flagella. Mol. Biol. Cell. 10:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., B.L. Dickert, Y. Vucica, E.S. Seeley, J.L. Rosenbaum, G.B. Witman, and D.G. Cole. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151:709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., N. Agrin, J. Leszyk, and G.B. Witman. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P. 2006. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics. 22:1031–1035. [DOI] [PubMed] [Google Scholar]
- Rupp, G., E. O'Toole, and M.E. Porter. 2001. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell. 12:739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager, R., and S. Granick. 1953. Nutritional studies with Chlamydomonas reinhardi. Ann. N. Y. Acad. Sci. 56:831–838. [DOI] [PubMed] [Google Scholar]
- Sapiro, R., I. Kostetskii, P. Olds-Clarke, G.L. Gerton, G.L. Radice, and I.J. Strauss. 2002. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell. Biol. 22:6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir, P., and T. Matsuoka. 1989. Splitting the ciliary axoneme: implications for a “switch-point” model of dynein arm activity in ciliary motion. Cell Motil. Cytoskeleton. 14:345–358. [DOI] [PubMed] [Google Scholar]
- Schloss, J.A. 1990. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 221:443–452. [DOI] [PubMed] [Google Scholar]
- Schrander-Stumpel, C., and J.P. Fryns. 1998. Congenital hydrocephalus: nosology and guidelines for clinical approach and genetic counselling. Eur. J. Pediatr. 157:355–362. [DOI] [PubMed] [Google Scholar]
- Silflow, C.D., M. LaVoie, L.W. Tam, S. Tousey, M. Sanders, W. Wu, M. Borodovsky, and P.A. Lefebvre. 2001. The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironen, A., B. Thomsen, M. Andersson, V. Ahola, and J. Vilkki. 2006. An intronic insertion in KPL2 results in aberrant splicing and causes the immotile short-tail sperm defect in the pig. Proc. Natl. Acad. Sci. USA. 103:5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E.F., and P. Yang. 2004. The radial spokes and central apparatus: mechano- chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton. 57:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo, M.J., E.E. Dymek, and E.F. Smith. 2005. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J. Cell Sci. 118:4655–4665. [DOI] [PubMed] [Google Scholar]
- Wilkerson, C.G., S.M. King, A. Koutoulis, G.J. Pazour, and G.B. Witman. 1995. The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280–290. [DOI] [PubMed] [Google Scholar]
- Witman, G.B., K. Carlson, J. Berliner, and J.L. Rosenbaum. 1972. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54:507–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B., J. Plummer, and G. Sander. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, R., E. O'Toole, S. Ghosh, and D.R. Mitchell. 2004. Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA. 101:17398–17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala, M.A., M.R. Knowles, and H. Omran. 2006. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed]
- Zhang, H., and D.R. Mitchell. 2004. Cpc1, a Chlamydomonas central pair protein with an adenylate kinase domain. J. Cell Sci. 117:4179–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]