Abstract
COUP-TF-interacting protein 2 (CTIP2), also known as Bcl11b, is a transcriptional regulatory protein that is highly expressed in and plays a critical role(s) during development of T lymphocytes and the central nervous system. We demonstrate herein that CTIP2 is also highly expressed in mouse skin during embryogenesis and in adulthood as revealed by immunohistochemical analyses. CTIP2 expression in the ectoderm was first detected at embryonic day 10.5 (E10.5), and became increasingly restricted to proliferating cells of the basal cell layer of the developing epidermis in later stages of fetal development and in adult skin. In addition, CTIP2 expression was also detected in some cells of the suprabasal layer of the developing epidermis, as well as in developing and mature hair follicles. Relatively fewer cells of the developing dermal component of skin were found to express CTIP2, and the adult dermis was devoid of CTIP2 expression. Some, but not all, of the cells present within hair follicle bulge were found to co-express CTIP2, keratin K15, and CD34, indicating that a subset of K15+ CD34+ skin stem cells may express CTIP2. Considered together, these findings suggest that CTIP2 may play a role(s) in skin development and/or homeostasis.
Keywords: COUP-TF-interacting protein 2, Bcl11b, nuclear receptor, Transcription, expression, zinc finger, skin, epidermis, dermis, ectoderm, mesoderm, keratinocyte, stratification, development, stem cells, proliferation, differentiation, morphogenesis, basal cell, suprabasal cell, hair bulb, hair follicle, hair bulge, immunohistochemistry, in situ hybridization, marker, mouse, embryo, K10, K14, K15, Ki67, CD34
1. Results and discussion
CTIP2 (Chicken ovalbumin upstream promoter transcription factor (COUP-TF)–interacting protein 2), also known as Bcl11b, is a C2H2 zinc finger protein (Avram et al., 2000) that has been shown to repress transcription though interaction with COUP-TF nuclear receptor proteins as well as through direct, sequence-specific DNA binding (Avram et al., 2002). CTIP2 is required for normal T cell development and CTIP2-null mice exhibit arrested thymocyte development (Wakabayashi et al., 2003b). Additionally, deregulation of CTIP2 may be implicated in immune system malignant transformation (Wakabayashi et al., 2003a; Bezrookove et al., 2004; Kamimura et al., 2007). It was shown that CTIP2 is also expressed in layer V of cerebral cortex and plays a critical role in the establishment of connections of corticospinal motor neurons to the spinal cord (Arlotta et al., 2005).
Mouse epidermis develops from a single-layered embryonic ectoderm (Mack et al., 2005). Subsequent stratification events lead to the formation of the periderm (around E9-E12), which overlies the ectoderm (Byrne et al., 2003; Mack et al., 2005). Cells of this two-layered epidermis then undergo a series of proliferation and terminal differentiation events which results in the formation of new cell layers of the future epidermis. Formation of the mature epidermis is completed by E18, at which the epidermis consists of four layers: the basal, spinous, granular and cornified layer (Mack et al., 2005).
Epidermis undergoes constant renewal, which is required to maintain normal tissue homeostasis. This is possible due to the presence of two populations of proliferating cells: transit amplifying cells with limited proliferative potential and keratinocyte stem cells, which are slow-cycling cells with high proliferative capacity (Lavker et al., 1993; Slack, 2000).
Previous RNA in situ hybridization using a CTIP2 RNA probe performed in our laboratory demonstrated that CTIP2 was highly expressed in developing and mature central nervous system and spinal cord as well as in the thymus (Leid et al., 2004). The epidermis was not specifically identified as a site of CTIP2 expression in the previous in situ hybridization studies, although CTIP2 mRNA is found in the skin (see Fig. 1G and I from Leid et al., 2004). Preliminary attempts to define CTIP2 expression pattern during mouse embryogenesis using a CTIP2-specific monoclonal antibody revealed high-level expression of CTIP2 in developing skin. To our knowledge this is the first evidence for expression of CTIP2 in skin, during development or in the adulthood, and it therefore provided a rationale to perform additional analyses.
Figure 1. Expression of CTIP2 in the mouse fetal skin.
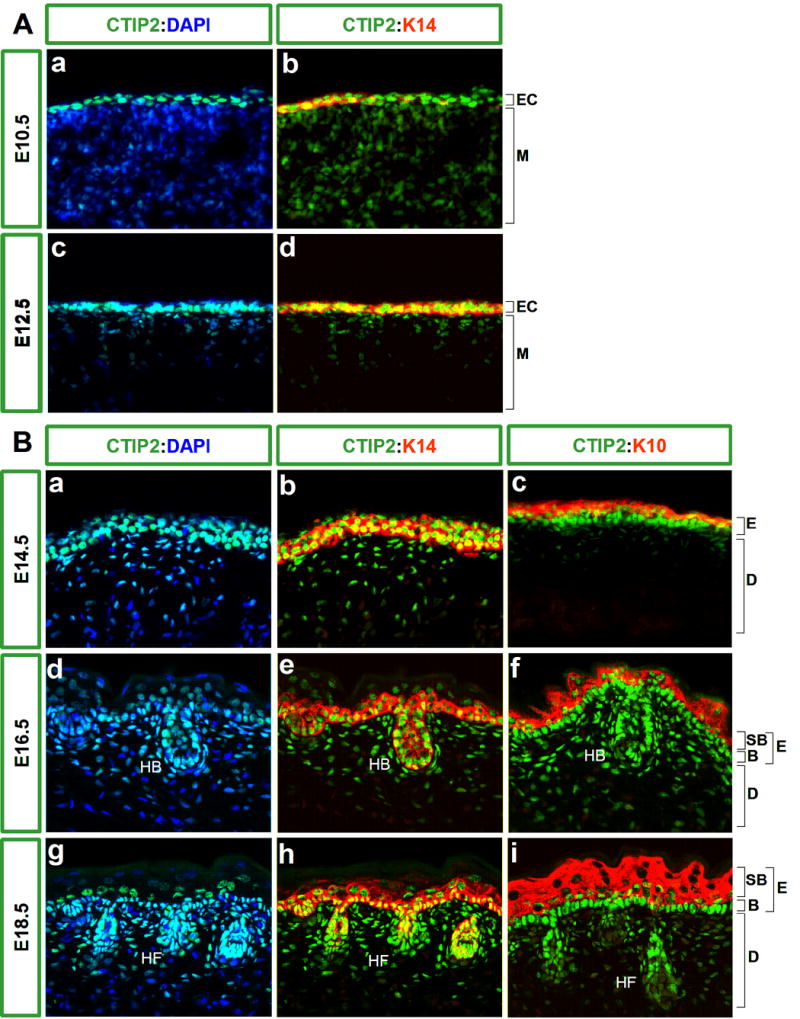
Immunohistochemistry was performed on 10 μm-thick frozen sections of wild type embryos using antibodies directed against CTIP2, K14 and K10. A, CTIP2 (in green) is highly expressed in the ectoderm at E10.5 (upper panel) and E12.5 (lower panel) and is co-localized with the expression of K14 (in red). B, high expression of CTIP2 was observed in the basal cells and upper layers of the epidermis of E14.5 (upper panel), E16.5 (middle panel) and E18.5 embryos (lower panel). K14 and K10 staining (in red) were used to label basal cells and suprabasal layers, respectively. E16.5 and E18.5 stages of development show high expression of CTIP2 in the basal layer of epidermis as well as in the dermis and hair follicles. All sections were counterstained with DAPI (in blue). Images were taken at 40X magnification. Abbreviation: EC- ectoderm, M-mesoderm, E-epidermis, D-dermis, B-basal cell layer, SB- suprabasal cell layers, HB-hair bulb, HF-hair follicle.
1.1 Expression of CTIP2 during epidermal morphogenesis
To characterize the expression profile of CTIP2 during mouse skin ontogenesis, we performed immunohistochemistry at different stages of development using an anti-CTIP2 monoclonal antibody, which has been previously described (Senawong et al., 2003; Arlotta et al., 2005; Topark-Ngarm et al., 2006).
CTIP2 expression was detected in the ectoderm as early as embryonic day 10.5 (E10.5), where CTIP2-positive cells were found in the outermost layer of the cross-section of a developing fetus (Fig.1A, a). Some of these cells were already expressing the basal cell marker keratin 14 (K14) (Fig.1A, b), which signifies a commitment of these cells to give rise to stratified epithelia, (Byrne et al., 1994).
Expression of CTIP2 persisted at E12.5 (Fig.1A, c). This stage of skin development is marked by formation and stratification of the embryonic basal layer and all cells of that layer were found to be positive for CTIP2 expression (Fig.1A). CTIP2 expression precisely co-localized with basal cell marker K14 (Vassar et al., 1989) at E12.5 (Fig.1A, d).
Strong expression of CTIP2 was detected in the rapidly dividing basal cell layer at E14.5, and expression of CTIP2 appeared to be co-localized with that of K14 (Fig.1B, a and b). At this stage, the early differentiating layers started to form, and were identified using differentiation marker keratin 10 (K10) (Byrne et al., 1994). CTIP2 expression also extended to some cells in the differentiating suprabasal cell layer at E14.5, as seen by co-localization of CTIP2 and K10 staining (Fig.1B, c). However, the level of CTIP2 expression in the suprabasal cell appeared to be lower than that of basal cells (Fig.1B, compare b and c). In addition, some cells of the future dermis were also found to express CTIP2 at this developmental stage (Fig.1B, a).
CTIP2 expression was further investigated at E16.5 and E18.5 (Fig. 1B, d-i). High levels of CTIP2 expression were consistently observed in the basal layer of the epidermis at these two stages (Fig.1B d, e, g, h). Some CTIP2 positive cells were observed in the suprabasal layers that expressed high levels of K10 at E16.5 (Fig.1B, f), whereas the suprabasal expression of CTIP2 at E18.5 was primarily restricted to a small number of K10-positive cells that were adjacent to the basal cells (Fig.1B, I; see supplemental Fig. 1).
Later stages of skin morphogenesis were marked by the formation of hair follicles, and anti-CTIP2 antibody robustly stained hair bulbs and follicles at E16.5 and E18.5, respectively (Fig.1B, d and g). These stages were also characterized by expansion of the dermal compartment of the skin, and some of these dermal cells were also found to express CTIP2 (Fig.1B, d and g, and data not shown).
1.2 CTIP2 expression in proliferating cells
To evaluate the proliferation status of CTIP2-expressing cells we performed co-labeling studies using anti-CTIP2 and an antibody against the proliferation marker Ki67 (Schluter et al., 1993) at different stages during epidermal morphogenesis (Fig.2). At early stages of development (E10.5), almost all cells of the fetus were positive for Ki67 expression (Fig.2, b and data not shown). This was expected as early stages of fetal development are marked by rapid and ubiquitous proliferation. All cells of the ectodermal compartment, as well as those of the underlying mesenchyme were found to express Ki67 (Fig.2, b). Two days later (at E12.5) virtually all cells of the ectoderm were positive for Ki67 expression (Fig.2, e). Most, but not all, cells of mesenchymal origin were negative for Ki67 staining at this developmental stage (Fig.2, f).
Figure 2. CTIP2 expression in proliferating cells.
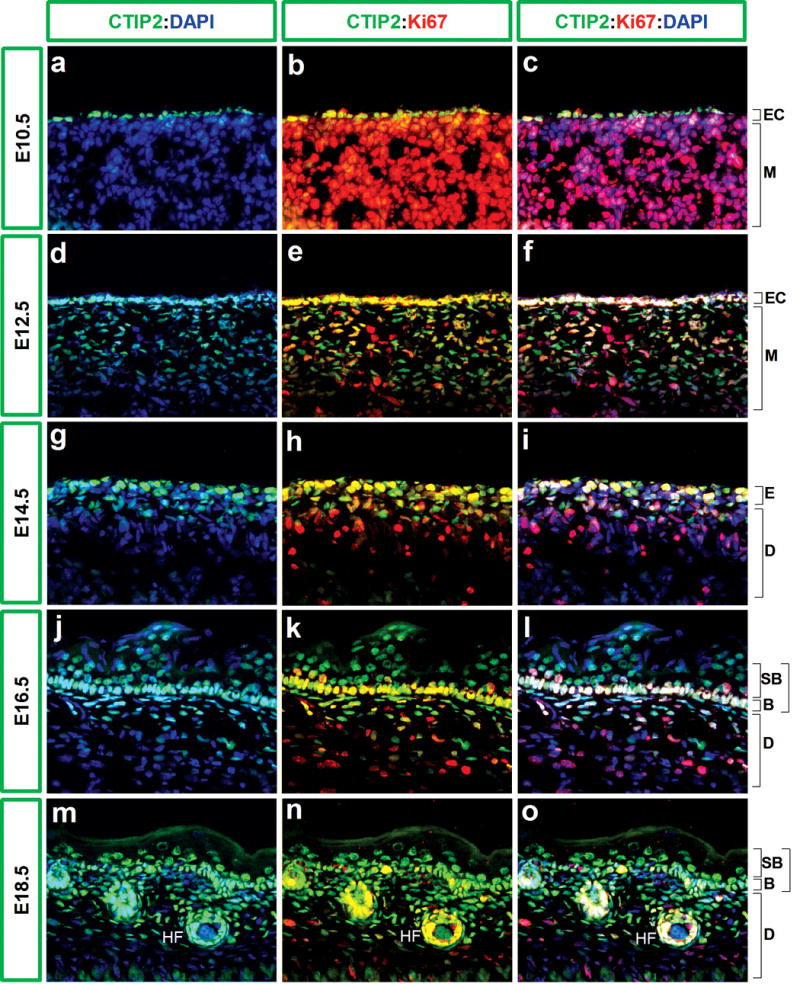
10 μm-thick frozen section of wild type fetuses at different stages of development were analyzed immunohistochemically using antibodies directed against CTIP2 (green) and Ki67 (red) and counterstained with DAPI (blue). Images were taken at 40X magnification. Abbreviations are described in the legend of Fig. 1.
The basal cell layer of epidermis is highly proliferative at E14.5 (Mack et al., 2005), which is reflected in the presence of multiple Ki67-positive cells (Fig.2, h-i). A smaller fraction of cells of the future dermis were positive for Ki67 staining at this stage (Fig.2, h-i), and all layers of the developing skin became less proliferative with developmental progression. For example, only a fraction of basal cells, as well as those cells of the dermal compartment, were positive for Ki67 staining at E16.5 (Fig.2, k-l). By E18.5 only a few cells were still proliferating in the basal cell layer, and the dermis was mostly non-proliferative. Interestingly, epithelia of the developing hair follicles, which were invaginating into the dermis, still harbored a considerable number with Ki67-positive cells (Fig.2, n).
At early, highly proliferative stages of development (E10.5-E14.5) almost all of the CTIP2-positive epidermal cells were found to be dividing as indicated by co-localization of CTIP2 and Ki67 staining (Fig.2, a-i). The total number of Ki67-positive cells was reduced in the epidermis at E16.5 but, in general, cells that were positive for Ki67 staining were also found to be positive for CTIP2 expression (Fig.2 j-l). By E18.5, the number of proliferating cells in fetal skin was comparatively reduced, but again a correspondence was observed between the Ki67 staining and that of CTIP2, indicating that majority of proliferating cells express CTIP2. Similarly, some CTIP2-positive cells were detected in the developing hair follicle epithelium, and these cells were also positive for Ki67 expression (Fig.2, m-o).
1.3 Expression of CTIP2 in adult skin
Immunohistochemistry was performed on 8-10-week old adult mouse skin using anti-CTIP2, -K14 and -K10 antibodies to evaluate the possibility that CTIP2 may be expressed in adult skin (Fig. 3). In the adult skin, CTIP2 expression was found in the majority, if not all cells of the basal layer (Fig. 3, a and b), hair follicles and some cells of the suprabasal K10-positive layers (Fig. 3, c), which is similar to the results that we obtained during embryogenesis (Fig. 1). In contrast to the developing dermis, CTIP2 expression was not detected in the dermal compartment of the adult skin (compare Fig. 3,a and Fig. 1B, g). Immunoblot analysis of CTIP2 in protein extracts from dorsal skin biopsies confirmed expression of CTIP2 in fetal (E18.5) and adult skin (Fig. 4, top panel).
Figure 3. Expression of CTIP2 in the adult mouse skin.
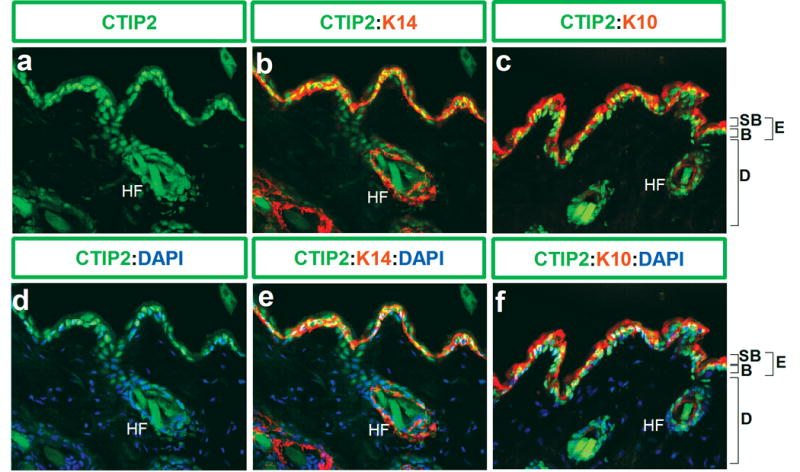
10 μm-thick frozen skin sections obtained from dorsal skin biopsies of 8-10-week old C7/BL6 mice were stained with anti-CTIP2 (green), -K14 (red), -K10 (red) antibodies and counterstained with DAPI (blue), and photographed using 40X objective. Abbreviation are described in the legend of Fig. 1.
Figure 4. Immunoblot analysis of CTIP2 expression in fetal and adult skin.
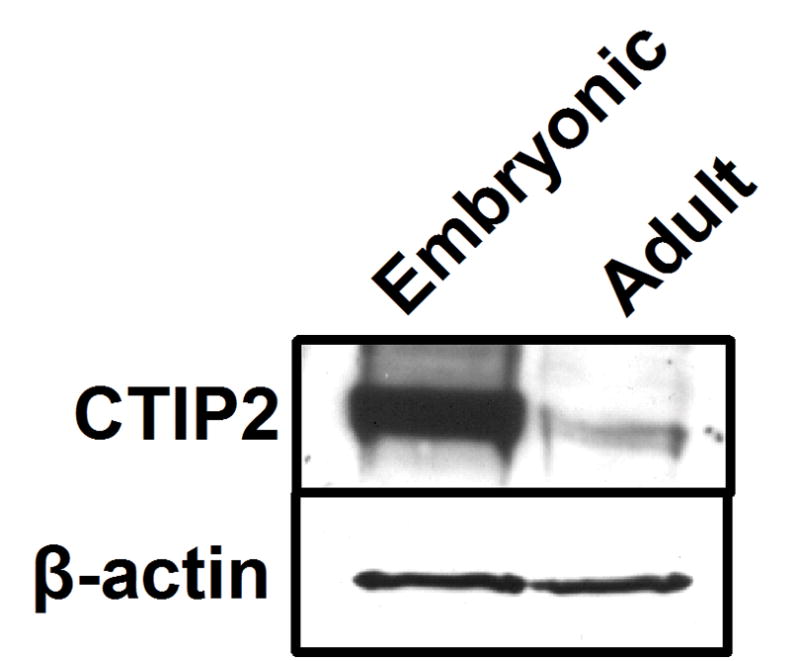
Whole skin protein extracts were prepared from adult (8-week old) and fetal (E18.5) dorsal skin biopsies and analyzed using CTIP2-specific antibody (upper panel). β-actin was used as an internal loading control (lower panel).
Expression of CTIP2 in whole skin, protein extracts of the E18.5 embryo was found to be significantly higher that that of the adult skin. This could be partially explained by our observation that CTIP2 expression is maintained in the epidermal basal and suprabasal cells throughout development, and into the adulthood, whereas the adult dermal compartment, which is dramatically expanded relative to fetal skin, does not express significant amounts of CTIP2.
1.4 CTIP2 expression in stem cells present in the bulge of the adult hair follicle
The “bulge” in the outer root sheath (ORS) of the hair follicle is believed to be the site of keratinocyte epithelial stem cells that contributes to morphogenesis of the hair follicle, and adjacent interfollicular epidermis (Lavker and Sun, 2000; Khavari, 2004). Two markers of bulge stem cells are known to date: keratin 15 (K15) (Liu et al., 2003), and CD34 (Trempus et al., 2003), which is also a hematopoietic stem cell marker (Krause et al., 1996). As our immunohistochemistry data in the adult tissue revealed that CTIP2 was highly expressed in the hair follicle in adult skin, we performed co-staining of CTIP2 with K15 and CD34 to investigate if CTIP2 is expressed in stem cells of the hair bulge. As expected, K15 and CD34 specifically labeled keratinocytes in the bulge region of adult mouse hair follicle (Fig. 5A and B). CTIP2 was found to be co-expressed in some, but not all, of the keratinocytes in the bulge region with K15 and CD34 expression. In particular, keratinocytes of the outermost cell layer and those located in lower part of the “bulge” stained positively for CTIP2 (Fig. 5A and B, white arrows), while other K15+CD34+ cells located more medially within the hair bulge did not. Our findings support the idea that hair follicle “bulge” consists of a heterogeneous cell population of keratinocyte stem and/or progenitor cells (Cotsarelis et al., 1990; Morris and Potten, 1994; Lavker and Sun, 2000). The positive staining of CTIP2 in the peripheral cells of the bulge suggests that CTIP2 might label the cells as they exit the stem/progenitor population. Further studies will be required to validate this hypothesis.
Figure 5. Expression of CTIP2 in stem cells of the bulge region of the mouse hair follicle.
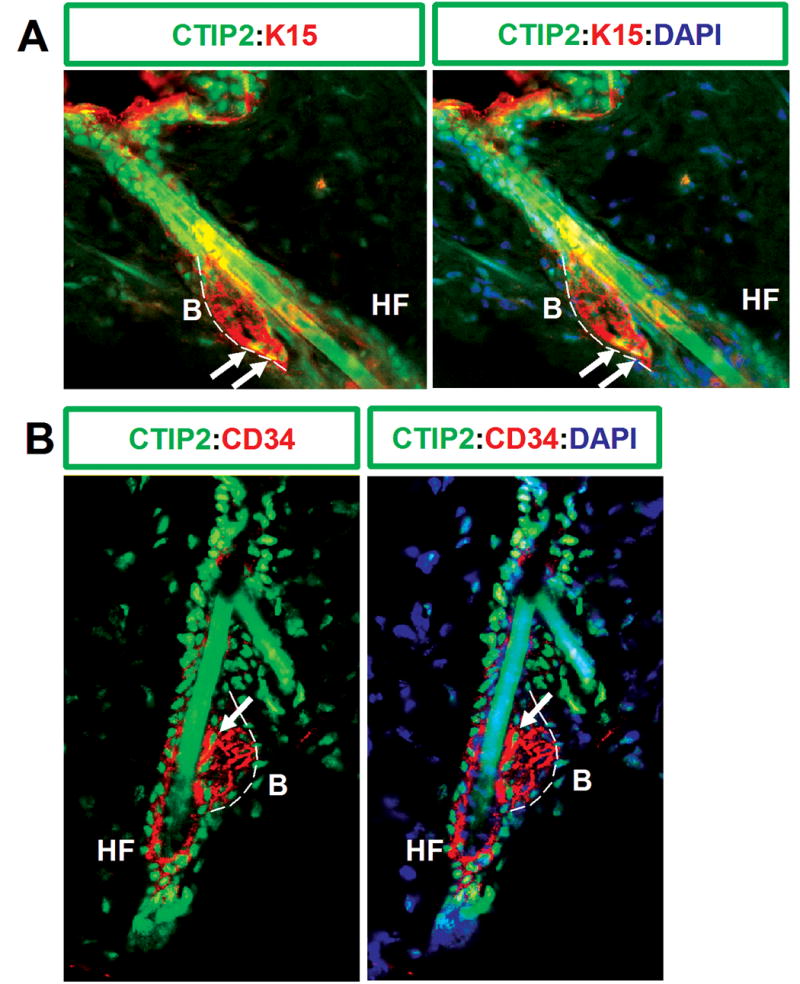
10 μm-thick frozen skin sections obtained from dorsal skin biopsies 8-week old mice were co-stained with CTIP2 (green) and K15 (red: A) or CTIP2 (green) and CD34 (red: B) and images of hair follicles in the bulge region were taken at 40X magnification. Sections were counterstained with DAPI. White arrows indicate CTIP2 positive cells in the bulge region that are also positive for K15 or CD34. Abbreviation: B-bulge, HF-hair follicle.
1. Experimental procedures
2.1. Sample preparation
Wild type fetuses from CD1 outbred mice strain (ICR or Swiss Mice from Charles River) were collected at stages E10.5, E12.5, E14.5, E16.5, E18.5, rinsed with PBS and fixed with 4% paraformaldehyde at 4°C. Fixed embryos were then washed with PBS overnight, cryopreserved in 30% sucrose, frozen in Optimum Cutting Temperature embedding compound (OCT) and stored at −80°C until used. For the purpose of developmental timing, 12:00 pm on the day after the night of mating was considered as E0.5. For adult skin studies, dorsal skin biopsies were taken from 8-10 week-old shaved mice and processed as described for fetal skin biopsies.
1.2. Immunohistochemistry
OCT-embedded embryos and adult skin samples were cross-sectioned at 10 μm intervals. Sections were mounted on “superfrost” slides and allowed to air dry. Sections were rinsed with PBS three times (5 min each) and permeabilized with ice-cold methanol for 3 min. Sections were allowed to dry and nonspecific binding of antibody was blocked with blocking buffer (0.3% Boehringer Mannheim Blocking reagent, 5% horse serum, 5% fetal Calf serum, 0.1 % triton X-100 in PBS) for 1 h. Slides were then incubated with rat anti-CTIP2 primary monoclonal antibody (Abcam product number 18465; clone 25B6) overnight in a humidified chamber. Primary antibody incubation was followed by three washes with PBST and incubation with fluorescently-labeled [Cy2 (1:250) or Cy3(1:500); Jackson ImmunoResearch] secondary antibody for 2 hours. Nuclei were counterstained with DAPI. Finally, sections were rinsed with PBST, dehydrated through sequential washes in 50, 70, 95 and 100% ethanol and then cleared in xylene. Slides were mounted with DPX mounting media and allowed to dry overnight. Images were captured at 40X magnification using Leica DMRA fluorescent microscope and Hamamatsu C4742-95 digital camera and processed using OpenLab software and Adobe Photoshop 7.0.
2.3 Whole skin protein extract preparation and Immunoblot analysis
Dorsal skin biopsies from E18.5 fetuses or adult mice were homogenized in RIPA buffer (50 mM Tris pH7.5, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1 %SDS, 150 mM NaCl, 5mM EDTA, proteinase inhibitors) using tissue grinder and cleared by centrifugation. Supernatants were collected and equal amounts of protein (determined using the Bio-Rad Protein Assay Kit) were analyzed by immunoblotting using antibodies against CTIP2 and β-actin as a loading control.
2.4 Antibodies
The following antibodies and their dilutions were used for immunohistochemical studies: anti-CTIP2 (Abcam, 1:300), -K14 (Covance, 1:1000), -K10 (Covance, 1:1000), -Ki67 (Abcam, 1:500), -K15 (Covance, 1:500), -CD34 (Santa Cruz, 1:50). Antibodies used for Western blot analysis: anti-CTIP2 (Abcam, 1:1000), -β-actin (Abcam, 1:4000).
Supplimental Section
10 μm-thick frozen sections of wild-type fetuses at E18.5 were analyzed immunohistochemically using antibodies directed against CTIP2 (green, panel a-c), K14 (red, middle panel b) and K10 (red, lower panel c) and counterstained with DAPI (blue). K10+ cells were detected in the suprabasal layers that stained negative for basal cell marker K14. Images were taken at 100X magnification. B, basal; S, suprabasal; D, dermis.
Acknowledgments
These studies were supported by a grant from the National Institutes of Health (GM60852) to ML and by a NIEHS Center grant (ES00210) to the Oregon State University Environmental Health Sciences Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezrookove V, van Zelderen-Bhola SL, Brink A, Szuhai K, Raap AK, Barge R, Beverstock GC, Rosenberg C. A novel t(6;14)(q25-q27;q32) in acute myelocytic leukemia involves the BCL11B gene. Cancer Genet Cytogenet. 2004;149:72–76. doi: 10.1016/s0165-4608(03)00302-9. [DOI] [PubMed] [Google Scholar]
- Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- Byrne C, Hardman M, Nield K. Covering the limb--formation of the integument. J Anat. 2003;202:113–123. doi: 10.1046/j.1469-7580.2003.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Kamimura K, Ohi H, Kubota T, Okazuka K, Yoshikai Y, Wakabayashi Y, Aoyagi Y, Mishima Y, Kominami R. Haploinsufficiency of Bcl11b for suppression of lymphomagenesis and thymocyte development. Biochem Biophys Res Commun. 2007;355:538–542. doi: 10.1016/j.bbrc.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Khavari PA. Profiling epithelial stem cells. Nat Biotechnol. 2004;22:393–394. doi: 10.1038/nbt0404-393. [DOI] [PubMed] [Google Scholar]
- Krause DS, Fackler MJ, Civin CI, May WS. CD34: structure, biology, and clinical utility. Blood. 1996;87:1–13. [PubMed] [Google Scholar]
- Lavker RM, Miller S, Wilson C, Cotsarelis G, Wei ZG, Yang JS, Sun TT. Hair follicle stem cells: their location, role in hair cycle, and involvement in skin tumor formation. J Invest Dermatol. 1993;101:16S–26S. doi: 10.1111/1523-1747.ep12362556. [DOI] [PubMed] [Google Scholar]
- Lavker RM, Sun TT. Epidermal stem cells: properties, markers, and location. Proc Natl Acad Sci U S A. 2000;97:13473–13475. doi: 10.1073/pnas.250380097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lyle S, Yang Z, Cotsarelis G. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J Invest Dermatol. 2003;121:963–968. doi: 10.1046/j.1523-1747.2003.12600.x. [DOI] [PubMed] [Google Scholar]
- Mack JA, Anand S, Maytin EV. Proliferation and cornification during development of the mammalian epidermis. Birth Defects Res C Embryo Today. 2005;75:314–329. doi: 10.1002/bdrc.20055. [DOI] [PubMed] [Google Scholar]
- Morris RJ, Potten CS. Slowly cycling (label-retaining) epidermal cells behave like clonogenic stem cells in vitro. Cell Prolif. 1994;27:279–289. doi: 10.1111/j.1365-2184.1994.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, Leid M. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J Biol Chem. 2003;78(44):43041–50. doi: 10.1074/jbc.M307477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter C, Duchrow M, Wohlenberg C, Becker MH, Key G, Flad HD, Gerdes J. The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM. Stem cells in epithelial tissues. Science. 2000;287:1431–1433. doi: 10.1126/science.287.5457.1431. [DOI] [PubMed] [Google Scholar]
- Topark-Ngarm A, Golonzhka O, Peterson VJ, Barrett B, Jr, Martinez B, Crofoot K, Filtz TM, Leid M. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J Biol Chem. 2006;281(43):32272–83. doi: 10.1074/jbc.M602776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, Tennant RW. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci U S A. 1989;86:1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Inoue J, Takahashi Y, Matsuki A, Kosugi-Okano H, Shinbo T, Mishima Y, Niwa O, Kominami R. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem Biophys Res Commun. 2003a;301:598–603. doi: 10.1016/s0006-291x(02)03069-3. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003b;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
10 μm-thick frozen sections of wild-type fetuses at E18.5 were analyzed immunohistochemically using antibodies directed against CTIP2 (green, panel a-c), K14 (red, middle panel b) and K10 (red, lower panel c) and counterstained with DAPI (blue). K10+ cells were detected in the suprabasal layers that stained negative for basal cell marker K14. Images were taken at 100X magnification. B, basal; S, suprabasal; D, dermis.


