Abstract
We have used isoform-specific RNA interference knockdowns to investigate the roles of myosin IIA (MIIA) and MIIB in the component processes that drive cell migration. Both isoforms reside outside of protrusions and act at a distance to regulate cell protrusion, signaling, and maturation of nascent adhesions. MIIA also controls the dynamics and size of adhesions in central regions of the cell and contributes to retraction and adhesion disassembly at the rear. In contrast, MIIB establishes front–back polarity and centrosome, Golgi, and nuclear orientation. Using ATPase- and contraction-deficient mutants of both MIIA and MIIB, we show a role for MIIB-dependent actin cross-linking in establishing front–back polarity. From these studies, MII emerges as a master regulator and integrator of cell migration. It mediates each of the major component processes that drive migration, e.g., polarization, protrusion, adhesion assembly and turnover, polarity, signaling, and tail retraction, and it integrates spatially separated processes.
Introduction
Cell migration is a highly regulated and coordinated process. It is comprised of several coupled steps that include polarization, protrusion, adhesion formation and turnover at the cell front, and adhesion disassembly and tail retraction at the cell rear. Many of the major regulatory pathways that control these processes are known (Zamir and Geiger, 2001; Ridley et al., 2003; Carragher and Frame, 2004; Webb et al., 2004). Most converge on Rho family GTPases, which in turn activate kinases like PAK or ROCK (Bokoch, 2003; Riento and Ridley, 2003). Recent studies point to other, analogous pathways that control protrusion, adhesion dynamics, and cell polarity. Cdc42 acting on MRCK, which is a kinase that phosphorylates MLC, regulates nuclear positioning in migrating cells (Gomes et al., 2005). In addition, PAK localizes to the centrosome, where it plays an essential role in MTOC positioning (Zhao et al., 2005). MII is a common effector for all of these pathways, and thus it is implicated as a key regulator of cell migration.
MII is a bipolar, contractile protein composed of two myosin heavy chains (MHCs), two regulatory myosin light chains (MLCs), and two essential MLCs. Each MHC contains an N-terminal globular motor domain that moves actin as it hydrolyzes ATP and a C-terminal tail region that binds to the other MHC (Landsverk and Epstein, 2005). MLC phosphorylation regulates the ATPase activity of MHC (Adelstein and Conti, 1975; Scholey et al., 1980). In addition to its contractile properties, MII also cross-links, and thus stabilizes, actin through its bivalent binding to actin filaments (Siddique et al., 2005).
In fibroblasts, two major isoforms of MHC-II have been described, MHC-IIA and -IIB. It is likely that they serve different roles in the regulation of the actin cytoskeleton because of their different ATPase activities, contraction rates, and subcellular localization (Kolega, 1998; Kim et al., 2005). Both MIIA and MIIB mediate stress fiber formation (Wei and Adelstein, 2000; Bao et al., 2005). MIIB contributes to cell migration by controlling protrusion stability (Lo et al., 2004), and MIIA is implicated in the regulation of actin retrograde flow (Cai et al., 2006).
Although these reports point to the participation of MII and its isoforms in migration, the mechanisms by which it controls and integrates its component processes are unclear. In this report, we reveal the integrative role of MII in migration and parse its isoform-dependent and contraction-independent activities. From these studies, MII emerges as a central, regulatory molecule that serves to integrate and coordinate diverse migration-related phenomena that comprise migration.
Results and discussion
MIIA and MIIB exert differential effects on polarity and tail retraction
Previous observations have shown the differential cellular localization of MII isoforms. In general, MIIA is present in regions distal to MIIB, and MII is largely absent from the lamellipodium of epithelial cells (Kolega, 1998; Gupton and Waterman-Storer, 2006). We have confirmed these observations using migrating CHO.K1 cells and reveal novel details (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1), as follows: (a) the two isoforms often decorate the same actin filaments in a stippled manner, suggesting that some functions might result from additive activities; (b) MIIA and MIIB likely mediate distinct functions because the two isoforms also occupy distinct areas, and therefore do not readily form cofilaments; and (c) MII resides well away from nascent adhesions; therefore, any effect on adhesion dynamics would result from an indirect rather than a local effect.
To determine whether the spatial segregation of MIIA and MIIB results in different roles during cell migration, we generated knockdown vectors that inhibit MIIA and MIIB expression with high specificity (Fig. 1, A and B). For both isoforms, down-regulation was comparable and maximal 96 h after transfection, where it averaged 75–95% by immunoblotting, depending on transfection efficiency (Fig. 1 A). Immunofluorescence revealed >95% knockdown in individual cells (Fig. 1 B).
Figure 1.
Knockdown of MIIA or MIIB differentially alters cell polarity. (A) CHO.K1 cells were transfected with pSUPER-GFP vector or pSUPER-GFP-RNAi against MIIA or MIIB, and blotted for MIIA or MIIB. GIT1 is a loading control. (B) Representative images of MIIA- (top) and MIIB-depleted cells (bottom) stained for MIIA or MIIB, respectively. Arrows point to transfected cells. (C–E) Time-lapse series of MIIA-depleted (C; Video 1), MIIB-depleted (D; Video 2), or control cells (E; Video 3). In C, arrowheads point to the defect in tail retraction. (D) Arrows point to transfected, unpolarized cells. Videos are representative of >25 cells in 6 independent experiments. (F) Fluorescence images depicting the enlargement of MIIB-deficient cells. Images are representative of >300 cells. (G; top) Effect of MIIB knockdown on cell area. Data are the mean ± the SD of 4 independent experiments comprising >300 cells/experiment. (bottom) Cell areas in control and MIIB-depleted cells. (H) MIIB-deficient cell showing clockwise rotation of the nucleus. Arrowhead points to nucleolus; top-right indicator, clockwise angular displacement (Video 4). (I) Depolarization of the Golgi is observed in MIIB-deficient cells (arrows) and not in nontransfected cells (arrowheads). Bars: (C) 40 μm; (D–F and H) 50 μm. Videos 1–4 are available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1.
When plated using migration-promoting conditions (see Materials and methods), MIIA-deficient cells exhibited broader lamellipodia than control cells and did not retract their trailing edge (Fig. 1 C and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1). This resulted in cells with extended tails (Fig. 1 C, arrowheads). This phenotype is reminiscent of the effect of Rho-kinase inhibitors in macrophages (Worthylake et al., 2001) and overexpression of a paxillin mutant with the LD4 domain deleted (West et al., 2001).
In contrast, MIIB-deficient cells were round and occupied a large area without distinguishable leading and trailing edges (Fig. 1, D and F–H; and Videos 2 and 4, available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1), e.g., front–back polarity. Depletion of MIIA or MIIB in Rat2 fibroblasts yielded similar results (Fig. S2). This phenotype differs somewhat from that reported for MEFs from MIIB−/− knockout mice (Lo et al., 2004). Although both showed inhibited migration, the MIIB−/− MEFs also showed long, unstable protrusions. This could arise either from incomplete ablation of MIIB by the RNAi knockdown or an uncharacterized adaptation. Off-target effects of the RNAi seem unlikely because our phenotypes were rescued by ectopic expression of RNAi-insensitive MIIA or MIIB, respectively (Fig. 2 E and not depicted).
Figure 2.
MIIA and IIB regulate protrusion and differentially control adhesion turnover. (A; top) Kymographs from control (pSUPER), MIIA-depleted (pSUP-IIA), and MIIB-depleted (pSUP-IIB) cells. (bottom) Overlay of periodicity and slope from the kymographs. (B) Protrusion rates from A. At least 12 cells (3–5 protrusions/cell) from four independent experiments were analyzed. Data is presented as the mean ± the SEM. (C) Image sequence of control (top; Video 7) and MIIA- (middle; Video 6) and MIIB-depleted cells (bottom; Video 8) cotransfected with paxillin-GFP. (D) Color-inverted image sequence of MIIA-depleted cell expressing paxillin-GFP (Video 9). Time is shown in seconds. (E) Image sequence of paxillin-GFP–expressing, MIIA- (top) or MIIB-depleted (bottom) cells cotransfected with mChe-MIIA or mChe-MIIB, respectively (not depicted). (F) Image sequence of paxillin-GFP–expressing, MIIA-depleted cell. Arrowheads point to central adhesions; arrows point to impaired disassembly at the trailing edge (Video 10). (G) Image sequence of paxillin-GFP in a MIIB-depleted cell (left) expressing mChe-IIA (not depicted) and a MIIA-depleted cell (right) expressing mChe-IIB (not depicted). Bars: (C) 5 μm; (D) 3 μm; (E) 5 μm; (F) 20 μm; (G) 5 μm. Videos 6–10 are available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1.
In addition to the round morphology, the MIIB-deficient cells also showed a defect in nuclear, centrosomal, and Golgi anchoring. More than 95% of the nuclei in the knockdown cells rotated clockwise (∼2 h/cycle; Fig. 1 H and Video 4). The centrosome accompanied this rotation, and the Golgi apparatus was distributed around the nucleus rather than polarized, as observed in nontransfected, migrating cells (Fig. 1 I and not depicted), suggesting a more general role of MIIB in cell polarization. Although the origin of this nuclear rotation is not known, it suggests that MIIB is part of a balanced mechanism of nuclear anchoring.
MII depletion increases cell protrusion and inhibits maturation of nascent adhesions at the leading edge
The increased area of MIIB-deficient cells and the broader lamellipodia in MIIA-deficient cells pointed to alterations in protrusion. By kymography, MIIA- and MIIB-deficient cells exhibited 2–3-fold increased rates of protrusion (Fig. 2, A–B). In addition, the protrusion was continuous, resulting in kymograms that showed a near linear, uninterrupted slope (Fig. 2 A, bottom). In contrast, wild-type cells often showed an interrupted, stepwise pattern, as previously reported (Giannone et al., 2004). Interestingly, during these periodic interruptions in protrusion, the adhesions stabilized and grew as MII began to localize in the previously protrusive region (Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1), suggesting a causal link. Finally, when the protrusions in MIIA- and MIIB-deficient cells stopped advancing, they did not retract efficiently (Videos 6 and 8), suggesting that both MII isoforms regulate retraction of the lamellipodium. Thus, both MII isoforms control the speed, stepwise pattern of extension, and retraction of protrusions.
To determine whether the abnormal protrusion is accompanied by alterations in adhesion dynamics, we knocked down MIIA or MIIB in paxillin-GFP– (Fig. 2 C and Videos 6–8, available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1) or vinculin-GFP–expressing cells (not depicted). Control cells showed numerous well-defined adhesions in protrusions, as well as some small adhesions near the leading edge (Fig. 2 C, top; and Video 7) that assembled and turned over as described previously (Webb et al., 2004). In contrast, MIIA- and MIIB-deficient cells showed few discrete adhesions in the protrusions; instead, there was a nearly continuous band of adhesions very close to the leading edge (Fig. 2 C, and Videos 7 and 8). The small individual adhesions that comprise this band were readily apparent at higher magnification (Fig. 2 D, Fig. 4, and Video 9). These adhesions disassembled and reformed rapidly (t 1/2 < 15 s) as the leading edge progressed (Fig. 2, C and D; and Videos 7–9). More importantly, they did not evolve into larger adhesions when lamellipodial growth halted. The defects were rescued when RNAi-insensitive mCherry (mChe)-MIIA or -MIIB were expressed in MIIA- or MIIB-deficient cells, respectively (Fig. 2 E).
Figure 4.
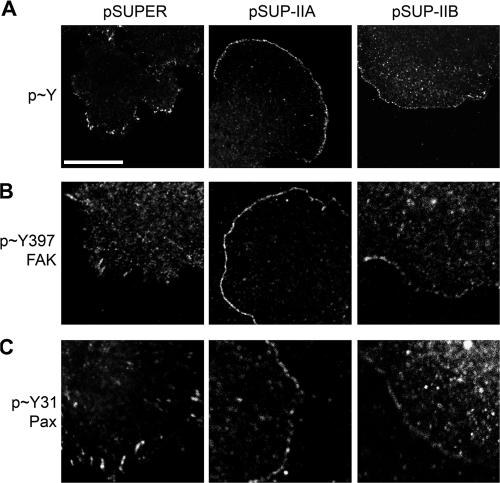
Adhesive signaling near the leading edge of MII-depleted cells. MIIA- or MIIB-depleted or control cells were plated on fibronectin and then fixed and stained for phosphotyrosine (A), phosphoTyr397-FAK (B), and phosphoTyr31-paxillin (C). Bar, 10 μm.
It is interesting that the phenotypes of MIIA- and MIIB-deficient cells on protrusion and the dynamics of adhesions in protrusions were almost indistinguishable because MIIA and MIIB occupy different regions of the cell, and neither is present in protrusions. This suggests that MII “acts at a distance”; that is, MII activity at the base of the lamellipodium or in central regions is transmitted to the leading edge, presumably through actin filaments, and generates periodic contractions that coincide with cessation of protrusion and retraction, adhesion maturation, and the movement (sliding) of larger adhesions toward the center of the cell. It also suggests that myosin activity regulates the behavior of adhesions at the leading edge, regardless of the isoform. Finally, these observations support the notion that protrusion and adhesion turnover are coupled.
MIIA promotes the growth of adhesions in central regions and disassembly at the trailing edge
Because both isoforms of MII regulate adhesion dynamics at the leading edge, but only MIIA inhibits rear retraction, we investigated the effect of MIIA and MIIB knockdowns on adhesions in other cellular regions. MIIB-deficient cells exhibit central adhesions comparable to those in control cells (unpublished data). In contrast, MIIA-deficient cells showed abnormally small, but static, adhesions in the central region of the cell (Fig. 2 F and Video 10, available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1). At the cell rear, where MIIA inhibits retraction, adhesion disassembly is greatly inhibited, e.g., the adhesions slide slowly and do not disassemble (Fig. 2 F and Video 10), thereby showing that adhesion sliding and disassembly are coordinately regulated by MIIA.
MIIA is required for the effects of MIIB on nascent, but not central, adhesions
Because both MIIA and MIIB mediate contraction and actin bundling (Kelley et al., 1996), we used cross-rescue experiments to determine whether their functions were overlapping in the regulation of adhesion assembly and disassembly. First, mChe-MIIA was expressed in MIIB-depleted cells coexpressing paxillin-GFP. mChe-MIIA localized in regions very similar to those in unperturbed cells (not depicted), and it restored the maturation of nascent adhesions (Fig. 2 G, left). However, the polarity defects and the appearance of multiple protrusions around the cell periphery remained (not depicted).
We then expressed mChe-IIB in MIIA-deficient cells. mChe-MIIB localized in the central areas of the cell, as it does in unperturbed cells. However, it did not rescue the inhibited maturation of nascent adhesions induced by the MIIA knockdown. Instead, a band of small, dynamic adhesions remained near the leading edge, as in MIIB knockdowns (Fig. 2 G, right). In contrast, mChe-MIIB rescued the effect on the central adhesions, i.e., they were larger (Fig. 2 G, top right).
Thus, increased MIIB in central areas (where endogenous MIIB resides) of MIIA knockdown cells rescues the maturation of adhesions in the central regions of the cell, but not the nascent adhesions at the cells periphery. In contrast, increased MIIA in MIIB knockdown cells rescues the maturation of adhesions at the leading edge. This points to a mechanism in which the central MIIB activity requires MIIA for its translation to the periphery, perhaps by organizing the actin so that tension produced in the middle of the cell propagates into protrusions. This suggestion is supported by our observation that overexpressed MIIA in wild- type cells localizes primarily in actin bundles and produces more and larger adhesions (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200612043/DC1), indicating that MII activity can dial up or down adhesion assembly, depending on its expression level.
Adhesion maturation at the leading edge depends on the ATPase activity of MIIA and MIIB
To separate the bundling from the contractile functions of MII on cell migration, we produced ATPase-inhibited mutants of MIIA and MIIB fused to GFP and mChe. The ATPase activity of N93K-MIIA and R709C-MIIB are inhibited 80 and 75%, respectively, in vitro (Heath et al., 2001; Ma et al., 2004). However, both mutants bind and cross-link, but do not move, actin filaments in vitro (Kim et al., 2005).
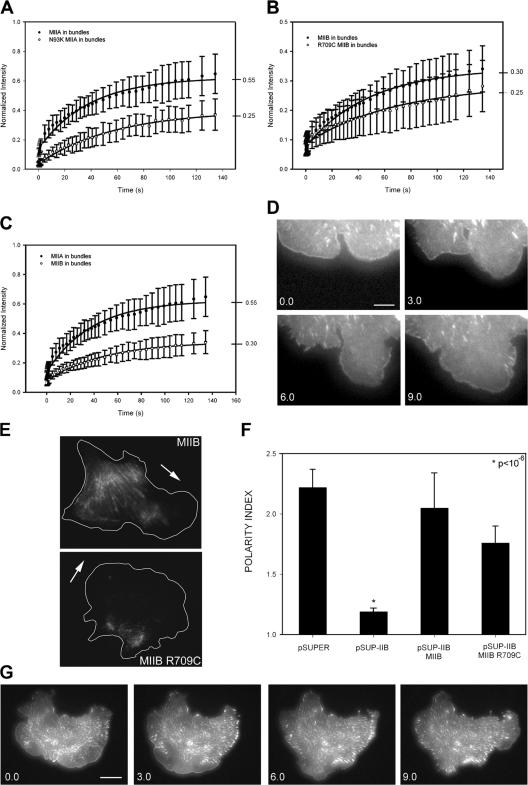
We used FRAP to show that the mutants exhibit increased time in the actin-bound state, as expected from their inhibited ATPase cycling. Both MIIA N93K and MIIB R709C showed decreased rates and fractional recoveries (Fig. 3, A–B). The fractional recovery observed for both mutants was the same, suggesting that the two isoforms bind actin similarly. Interestingly, wild-type MIIA exhibited faster and higher fractional recovery than MIIB (Fig. 3 C). This suggests that MIIA is more active than MIIB, and therefore binds actin strongly in a smaller fraction of time, as shown previously in vitro (Kelley et al., 1996). It also points to the use of FRAP as a method to determine MII activity in living cells.
Figure 3.
Contractility-deficient mutants of MIIA and MIIB exhibit differential rescue of actin bundling, protrusion, and adhesion dynamics in MIIA- and MIIB-deficient cells. Inhibited FRAP of GFP-MIIA N93K (A) and GFP-MIIB R709C (B) in actin bundles. Data are the mean ± the SD of >20 individual measurements from four independent experiments. (C) Differential FRAP of GFP-MIIA-WT and GFP-MIIB-WT in actin bundles. (D) A MIIB-depleted cell cotransfected with paxillin-GFP and mChe-MIIB R709C (not depicted). (E) Localization of GFP-MIIB and GFP-MIIB R709C. Arrows show the direction of migration. (F) Polarity index of migrating CHO.K1 cells. Data are the mean ± the SD of >100 cells analyzed/condition. (G) Time-lapse series of a MIIA-depleted cell expressing paxillin-GFP and mChe-MIIA (not depicted). Bars: (D) 3 μm; (G) 15 μm.
When expressed in MIIB-deficient cells, MIIB R709C did not effectively restore adhesion maturation (Fig. 3 D). However, MIIB R709C partially restored the front–back polarity and localized at the back of the cell (Fig. 3, E and F). Thus, maturation of adhesions at the leading edge requires MIIB activity; but its role in determining front– back polarity suggests a cross-linking contribution.
In contrast, MIIA N93K localized like its wild-type counterpart (unpublished data), rather than the rearward localization of MIIB R709C, and did not rescue the increased protrusiveness observed in MIIA-deficient cells. However, it did restore leading edge retraction and the concomitant growth of adhesions in protrusions pointing to its actin-binding function in these activities (Fig. 3 G).
MII regulates adhesive signaling
Adhesive signaling through integrin receptors both stimulates and responds to tension through Rho GTPases, thus constituting a feedback loop connecting adhesion and contraction through MII regulation (Chrzanowska-Wodnicka and Burridge, 1996). The phosphorylation of paxillin on Y31, Y118, and S273 and the phosphorylation of FAK on Y397 are part of this signaling mechanism and serve as markers for the activation of this pathway (Katz et al., 2003; Zaidel-Bar et al., 2003; Nayal et al., 2006).
The small dynamic adhesions near the leading edge of MIIA- and MIIB-depleted cells were prominently phosphorylated on tyrosine (Fig. 4, A–C). They also stained positively for Y397-FAK and Y31-paxillin, defining an almost continuous band of adhesions (Fig. 4, B and C). The staining of these phosphomarkers decreased in the stable adhesions that reside in regions removed from the leading edge (Fig. 4 B). Thus, depletion of myosin function at the lamellipodium enhances an adhesive signaling pathway that regulates adhesion turnover and MII activity, providing a mechanistic link between myosin-generated tension in the control of adhesion maturation at the leading edge.
Conclusions
The complex interplay between myosin-mediated contraction, protrusion, adhesion, and polarization underscores the central role of MII and its integrative properties in cell migration. Although the protrusion rate is determined by factors that regulate actin polymerization (Pollard and Borisy, 2003), it is also influenced by the rate of retrograde flow, which, in turn, is regulated by MII activity and serves to counterbalance actin polymerization (Lin and Forscher, 1995; Mitchison and Cramer, 1996). The retrograde flow rate is also influenced by adhesion, through a clutch-like mechanism, which links actin filaments to the substratum and can inhibit retrograde flow (Mitchison and Kirschner, 1988; Lin et al., 1994; Smilenov et al., 1999). The net protrusion rate is also influenced by cycles of retraction and adhesion maturation at the leading edge. Highly motile cells protrude and move nearly continuously (Bear et al., 2002; Jurado et al., 2005), whereas other cells can show cycles of protrusion and retraction (Giannone et al., 2004).
MII is also involved in a feedback loop that links adhesion, protrusion, and tension. Adhesion initiates signaling through Rho family GTPases that leads to the formation of adhesions and protrusions and generates tension. Tension also acts on adhesions to promote their maturation and the formation of actin filament bundles (Bershadsky et al., 2006). Highly motile cells tend to have small, highly dynamic adhesions that turnover rapidly, whereas the adhesions in slower moving cells stabilize and grow in response to increased tension before turning over (Nayal et al., 2006). Signaling components, such as phosphorylated paxillin and PAK, localize in the small, dynamic adhesions near the leading edge, where they function in a signaling pathway that inhibits adhesion maturation and promote protrusion (Nayal et al., 2006). Interestingly, in retracting regions, MII mediates the disassembly, rather than the assembly, of adhesions.
Finally, MII polarizes and connects spatially segregated activities. Myosin acts “at a distance” in regulating protrusion and adhesion. It also contributes to the overall polarity of the migrating cell and establishes front and rear. The former is through the role of MII in orienting microtubules, Golgi, and the nucleus, and the latter is through actin bundling at the rear and sides (Xu et al., 2003).
Thus, MII functions as a master regulator of cell migration. It can integrate spatially separated processes, and it is a key effector of signaling pathways that regulate each of the major component processes that drive migration.
Materials and methods
Plasmids
To generate MIIA and MIIB siRNA, the oligonucleotides GATCTGAACTCCTTCGAGC (IIA) and GGATCGCTACTATTCAGGA (IIB) were inserted into the appropriate pSUPER cassette according to the vector manufacturer's instructions (OligoEngine). The siRNA sequences correspond to nt 1,396–1,414 and 506–524 of rat MIIA (NM_013194) and MIIB (NM_031520), respectively.
GFP-MIIA and GFP-MIIB were gifts from R.S. Adelstein (National Institutes of Health, Bethesda, MD; Wei and Adelstein, 2000). Where indicated, GFP was replaced by mChe, which was obtained from R. Tsien (University of California, San Diego, La Jolla, CA; Shaner et al., 2004). siRNA-insensitive MIIB was generated by site-directed mutagenesis (QuickChange kit; Stratagene) introducing a silent mutation (TCA → AGC = Ser → Ser) in the RNAi target region of human MIIB. The MIIB R709C and MIIA N93K mutants were generated by site-directed mutagenesis using the appropriate primers.
Antibodies and reagents
The following antibodies were used: MIIA, MIIB, and GIT1 (rabbit, pAb) were purchased from Covance; paxillin (mouse, IgG1) was obtained from BD Biosciences; α-actinin (mouse, IgM) was purchased from Sigma- Aldrich; phosphotyrosine 4G10 (mouse, Ig2b) was obtained from Millipore; phosphoTyr31-paxillin (rabbit, pAb) was purchased from BioSource; and phosphoTyr397-FAK (rabbit, pAb) was obtained from CHEMICON International, Inc. Bodipy FL C5-ceramide (for Golgi detection) was obtained from Invitrogen and used as described by the manufacturer.
Cell culture and transfection
CHO-K1 cells and Rat2 cells were cultured in standard conditions and transfected using Lipofectamine (Invitrogen). For cotransfection experiments, plasmids containing the siRNA sequences were used in 10:1 excess to GFP or mChe-containing plasmids to ensure knockdown in fluorescence-positive cells.
Immunofluorescence
Cells were allowed to adhere to 2 μg/ml fibronectin-coated coverslips for 60 min, fixed using 4% paraformaldehyde, and permeabilized with either 0.5% Triton X-100 for 5 min or ice-cold methanol for 10 min. Coverslips were incubated with primary antibodies and a species-appropriate secondary antibody coupled to either Alexa Fluor 488 or 568 (Invitrogen).
Microscopy and image processing
Cells were plated on 2 μg/ml fibronectin–coated glass-bottomed dishes (migration-promoting conditions) in CCM1 for 1 h and maintained at 37°C at pH 7.4. For phase analyses, time-lapse images were captured at 10 min (NA 0.50; Nikon) with a charge-coupled device camera (Orca II; Hamamatsu) attached to an inverted microscope (TE-300; Nikon) using Metamorph software (Universal Imaging Corp.). Time is in minutes unless otherwise indicated. For assessment of cell polarity, the polarity index was calculated as the length of the major migration axis (parallel to the direction of movement) divided by the length of the perpendicular axis that intersects the center of the cell nucleus.
Confocal images were collected on a FluoView 300 system (60×/1.45 NA [oil] PlanApo 60× OTIRFM objective [all Olympus]). GFP and RFP were excited using the 488-nm laser line of an Ar ion laser and the 543-nm laser line of a He-Ne laser (Melles Griot), respectively. A Q500LP dichroic mirror (Chroma Technology Corp.) was used for GFP-labeled cells. For dual-color GFP-RFP imaging, a green–red cube (488/543/633) with a DM570 dichroic mirror (Chroma Technology Corp.) was used. Fluorescence and differential interference contrast images were acquired using FluoView software (Olympus).
TIRF images were acquired in an inverted microscope (IX70; Olympus). The excitation laser lines used were as described for confocal microscopy. A dichroic mirror (HQ485/30) was used for GFP-labeled cells. For dual GFP-RFP, a dual-emission filter (z488/543) was used. Images were acquired with a charge-coupled device camera (Retiga Exi; Qimaging) and analyzed using Metamorph.
Kymography
Protrusion parameters were quantified using kymography (Hinz et al., 1999). Images were captured every 5 s for 3 min. Kymographs were generated using Metamorph software along 1-pixel-wide regions oriented in the protrusion direction and perpendicular to the lamellipodial edge.
FRAP
Confocal images for FRAP analysis were acquired using the FluoView system. Initially, a cellular area (34.72 μm2) that contained GFP-MII–decorated stress fibers was scanned 3 times and bleached using 15 scans at 100% laser power. To image the FRAP, we did 15 scans every 0.1 s, 15 scans every 3 s, 14 scans every 5 s, and 2 scans every 10 s. Background subtraction and normalization were calculated, and normalized intensity versus times were fitted by a single exponential equation (R2 > 0.98).
Online supplemental material
Fig. S1 shows the spatial localization of MIIA and MIIB in a migrating cell. Fig. S2 shows the migratory phenotypes of MIIA-depleted, MIIB-depleted, and control Rat2 fibroblasts. Fig. S3 shows that MIIA expression levels affect the number of adhesions. Video 1 (corresponding to Fig. 1 C) is a time-lapse video of MIIA-deficient CHO.K1 cells. Video 2 (corresponding to Fig. 1 D) is a time-lapse video of MIIB-deficient CHO.K1 cells. Video 3 (corresponding to Fig. 1 E) is a time-lapse video of pSUPER-transfected (control) CHO.K1 cells. Video 4 (corresponding to Fig. 1 H) is a time-lapse video of a MIIB-deficient CHO.K1 cell, highlighting nuclear spinning. Video 5 is a dual-color TIRF time-lapse video of a protrusion of a MIIB-deficient CHO.K1 cell cotransfected with mChe-MIIB (magenta) and paxillin-GFP (green). Video 6 (corresponding to Fig. 2 C) is a TIRF time-lapse video of a protrusion of a MIIA-deficient CHO.K1 cell cotransfected with paxillin-GFP. Video 7 (corresponding to Fig. 2 C) is a TIRF time-lapse video of a protrusion of a pSUPER-transfected control CHO.K1 cell cotransfected with paxillin-GFP. Video 8 (corresponding to Fig. 2 C) is a TIRF time-lapse video of a protrusion of a MIIB-deficient CHO.K1 cell cotransfected with paxillin-GFP. Video 9 (corresponding to Fig. 2 D) is a TIRF time-lapse video of a protrusion of a MIIA-deficient CHO.K1 cell cotransfected with paxillin-GFP. Video 10 (corresponding to Fig. 2 F) is a TIRF time-lapse video of a MIIA-deficient CHO.K1 cell cotransfected with paxillin-GFP (shown).
Acknowledgments
We thank Bob Adelstein and Roger Tsien for providing reagents, and Bob Adelstein, Meg Titus, and Rex Chisholm for useful discussions. We also thank Bob Adelstein for valuable comments on the manuscript.
This work was supported by National Institutes of Health grant GM23244.
Abbreviations used in this paper: MHC, myosin heavy chain; MII, myosin II; MLC, myosin light chain.
References
- Adelstein, R.S., and M.A. Conti. 1975. Phosphorylation of platelet myosin increases actin-activated myosin ATPase activity. Nature. 256:597–598. [DOI] [PubMed] [Google Scholar]
- Bao, J., S.S. Jana, and R.S. Adelstein. 2005. Vertebrate nonmuscle myosin II isoforms rescue small interfering RNA-induced defects in COS-7 cell cytokinesis. J. Biol. Chem. 280:19594–19599. [DOI] [PubMed] [Google Scholar]
- Bear, J.E., T.M. Svitkina, M. Krause, D.A. Schafer, J.J. Loureiro, G.A. Strasser, I.V. Maly, O.Y. Chaga, J.A. Cooper, G.G. Borisy, and F.B. Gertler. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521. [DOI] [PubMed] [Google Scholar]
- Bershadsky, A., M. Kozlov, and B. Geiger. 2006. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr. Opin. Cell Biol. 18:472–481. [DOI] [PubMed] [Google Scholar]
- Bokoch, G.M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72:743–781. [DOI] [PubMed] [Google Scholar]
- Cai, Y., N. Biais, G. Giannone, M. Tanase, G. Jiang, J.M. Hofman, C.H. Wiggins, P. Silberzan, A. Buguin, B. Ladoux, and M.P. Sheetz. 2006. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys. J. 91:3907–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher, N.O., and M.C. Frame. 2004. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol. 14:241–249. [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka, M., and K. Burridge. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone, G., B.J. Dubin-Thaler, H.G. Dobereiner, N. Kieffer, A.R. Bresnick, and M.P. Sheetz. 2004. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 116:431–443. [DOI] [PubMed] [Google Scholar]
- Gomes, E.R., S. Jani, and G.G. Gundersen. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463. [DOI] [PubMed] [Google Scholar]
- Gupton, S.L., and C.M. Waterman-Storer. 2006. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 125:1361–1374. [DOI] [PubMed] [Google Scholar]
- Heath, K.E., A. Campos-Barros, A. Toren, G. Rozenfeld-Granot, L.E. Carlsson, J. Savige, J.C. Denison, M.C. Gregory, J.G. White, D.F. Barker, et al. 2001. Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am. J. Hum. Genet. 69:1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz, B., W. Alt, C. Johnen, V. Herzog, and H.W. Kaiser. 1999. Quantifying lamella dynamics of cultured cells by SACED, a new computer-assisted motion analysis. Exp. Cell Res. 251:234–243. [DOI] [PubMed] [Google Scholar]
- Jurado, C., J.R. Haserick, and J. Lee. 2005. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol. Biol. Cell. 16:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, B.Z., L. Romer, S. Miyamoto, T. Volberg, K. Matsumoto, E. Cukierman, B. Geiger, and K.M. Yamada. 2003. Targeting membrane-localized focal adhesion kinase to focal adhesions: roles of tyrosine phosphorylation and SRC family kinases. J. Biol. Chem. 278:29115–29120. [DOI] [PubMed] [Google Scholar]
- Kelley, C.A., J.R. Sellers, D.L. Gard, D. Bui, R.S. Adelstein, and I.C. Baines. 1996. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J. Cell Biol. 134:675–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.Y., M. Kovacs, S. Kawamoto, J.R. Sellers, and R.S. Adelstein. 2005. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J. Biol. Chem. 280:22769–22775. [DOI] [PubMed] [Google Scholar]
- Kolega, J. 1998. Cytoplasmic dynamics of myosin IIA and IIB: spatial ‘sorting’ of isoforms in locomoting cells. J. Cell Sci. 111:2085–2095. [DOI] [PubMed] [Google Scholar]
- Landsverk, M.L., and H.F. Epstein. 2005. Genetic analysis of myosin II assembly and organization in model organisms. Cell. Mol. Life Sci. 62:2270–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.H., and P. Forscher. 1995. Growth cone advance is inversely proportional to retrograde F-actin flow. Neuron. 14:763–771. [DOI] [PubMed] [Google Scholar]
- Lin, C.H., C.A. Thompson, and P. Forscher. 1994. Cytoskeletal reorganization underlying growth cone motility. Curr. Opin. Neurobiol. 4:640–647. [DOI] [PubMed] [Google Scholar]
- Lo, C.M., D.B. Buxton, G.C. Chua, M. Dembo, R.S. Adelstein, and Y.L. Wang. 2004. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Biol. Cell. 15:982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X., S. Kawamoto, Y. Hara, and R.S. Adelstein. 2004. A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol. Biol. Cell. 15:2568–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T., and M. Kirschner. 1988. Cytoskeletal dynamics and nerve growth. Neuron. 1:761–772. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J., and L.P. Cramer. 1996. Actin-based cell motility and cell locomotion. Cell. 84:371–379. [DOI] [PubMed] [Google Scholar]
- Nayal, A., D.J. Webb, C.M. Brown, E.M. Schaefer, M. Vicente-Manzanares, and A.R. Horwitz. 2006. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J., M.A. Schwartz, K. Burridge, R.A. Firtel, M.H. Ginsberg, G. Borisy, J.T. Parsons, and A.R. Horwitz. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. [DOI] [PubMed] [Google Scholar]
- Riento, K., and A.J. Ridley. 2003. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4:446–456. [DOI] [PubMed] [Google Scholar]
- Scholey, J.M., K.A. Taylor, and J. Kendrick-Jones. 1980. Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature. 287:233–235. [DOI] [PubMed] [Google Scholar]
- Shaner, N.C., R.E. Campbell, P.A. Steinbach, B.N. Giepmans, A.E. Palmer, and R.Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572. [DOI] [PubMed] [Google Scholar]
- Siddique, M.S., G. Mogami, T. Miyazaki, E. Katayama, T.Q. Uyeda, and M. Suzuki. 2005. Cooperative structural change of actin filaments interacting with activated myosin motor domain, detected with copolymers of pyrene-labeled actin and acto-S1 chimera protein. Biochem. Biophys. Res. Commun. 337:1185–1191. [DOI] [PubMed] [Google Scholar]
- Smilenov, L.B., A. Mikhailov, R.J. Pelham, E.E. Marcantonio, and G.G. Gundersen. 1999. Focal adhesion motility revealed in stationary fibroblasts. Science. 286:1172–1174. [DOI] [PubMed] [Google Scholar]
- Webb, D.J., K. Donais, L.A. Whitmore, S.M. Thomas, C.E. Turner, J.T. Parsons, and A.F. Horwitz. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161. [DOI] [PubMed] [Google Scholar]
- Wei, Q., and R.S. Adelstein. 2000. Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol. Biol. Cell. 11:3617–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, K.A., H. Zhang, M.C. Brown, S.N. Nikolopoulos, M.C. Riedy, A.F. Horwitz, and C.E. Turner. 2001. The LD4 motif of paxillin regulates cell spreading and motility through an interaction with paxillin kinase linker (PKL). J. Cell Biol. 154:161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake, R.A., S. Lemoine, J.M. Watson, and K. Burridge. 2001. RhoA is required for monocyte tail retraction during transendothelial migration. J. Cell Biol. 154:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., F. Wang, A. Van Keymeulen, P. Herzmark, A. Straight, K. Kelly, Y. Takuwa, N. Sugimoto, T. Mitchison, and H.R. Bourne. 2003. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 114:201–214. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar, R., C. Ballestrem, Z. Kam, and B. Geiger. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613. [DOI] [PubMed] [Google Scholar]
- Zamir, E., and B. Geiger. 2001. Components of cell-matrix adhesions. J. Cell Sci. 114:3577–3579. [DOI] [PubMed] [Google Scholar]
- Zhao, Z.S., J.P. Lim, Y.W. Ng, L. Lim, and E. Manser. 2005. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol. Cell. 20:237–249. [DOI] [PubMed] [Google Scholar]