Abstract
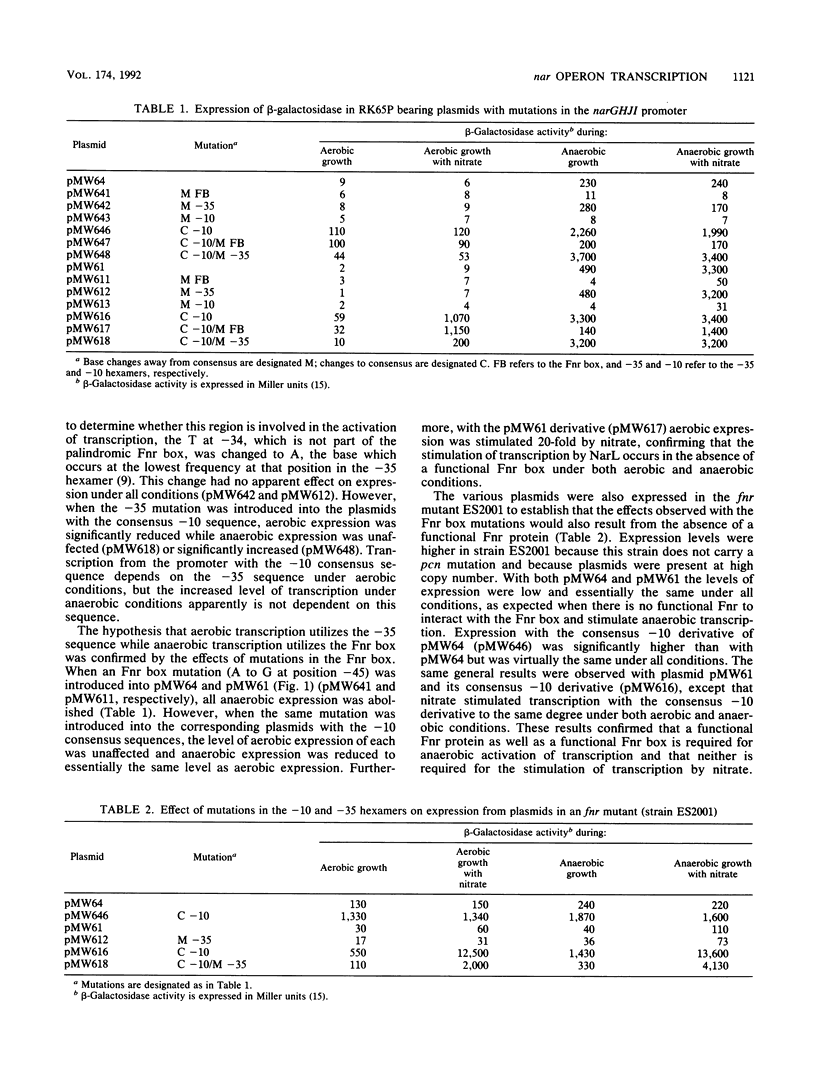
The effects of mutations in the -10, -35, and Fnr box regions of the narGHJI promoter of Escherichia coli were determined by assaying the expression of beta-galactosidase from narG::lacZ fusion plasmids under aerobic and anaerobic conditions. A 1-base change in the -10 hexamer completely abolished expression, whereas a 3-base change to create the consensus TATAAT resulted in significant aerobic as well as anaerobic expression. A mutation in the putative -35 hexamer did not affect anaerobic expression but reduced aerobic expression from the construction with the -10 consensus sequence. A mutation in the Fnr box severely reduced anaerobic expression but did not affect aerobic expression. When the complete 5' region of the nar operon including the NarL box was present, nitrate stimulated both aerobic and anaerobic expression. Stimulation of expression by nitrate occurred in an fnr mutant but not in a narL mutant. We conclude that the rate of transcription of the nar operon is dependent on two distinct modes of transcription. One mode, which occurs at low levels, depends on the -10 and -35 hexamer sequences and is dramatically enhanced by changing the -10 sequence to the consensus TATAAT. The second depends on the -10 and Fnr box sequences but is independent of the -35 sequence. This second mode occurs at a very high level under anaerobic conditions when Fnr is activated and is also enhanced by changing the -10 sequence to the consensus TATAAT. NarL, activated by nitrate, stimulated both modes of transcription, indicating that it does not act through Fnr but that it directly affects the interaction of RNA polymerase with the promoter.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Garges S. Positive control. J Biol Chem. 1990 Jul 5;265(19):10797–10800. [PubMed] [Google Scholar]
- Bonnefoy V., Pascal M. C., Ratouchniak J., Chippaux M. Alteration by mutation of the control by oxygen of the nar operon in Escherichia coli. Mol Gen Genet. 1986 Nov;205(2):349–352. doi: 10.1007/BF00430449. [DOI] [PubMed] [Google Scholar]
- Bonnefoy V., Pascal M. C., Ratouchniak J., Chippaux M. Autoregulation of the nar operon encoding nitrate reductase in Escherichia coli. Mol Gen Genet. 1986 Jul;204(1):180–184. doi: 10.1007/BF00330207. [DOI] [PubMed] [Google Scholar]
- Chan B., Spassky A., Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus -35 region sequences. Biochem J. 1990 Aug 15;270(1):141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux M., Bonnefoy-Orth V., Ratouchniak J., Pascal M. C. Operon fusions in the nitrate reductase operon and study of the control gene nir R in Escherichia coli. Mol Gen Genet. 1981;182(3):477–479. doi: 10.1007/BF00293938. [DOI] [PubMed] [Google Scholar]
- Chippaux M., Giudici D., Abou-Jaoudé A., Casse F., Pascal M. C. Laboratoire de Chimie Bactérienne C.N.R.S., Marsielle, France. Mol Gen Genet. 1978 Apr 6;160(2):225–229. doi: 10.1007/BF00267485. [DOI] [PubMed] [Google Scholar]
- Eiglmeier K., Honoré N., Iuchi S., Lin E. C., Cole S. T. Molecular genetic analysis of FNR-dependent promoters. Mol Microbiol. 1989 Jul;3(7):869–878. doi: 10.1111/j.1365-2958.1989.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Gaston K., Bell A., Kolb A., Buc H., Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990 Aug 24;62(4):733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman P. S., Gaston K. L., Cole J. A., Busby S. J. The nirB promoter of Escherichia coli: location of nucleotide sequences essential for regulation by oxygen, the FNR protein and nitrite. Mol Microbiol. 1988 Jul;2(4):527–530. doi: 10.1111/j.1365-2958.1988.tb00059.x. [DOI] [PubMed] [Google Scholar]
- Lambden P. R., Guest J. R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976 Dec;97(2):145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- Li S. F., DeMoss J. A. Location of sequences in the nar promoter of Escherichia coli required for regulation by Fnr and NarL. J Biol Chem. 1988 Sep 25;263(27):13700–13705. [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989 Mar;171(3):1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman B. M., Cole J. A. The chromosomal location and pleiotropic effects of mutations of the nirA+ gene of Escherichia coli K12: the essential role of nirA+ in nitrite reduction and in other anaerobic redox reactions. J Gen Microbiol. 1978 May;106(1):1–12. doi: 10.1099/00221287-106-1-1. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Shaw D. J., Rice D. W., Guest J. R. Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J Mol Biol. 1983 May 15;166(2):241–247. doi: 10.1016/s0022-2836(83)80011-4. [DOI] [PubMed] [Google Scholar]
- Sodergren E. J., DeMoss J. A. narI region of the Escherichia coli nitrate reductase (nar) operon contains two genes. J Bacteriol. 1988 Apr;170(4):1721–1729. doi: 10.1128/jb.170.4.1721-1729.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S., Gaston K. L., Bell A. I., Roberts R. E., Busby S. J., Guest J. R. Interconversion of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol. 1990 Nov;4(11):1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. Regulation and over-expression of the fnr gene of Escherichia coli. J Gen Microbiol. 1987 Dec;133(12):3279–3288. doi: 10.1099/00221287-133-12-3279. [DOI] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trageser M., Spiro S., Duchêne A., Kojro E., Fahrenholz F., Guest J. R., Unden G. Isolation of intact FNR protein (Mr 30,000) of Escherichia coli. Mol Microbiol. 1990 Jan;4(1):21–27. doi: 10.1111/j.1365-2958.1990.tb02011.x. [DOI] [PubMed] [Google Scholar]
- Unden G., Guest J. R. Isolation and characterization of the Fnr protein, the transcriptional regulator of anaerobic electron transport in Escherichia coli. Eur J Biochem. 1985 Jan 2;146(1):193–199. doi: 10.1111/j.1432-1033.1985.tb08638.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Holst B., Søgaard-Andersen L., Martinussen J., Nesvera J., Douthwaite S. R. Design of cAMP-CRP-activated promoters in Escherichia coli. Mol Microbiol. 1991 Feb;5(2):433–437. doi: 10.1111/j.1365-2958.1991.tb02126.x. [DOI] [PubMed] [Google Scholar]
- Walker M. S., DeMoss J. A. Promoter sequence requirements for Fnr-dependent activation of transcription of the narGHJI operon. Mol Microbiol. 1991 Feb;5(2):353–360. doi: 10.1111/j.1365-2958.1991.tb02116.x. [DOI] [PubMed] [Google Scholar]



