Abstract
Nucleosomes containing the centromere-specific histone H3 variant centromere protein A (CENP-A) create the chromatin foundation for kinetochore assembly. To understand the mechanisms that selectively target CENP-A to centromeres, we took a functional genomics approach in the nematode Caenorhabditis elegans, in which failure to load CENP-A results in a signature kinetochore-null (KNL) phenotype. We identified a single protein, KNL-2, that is specifically required for CENP-A incorporation into chromatin. KNL-2 and CENP-A localize to centromeres throughout the cell cycle in an interdependent manner and coordinately direct chromosome condensation, kinetochore assembly, and chromosome segregation. The isolation of KNL-2–associated chromatin coenriched CENP-A, indicating their close proximity on DNA. KNL-2 defines a new conserved family of Myb DNA-binding domain–containing proteins. The human homologue of KNL-2 is also specifically required for CENP-A loading and kinetochore assembly but is only transiently present at centromeres after mitotic exit. These results implicate a new protein class in the assembly of centromeric chromatin and suggest that holocentric and monocentric chromosomes share a common mechanism for CENP-A loading.
Introduction
Centromeric chromatin, in which centromere protein A (CENP-A) replaces histone H3, is organized during mitosis to form a surface for kinetochore assembly (Palmer et al., 1991; Sullivan et al., 2001; Cleveland et al., 2003). Studies of neocentromeres, which spontaneously form at low frequency on acentric chromosome fragments, have suggested that CENP-A nucleosomes rather than specific DNA sequences define centromere activity (Choo, 2000). The mechanisms that load CENP-A nucleosomes onto chromatin and maintain their restricted distribution are topics of active investigation (Henikoff and Dalal, 2005). Work in budding and fission yeasts has identified proteins that contribute to CENP-A localization (Takahashi et al., 2000; Mellone and Allshire, 2003; Hayashi et al., 2004), including the general chromatin-remodeling complex CAF-1. However, because CAF-1 inhibition destabilizes CENP-A protein levels, it is unclear whether CAF-1 has a direct role in CENP-A loading (Hayashi et al., 2004).
To identify proteins that are required to localize CENP-A, we took an unbiased functional genomic approach in the nematode Caenorhabditis elegans. C. elegans has holocentric chromosomes in which kinetochores form along the entire length of each sister chromatid instead of being confined to a localized chromosomal region. Fundamental similarities in the mitotic kinetochores of holocentric and monocentric chromosomes are indicated by both high resolution ultrastructural studies and conservation of the constituent proteins, from CENP-A on the DNA to components of the spindle microtubule interface (for review see Maddox et al., 2004). In both types of chromosome architectures, CENP-A is restricted to a subset of chromatin organized on opposing faces of sister chromatids to geometrically constrain kinetochore assembly and ensure correct attachment to the spindle (Buchwitz et al., 1999; Oegema et al., 2001). These similarities suggest that the mechanism of CENP-A loading is likely to be conserved between holocentric chromosomes of C. elegans and monocentric chromosomes of vertebrates. In this study, we provide evidence for this assertion by identifying and characterizing a protein family with a Myb-like DNA-binding domain that is specifically required for CENP-A loading in both nematodes and mammalian cells. Identification of this protein class provides a starting point for understanding the mechanism of this critical step in genome inheritance.
Results and discussion
In C. elegans, RNAi-mediated protein depletion is highly penetrant and largely independent of intrinsic protein turnover, making it possible to generate oocytes that are >95% depleted of a target protein (Oegema and Hyman, 2006). Fertilization of depleted oocytes triggers the first embryonic mitosis, which is highly stereotypical and, therefore, amenable to quantitative live imaging assays (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). In CeCENP-A–depleted embryos, kinetochores fail to form, resulting in a distinctive kinetochore-null (KNL) phenotype characterized by clustering of chromosomes from each pronucleus, premature spindle pole separation, defective chromosome alignment, and failure of chromosome segregation (Fig. 1 A and Video 2; Oegema et al., 2001).
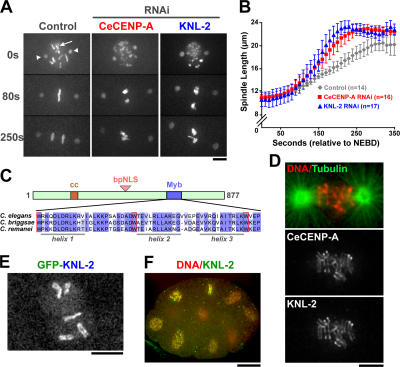
Figure 1.
KNL-2 is a novel Myb domain protein whose depletion results in a CeCENP-A loss of function phenotype. (A) Images of control, CeCENP-A, and KNL-2–depleted embryos expressing GFP-histone H2b (arrow) to mark chromosomes and GFP–γ-tubulin (arrowheads) to mark the spindle poles. Time is given in seconds relative to nuclear envelope breakdown (NEBD). (B) Spindle pole separation kinetics in control, CeCENP-A depletion, and KNL-2 depletion. Error bars represent the SEM with a confidence interval of 0.95. (C) Primary sequence features of KNL-2. The divergent Myb domain (amino acids 624–677) is shown on the bottom for C. elegans, C. briggsae, and C. remanei KNL-2. The predicted helical secondary structure is shown under the sequences, and the conserved tryptophan (W) residues are boxed in red. (D) KNL-2 and CeCENP-A colocalize at kinetochores during mitosis. (E) GFP–KNL-2 exhibits kinetochore localization. (F) KNL-2 localizes to kinetochores in later embryonic divisions. A multicellular embryo in which DNA (red) and KNL-2 (green) are labeled is shown. Bars (A, D, and E), 5 μm; (F) 10 μm.
To systematically identify the set of C. elegans proteins whose depletion results in a KNL phenotype, we used a fluorescence microscopy–based assay to rescreen a collection of ∼250 genes implicated in chromosome segregation by a previous comprehensive genome-wide screen. In the initial screen that targeted 98% of the ∼19,000 predicted C. elegans genes (Sonnichsen et al., 2005), embryos individually depleted of each of the ∼1,700 gene products required for embryonic viability were filmed by differential interference contrast (DIC) microscopy. Cluster analysis of the DIC video data revealed a set of ∼250 genes that are generally required for chromosome segregation (Sonnichsen et al., 2005). Because the KNL phenotype is not discernable by DIC, we used high resolution fluorescence time-lapse imaging to analyze living embryos coexpressing GFP-histone H2b and either GFP–γ-tubulin or GFP–α-tubulin that were depleted of each of these 250 gene products (unpublished data). This approach, which is expected to uncover all nonredundant gene products whose inhibition results in a KNL phenotype, identified five proteins: CeCENP-A, CeCENP-C, KNL-1, KNL-2, and KNL-3. Three of these, CeCENP-C, KNL-1, and KNL-3, function downstream of CeCENP-A because they require CeCENP-A to localize to kinetochores, and their depletion does not prevent CeCENP-A targeting (Oegema et al., 2001; Desai et al., 2003; Cheeseman et al., 2004). The remaining KNL protein, KNL-2 (K06A5.4), specifically targets CeCENP-A to chromatin (see Fig. 2) and represents the only such protein identified by this comprehensive strategy.
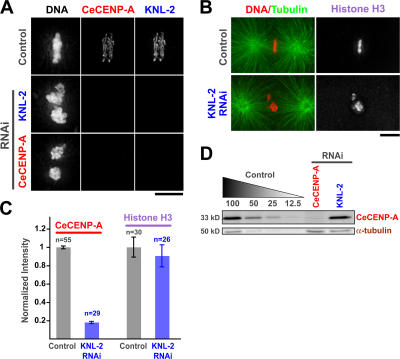
Figure 2.
KNL-2 is required for CeCENP-A but not histone H3 targeting to chromatin. (A) CeCENP-A and KNL-2 localization in wild-type, KNL-2–depleted, or CENP-A– depleted embryos. (B) KNL-2 depletion does not prevent histone H3 localization. Note that GFP-histone H2b levels on chromatin were also unaffected (Fig. 1 A). (C) Quantitation of CeCENP-A and histone H3 localization. For CeCENP-A analysis, KNL-2 was depleted in embryos expressing GFP-histone H2b, and the fluorescence intensity ratio of CeCENP-A to GFP-histone H2b was calculated in deconvolved images of fixed embryos. For histone H3, the signal on chromatin was directly quantified. Error bars represent the SEM with a confidence interval of 0.95. (D) KNL-2 depletion does not affect CeCENP-A protein levels. Immunoblot comparing serially diluted control C. elegans extract to CeCENP-A–and KNL-2–depleted C. elegans extracts is shown. α-Tubulin was used as a loading control. Bars, 5 μm.
The depletion of KNL-2 resulted in a defect in mitotic chromosome segregation that was essentially identical to that of CeCENP-A–depleted embryos (Fig. 1 A and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). Premature separation of spindle poles, indicating the absence of kinetochore-microtubule attachments, was quantitatively similar for the two depletions (Fig. 1 B). In contrast to CeCENP-A depletions (Video 2; Monen et al., 2005), KNL-2 depletion also resulted in a meiotic chromosome segregation defect that is evident from the aberrant nature of the oocyte pronucleus (on the embryo anterior/left side in Video 3). The meiotic role of KNL-2 in the segregation of holocentric C. elegans chromosomes is not discussed further here and will be the subject of a separate study. KNL-2 is an ∼103-kD basic protein with a short coiled-coil stretch and a bipartite nuclear localization sequence. Sequence analysis revealed the presence of a divergent version of a DNA-binding domain at its C terminus, which was first defined in the protooncogene Myb (Fig. 1 C). Myb domains as well as the related SANT domains are present in a large number of proteins implicated in chromatin dynamics, including transcription factors and subunits of chromatin-remodeling enzymes that interact with histones (Aasland et al., 1996; Lipsick, 1996; Mo et al., 2005). Homologues of KNL-2 were present in nematodes closely related to C. elegans, but a clear link to Myb domain proteins in other organisms was not evident in initial bioinformatic analysis (see Fig. 4 for more on this topic).
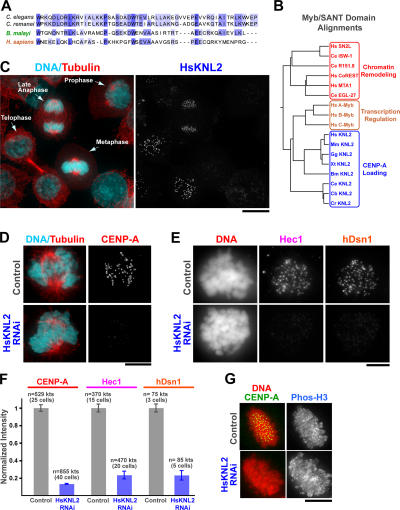
Figure 4.
KNL-2 is the founding member of a widely conserved protein family required for CENP-A loading in both invertebrates and vertebrates. (A) The Myb domain of the B. malayi KNL-2 homologue, which was aligned with two of the rhabditid KNL-2 Myb domains, was used to identify a putative human KNL2 homologue (c14orf106). Shading indicates degrees of conservation. (B) Tree of a small subset of the Myb/SANT domain protein superfamily aligned using Myb/SANT domain sequences. SANT domain proteins (red) are implicated in chromatin remodeling activities. The founding canonical Myb protooncogene (A–C refer to three isoforms in humans; gold) is a transcription factor that directly binds specific DNA sequences. A new subfamily (blue) is defined by the KNL-2 Myb domain and includes, in addition to the nematode members, a clear homologue in every sequenced vertebrate. KNL-2 homologues were not evident by sequence in fungi or Drosophila. GenBank/EMBL/DDBJ accession nos. for the putative vertebrate KNL2s are as follows: Hs, human c14orf106; Mm, mouse C79407; Gg, chicken XP_421481; and Xt, frog AL873597.2. (C) Localization of HsKNL2 in HeLa cells. The chosen field includes cells in different mitotic stages and highlights the striking increase in centromere staining in the late anaphase/telophase stage of the cell cycle. (D–F) HsKNL2 depletion dramatically reduces CENP-A localization at centromeres and equivalently reduces targeting of medial (hDsn1) and outer (Hec1) kinetochore proteins. Anticentromere antibody staining was used to identify centromere regions in all cases but is not shown for brevity. The total number of kinetochores (kts) and cells in which fluorescence intensities were measured is indicated for each condition. As expected from the kinetochore assembly defect, chromosome segregation is severely perturbed in HsKNL2-depleted cells (Fig. S4 and Video 10, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). Error bars represent SEM. (G) HsKNL2 depletion does not affect phosphohistone (S10) H3 staining. Bars (C), 10 μm; (D, E, and G) 5 μm.
To determine the subcellular localization of KNL-2, we generated an affinity-purified antibody and a stable C. elegans strain expressing a GFP fusion with KNL-2 (Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). Both tools revealed that KNL-2 localizes to the centromere/kinetochore region throughout mitosis (Fig. 1, D and E). The specificity of antibody localization was established using RNAi-mediated protein depletion (Fig. 2 A). Because our screening strategy focused on the first embryonic division, it was possible that the function of KNL-2 was limited to the specialized changes in genome architecture that immediately follow fertilization. However, KNL-2 was observed at kinetochores throughout embryogenesis (Fig. 1 F), and an RNAi-based strategy in which KNL-2 is not inhibited until after early embryogenesis (Maddox et al., 2005) indicated a continuous requirement for KNL-2 during development (Fig. S1).
The depletion of KNL-2 prevented the localization of CeCENP-A to chromatin (n = 29 first-division embryos; Fig. 2, A and C) but did not affect the stability of CeCENP-A protein (Fig. 2 D). The chromosomal targeting of KNL-2, in turn, required CeCENP-A (Fig. 2 A), indicating that these two proteins are interdependent for their localization. In contrast to its effect on CeCENP-A localization, the depletion of KNL-2 did not affect the chromosomal targeting of histone H3 (Fig. 2, B and C) or GFP-histone H2b (Fig. 1 A), indicating that KNL-2 is specifically required for loading CeCENP-A. The interdependence of KNL-2 and CeCENP-A localization was further confirmed using the live imaging of embryos expressing GFP–KNL-2 or GFP–CeCENP-A (Videos 4–7, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1).
Reciprocal depletion and localization experiments place CeCENP-A at the top of the kinetochore assembly hierarchy (Oegema et al., 2001; Desai et al., 2003; Cheeseman et al., 2004). The interdependence of KNL-2 and CeCENP-A for chromosomal targeting (Fig. 2 A) predicted that KNL-2, like CeCENP-A, should be upstream of other kinetochore components. To test this prediction, we examined the recruitment of three widely conserved proteins from distinct positions within the substructure of the mitotic kinetochore: CeCENP-C (inner), KNL-1 (medial), and BUB-1 (outer). As expected, all three proteins failed to localize to chromosomes in KNL-2–depleted embryos (Fig. 3 A). In reciprocal depletions, GFP–KNL-2 was present on chromosomes in embryos depleted of CeCENP-C, KNL-1, or BUB-1 (Fig. 3 B and Videos 8 and 9, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1), placing KNL-2 with CeCENP-A at the top of the kinetochore assembly hierarchy (Fig. 3 E).
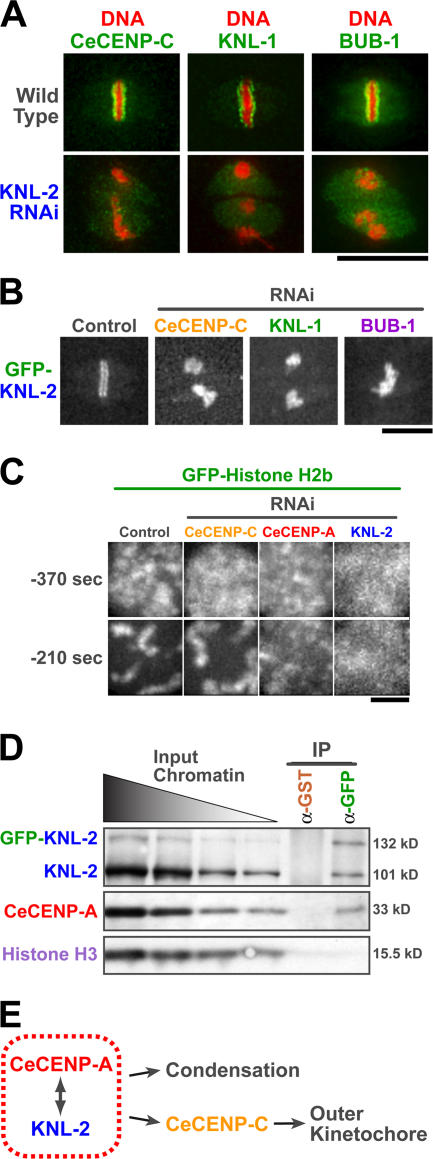
Figure 3.
KNL-2 and CeCENP-A make functionally equivalent contributions to kinetochore assembly and chromosome condensation and are physically proximal on chromatin. (A) Like CeCENP-A, KNL-2 is required for the localization of CeCENP-C, KNL-1, and BUB-1 to kinetochores. (B) Depletion of CeCENP-C, KNL-1, or BUB-1 does not prevent GFP–KNL-2 localization to chromatin. (C) CeCENP-A–and KNL-2–depleted embryos exhibit a similar chromosome condensation defect. Representative images of the sperm pronucleus at −370 and −210 s before nuclear envelope breakdown (for quantitation see Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). (D) KNL-2 and CeCENP-A are physically proximal on chromatin. Nuclei purified from C. elegans embryos expressing GFP–KNL-2 were sonicated to generate chromatin fragments of ∼500–1,500 bp. The sonicated chromatin was used as the input for anti-GST (control) and anti-GFP immunoprecipitations. Dilutions of the input chromatin and immunoprecipitates were blotted for KNL-2, CeCENP-A, and histone H3. Chromatin fragments enriched for KNL-2 were coenriched for CeCENP-A but not for histone H3 (Fig. S3). (E) CeCENP-A and KNL-2 coordinately direct kinetochore assembly and chromosome condensation. Bars (A and B), 10 μm; (C) 2 μm.
CENP-A containing nucleosomes play an important structural role in the condensation of mitotic C. elegans chromosomes that is independent of their role in directing kinetochore assembly (Maddox et al., 2006). To determine whether KNL-2 is also required for chromosome condensation during mitotic entry, we used a recently developed quantitative assay to compare condensation after the depletion of CeCENP-A, KNL-2, or CeCENP-C. The depletion of KNL-2 resulted in a severe condensation defect that was essentially identical to that observed after the depletion of CeCENP-A and distinct from the more subtle effects of CeCENP-C depletion (Fig. 3 C and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). Condensation analysis is performed solely on chromatin in the sperm pronucleus, which is formed before introduction of the inhibitory double-stranded RNA (dsRNA) and is consequently free of complications arising from meiotic defects (Maddox et al., 2006). This result further confirms that KNL-2 is specifically required for the assembly of CENP-A chromatin.
The aforementioned results suggest that KNL-2 physically associates with CeCENP-A nucleosomes to facilitate their loading and function. To test this idea, we isolated embryonic interphase nuclei from a strain expressing GFP–KNL-2, sonicated the chromatin to 500–1,500-bp fragments, and immunoprecipitated the KNL-2 fusion protein with an anti-GFP antibody (Fig. 3 D). Analysis of the immunoprecipitates revealed that CeCENP-A but not histone H3 was coenriched with KNL-2 by this procedure (Fig. 3 D). Complementing this biochemical approach, high resolution imaging revealed the partial colocalization of KNL-2 with dispersed CeCENP-A foci in interphase nuclei (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). CeCENP-A and KNL-2 are the only centromere/kinetochore proteins identified in C. elegans to date that are present in nuclei throughout the cell cycle. Cumulatively, these results indicate that KNL-2 and CeCENP-A are in close physical proximity on chromatin (and may associate directly), where they coordinately maintain centromere structure, initiate kinetochore assembly, and direct chromosome segregation.
Our initial efforts to identify KNL-2 homologues outside of nematodes were unsuccessful because the three nematodes with sequenced genomes are closely related rhabditids. Searches of the recently sequenced genome of the parasitic filarial nematode Brugia malayi (http://www.tigr.org/tdb/e2k1/bma1/) revealed a more distant nematode KNL-2 homologue (Fig. 4 A). By aligning the Myb domain sequence from the filarial KNL-2 with the analogous region of the three rhabditid proteins, we generated a profile that allowed us to identify a KNL-2–related subfamily of the Myb/SANT domain–containing protein superfamily with members in all sequenced vertebrates (Fig. 4 B). The human homologue of KNL-2 was independently discovered in a recent study based on its physical association with Mis18 (Fujita et al., 2007). Mis18 was first found in fission yeast, and both fungal Mis18 and its human homologue are implicated in CENP-A loading (Hayashi et al., 2004; Fujita et al., 2007). No homologue of Mis18 is evident in nematodes, and no KNL-2 homologue is recognizable in fungi.
To determine whether the human homologue of KNL-2 (which we refer to as HsKNL2) is required for CENP-A loading, we characterized its localization using an affinity-purified antibody generated against amino acids 661–899 and performed siRNA-mediated depletion. HsKNL2 localizes to the centromere region of chromosomes prominently between late anaphase/telophase and early G1 phase (Fig. 1 C), which coincides with the time of CENP-A loading in HeLa cells (see Jansen et al. on p. 795 of this issue). The reason for the difference in the localization timing of KNL2 between nematode embryos and mammalian cells in culture is currently unclear. Most importantly, the depletion of HsKNL2 reduced centromeric levels of CENP-A as well as levels of the core kinetochore components hDsn1 (a subunit of the human Mis12 complex; Kline et al., 2006) and Hec1 (the Ndc80-like subunit of the Ndc80 complex; Fig. 4, D–F; DeLuca et al., 2005). In contrast, phosphohistone H3 (S10) staining was unaffected (Fig. 4 G). HsKNL2 depletion did not affect CENP-A protein levels (not depicted), which is in agreement with the findings in C. elegans embryos (Fig. 2 D). HsKNL2-depleted cells exhibited severe chromosome segregation defects, which is consistent with their dramatic kinetochore assembly defect (Fig. S4 and Video 10, available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1). These results establish that involvement of the KNL2 protein family in CENP-A chromatin assembly is conserved between the holocentric chromosomes of nematodes and the monocentric chromosomes of vertebrates.
In summary, by using a functional genomic approach in C. elegans based on the signature CENP-A loss of the function phenotype during the first embryonic division, we identified a conserved Myb domain–containing protein family that is critical for CENP-A loading. The depletion of KNL-2 results in all of the defects expected for a CENP-A–loading factor, and, after extensive functional genomics analysis, KNL-2 is the only protein identified to date in C. elegans that meets this essential criterion. The fact that KNL2 contains a Myb domain raises the exciting possibility that recognition of short, specific DNA sequences may play a role in CENP-A deposition. Although studies of neocentromeres have suggested that centromere identity is not strictly defined by DNA sequence (Choo, 2000; Cleveland et al., 2003), it remains possible that short sequence stretches bias centromeric chromatin assembly. Further investigation of how the Myb domain of the KNL2 protein family contributes to CENP-A loading will help elucidate the mechanism by which this widely conserved protein class directs centromeric chromatin formation.
Materials and methods
RNAi and antibody production
RNAi was performed by injection of dsRNA against the target gene as described previously (Desai et al., 2003). Embryos were analyzed from injected adults that were kept for 48 h at 20°C after injection. For KNL-2, oligonucleotides AATTAACCCTCACTAAAGGTCGACTTGGTCGGACAGATT and TAATACGACTCACTATAGGTGCGATATGTGGCGTTATGT were used to create an ∼1,000-bp dsRNA by standard methods; all other dsRNAs were previously described (Desai et al., 2003). siRNA treatment was performed as described previously (Kline et al., 2006) using a cocktail of oligonucleotides purchased from Dharmacon. Polyclonal antibodies were generated to amino acids 2–153 of KNL-2 and amino acids 661–899 of HsKNL2 fused to GST and were affinity purified. Immunofluorescence was performed as described previously using directly labeled polyclonal antibodies (Oegema et al., 2001). All immunofluorescence images were acquired using a DeltaVision-modified inverted microscope (IX70; Olympus) and Softworx software (Applied Precision) and were deconvolved. Quantification of immunofluorescence was performed in MetaMorph software (Molecular Devices) on images acquired using fixed exposure conditions.
HeLa and C. elegans culture
HeLa cells were cultured in DME as previously described (Kline et al., 2006). C. elegans was grown using standard conditions (Oegema et al., 2001).
Live imaging and GFP fusions
All live images were acquired via a spinning disk confocal microscope (CSU10; McBain Instruments) mounted on an inverted microscope (TE2000e; Nikon). Strain TH32 coexpressing GFP-histone H2b and GFP–γ-tubulin was imaged using a 60× 1.4 NA plan Apo objective with 1.5× auxiliary magnification and a cooled CCD camera (Orca ER; Hamamatsu) binning 2 × 2. Live imaging of HeLa cells was performed using similar imaging conditions at 37°C. Strain OD31 expressing GFP–KNL-2 was imaged in the same manner without the 1.5× auxiliary magnification. To construct strain OD31, oligonucleotides CGCTTCCACTAGTGGTGATACGGAAATTGTTCCTC and CGCTTCCACTAGTTTAGTAGATGGATGTGTCTTCTTCA were used to PCR KNL-2 from cDNA (provided by Y. Kohara, National Institute of Genetics, Shizuoka, Japan), and the resulting fragment was digested with SpeI and cloned into pIC26 to create pPM3. pPM3 was biolistically transformed into C. elegans to generate a stable integrated strain. The HCP-3–GFP strain OD101 expressed HCP-3 with an internal GFP fused between the N-terminal tail and the histone core of HCP-3. The DNA construct for HCP-3–GFP bombardment was made by digesting a GFP PCR product (forward oligonucleotide GCGCGGAGCTCCATGAGTAAAGGAGAAGAACTTTTCAC and reverse oligonucleotide CGCGCGAGCTCGCTTTGTATAGTTCATCCATGCCAT) with Sac-I and inserting at a Sac-I restriction site within the HCP-3 coding region at residue 173. The HCP-3–GFP construct was then inserted at the Bam-HI site of pAZ132 and biolistically transformed into C. elegans to generate a stable, integrated strain. The YFP–CENP-A clonal cell line was a gift from D. Foltz (Ludwig Institute for Cancer Research, San Diego, CA; Foltz et al., 2006).
Biochemistry
Nuclei were isolated from adult OD31 C. elegans. In brief, synchronous liquid cultures of OD31 were grown until 5–10 embryos were present per adult. Embryos were isolated by bleaching, and blastomeres were dissociated by treatment with chitinase (0.5-μg/ml final concentration; 30 min at RT; Sigma-Aldrich). Nuclei were isolated from blastomeres by dounce homogenization in a low ionic strength buffer (0.35 M sucrose, 15 mM Hepes-KOH, pH 7.6, 0.5 mM EGTA, 5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, and protease inhibitor cocktail) with digitonin (0.125% final concentration; Sigma-Aldrich). Nuclei were washed in lysis buffer (50 mM Hepes, pH 7.4, 5 mM EGTA, 1 mM MgCl2, 300 mM KCl, 10% glycerol, 0.1% Triton X-100, 0.05% NP-40, and protease inhibitors) and sonicated to create chromatin fragments. The sonicated mixture was clarified, and the supernatant was used for immunoprecipitations as described previously (Cheeseman et al., 2004). Western blots were performed using standard methods.
Condensation assay
Quantification of mitotic chromosome condensation was performed as described previously (Maddox et al., 2006) using strain TH32 and the imaging conditions described in the Live imaging and GFP fusions section.
Online supplemental material
Fig. S1 shows that KNL-2 function is not restricted to the first embryonic division of C. elegans. Fig. S2 shows that KNL-2 makes an equivalent contribution to CeCENP-A in condensation and partially colocalizes with CeCENP-A in interphase nuclei of C. elegans embryos. Fig. S3 shows that HsKNL2 is required for chromosome segregation in human cells. Video 1 shows chromosome segregation in a control C. elegans embryo coexpressing GFP-histone H2b and GFP–γ-tubulin. Videos 2 and 3 show chromosome segregation in CeCENP-A– (Video 2) and KNL-2–depleted (Video 3) C. elegans embryos coexpressing GFP-histone H2b and GFP–γ-tubulin. Video 4 shows GFP–KNL-2 in a control C. elegans embryo, and Video 5 shows GFP–KNL-2 in a CeCENP-A–depleted C. elegans embryo. Video 6 shows GFP–CeCENP-A in a control C. elegans embryo, and Video 7 shows GFP–CeCENP-A in a KNL-2–depleted C. elegans embryo. Videos 8 and 9 show GFP–KNL-2 in CeCENP-C– (Video 8) and KNL-1–depleted (Video 9) C. elegans embryos. Video 10 shows DIC and YFP–CENP-A time lapse of control (top) and HsKNL2-depleted (bottom) mitotic HeLa cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200701065/DC1.
Acknowledgments
We thank Tony Hyman for sharing the functional genomics screen data and for his support, Yuji Kohara for cDNAs, Iain Cheeseman and Amy Shaub Maddox for critical reading, Dan Foltz for the YFP–CENP-A cell line, Lars Jansen and Don Cleveland for sharing results before publication, and members of the Oegema and Desai laboratories for helpful discussions.
P.S. Maddox is the Fayez Sarofim Fellow of the Damon Runyon Cancer Research Foundation (DRCRF; grant DRG-1808-04), K. Oegema is a Pew Scholar in the Biomedical Sciences, and A. Desai is the Connie and Bob Lurie Scholar of the DRCRF (grant DRS 38-04). This work was supported by a grant from the National Institutes of Health to A. Desai (R01-GM074215) and by salary support from the Ludwig Institute for Cancer Research to K. Oegema and A. Desai.
Abbreviations used in this paper: CENP-A, centromere protein A; DIC, differential interference contrast; dsRNA, double-stranded RNA; KNL, kinetochore null.
References
- Aasland, R., A.F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87–88. [PubMed] [Google Scholar]
- Buchwitz, B.J., K. Ahmad, L.L. Moore, M.B. Roth, and S. Henikoff. 1999. A histone-H3-like protein in C. elegans. Nature. 401:547–548. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Niessen, S. Anderson, F. Hyndman, J.R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H. 2000. Centromerization. Trends Cell Biol. 10:182–188. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., Y. Dong, P. Hergert, J. Strauss, J.M. Hickey, E.D. Salmon, and B.F. McEwen. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Shevchenko, A. Hyman, and K. Oegema. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz, D.R., L.E. Jansen, B.E. Black, A.O. Bailey, J.R. Yates III, and D.W. Cleveland. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. [DOI] [PubMed] [Google Scholar]
- Fujita, Y., T. Hayashi, T. Kiyomitsu, Y. Toyoda, A. Kokubu, C. Obuse, and M. Yanagida. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 12:17–30. [DOI] [PubMed] [Google Scholar]
- Hayashi, T., Y. Fujita, O. Iwasaki, Y. Adachi, K. Takahashi, and M. Yanagida. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 118:715–729. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Y. Dalal. 2005. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15:177–184. [DOI] [PubMed] [Google Scholar]
- Jansen, L.E.T., B.E. Black, D.F. Foltz, and D.W. Cleveland. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline, S.L., I.M. Cheeseman, T. Hori, T. Fukagawa, and A. Desai. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick, J.S. 1996. One billion years of Myb. Oncogene. 13:223–235. [PubMed] [Google Scholar]
- Maddox, P.S., K. Oegema, A. Desai, and I.M. Cheeseman. 2004. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 12:641–653. [DOI] [PubMed] [Google Scholar]
- Maddox, A.S., B. Habermann, A. Desai, and K. Oegema. 2005. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 132:2837–2848. [DOI] [PubMed] [Google Scholar]
- Maddox, P.S., N. Portier, A. Desai, and K. Oegema. 2006. Molecular analysis of mitotic chromosome condensation using a quantitative time- resolved fluorescence microscopy assay. Proc. Natl. Acad. Sci. USA. 103:15097–15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone, B.G., and R.C. Allshire. 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13:191–198. [DOI] [PubMed] [Google Scholar]
- Mo, X., E. Kowenz-Leutz, Y. Laumonnier, H. Xu, and A. Leutz. 2005. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 19:2447–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen, J., P.S. Maddox, F. Hyndman, K. Oegema, and A. Desai. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 7:1248–1255. [DOI] [PubMed] [Google Scholar]
- Oegema, K., and A.A. Hyman. 2006. Cell division. In WormBook. The C. elegans Research Community, editors. Doi: 10.1895/wormbook.1.72.1 http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D.K., K. O'Day, H.L. Trong, H. Charbonneau, and R.L. Margolis. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA. 88:3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., L.B. Koski, A. Walsh, P. Marschall, B. Neumann, M. Brehm, A.M. Alleaume, J. Artelt, P. Bettencourt, E. Cassin, et al. 2005. Full- genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469. [DOI] [PubMed] [Google Scholar]
- Sullivan, B.A., M.D. Blower, and G.H. Karpen. 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2:584–596. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., E.S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 288:2215–2219. [DOI] [PubMed] [Google Scholar]